Abstract
Protein-protein interactions usually involve a large number of residues; thus it is difficult to elucidate functional and structural roles of specific residues located in the interface. This problem is particularly challenging for ankyrin repeat proteins (ARs), which consist of linear arrays of small repeating units and play critical roles in almost every life process via protein-protein interactions, because the residues involved are discontinuously dispersed in both the ARs and their partners. Our previous studies showed that while both P16 and gankyrin bind to CDK4 in similar fashion, only P16 inhibits the kinase activity of CDK4. While this could explain why P16 is a tumor suppressor and gankyrin is oncogenic, the structural basis of these contrasting properties was unknown. In this study we show that a double mutant of gankyrin, L62H/I79D, inhibits the kinase activity of CDK4, similar to P16, and such CDK4-inhibtory activity is associated with the I79D but not L62H mutation. In addition, mutations at I79 and L62 bring about moderate decrease in the stability of gankyrin. Further structural and biophysical analyses suggest that the substitution of Ile 79 with Asp leads to local conformational changes in loops I–III of gankyrin. Taken together, our results allow the dissection of the “protein-protein binding” and “CDK4 inhibition” functions of P16, show that the difference between tumor suppressing and oncogenic functions of P16 and gankyrin, respectively, mainly resides in a single residue, and provide structural insight to the contrasting biological functions of the two AR proteins.
Introduction
Ankyrin repeat (hereafter, AR) proteins are a class of proteins in which multiple conserved repeats, namely ankyrin repeats, stack in a linear array to form a repetitive skeletal architecture with variable molecular surfaces1–3. They are involved in numerous physiological processes, including cell signaling, cytoskeleton integrity, transcription, cell cycle control, apoptosis, inflammatory response, development and differentiation, and transport, through mediating protein-protein interactions. While protein interactions such as SH2 or SH3 binding to their target proteins usually involves a localized region of SH2 or SH3 and a very short motif in the target (phosphorylated Tyr and polyproline stretches, respectively), AR proteins are not known to bind to a specific motif in the binding partner3. It is thus a very intriguing question as to how AR proteins control their functional specificity.
P16 and gankyrin are two AR proteins (consisting of 4 and 7 ARs, respectively) with opposite biological functions. P16 specifically binds to CDK4 and inhibits the latter’s Rb-phosphorylating activity4, 5. Such inhibition precludes the release of transcription factors E2Fs from incompetent Rb/E2F complexes, thus blocking the transactivation of downstream genes required for entry into the S phase. It has been well established that P16 is a tumor suppressor whose genetic inactivation through deletion, methylation, or mutation has been found in almost every type of human cancers5. In contrast, binding of gankyrin to CDK4 renders the kinase resistant to P16 inhibition6, 7, which results in enhanced Rb-phosphorylation and stimulation of E2F transcription activity, thus effecting cell cycle progression. Gankyrin is also able to directly bind Rb and facilitate Rb phosphorylation and degradation8. Furthermore, gankyrin interacts with MDM2 and promotes the ubiquitination and targeting of P53 to the proteasome for degradation9. Therefore, gankyrin functions as a negative regulator of three prominent tumor suppressors, P16, Rb, and P5310, 11, and overexpression of gankyrin leads to cell transformation8. In addition, it has been reported that gankyrin is competent in binding to S6 ATPase of the 26S proteasome6 and MAGE A412, and gankyrin is overexpressed in most of human hepatocellular carcinomas8, 13, 14. Taken together, these findings indicate that gankyrin is an oncogenic protein involved in the development of human cancers through down-regulation of CDK4, Rb, and P53.
The goal of this study was to understand the structural basis of the contrasting properties of P16 and gankyrin, and to use these studies to further understand how AR proteins control their biological specificity. As revealed in the crystal structure of P16/CDK615 (a close homolog of CDK4), and other biochemical studies16–18 (Figure 1), contacts between P16 (or other INK4 proteins, in general) and CDK4 occur in discontinuous patches, and a number of residues located in both loop and helical regions of P16 contribute to CDK4 binding through electrostatic, hydrogen bonding, and van der Waals interactions3. While mutations of most of CDK4-interacting P16 residues only lead to mild decrease in its inhibitory activity, the D84H mutant of P16 (a mutant found in human cancers), or the corresponding mutant of P18, D76A, loses all inhibitory activity even though these two mutants are still able to bind to CDK416, 18. On the other hand, it has been reported that CDK4 R24C, a mutant frequently found in human cancers, retains its kinase activity but is resistant to P16 inhibition19, 20. Further analysis of the crystal structure of the P16/CDK6 complex indicates that there is a strong electrostatic interaction between the side chains of D84 of P16 (D76 of P18) and R24 of CDK4 (R26 of CDK6) at the kinase active site, and mutation in either D84 of P16 or R24 of CDK4 disrupts this interaction, thus abolishing P16 inhibition.
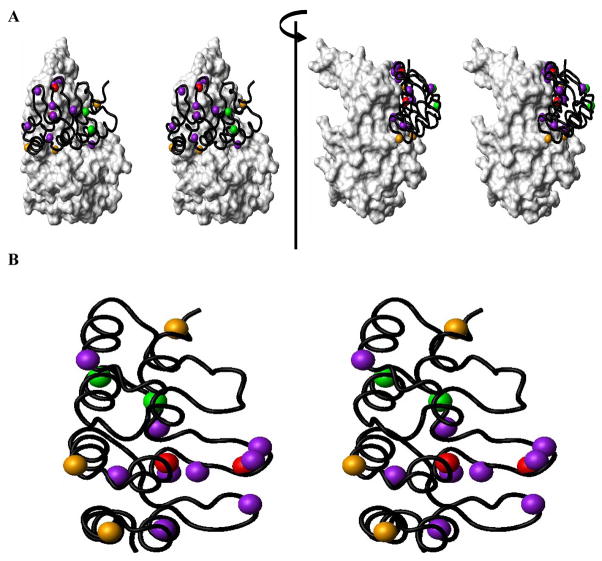
Figure 1. Structural Basis of p16/CDK6 (or CDK4) Interaction.
(A) Structural positioning of the functionally important residues of P16 in contact with CDK6 is shown using the crystal structure of the P16/CDK6 complex15 (PDB code: 1BI7). (B) Quantitative contributions of functionally important residues of P16. Residues are presented in different colors based on changes in the values of IC50 when mutated16, 17. Residues with >20 fold increase in IC50 when mutated are indicated in red (L78 and D84); 10–20 fold, orange (W15, D92 and R124); 5–10 fold, green (H66 and E69), and 3–5 fold, purple (E26, N71, P76, A77, T80, H83, F90, W110, and L121).
Gankyrin competes with P16 for CDK4 binding7, but it does not influence the kinase activity of CDK4, suggesting that gankyrin acts as a P16 blocker rather than a CDK4 inhibitor or activator. The structure of gankyrin in complex with CDK4 is not available, although the structure of free gankyrin has been solved by both NMR21 and X-ray22–24. Our structure-based sequence homology analysis showed that the residue corresponding to D84 of P16 is I79 in gankyrin (Figure 2). Using site-directed mutagenesis, we show that I79D mutant of gankyrin, as well as a double mutant L62H/I79D, binds to CDK4 and inhibits its kinase activity similar to P16. Further structural analyses suggest that these substitutions bring about notable local conformational changes in gankyrin, most likely facilitating the positioning of a negatively-charged residue into the proximity of positively-charged R24 of CDK4. Analysis of conformational stability also suggests that the CDK4-inhibitory activity likely comes at the expense of stability. Overall, our studies provide insights into the molecular mechanisms underlying interactions between AR proteins and their partners.
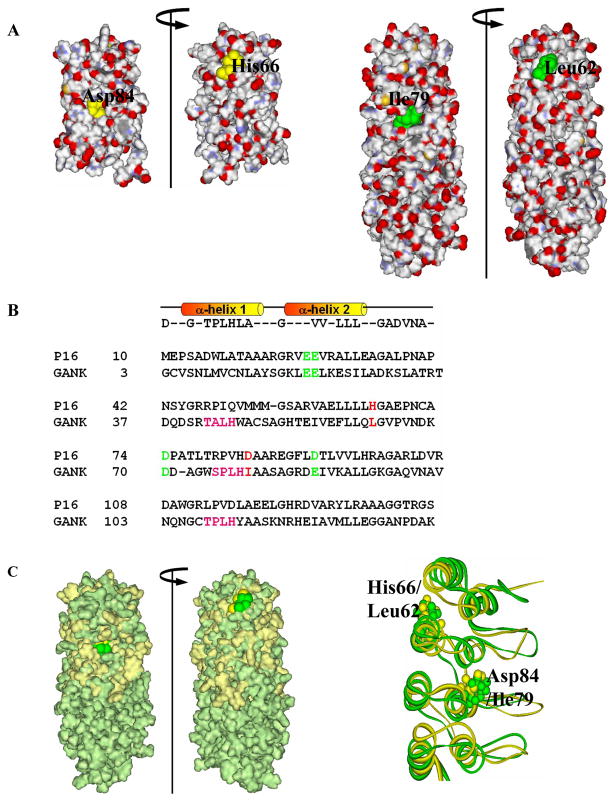
Figure 2. Structure-based sequence homology between P16 and gankyrin.
(A) Surface charge distribution of P16 (PDB ID: 1DC2; left) and gankyrin (PDB ID: 1TR4; right) indicating the location of the two residues of interest. (B) Structure-based sequence alignment of human P16 and the first four ARs of gankyrin. Four negatively charged CDK4-binding residues, E28, E29, D74, and D92 of P16 (highlighted in green) are conserved in gankyrin as E20, E21, D70, and E87, respectively (green). Two residues of interest, H66 and D84 of P16, and corresponding gankyrin residues L62 and I79, are highlighted in red. (C) Surface-fitted overlay of P16 and gankyrin (left) and the ribbon drawing of best fit superimposition of the backbone (N, Cα, and C) atoms of the NMR structures of P16 (gold) and the first four ankyrin repeats of gankyrin (green). Two residues of interest, H66 and D84 of P16 (gold) as well as their corresponding residues, L62 and I79 (green) in gankyrin are highlighted.
Results
Structure-based protein engineering
Previous studies in our as well as other laboratories showed that gankyrin binds to CDK4 in a way similar to that of P16 in vitro and in vivo, and this binding counteracts the CDK4-inhibiting activity of P166, 7. Further fragmentation experiments demonstrated that the first four ARs of gankyrin are required and sufficient for CDK4 binding and P16 counteraction7. To understand the differences between the functions of gankyrin and P16, structural and sequence alignments of the two proteins were performed as shown in Figure 2 (A and B, respectively). As shown in Figure 2B, four negatively-charged CDK4-binding residues, E26, E27, D74, and D92 of P16 align perfectly with E20, E21, D70, and E87, respectively, of gankyrin thus justifying this structure-based sequence homology analysis (Figure 2C). These residues are very likely involved in the binding of gankyrin with CDK421.
One interesting notion of this analysis is that D84 of P16 corresponds to I79 in gankyrin. Apparently, the hydrophobic side chain of I79 is incapable of interacting via a salt bridge or a hydrogen bond with the positively charged side chain of R24 at the active site of CDK4, although a much weaker nonpolar contact could not be ruled out. Therefore, this residue is an interesting candidate to explore the functional role and to see if introducing a negatively charged Asp residue in place of I79 of gankyrin could rescue its CDK4-inhibiting activity. H66 is another important residue in P16 which is not conserved between P16 and gankyrin. The crystal structure of the P16/CDK6 complex showed that this residue does not directly contact CDK4, however, mutations of this residue led to the loss of 80% of the CDK4-inhibitory activity of P1616, suggesting that H66 contributes indirectly to inhibition, likely through stabilizing the structure of P16 and facilitating the positioning of D84. The residue corresponding to H66 in P16 is L62 in gankyrin, and it can be reasoned that introducing a L62H mutation in gankyrin may influence the stability of gankyrin but not its inhibitory ability, as long as the critical Asp is absent at position 79 of gankyrin. To confirm the above notion, two single mutants, L62H and I79D, and one double mutant, L62H/I79D of gankyrin were generated by site-directed mutagenesis, and their biochemical and biophysical properties were subsequently evaluated.
I79D and L62H/I79D of gankyrin bind and inhibit CDK4
The binding of these gankyrin mutants to CDK4 was first investigated using pull-down assays with glutathione-S-transferase (GST)-tagged gankyrin proteins and the CDK4/cyclin D2 holoenzyme. Instead of CDK4, the CDK4/cyclin D2 holoenzyme was used in this assay mainly due to the fact that the holoenzyme is the biological active form and there is no interaction between gankyrin and cyclin D27. As shown in Figure 3A, CDK4 was detected in the reaction mixtures containing GST-gankyrin wild type (lane 3) and mutant proteins (lanes 4–6) as well as in the reaction mixture containing GST-P16 (lane 2), which was used as positive control in this assay, suggesting that all three gankyrin mutants bind to CDK4. Even though this assay should not be used to quantitatively evaluate protein/protein interaction, the amounts of CDK4 in the pull-down products were comparable to each other, indicating that there are no significant differences among the wild type and the mutant gankyrin proteins in their CDK4-binding affinity.
Figure 3. Interactions between gankyrin proteins and CDK4 as evaluated using pull-down assays and in vitro kinase assays.
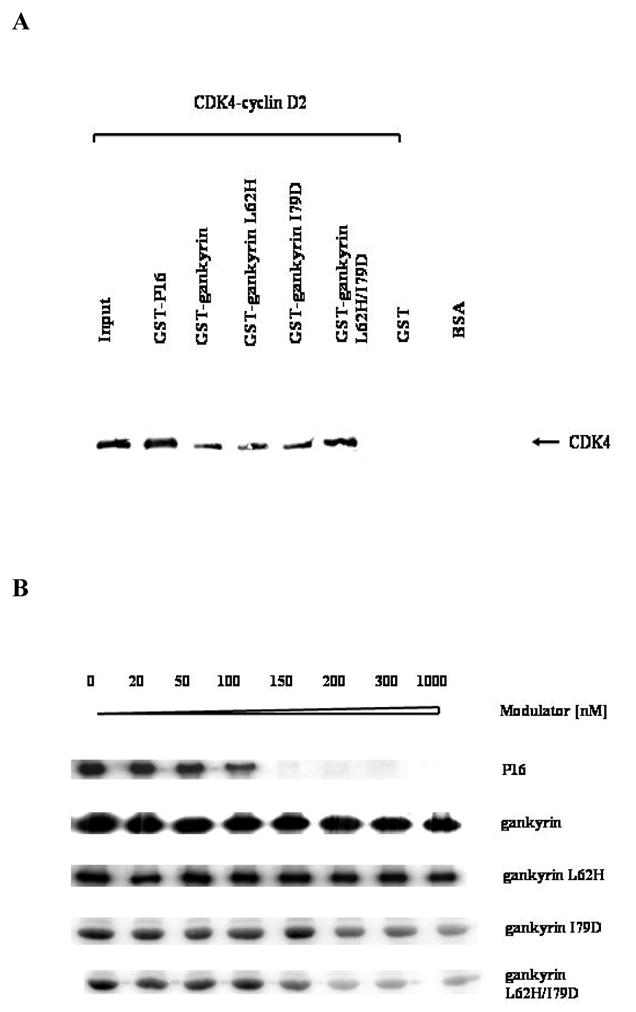
(A) Pull-down assays. The reaction mixtures containing GST-tagged proteins and the CDK4-cyclin D2 holoenzyme were incubated with reduced glutathione-agarose, and after elution with reduced glutathione the bound proteins were separated by SDS-PAGE, and blotted against anti-human CDK4 antibody (Santa Cruz, C-22). Lanes: 1, the input only containing 5% of the amount of purified CDK4-cyclin D2 in other lanes; 2, positive control, GST-P16 (0.5 μM)/CDK4-cyclin D2 (0.1 μM); 3, GST-gankyrin wild type (0.5 μM)/CDK4-cyclin D2 (0.1 μM); 4, GST-gankyrin L62H (0.5 μM)/CDK4-cyclin D2 (0.1 μM); 5, GST-gankyrin I79D (0.5 μM)/CDK4-cyclin D2 (0.1 μM); 6, GST-gankyrin L62H/I79D (0.5 μM)/CDK4-cyclin D2 (0.1 μM); 7, mock lane, GST (0.5 μM)/CDK4-cyclin D2 (0.1 μM); 8, BSA, as a negative control in Western Blot. (B) in vitro kinase assays. Each reaction mixture included 3 units of CDK4-cyclin D2 holoenzyme (about 0.3 μg), 50 ng GST-Rb791-928, 5 μCi [γ-32P] ATP, and varying amounts of effector proteins. After incubation at 30 °C for 15 minutes, the reaction mixtures were separated by 10% SDS-PAGE, dried, and analyzed by autoradiography.
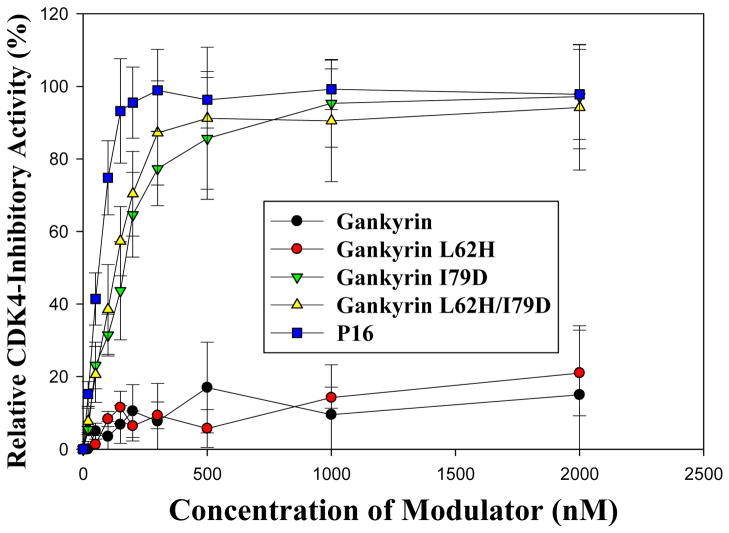
The CDK4-modulating activities of gankyrin and the three mutants were then assessed using an in vitro kinase assay as previously described7. As shown in Figure 3B, neither gankyrin nor L62H mutant brought about any significant change in CDK4-mediated phosphorylation of Rb, suggesting that wild type gankyrin and L62H mutant do not influence the activity of CDK4. However, in the reaction mixtures containing mutant I79D or L62H/I79D, as the concentration of I79D or L62H/I79D increased, CDK4-mediated phosphorylation of Rb decreased, indicating that unlike wild type gankyrin, both I79D and L62H/I79D mutants inhibit the kinase activity of CDK4. As a positive control, increasing amounts of P16 in the reaction mixtures also led to a decrease in the kinase activity of CDK4. Further quantitative analyses yielded IC50 values of 170 ± 32 nM and 120 ± 27 nM for I79D and L62H/I79D mutants, respectively (Figure 4), only slightly higher than that for P16 (72 ± 15 nM), suggesting that the CDK4-inhibitory activities of I79D and L62H/I79D are comparable to that of P16. In contrast, the IC50 values for wild type gankyrin and L62H mutant were higher than 3.0 μM, indicating that gankyrin wild type and L62H mutant do not inhibit CDK4 under the experimental conditions used. Interestingly, our previous studies16 showed that while mutations at D84 of P16 completely abolished its CDK4-inhibitory activity, mutations at H66 of P16 also led to an 8-fold decrease in the CDK4-inhibitory activity suggesting that H66 of P16 is able to affect the CDK4-inhibitory activity through certain ways. In contrast, gankyrin L62H mutation does not exhibit any detectable CDK4-inhibitory activity, and the IC50 values for gankyrin I79D and L62H/I79D are almost identical. Taken together, an Ile → Asp substitution at position 79 of gankyrin does not significantly affect the binding of gankyrin to CDK4 but enables gankyrin to inhibit the kinase activity of CDK4. In comparison, a Leu → His mutation at position 62 apparently affects neither the binding nor the inhibition.
Figure 4. Quantitative measurement of the CDK4-inhibitory activities of gankyrin proteins.
The in vitro kinase assays were performed as described in Figure 3B, and the incorporation of 32P into the Rb substrate was quantitated using ImageQuant (Molecular Dynamics). IC50 was defined as the inhibitor concentration at which half of the maximum inhibition can be obtained. The experimental error for this assay was estimated to be ± 30%. All assays were performed in triplicate, and P16 was used as a positive control.
I79D and L62H/I79D mutations bring about substantial perturbation to the local conformation of gankyrin
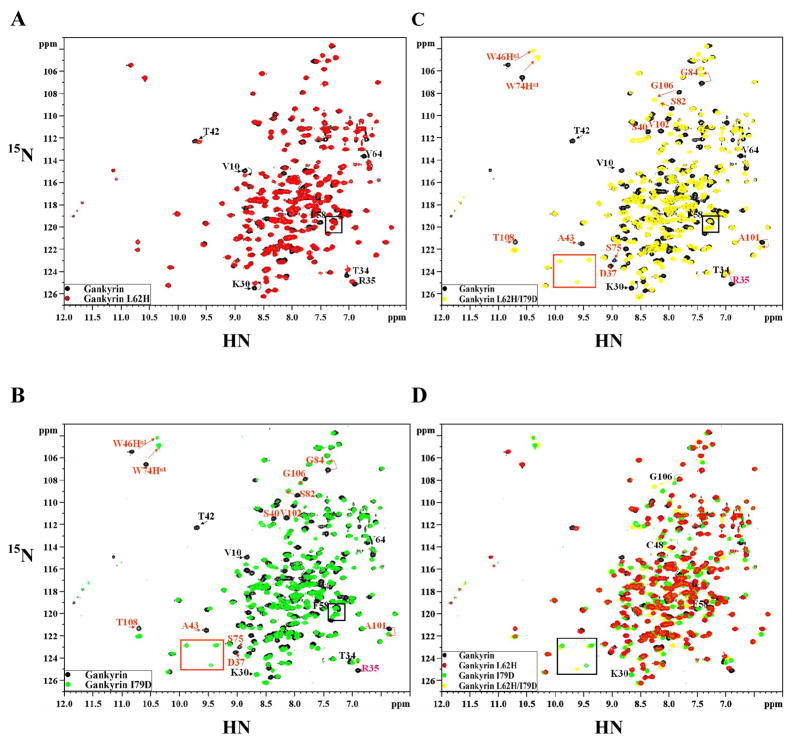
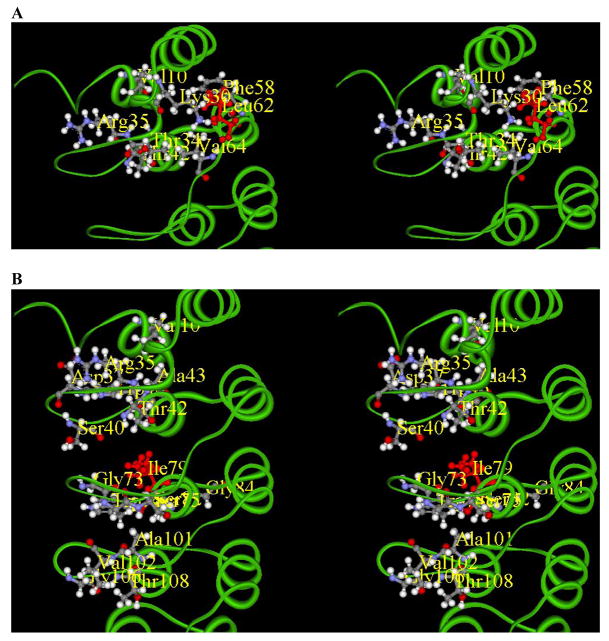
2D 1H-15N heteronuclear single-quantum coherence (HSQC) NMR spectroscopy was subsequently used to investigate the possible structural perturbation caused by the above gankyrin mutations. As shown in Figure 5A, the spectrum of L62H is almost identical to that of wild type gankyrin. The few notable chemical shift changes are mapped to residues Lys30, Lys35, Thr34, Thr42, Phe58, and Val64, all of which are located in the first two ARs, and thus in close vicinity of L62H mutation (Figure 6A). Hence, L62H mutation brings about only minor changes in local environments rather than any significant change in the global structure of gankyrin. In contrast, there are considerable and extensive changes in the spectra of I79D and L62H/I79D in comparison to the spectrum of wild type gankyrin (Figures 5B and 5C). In addition, the spectra of I79D and L62H/I79D are virtually superimposable, further supporting our previous observation that L62H mutation does not cause any significant change in the global structure of gankyrin.
Figure 5. Comparison of 15N-1H HSQC spectra of wild type gankyrin and its mutants .
The HSQC spectra were obtained in D2O solution using a 600 MHz spectrometer. (A) L62H (red) and wild type (black); (B) I79D (blue) and wild type (black); (C) L62H/I79D (green) and wild type (black); (D) superimposition of spectra in A, B, and C. L62 and I79 peaks in the spectra of wild type gankyrin are in overlapping regions, hence, cannot be precisely pointed at in the respective mutant data. The peaks that show larger chemical shifts are labeled with arrows.
Figure 6. Mapping of chemical shift changes on the gankyrin structure.
(A) L62H (highlighted in red) mutation and the residues affected as a consequence are localized in AR1 and AR2. (B) I79D (highlighted in red) mutation and the residues affected as a consequence are localized in loop regions between AR1-2, AR2-3 and AR3-4.
On the basis of 3D 15N-edited NOESY recorded on a 15N-labeled I79D sample together with the previous assignments on wild type (WT)21, a total of 168 backbone amides have been assigned, among which the residues in the AR2 and AR3 are least assigned due to the largest chemical shift perturbations. It can be concluded that while the residues in AR5-AR7 retain virtually the same chemical shifts, the residues perturbed as a result of the Ile → Asp substitution are located in the flexible loop regions within the first four ARs, including Val10, Arg35, Arg37, Ser40, Thr42, Ala43 (the first loop), Gly73, Trp74, Ser75, Ser82 (the second loop), Ala101, Val102, Gly106, and Thr108 (the third loop) (Figure 6B). Interestingly, all these residues are part of the TPLH interaction network of central ankyrin repeats of gankyrin21. These results suggest that the conformations of loops I to III are perturbed in the I79D mutant, and raise the possibility that the observed functional change upon this mutation may not be a simple effect of a side chain substitution. Instead, the I79D substitution possibly leads to local structural adjustments that help to position the side chain for the inhibitory function.
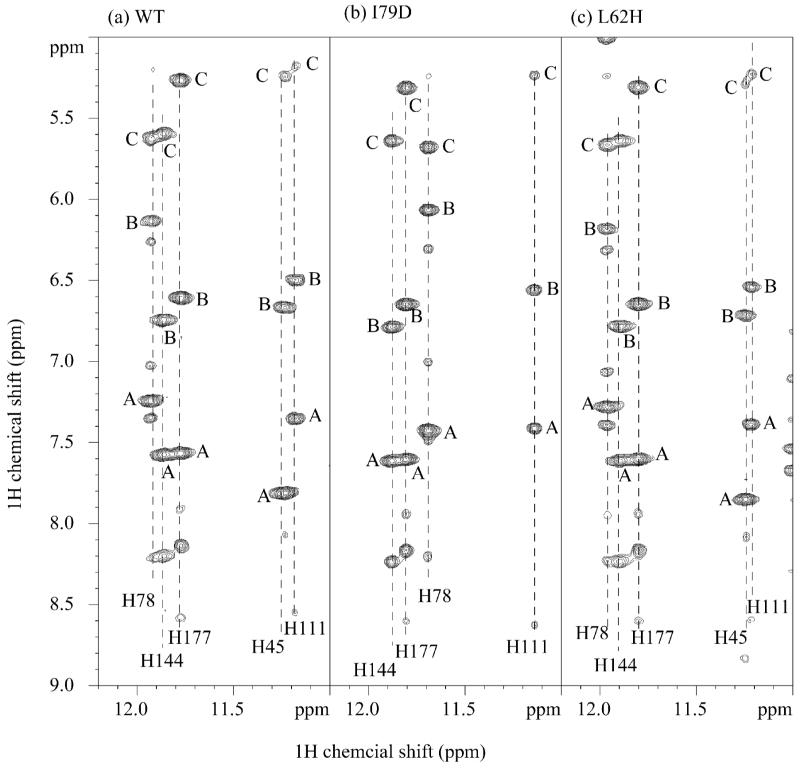
This concept of full integration of structure and function is further supported by some preliminary NOE analysis on the aforementioned 3D 15N-edited NOESY as well as parallel 2D 1H homonuclear NOESY recorded on WT, I79D, and L62H. Firstly, in 3D NOESY data set relative NOE intensity changes have been observed for the limited number of residues that have been assigned in the second and third loop. For example, the NOE between Hγ2/T37 and HN/S40 is becoming much stronger whereas the sequential NOE between Hε2/N67 and HN/D68 is comparatively weaker in I79D mutant. Secondly, in the downfield region of 2D NOESY recorded in H2O (Figure 7), I79D mutant shows contrasting result compared with L62H mutant concerning the Hε2 of the histidine residue in a TPLH motif. The significant change in I79D is highlighted by the large perturbation on Hε2/H78 resonance and more importantly, the disappearance of the Hε2/H45 signal despite extensive search aided by 2D 1H-15N HSQC and 2D 1H-15N heteronuclear multiple-bond correlation (HMBC) experiments on a 15N-labeled I79D. Since Hε2/H45 is the bridging proton of the inter-AR hydrogen bond between H45 imidazole ring and the backbone oxygen of W74, its disappearance could be indicative of the disruption of the inter-AR interaction and the local conformational changes in both AR2 and AR3.
Figure 7. 2D NOESY spectra on WT (a), I79D (b), and L62H (c) showing the proton downfield region.
The cross peaks designated A–C are NOEs from Hε2/His in a TPLH motif to Hε1 and Hδ2 of the same histidine residue, and Hα/(His+30) residue (Hα/T207 for H177), respectively. The latter is a supporting evidence of inter-ankyrin repeat hydrogen bond between Nε2/His and O/(His+29) (e.g. Nε2/H177 and O/K206). It is clear from the comparisons that L62H mutant retains the TPLH features from AR2 to AR6, while I79D has large perturbation in TPLH of AR3 (H78) and ANK4 (H111), and likely disruption in 42TALH45 evidenced by the disappearance of Hε2/H45 signal.
I79D and L62H/I79D destabilize the global structure of gankyrin
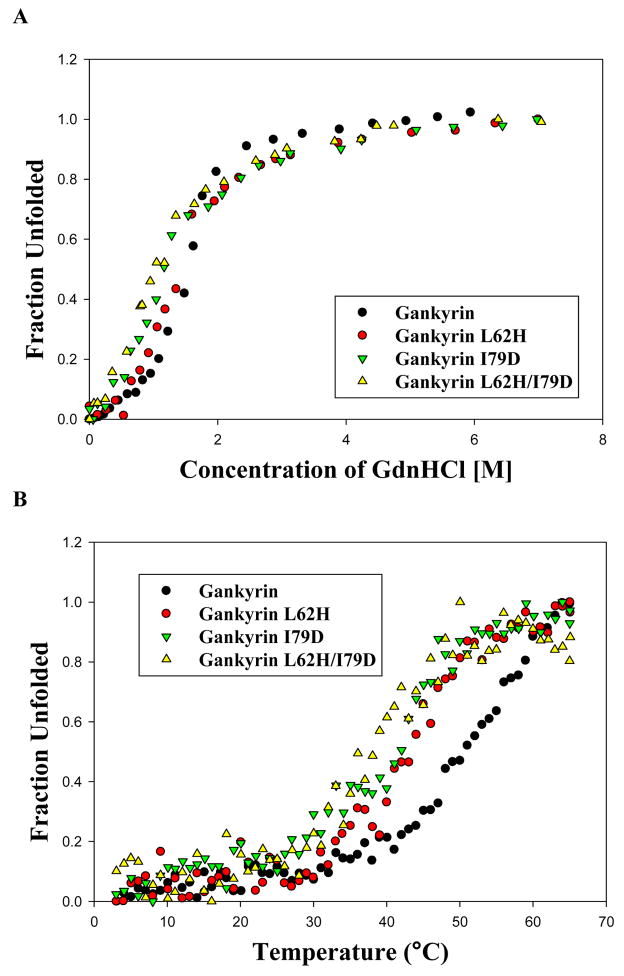
The effect of the above gankyrin mutations on the conformational stability of gankyrin was evaluated using far-UV Circular Dichroism (CD) spectroscopy. In guanidinium hydrochloride (GdnHCl)-induced unfolding, changes in the ellipticity at 222 nm, indicative of changes in the α-helical content, were monitored. The unfolded fraction was derived from the raw data and plotted against the concentration of denaturant (Figure 8A). The unfolding curves of all three mutants as well as the wild type gankyrin can be fitted well to a model with a two-state transition between native and unfolded proteins25; the resultant values of ΔGdwater (the denaturation free energy in water), D1/2 (the denaturant concentration at the midpoint of transition), and the slope m are listed in Table 1. Since the m values of the mutants and the wild type gankyrin are almost identical, differences in the values of ΔGdwater can be directly interpreted as their differences in conformation stability25. Compared to wild type gankyrin, both L62H and I79D mutations have moderately destabilized the conformation by 0.51 and 0.85 kcal*mol−1, respectively. The double mutant, L62H/I79D, which possesses the highest CDK4-inhibitory activity, has additional 0.34 kcal*mol−1 loss of conformational stability from I79D. The relative stability has been further confirmed in a heat-induced unfolding experiment (Figure 8B), in which the temperatures at the midpoint of two-state unfolding transition26, 27, Tm values, were extracted (Table 1). Compared to gankyrin WT, all three mutants have decreased Tm values, and the double mutant has the lowest Tm. In conclusion, it appears that the relative conformation stability of the three mutants coincides with the relative perturbations in HSQC, and the CDK4-inhibitory activity via mutation likely comes at the expense of stability.
Figure 8. Chemical- and heat-induced unfolding of gankyrin mutants monitored by far-UV CD.
(A) GdnHCl-induced unfolding. Samples containing 7.5–10.0 μM proteins were incubated with different amounts of GdnHCl on ice overnight and the ellipticity at 222 nm was monitored by far-UV CD (190–260nm) at 25 °C. The fraction unfolded, defined as (the ellipticity at 222 nm at a denaturant concentration-the ellipticity at 222 nm at the native state)/(the ellipticity at 222 nm at the fully unfolded state-the ellipticity at 222 nm at the native state), was plotted against the GdnHCl concentration37. The unfolding curve was fitted to a model of two-state approximation25. (B) Thermal melting spectra were recorded at 222nm by heating from 3 °C to 65 °C with a rate of 1 °C per minute and a 1 °C interval. Tm, the temperature at the midpoint of transition, was obtained through fitting the melting curve to a two-state transition model.
Table 1.
Biochemical and biophysical parameters of gankyrin and its mutant proteins
| Protein | In vitro CDK4 inhibitiona | Chemical-induced unfoldingb | Heat-induced unfoldingc | ||
|---|---|---|---|---|---|
| IC50 (nM) | ΔGdwater (kcal*mol−1) | D1/2 (M) | m (kcal*mol−1 *M−1) | Tm (°C) | |
| gankyrin | > 3000 | 2.77 | 1.54 | 1.80 | 51.5 |
| gankyrin L62H | > 3000 | 2.26 | 1.38 | 1.64 | 44.5 |
| gankyrin I79D | 170 ± 32 | 1.92 | 1.14 | 1.68 | 42.0 |
| gankyrin L62H/I79D | 120 ± 27 | 1.58 | 1.01 | 1.57 | 38.5 |
IC50 was defined as the concentration of gankyrin proteins required for 50% of the maximum inhibition. The highest concentration of gankyrin proteins tested in such in vitro kinase assay was 3.0 μM, and an IC50 value higher than 3.0 μindicated that at the tested concentration range of gankyrin proteins, the CDK4 kinase activity was higher than 50% of that in the absence of any gankyrin protein in the reaction mixture. The estimated error in the determination of IC50 is ± 20%, and the experiments were performed in triplicate28.
ΔGdwater, D1/2, and m values were calculated according to a two-state model, and the error limit in ΔGdwater is estimated to be ± 0.5 kcal/mol.36
Tm was defined as the temperature at the midpoint of transition27, and the error is estimated to be ± 0.5 °C.
Discussion
Possible conformational adjustments of local loops upon I79D mutation of gankyrin
In spite of the striking similarity between the skeletal structures of P16 and the first four ARs of gankyrin, available structures indicate that the microenvironments around the “effector” residues, i.e. D84 and I79 for P16 and gankyrin, respectively, are different from each other. In P16, D84 is located at the beginning of the first helix of AR3 and structurally, it has more freedom than the residues in the middle of the helix16. Hence, even though D84 is only slightly exposed on the surface of the free protein (Figure 2A), the conformationally flexible loop (loop 2) preceding D84 could influence the microenvironment around D84 and make this negatively charged residue accessible to the solvent or the positively-charged side chain of R24 at the active site of CDK4. In the crystal structure of the P16/CDK6 complex, negatively charged side chain of D84 is in the vicinity of a positively charged environment (Figure 9). In gankyrin, I79 structurally and sequentially corresponds to D84 of P16. While I79 is located at the beginning of the first helix of AR3, its bulky, aliphatic side chain presumably makes it less accessible to the aqueous surrounding. As shown in the docking model of gankyrin and CDK6 (Figure 9), nonpolar I79 is positioned in an unfavorable electrostatic surrounding at the active site of CDK6. Additionally, the preceding H78 or other nearby residues are unable to provide any assistance in stabilizing the aliphatic side chain of I79. Therefore, after introduction of Asp residue at position 79, the microenvironments surrounding this residue need to be adjusted in order to position the negatively-charged side chain of Asp in the proximity of the active site of CDK4 and facilitate the kinase inhibition. Such structural adjustment in gankyrin I79D mutant is supported by the substantial chemical shift changes in the HSQC spectra of gankyrin I79D and L62H/I79D mutants and the discernable changes in NOE pattern. While large chemical shift changes were observed mostly for residues located in the flexible loops connecting AR2, AR3, and AR4, significant changes were also observed for the residues constituting the TPLH network. The residues involved in the TPLH network include, for example, T42 and A43 of 42TALH45 (AR2), S75 of 75SPLH78 (AR3) and T108 of 108TPLH111 (AR4), as well as the residues preceding the TPLH sequence, such as S40, G74, W75, and G106. Most of these residues are located on the concave surface of gankyrin facing CDK4 (Figure 6). The 2D NOESY have provided more insights about the structural perturbation in TPLH motifs, 42TALH45 in particular, induced by I79 mutation. The observation is not surprising, considering that the side chain of I79 is pointing to AR2 and directly interacts with H45 evidenced by a NOE assigned previously between Hδ1/I79 and Hδ2/H4521. It is expected that such interaction between these two side chains helps to orient the histidine imidazole ring and restrict its flexibility, enabling H45 to adopt an appropriate Nε2-H tautomeric form and engage in several hydrogen bonds. A modeling structure of I79D (Figure 10) shows that the Asp side chain is incapable of such an interaction without significant structural adjustment. Since P16 does not have the TPLH motif or its close variant, and specifically, 46RPIQ49 of P16 is aligned with 42TALH45 of gankyrin, the destabilization in TPLH motifs induced by I79 mutation likely makes the local conformation and dynamics at the binding site mimic the counterpart in P16.
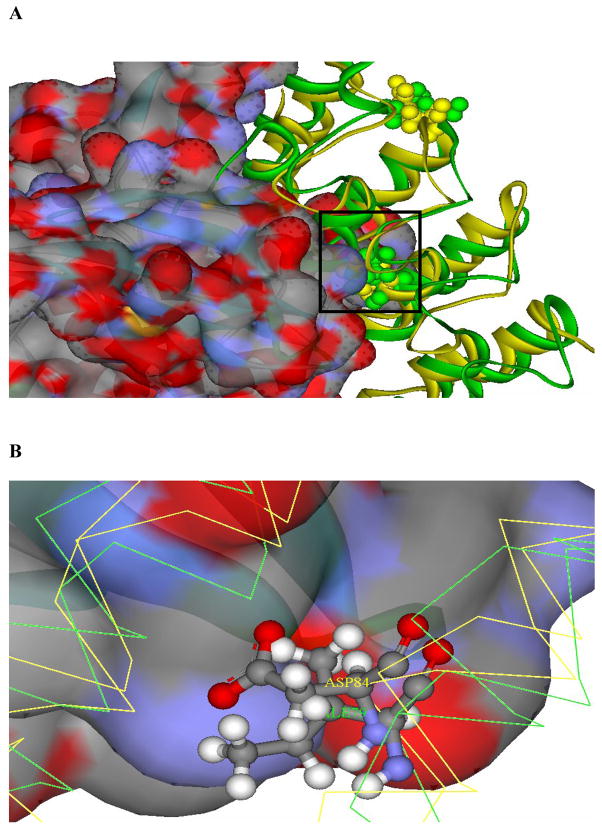
Figure 9. Docking model showing the interactions of I79 of gankyrin with the active site of CDK6.
(A) Docking model of the putative gankyrin/CDK6 complex. This model was constructed by superimposing the first four ARs of gankyrin (ribbon diagram, green) onto P16 (ribbon diagram, golden) in the crystal structure of the P16/CDK6 complex15 (PDB ID: 1BI7; CDK6 is shown in solid-filled diagram). Since no structure of CDK4 is available, crystal structure of CDK6, a close homologue of CDK4, is used in this modeling. H66 and D84 of P16, as well as L62 and I79 of gankyrin are highlighted. Surface residues of CDK6 with positive and negative charges are shown in red and blue, respectively, in the solid-filled diagram. The active site of CDK6 is framed and analyzed in (B). (B) Interactions between D84 of P16 (corresponding to I79) and the active site of CDK6. P16 is in golden, and gankyrin is in green. Apparently, interactions with the charged surface of R26 in the active site of CDK6 (R24 in CDK4) depend on the negatively charged Asp of P16.
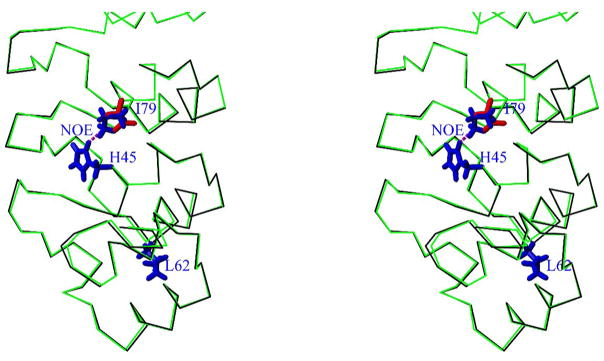
Figure 10. Stereoview showing the overlay of Cα trace of gankyrin WT (black) and modeled I79D mutant (green).
Only the AR1-AR3 repeats are shown. The side chains of the following residues are highlighted: H45, L62 and I79 of gankyrin WT (in blue), and D79 of I79D mutant (in red). In previous NMR studies an NOE between Hδ2/H45 and Hδ1/I79 was observed (dashed line in magenta) 21. Apparently the modeling I79D structure indicates that such side-chain interactions would be impossible without major structural adjustment. Thus the local structures around H45 including the binding residues may be destabilized by this mutation.
In conclusion, our results suggest a structural adjustment in I79D, especially in the loop regions, and that such adjustment may facilitate the positioning of negatively-charged D79 for an electrostatic interaction with R24 of CDK4. Moreover, this conformational adjustment was observed for free gankyrin I79D mutant under physiological condition. Since structural analyses show that there is no significant structural change on P16 or P18 upon binding to CDK4 (data not shown), i.e. no induced-fit event happens on the AR protein part after binding to CDK4, like P16 or P18, free gankyrin I79D mutant is present in a functional conformation (in regard to CDK4 binding and inhibition).
Structural basis for the decreased stability of the gankyrin mutants
As demonstrated by our GdnHCl- and heat-induced unfolding experiments, the conformation of gankyrin mutants was destabilized in the order L62H < I79D < L62H/I79D. One structural difference between P16 and gankyrin is that there is a TPLH stretch, or its variant, present in six out of the seven ankyrin repeats of gankyrin but none as highly matched TPLH variant is present in any of the four P16 ankyrin repeats. These TPLH stretches are located within neighboring loops and form an intra- and inter-AR interaction network that stabilizes the global structure of gankyrin2, 17, 21. Besides the potential electrostatic and steric disturbance caused by the above mutations, impairment to the TPLH interaction network could be the major cause for structural destabilization of gankyrin mutants. As discussed earlier, an Ile → Asp substitution at position 79 brings about considerable conformational changes in residues within/around these TPLH stretches which perturb the TPLH interaction network and subsequently destabilize the global structure of gankyin. As for L62H, such mutation only caused “minor” conformational changes in residues within the first two ARs (including the first TPLH stretch), and the “impairment” to the TPLH network is minor and local (Figure 6A and Figure 7C). This can explain why L62H mutation destabilizes the conformation to a lesser extent and does not affect the function in CDK4 binding and modulation. In contrast, due to the absence of the TPLH interaction network, the corresponding H66Y mutation in P16 with marginal stability could strongly destabilize the structure, which consequently can impair its CDK4-inhibitory activity as evidenced by a loss of 80% of CDK4-inhibitory activity of this mutant16.
Structural basis for the functional diversity of the AR proteins
The residues in P16 could play three major roles: (1) maintaining the skeletal structure of P16, e.g. L63 and L64, which are conserved in almost all AR motifs1, 28; (2) CDK4 binding (but not inhibition), e.g. E28 and E2916; and (3) inhibition (may also contribute to binding), e.g. D84. Our studies suggest that these three functions are inter-related. For example, residues in groups 1 and 2 that are responsible for generating the skeletal structure, may facilitate the positioning of D84 to R24 at the active site of CDK4, thus inhibiting its kinase activity (Figure 9). Therefore, mutations in group 1 and 2 residues could affect the inhibition by destabilizing the global structure and/or impairing CDK4 binding, whereas mutation of D84 directly abolishes the CDK4-inhibtory activity. On the other hand, binding of gankyrin to CDK4 brings the extremely hydrophobic I79 residue in close proximity of R24 of CDK4 (Figure 9) which precludes any electrostatic interaction between the side chains of these two residues and as a result gankyrin just acts as a P16 blocker rather than a CDK4 inhibitor. However, when a negatively-charged Asp residue is introduced at position 79 of gankyrin, following certain adjustments in the local environment, its electrostatic interaction with R24 of CDK4 resumes, and so does the inhibition.
In regard to the modular and repetitive nature of AR proteins, one might assume that the AR proteins are structurally very rigid. However, the results presented in this and previous reports suggest that AR proteins are highly tolerant of structural variations, which could be the basis for its functional diversity since structural and functional properties are intimately related. Some of these properties, relating to the structural pliancy of AR proteins, are summarized here: First, there are significant structural variations in the loops linking neighboring ARs among AR proteins, even though the global topology, including the small curvature, is highly conserved1, 2. Second, some AR proteins can tolerate addition or deletion of AR repeats in the middle or at the end of AR proteins. For example, our previous biochemical studies demonstrated that the removal of up to three ARs at the C-terminus of gankyrin did not affect its structural topology and CDK4-binding ability7. It has also been reported that deletion of individual ARs from Notch AR domain, or insertion of various copies of ARs from Notch AR domain or from consensus-designed ARs, did not affect chemical- and heat-induced unfolding of Notch AR domain29–31. Last but not least, our results from the gankyrin mutants showed that when negatively-charged Asp was introduced to the site originally occupied by Ile, the local conformation of the loops adjusted accordingly (accompanied by partial loss of stability), likely to help position the Asp residue for the CDK4-inhibiting function. These properties suggest that the AR proteins are structurally pliant and are capable of conformational adjustments as a result of relatively minor changes in the sequence. This, in turn, can introduce the functional diversity seen in many of the AR proteins.
Potential biological significance
In its natural form, gankyrin is bi-functional in regulating the CDK4-Rb pathway: it counteracts P16 inhibition of CDK47, and facilitates the ubiquitin-mediated degradation of Rb, both of which result in forcing the cells into cell cycle progression8, 11. Through structure-based protein engineering, we have transformed gankyrin from a simple competitor of P16 in CDK4 binding into a potent CDK4 inhibitor. In addition, it is plausible that for the gankyrin I79D mutant its Rb-binding and promoting function may remain intact since the conformational adjustments upon this mutation are limited to loops I–III, and results from our previous studies showed that the three C-terminal ARs are responsible for Rb binding, and this binding is independent of CDK4 binding. Therefore, gankyrin I79D mutant likely remains bi-functional, but the outcomes of these two functions are now in conflict: while its Rb-binding and promoting role assists cell cycle progression, its CDK4-inhibitory role acts against cell cycle progression. It will be very interesting to see which of these two opposing functions is dominant when gankyrin I79D is overexpressed in cells. Results from such studies could further our understanding of how nature balances the two functions of gankyrin, or the contrasting functions between gankyrin and P16 in relation to cell cycle and cancer.
While this manuscript was in preparation, the crystal structure of the complex between mouse gankyrin and the C-terminal domain of S6 ATPase of the 26S proteasome was reported32. In this complex, most of S6 ATPase-interacting residues are located in the concave surface of mouse gankyrin, especially in loops II and III, which also contribute to binding to CDK4 as demonstrated in previous studies (reviewed in Reference 3) as well as in our current study. Furthermore, mouse gankyrin binds to S6 ATPase in the presence and absence of Rb, indicating that like CDK4 binding, S6 ATPase binding is independent of Rb binding. These findings suggest that CDK4 and S6 ATPase may compete with each other for gankyrin binding in cells. From this perspective, the similarities between CDK4 and S6 ATPase in binding to gankyrin make our question about the molecular mechanisms in which gankyrin balances and coordinates its versatile functions in cells more interesting and challenging. The significance of our current study, however, goes beyond binding – it dissects the binding and inhibition functions of gankyrin toward CDK4.
Materials and Methods
Cloning, expression, and purification of human gankyrin and its mutants
As previously described7, human gankyrin cDNA was cloned into pGEX-6p-2 vector (Amersham) and expressed in Escherichia coli BL21 (DE3) Codon plus cells (Novagen) as glutathione-S-transferase-fusion proteins (GST) upon IPTG induction. GST-fusion gankyrin protein was purified from the cell lysate using a reduced glutathione-agarose column (about 15 mL of G beads, Sigma). After washing with TES buffer (10 mM Tris-HCl-1 mM EDTA-1 mM β-mercaptoethanol-0.15 M NaCl, pH 7.5), 2.0 mL of protein-bound G beads were washed with 10 mL of TES-50 mg/mL reduced glutathione (pH 7.5) and the elute was further purified using a Q Fastflow column (Pharmacia) to get pure GST-gankyrin. To obtain gankyrin without the GST tag, the remaining protein-bound G beads (about 13 mL) were suspended in 30 mL of TES in a 50 mL tube, and 100 units of PreScission protease (2 units/μL, Amersham) were added into the tube. After incubation at 4 °C for 24 hours, the G beads were re-packed on a column and the flowthrough was further purified by an S100 column (Pharmacia) equilibrated with 5 mM HEPES, 1 μM EDTA, and 1 mM DTT (pH 7.5). After SDS-PAGE analysis, fractions containing free gankyrin were pooled, concentrated, and lyophilized for further analyses.
All gankyrin mutants were generated through PCR-based Quickchange site-directed mutagenesis (Stratagene), and were expressed and purified as wild type.
Pull-down assay
To investigate the binding of gankyrin proteins to CDK4, 25 μg of GST-gankyrin proteins were incubated with 10 μg of recombinant CDK4-cyclin D2 complex (see below) and were incubated at 4 °C in 250 μL of TES (pH 7.5) for 2 hours7. The concentrations of the holoenzyme and GST-gankyrin proteins were 0.4 and 2.0 μM, respectively. Subsequently, 250 μL of fresh G beads were added into the reaction mixture, and after incubation at 4 °C for one hour, the reaction mixture was loaded onto a spin column (Fisher Scientific) and centrifuged at 4 °C, 1500 rpm for 3 minutes. After washing the G beads with TES 5 times, 1.0 mL each time, the beads were eluted with 200 μL of TES-50 mg/mL reduced glutathione, and the elute was further analyzed with Western Blot using rabbit polyclonal anti-human CDK4 antibody (Santa Cruz, C-22). In this assay, GST-P16 was used as positive control, and GST was used as negative control.
In vitro CDK4 kinase assay
The in vitro CDK4 activity assay was performed as previously described16. Briefly, each reaction mixture contained 0.3 μg of recombinant CDK4/cyclin D2 holoenzyme and varying concentrations of AR proteins in 15 μL of the kinase buffer, 50 mM HEPES, 10 mM MgCl2, 2.5 mM EGTA, 0.1 mM Na3VO4, 1 mM NaF, 10 mM β–glycerolphosphate, 1 mM DTT, 0.2 mM AEBSF, 2.5 mg/mL leupeptin, and 2.5 mg/mL aprotinin. After incubation at 30 °C for 30 minutes, 50 ng of GST-Rb791-928 and 5 μCi [γ-32P] ATP were added in the reaction mixture which was then incubated at 30 °C for another 15 minutes. Proteins in the reaction mixture were separated by SDS-PAGE, and the incorporation of 32P into GST-Rb791-92833 was quantitatively evaluated using a PhosphorImager (Molecular Dynamics). The concentrations of AR proteins were determined using absorbance at 280 nm in the kinase buffer and the molar extinction coefficients from ProtParam at ExPASY. The IC50 value was defined as the concentration of kinase inhibitor required for 50% of the maximal inhibition of CDK4, and measurements were repeated in triplicate.
Recombinant CDK4/cyclin D2 holoenzyme was expressed and purified as previously described7, 16. Briefly, human CDK4 and cyclin D2 cDNAs were cloned into pBacBAK8 and pBacBAK6 vectors, respectively. Of note, a Hisx6 tag was fused to the C-terminus of CDK4 to facilitate the following purification of the CDK4/cyclin D2 holoenzyme. Each construct was co-transfected into Spodoptera frugiperda SF-9 cells with Autographa California nuclear polyiruhedrosis virus BacPAK6/Bsu-361 DNA (BD Clontech) to generate baculovirus particles. Both baculovirus particles were co-transfected into HighFive insect cells (Invitrogen), and the CDK4/cyclin D2 holoenzyme was purified through affinity chromatography using Talon resin (BD Clontech). The final product was concentrated to approximately 0.3 mg/mL in the above kinase buffer, and aliquots were stored at −80 °C.
Circular dichroism (CD) analyses of gankyrin proteins
In GdnHCl-induced unfolding, recombinant gankyrin proteins were dissolved in 20 mM sodium borate buffer (pH 7.4) containing 40 μM DTT and dialyzed against this borate buffer at 4 °C overnight. Samples containing 7.5–10.0 μM proteins were incubated with different amounts of GdnHCl (in a stock solution of 8.5 M) on ice overnight and then equilibrated at 25 °C just prior to CD analysis. The rotation at 222 nm was measured on an AVIV far-UV spectropolarimeter using a quartz microcell (Helma) of 0.1 cm light pass length, and the exact concentrations of GdnHCl were determined using its refractive index, and three scans were averaged. In this study, the ellipticity at 222 nm, an indicator of the existence of α–helical secondary structure was taken as the measure of the degree of structure present in the protein at each GdnHCl concentration, and the free energy of protein denaturation in aqueous condition was obtained on the basis of two-state approximation25. To determine the protein concentration, absorbance at 280 nm was measured in the borate buffer, and the molar extinction coefficient for each protein was determined using ProtParam at ExPASY.
Heat-induced unfolding experiments were performed using 10 μM proteins in the borate buffer (pH 7.4) with 1 nm bandwidth and 10 second response time. Thermal melting spectra were recorded at 222nm by heating from 3 °C to 65 °C at the rate of 1 °C per minute and a 1 °C interval and then cooling down to 3 °C at the same rate. Tm was defined as the temperature at the midpoint of transition26. The heat-induced unfolding of these gankyrin proteins was highly reversible, and for each protein, more than 90% of the CD signal was recovered after returning to the initial temperature.
NMR analyses
Unlabeled WT, I79D, and L62H, and uniformly 15N-labeled I79D proteins were prepared as previously described21. The NMR samples contained 4 mM HEPES, 1 mM DTT, and 5 μM EDTA in 95% H2O/5% 2H2O at pH 7.5, and about 0.4 mM protein. All NMR experiments, including 2D 1H-15N HSQC, 2D 1H-15N HMBC, 2D 1H homonuclear NOESY, and 3D 15N-edited NOESY, were performed at 27 °C, on a Bruker DRX-600 or Bruker DRX-800 spectrometer equipped with cryoprobe. The NOE mixing time was 150 ms. Data were processed with NMRPipe34 and analyzed with NMRView35.
Bioinformatics analysis
All structural modeling and comparisons were performed using DS VISUALIZER (ACCELRYS) and MOLMOL. The molar extinction coefficient of gankyrin was calculated using ProtParam from ExPASY.
Acknowledgments
We thank Sandeep Kumar for critical reading of this manuscript. This work was supported by Research Grants from NIH (RO1 CA69472 to M.-D. T, and RO1 DE011943 and R21 DE016361 to C. M. W.).
Abbreviations
- AR
ankyrin repeat
- BSA
bovine serum albumin
- CD
circular dichroism
- CDK4
cyclin-dependent kinase 4
- D1/2
the denaturant concentration at the midpoint of transition during chemical-induced unfolding
- G bead
reduced glutathione-agarose (Sigma)
- GdnHCl
guanidinium hydrochloride
- GST
glutathione-S-transferase
- HMBC
heteronuclear multiple-bond correlation
- HSQC
heteronuclear single-quantum coherence
- INK4
inhibitors of CDK4
- IPTG
isopropyl-β-D-thiogalactopyranoside
- NMR
nuclear magnetic resonance spectroscopy
- NOE
nuclear overhauser enhancement
- NOESY
NOE spectroscopy
- P16
specific CDK4 inhibitor p16INK4A, also called CDKNA2 or MTS1
- Rb
retinoblastoma susceptible gene product
- Tm
the temperature at the midpoint of transition during thermal denaturation
- WT
wild type
Footnotes
This is Paper 3 in the Structure, stability and specificity of gankyrin series. For Papers 1 and 2, see References 7 and 20.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sedgwick SG, Smerdon SJ. The ankyrin repeat: a diversity of interactions on a common structural framework. Trends Biochem Sci. 1999;24:311–316. doi: 10.1016/s0968-0004(99)01426-7. [DOI] [PubMed] [Google Scholar]
- 2.Mosavi LK, Cammett TJ, Desrosiers DC, Peng Z-y. The ankyrin repeat as molecular architecture for protein recognition. Protein Sci. 2004;13:1435–1448. doi: 10.1110/ps.03554604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li J, Mahajan A, Tsai MD. Ankyrin repeat: a unique motif mediating protein-protein interactions. Biochemistry. 2006;45:15168–15178. doi: 10.1021/bi062188q. [DOI] [PubMed] [Google Scholar]
- 4.Sherr CJ, Roberts JM. Living with or without cyclins and cyclin-dependent kinases. Genes Dev. 2004;18:2699–2711. doi: 10.1101/gad.1256504. [DOI] [PubMed] [Google Scholar]
- 5.Ortega S, Malumbres M, Barbacid M. Cyclin D-dependent kinases, INK4 inhibitors and cancer. Biochim Biophys Acta. 2002;1602:73–89. doi: 10.1016/s0304-419x(02)00037-9. [DOI] [PubMed] [Google Scholar]
- 6.Dawson S, Apcher S, Mee M, Higashitsuji H, Baker R, Uhle S, Dubiel W, Fujita J, Mayer RJ. Gankyrin is an ankyrin-repeat oncoprotein that interacts with CDK4 kinase and the S6 ATPase of the 26 S proteasome. J Biol Chem. 2002;277:10893–10902. doi: 10.1074/jbc.M107313200. [DOI] [PubMed] [Google Scholar]
- 7.Li J, Tsai MD. Novel insights into the INK4-CDK4/6-Rb pathway: counter action of gankyrin against INK4 proteins regulates the CDK4-mediated phosphorylation of Rb. Biochemistry. 2002;41:3977–3983. doi: 10.1021/bi011550s. [DOI] [PubMed] [Google Scholar]
- 8.Higashitsuji H, Itoh K, Nagao T, Dawson S, Nonoguchi K, Kido T, Mayer RJ, Arii S, Fujita J. Reduced stability of retinoblastoma protein by gankyrin, an oncogenic ankyrin-repeat protein overexpressed in hepatomas. Nat Medicine. 2000;6:96–99. doi: 10.1038/71600. [DOI] [PubMed] [Google Scholar]
- 9.Higashitsuji H, Higashitsuji H, Itoh K, Sakurai T, Nagao T, Sumitomo H, Masuda T, Dawson S, Shimada Y, Mayer RJ, Fujita J. The oncoprotein gankyrin binds to MDM2/HDM2, enhancing ubiquitylation and degradation of p53. Cancer Cell. 2005;8:75–87. doi: 10.1016/j.ccr.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 10.Lozano G, Zambetti GP. Gankyrin: An intriguing name for a novel regulator of p53 and Rb. Cancer Cell. 2005;8:3–4. doi: 10.1016/j.ccr.2005.06.014. [DOI] [PubMed] [Google Scholar]
- 11.Dawson S, Higashitsuji H, Wilkinson AJ, Fujita J, Mayer RJ. Gankyrin: a new oncoprotein and regulator of pRb and p53. Trends Cell Biol. 2006;16:229–233. doi: 10.1016/j.tcb.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 12.Nagao T, Higashitsuji H, Nonoguchi K, Sakurai T, Dawson S, Mayer RJ, Itoh K, Fujita J. MAGE-A4 interacts with the liver oncoprotein gankyrin and suppresses its tumorigenic activity. J Biol Chem. 2003;278:10668–10674. doi: 10.1074/jbc.M206104200. [DOI] [PubMed] [Google Scholar]
- 13.Fu XY, Wang HY, Tan L, Liu SQ, Cao HF, Wu MC. Overexpression of p28/gankyrin in human hepatocellular carcinoma and its clinical significance. World J Gastroenterol. 2002;8:638–643. doi: 10.3748/wjg.v8.i4.638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lim IK. Spectrum of molecular changes during hepatocarcinogenesis induced by DEN and other chemicals in Fisher 344 male rats. Mech Ageing Dev. 2003;124:697–708. doi: 10.1016/s0047-6374(03)00010-1. [DOI] [PubMed] [Google Scholar]
- 15.Russo AA, Tong L, Lee J, Jeffery PD, Pavletich NP. Structural basis for inhibition of the cyclin-dependent kinase Cdk6 by the tumour suppressor p16INK4a. Nature. 1998;395:237–243. doi: 10.1038/26155. [DOI] [PubMed] [Google Scholar]
- 16.Byeon IJ, Li J, Ericson K, Selby T, Tevelev A, Kim HJ, O’Maille P, Tsai MD. Tumor suppressor p16INK4A: determination of solution structure and analyses of its interaction with cyclin-dependent kinase 4. Mol Cell. 1998;1:421–431. doi: 10.1016/s1097-2765(00)80042-8. [DOI] [PubMed] [Google Scholar]
- 17.Li J, Byeon I-J, Ericson K, Poi MJ, O’Maille P, Selby T, Tsai M-D. Tumor suppressor INK4: determination of the solution structure of p18INK4C and demonstration of the functional significance of loops in p18INK4C and p16INK4A . Biochemistry. 1999;38:2930–2940. doi: 10.1021/bi982286e. [DOI] [PubMed] [Google Scholar]
- 18.Li J, Poi MJ, Qin D, Selby T, Byeon IJ, Tsai MD. Tumor suppressor INK4: quantitative structure-function analyses of p18INK4C as an inhibitor of cyclin-dependent kinase 4. Biochemistry. 2000;39:649–657. doi: 10.1021/bi991281u. [DOI] [PubMed] [Google Scholar]
- 19.Sotillo R, Dubus P, Martin J, de la Cueva E, Ortega S, Malubres M, Barbacud M. Wide spectrum of tumors in knock-in mice carrying a Cdk4 protein insensitive to INK4 inhibitors. EMBO J. 2001;20:6637–6647. doi: 10.1093/emboj/20.23.6637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mettus RV, Rane SG. Characterization of the abnormal pancreatic development, reduced growth and infertility in Cdk4 mutant mice. Oncogene. 2003;22:8413–8421. doi: 10.1038/sj.onc.1206888. [DOI] [PubMed] [Google Scholar]
- 21.Yuan C, J Li, et al. Solution structure of the human oncogenic protein gankyrin containing seven ankyrin repeats and analysis of its structure--function relationship. Biochemistry. 2004;43:12152–12161. doi: 10.1021/bi049116o. [DOI] [PubMed] [Google Scholar]
- 22.Krzywda S, Brzozowski AM, Higashitsuhi H, Fujita J, Welchman R, Dawson S, Mayer RJ, Wilkinson AJ. The crystal structure of gankyrin, on oncoprotein found in complexes with cyclin-dependent kinase 4, a 19S proteasomal ATPase regulator, and the tumor suppressors Rb and P53. J Biol Chem. 2004;279:1541–1545. doi: 10.1074/jbc.M310265200. [DOI] [PubMed] [Google Scholar]
- 23.Padmanabhan B, Adachi N, Kataoka K, Horikoshi M. Crystal structure of the homolog of the oncoprotein gankyrin, an interactor of Rb and Cdk4/6. J Biol Chem. 2004;279:1546–1552. doi: 10.1074/jbc.M310266200. [DOI] [PubMed] [Google Scholar]
- 24.Manjasetty BA, Quedenau C, Sievert V, Bussow K, Niesen F, Delbruck H, Heinemann U. X-ray structure of human gankyrin, the product of a gene linked to hepatocellular carcinoma. Proteins. 2004;55:214–217. doi: 10.1002/prot.20028. [DOI] [PubMed] [Google Scholar]
- 25.Pace CN. Determination and analysis of urea and guanidine hydrochloride denaturation curves. Methods Enzymol. 1986;131:266–280. doi: 10.1016/0076-6879(86)31045-0. [DOI] [PubMed] [Google Scholar]
- 26.Zhang B, Peng Z-y. A minimum folding unit in the ankyrin repeat protein p16(INK4) J Mol Biol. 2000;299:1121–1132. doi: 10.1006/jmbi.2000.3803. [DOI] [PubMed] [Google Scholar]
- 27.Devi VS, Binz HK, Stumpp MT, Pluckthun A, Bosshard HR, Jelesarov I. Folding of a designed simple ankyrin repeat protein. Protein Sci. 2004;13:2864–2870. doi: 10.1110/ps.04935704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tevelev A, Byeon IJ, Selby T, Ericson K, Kim HJ, Kraynov V, Tsai MD. Tumor suppressor p16INK4A: structural characterization of wild-type and mutant proteins by NMR and circular dichroism. Biochemistry. 1996;35:9475–9487. doi: 10.1021/bi960211+. [DOI] [PubMed] [Google Scholar]
- 29.Mello CC, Barrick D. An experimentally determined protein folding energy landscape. Proc Natl Acad Sci USA. 2004;101:14102–14107. doi: 10.1073/pnas.0403386101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tripp KW, Barrick D. The tolerance of a modular protein to duplication and deletion of internal repeats. J Mol Biol. 2004;344:169–178. doi: 10.1016/j.jmb.2004.09.038. [DOI] [PubMed] [Google Scholar]
- 31.Tripp KW, Barrick D. Enhancing the stability and folding rate of a repeat protein through the addition of consensus repeats. J Mol Biol. 2007;365:1187–1200. doi: 10.1016/j.jmb.2006.09.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakamura Y, Nakanok K, Umechara T, Kimura M, Hayashizaki Y, Tanaka A, Horikoshi M, Padmanabhan B, Yokoyama S. Structure of the oncoprotein gankyrin in complex with S6 ATPase of the 26S proteasome. Structure. 2007;15:179–189. doi: 10.1016/j.str.2006.11.015. [DOI] [PubMed] [Google Scholar]
- 33.Li J, Joo S-H, Tsai MD. An NF-kappaB-specific inhibitor, IkappaBalpha, binds to and inhibits cyclin-dependent kinase 4. Biochemistry. 2003;42:13476–13483. doi: 10.1021/bi035390r. [DOI] [PubMed] [Google Scholar]
- 34.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 35.Johnson BA, Blevins RA. NMRView: A computer program for the visualization and analysis of NMR data. J Biomol NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- 36.Hondal RJ, Riddle SR, Kravchuk AV, Zhao Z, Liao H, Bruzik KS, Tsai MD. Phosphatidylinositol-specific phospholipase C: kinetic and stereochemical evidence for an interaction between arginine-69 and the phosphate group of phosphatidylinositol. Biochemistry. 1997;36:6633–6642. doi: 10.1021/bi962866g. [DOI] [PubMed] [Google Scholar]
- 37.Zeeb M, Rosner H, Wojciech Z, Canet D, Holak TA, Balbach J. Protein folding and stability of human CDK inhibitor p19INK4D. J Mol Biol. 2002;315:447–457. doi: 10.1006/jmbi.2001.5242. [DOI] [PubMed] [Google Scholar]