Abstract
We used deletion mutants to study β-catenin function in axon arborization of retinal ganglion cells (RGCs) in live Xenopus laevis tadpoles. A deletion mutant βcatΔARM consists of the N- and C-terminal domains of wild-type β-catenin that contain, respectively, α-catenin and postsynaptic density-95 (PSD-95)/discs large (Dlg)/zona occludens-1 (ZO-1) (PDZ) binding sites but lacks the central armadillo repeat region that binds cadherins and other proteins. Expression of ΔARM in RGCs of live tadpoles perturbed axon arborization in two distinct ways: some RGC axons did not form arbors, whereas the remaining RGC axons formed arbors with abnormally long and tangled branches. Expression of the N- and C-terminal domains of β-catenin separately in RGCs resulted in segregation of these two phenotypes. The axons of RGCs overexpressing the N-terminal domain of β-catenin developed no or very few branches, whereas axons of RGCs overexpressing the C-terminal domain of β-catenin formed arbors with long, tangled branches. Additional analysis revealed that the axons of RGCs that did not form arbors after overexpression of ΔARM or the N-terminal domain of β-catenin were frequently mistargeted within the tectum. These results suggest that interactions of the N-terminal domain of β-catenin with α-catenin and of the C-terminal domain with PDZ domain-containing proteins are required, respectively, to initiate and shape axon arbors of RGCs in vivo.
Keywords: β-catenin, α-catenin, PDZ proteins, lipofection, axon arborization, axon branching, retinal ganglion cells, Xenopus laevis
Introduction
In many types of neurons, axons branch or arborize when they reach their target tissue. Axon arborization is spatiotemporally correlated with neuron-target interactions (Sakaguchi and Murphey, 1985; Yates et al., 2001) and with synapse formation and function (Cantallops et al., 2000; Alsins et al., 2001), but in contrast to neuron-target interactions and synaptogenesis, we know very little about the molecules that control axon arborization, with a few notable exceptions (Cohen-Cory and Fraser, 1995; Wang et al., 1999; Zou and Cline, 1999).
β-catenin participates in several molecular interactions that could regulate axon arborization. First, β-catenin links the cadherin family of cell adhesion molecules to α-catenin and the cytoskeleton, thereby strengthening adhesion (for review, see Ivanov et al., 2001; Schneider et al., 2003). N-cadherin, β-catenin, and α-catenin are expressed in axons (Murphy-Erdosh, 1994; Riehl et al., 1996; Benson and Tanaka, 1998) and at synapses (Uchida et al., 1996; Benson and Tanaka, 1998; Miskevich et al., 1998). N-cadherin is required for normal axonal targeting and arborization (Inoue and Sanes, 1997; Lee et al., 2001), as well as for normal synapse formation and function (Tang et al., 1998; Togashi et al., 2002). We do not know currently whether β-catenin is also required for these processes. At the synapse, β-catenin interacts with at least two postsynaptic density-95 (PSD-95)/discs large (Dlg)/zona occludens-1 (ZO-1) (PDZ) proteins, Lin-7/Velis and synaptic scaffolding molecule (S-SCAM) (Perego et al., 2001; Nishimura et al., 2002). Together with cadherins, these proteins recruit and localize proteins at synaptic junctions (Perego et al., 2001; Nishimura et al., 2002). Here we use deletion mutants to examine how disrupting the interactions mediated by the N- and C-terminal domains of β-catenin, which interact, respectively, with α-catenin and synaptic PDZ proteins, specifically affects stages in axonal arborization.
We have studied β-catenin function in axonal arborization of RGCs in tecta of live Xenopus tadpoles. An advantage of this system is that the effects of β-catenin on arborization of single axons in vivo can be examined over several days (O'Rourke and Fraser, 1990; Cohen-Cory and Fraser, 1995; Zou and Cline, 1999). We first show using Xenopus brain sections that β-catenin is expressed in RGC axon arbors. We then express deletion mutants of β-catenin in RGCs and examine their effects on the formation of axonal arbors during development in vivo. These experiments suggest that interactions mediated by the N- and C-terminal domains of β-catenin, which contain α-catenin and PDZ interaction sites, respectively, initiate and shape the axonal arbors of RGCs in vivo.
Materials and Methods
Xenopus laevis tadpoles. Xenopus laevis embryos were generated by in vitro fertilization of eggs obtained from females primed with human chorionic gonadotrophin. Embryos were cultured in a 10% modified Ringer's solution (MMR) and staged according to Nieuwkoop and Faber (1956); 0.001% phenylthiocarbamide was added to the Ringer's solution to reduce pigmentation (Cohen-Cory and Fraser, 1995).
Antibody staining. Brains with and without green fluorescent protein (GFP)-expressing retinal ganglion cell (RGC) axon arbors were dissected from stage 45/46 tadpoles. Tadpoles were anesthetized in a 0.02% benzocaine solution before dissection. The dissected brains were fixed overnight at 4°C in a freshly made 4% paraformaldehyde solution and sunk in sucrose over 2 nights (15 and 30% sucrose in phosphate buffer). The brains were then embedded in an optimal cutting temperature compound mold, cryostat sectioned into 30-μm-thick horizontal sections, and mounted onto glass slides. Antibody staining was performed as follows. Sections were rinsed in PBS for 5 min, blocked with normal goat serum [5% in PBS plus 0.1% Triton X-100 (PBST)] for 30 min at room temperature, and incubated overnight at 4°C with a polyclonal rabbit anti-β-catenin antibody generated against Xenopus β-catenin (1:500 in PBST) (Kypta et al., 1996). Sections were then rinsed in PBS three times for 8 min and incubated with a Texas Red-tagged goat anti-rabbit secondary antibody (1:200 in PBST) (Molecular Probes, OR) for 1 hr at room temperature. Control sections underwent the same procedure but were incubated with only the Texas Red secondary antibody.
To determine whether β-catenin staining colocalized with GFP axon arbors, we followed a procedure similar to the one described previously by Pinches and Cline (1998). Antibody-stained tectal sections (30 μm) containing a single GFP arbor were imaged with a confocal microscope (Bio-Rad 600) using a 100× objective [Zeiss Plan Apochromat; numerical aperture (NA) = 1.40] and a z-series protocol (z-interval 0.5-1.0 μm). We then opened each optic slice individually in NIH image (Version 1.62) and traced the portion of the GFP arbor in that optic slice. We noted all yellow β-catenin puncta that colocalized with the GFP arbor in that particular optic slice. Two criteria were used to define a yellow spot as a countable puncta. First, in the image containing the green (GFP)/red (anti-β-catenin) overlay, we counted only yellow spots of >1 μm diameter. Second, we counted only those spots with intensity in the red channel that was twofold greater than that of the average intensity in the neuropil region of the corresponding control section (stained only with a secondary antibody). All of the individual tracings for a single tectal section were combined into one image, and thus the arbor and corresponding β-catenin staining were reconstructed. In all cases, this constituted a partial reconstruction of the total arbor, comprising only that portion of arbor present in the 15 μm of the tectal section that had optimal antibody penetration. In each of these reconstructed GFP arbors, we counted the total number of branches, measured the length of each of the branches, and counted the number of β-catenin puncta in each of the branches. From these data, we calculated the average number of β-catenin puncta per 10 μm of branch length. To confirm the specificity of overlap of red β-catenin puncta with the green GFP arbor branches, we mismatched the red and green images along the y-axis and then calculated the number of yellow puncta in the mismatched images. On average, mismatched images contained 33% of the number of yellow puncta as did the correctly matched images.
Lipofection. To transfect RGCs with DNA constructs, we used the lipofection protocol first developed for studying axon outgrowth by Xenopus RGCs (Holt et al., 1990; Riehl et al., 1996). Briefly, 24 hr after fertilization, at stages 20-23, we manually removed vitteline envelopes from embryos and transferred the embryos to a high salt solution (1× MMR). We back filled micropipettes with a mixture of DNA and DOTAP lipofection reagent (at a ratio of 1:3) and pressure injected 50-200 nl of the DNA-DOTAP solution into eye bud primordia. We then transferred embryos to low-salt 0.1× MMR containing 0.001% phenylthiocarbamide to inhibit pigmentation. Embryos were raised in the dark at 23°C for 4 d (until stage 45/46). Expression of proteins has been shown to peak 4 d after lipofection (Holt et al., 1990). This corresponds to stage 45/46, by which time RGC axons have reached the tectum and are forming arbors.
Arbor imaging. At stage 45/46, the heads of tadpoles are transparent, so that GFP-expressing axon arbors can be imaged in live animals using fluorescent illumination. For screening and confocal imaging of arbors, we anesthetized tadpoles in a 0.02% benzocaine solution and then mounted tadpoles in a chamber made of silicone and sealed with a coverslip. To screen tadpoles for GFP arbors, we used a 10× objective (Nikon Plan Apo; NA = 0.45) on an upright microscope (Nikon Microphot-FXA). For each tadpole that contained a GFP arbor we drew a rough sketch of the morphology of the arbor and its location in the tectum. We confirmed that GFP and GFPΔARM-expressing arbors originated from cells located in the contralateral retina by following several brighter axons back to the contralateral eye. Animals containing fluorescent arbors were taken to an inverted confocal microscope (Bio-Rad 600) and imaged using a 40× objective (Zeiss Plan Neofluor; NA = 0.75). Some of the arbors were confocal imaged at 24 hr intervals over a 2 or 3 d period. All of the arbors that we imaged [the thicker, unbranched axons expressing GFPΔARM and GFP tagged to the N-terminal domain of β-catenin (GFPNTERM), as well as the thinner, branched arbors expressing GFP, GFPΔARM and GFP tagged to the C-terminal domain of β-catenin (GFPCTERM)] were located at a relatively superficial depth within the tectum.
Arbor analysis. Confocal images were projected using the NIH Image (version 1.62) projection algorithm. From this projected image we counted the number of branches in the arbors and measured the total length of arbors. By using the projection algorithm in NIH image, we underestimate both the number of branch tips and the length of arbors, relative to other studies that manually reconstructed arbors by tracing each individual slice (Zou and Cline, 1996).
DNA plasmid construction. The β-catenin deletion mutant ΔARM contains amino acid residues 1-151 of β-catenin fused to residues 648-781. NTERM contains residues 1-151 of β-catenin, and CTERM contains residues 648-781 of β-catenin.
All constructs, including GFP, were subcloned into the Xenopus expression vector pCS2 [originally constructed by D. Turner (University of Michigan) and R. Rupp (Max-Planck-Institute, Tuebingen, Germany)]. ΔARM and CTERM were constructed previously in our laboratory from a full-length Xenopus β-catenin that had a myc-epitope tag fused to its C terminus and were subcloned into the expression vector pEGFP-C1 (Clontech) (Kypta et al., 1996). We subcloned ΔARM and CTERM (with GFP at their N termini and myc tags at their C termini) from pEGFP-C1 into pCS2 using NheI and XbaI. To make pCS2ΔARM without GFP we subcloned ΔARM (with the myc tag) from peGFP-C1 into pCS2 using EcoRI. To make CTERM without GFP or a myc tag, we performed PCR on a full-length PBS Xenopus β-catenin (without a myc tag) and subcloned the resulting PCR fragment into the pCR-Blunt II-TOPO vector. We then subcloned CTERM (no myc) from pCR Blunt II-TOPO into pCS2 using XbaI.
To make pCS2-GFPNTERM we digested peGFP-C1-β-catenin (full length) with XhoI and then subcloned the Xho fragment (containing the N-terminal domain of β-catenin) into peGFP-Cl. We then subcloned GFPNTERM out of pEGFP-C1 into PCS2 with NheI and XbaI.
Results
β-catenin is expressed in RGC axon arbors in the tectum
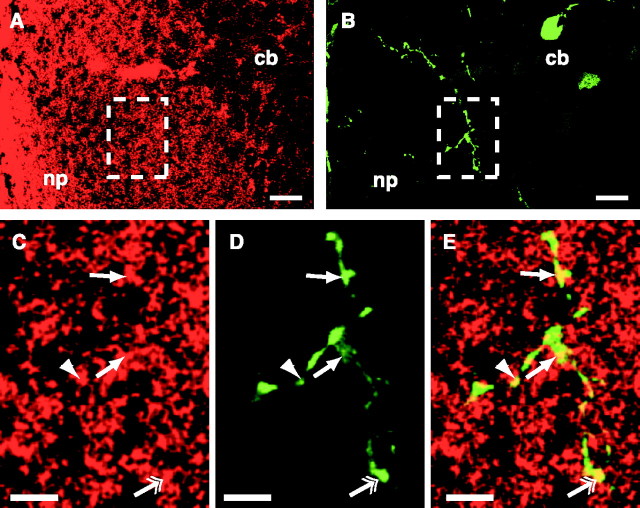
To determine whether β-catenin was expressed in RGC axon arbors in their target tissue, the tectal midbrain, sections from brains of stage 45/46 tadpoles were stained with anti-β-catenin. Results showed β-catenin expression throughout the neuropil region of the tectum (Fig. 1A) (tectal sections from six different brains showed similar staining patterns). Within the neuropil, β-catenin was expressed in a nonuniform pattern, with the strongest, most dense expression in the lateral neuropil (Fig. 1A). This corresponds to the outermost region of the tectum that is first invaded by RGC axons. In the more medial neuropil, the region of the tectum in which RGC axons form arbors, the expression of β-catenin appeared more sparse and punctate (Fig. 1A,C). N-cadherin is also expressed in a similar pattern in the neuropil region of the Xenopus tectum (Riehl et al., 1996).
Figure 1.
β-catenin is expressed in RGC axon arbors in the tectum. Confocal image (single optic section) of a tectal section taken from a stage 45/46 tadpole stained with anti β-catenin antibody (red) shows expression in the neuropil area, the target region for RGC axons (A). Confocal image of a GFP-expressing RGC axon arbor (green) in the same optic section shows that the axon arbor is in the neuropil region of the tectal section (B). Higher magnification zoom of boxed regions in A and B shows that β-catenin staining is punctate in the neuropil (C) and that the GFP-expressing RGC axon arbor contains β-catenin puncta (E, arrows, double arrow, arrowhead). β-catenin puncta are located at branch points (D, arrows), in growth cones (D, double arrow), and along branch shafts (D, arrowhead). np, Neuropil region of the tectum; cb, cell body region of the tectum. Scale bars: A, B,10 μm; C-E, 5 μm.
To examine directly whether RGC axon arbors in the tectum contained β-catenin puncta, single RGC axon arbors were labeled with GFP (Fig. 1B,D). Tectal sections containing a single GFP RGC axon arbor were then stained for β-catenin and imaged on a confocal microscope. Examination of these images revealed specific (yellow) puncta of overlap between the green GFP-labeled RGC arbors and red-stained β-catenin puncta (Figs. 1C-E). The density of β-catenin puncta in RGC axon arbors was 3.4 ± 0.85 β-catenin puncta per 10 μm of branch length (440 μm of arbor length analyzed, 100 μm or more per arbor). Within the RGC axon arbors, β-catenin puncta were found at branch points (Fig. 1E, arrows), along shafts of branches (Fig. 1E, arrowhead), and in branch tips (Fig. 1E, double arrow). To confirm the specificity of overlap of red β-catenin puncta with the green GFP arbor branches, we mismatched the red and green images along the y-axis and then compared the number of yellow puncta in the mismatched images with that in correctly matched red and green images. On average, mismatched images contained 33% of the number of yellow puncta as observed in the correctly matched images (see Materials and Methods). In summary, these data demonstrate that β-catenin is expressed in RGC axon arbors in the tectum. Thus, β-catenin may play a role in RGC axon arborization.
Perturbation of β-catenin function does not inhibit RGC axon outgrowth to the tectum
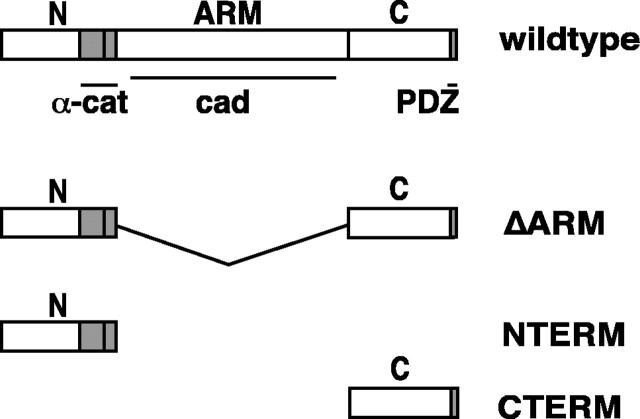
To perturb β-catenin function in RGCs, we used a deletion mutant of β-catenin named ΔARM that is a fusion of the N-terminal and C-terminal domains of β-catenin (Fig. 2). The N-terminal domain of β-catenin contains a highly conserved subdomain that mediates interactions with α-catenin that in turn associates with the F-actin cytoskeleton (Ivanov et al., 2001; Schneider et al., 2003). The interactions between β-catenin and α-catenin are required for strong cadherin-based cell-cell adhesion (Ivanov et al., 2001). The C-terminal domain contains a PDZ binding motif that has been shown to interact with two synaptic PDZ proteins (Perego et al., 2001; Nishimura et al., 2002). ΔARM lacks the central armadillo repeat region of β-catenin that contains interaction sites for cadherins and several proteins that mediate Wnt signaling, including APC (adenomatous polyposis coli) and TCF/LEF (T-cell factor/lymphoid enhancing factor)(Hulsken et al., 1994; Gottardi and Gumbiner, 2001; Pokutta and Weis, 2002). Previous studies have shown that ΔARM does not activate (or interfere with) Wnt signaling when expressed in cleavage stage Xenopus embryos (Funayama et al., 1995; Sehgal et al., 1997). Because ΔARM is expected to compete with endogenous, full-length β-catenin for binding to α-catenin and PDZ-containing proteins, we hypothesized that overexpression of ΔARM will interfere with recruitment of these proteins to cadherins, thereby perturbing cadherin-based adhesion and recruitment of PDZ-containing synaptic proteins (see Discussion).
Figure 2.
Schematic diagram of mutant β-catenin constructs. Wild-type β-catenin consists of three distinct functional and structural domains: an N-terminal domain, a central core consisting of 12 armadillo repeats, and a C-terminal domain. Within these three domains, β-catenin contains binding sites for α-catenin (N-terminal domain and armadillo repeats R1-R2; amino acid residues 120-150), cadherins (binding spans throughout armadillo repeats R3-R12), and PDZ-domain-containing proteins (C-terminal domain; amino acid residues 777-781). The deletion mutant ΔARM lacks almost the entire armadillo repeat region found in wild-type β-catenin; it consists of the N-terminal residues 1-151 of wild-type β-catenin fused to the C-terminal residues 648-781. Thus, ΔARM retains binding to α-catenin and PDZ-binding proteins but not to cadherin. The deletion mutants NTERM and CTERM are simply the N- and C-terminal domains of β-catenin that together constitute ΔARM. Each protein had GFP fused to its N terminus and was expressed using the Xenopus expression vector pCS2. ΔARM and CTERM also contained a myc-epitope tag at their C termini. We also confirmed the effects of ΔARM without an attached GFP tag and of CTERM without attached GFP or myc tags (see Results).
We first asked whether ΔARM inhibited RGC axon outgrowth from the retina to the tectum. To address this issue we examined whether GFPΔARM-expressing RGC axons were present in the tecta of stage 45/46 tadpoles. In wild-type animals, a significant number of RGC axons have reached the tectum by this stage (Holt, 1984). RGCs lipofected with GFPΔARM extended green fluorescent axons to the tectum at stage 45/46. To determine whether GFPΔARM- and GFP-expressing axons reached the tectum at the same frequency, we lipofected these two plasmids into two groups of 30 tadpoles. By stage 45/46, one-third of the tadpoles in each of the two groups showed green fluorescent RGC axons in the tectum (data not shown). In addition, embryos injected with ΔARM in the eyebud of a single, determined hemisphere extended axons only to the contralateral tectum, similar to embryos expressing GFP alone (eight embryos analyzed; data not shown). Thus, overexpression of ΔARM does not inhibit the extension of RGC axons from the retina to the tectum. These data are consistent with previous findings showing that β-catenin function is not essential for axon outgrowth (Loureiro and Peifer, 1998; Loureiro et al., 2001) and that overexpression of the region of the cytoplasmic tail of cadherin that interacts with β-catenin is not sufficient to inhibit growth of Xenopus RGC axons (Riehl et al., 1996).
βcatΔARM expression perturbs axon arborization of RGCs in vivo in two different ways
Is β-catenin function required for axon arborization of RGCs? Before examining the arbors of RGCs expressing ΔARM, we imaged and measured GFP-expressing RGC axon arbors in live tadpoles. At stage 45/46, arbors containing GFP were moderately branched, with an average branch number of ∼11 and total arbor length of 300 μm (Figs. 3A, 4A, 5A,B; Table 1). In a 24 hr period, these arbors increased their average branch number to 16 and their average total arbor branch length to 350 μm (Figs. 4A, 5B,C, Table 1). These axonal arborization parameters agree with values for wild-type RGC axon arbors at stage 45/46 reported in a previous study (O'Rourke and Fraser, 1990).
Figure 3.
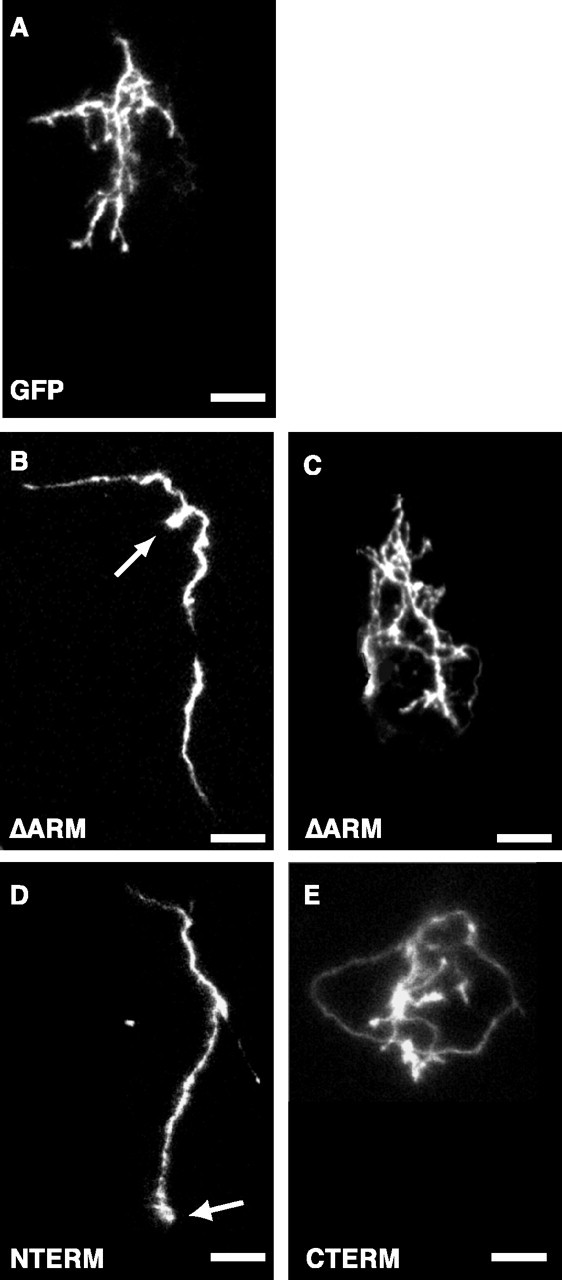
Axonal arborization phenotypes of RGCs expressing β-catenin deletion mutants. Projected confocal z-series images taken of axon arbors in live tadpoles show arbor morphologies for RGCs expressing GFP (A) or GFP-tagged-β-catenin deletion mutants (B-E). RGC axons that express the β-catenin deletion mutant ΔARM have two distinct phenotypes. They either do not arborize (B) or they arborize but contain longer, tangled branches (C). RGC axons expressing the β-catenin deletion mutant NTERM form severely reduced arbors (D) that are similar to the subpopulation of ΔARM arbors with no or few branches (B). RGC axons expressing CTERM form arbors with long, tangled branches (E) that are similar to the subpopulation of ΔARM arbors with effusive branching (C). RGC axons that express ΔARM and NTERM and form severely reduced arbors (B, D) also have bulbous endings at their axon/branch tips (B, D, arrows) and appear thicker than RGC arbors that express GFP, ΔARM, and CTERM (compare axon/branch thicknesses in B, D with those in A, C, E). Note that at the level of magnification of these projected confocal z-series (40×), all of the GFP-tagged-β-catenin deletion mutant constructs appear to be uniformly distributed within the axon terminals and arbors. Scale bars: A-E, 20 μm.
Figure 4.
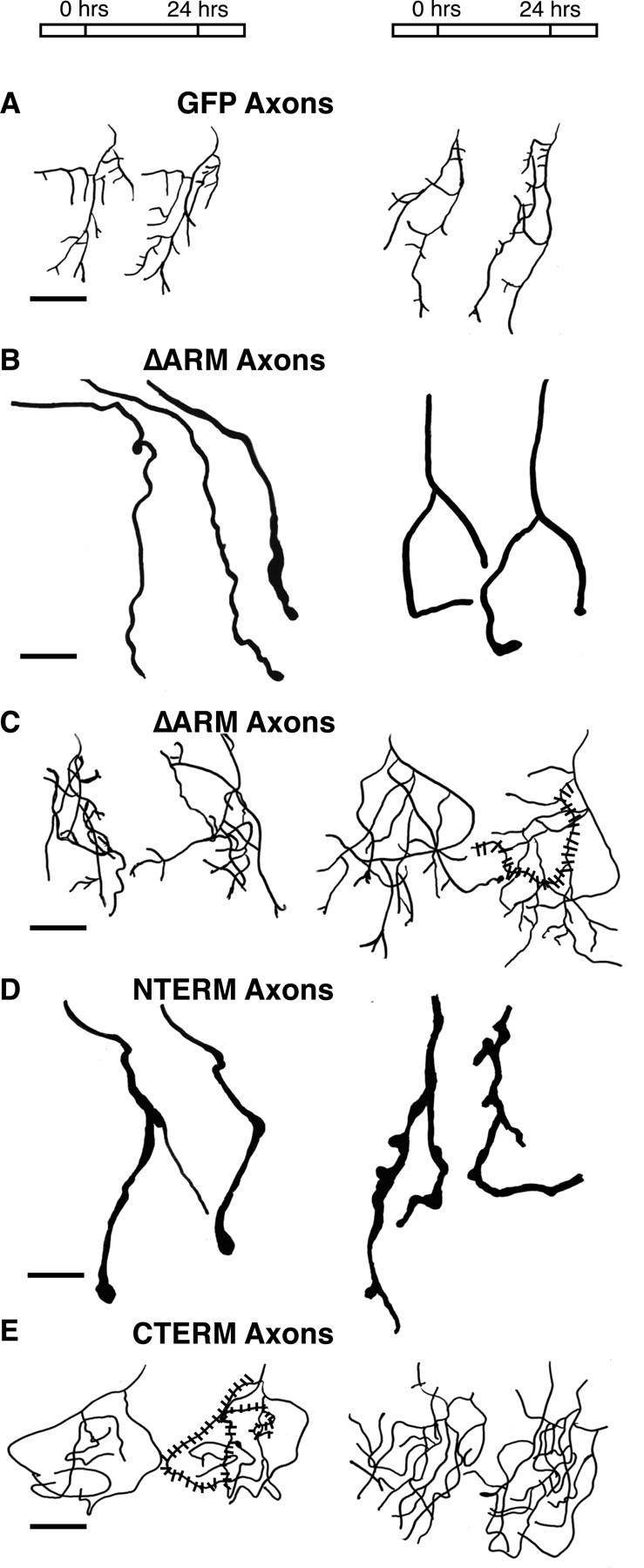
Tracings of projected confocal z-series of RGC axon arbors expressing β-catenin deletion mutants confirm morphological phenotypes. Compared with controls, RGC axons that express ΔARM have two distinct phenotypes. They either do not arborize (B) or they arborize but have abnormally long and tangled branches (C). NTERM-expressing arbors (D) are similar to the subpopulation of ΔARM arbors with no or few branches (B). CTERM-expressing arbors (E) are similar to the subpopulation of ΔARM arbors with abnormally long, tangled branches (C). For each condition (A-E), two examples of arbors are shown. In each example of an arbor, two (or three) (B, left) tracings are shown that correspond to confocal images taken ∼ 24 hr apart. The first tracing of each example corresponds to a confocal image taken at stage 45/46. The left most arbor tracing in A-E corresponds, respectively, to the actual confocal z-series projection image shown in Figure 3A-E. Stippling over a single arbor branch in C and E illustrates a particularly tangled branch. Rostral is to the top and lateral is to the right in all images. Scale bars, 20 μm (A-E).
Figure 5.
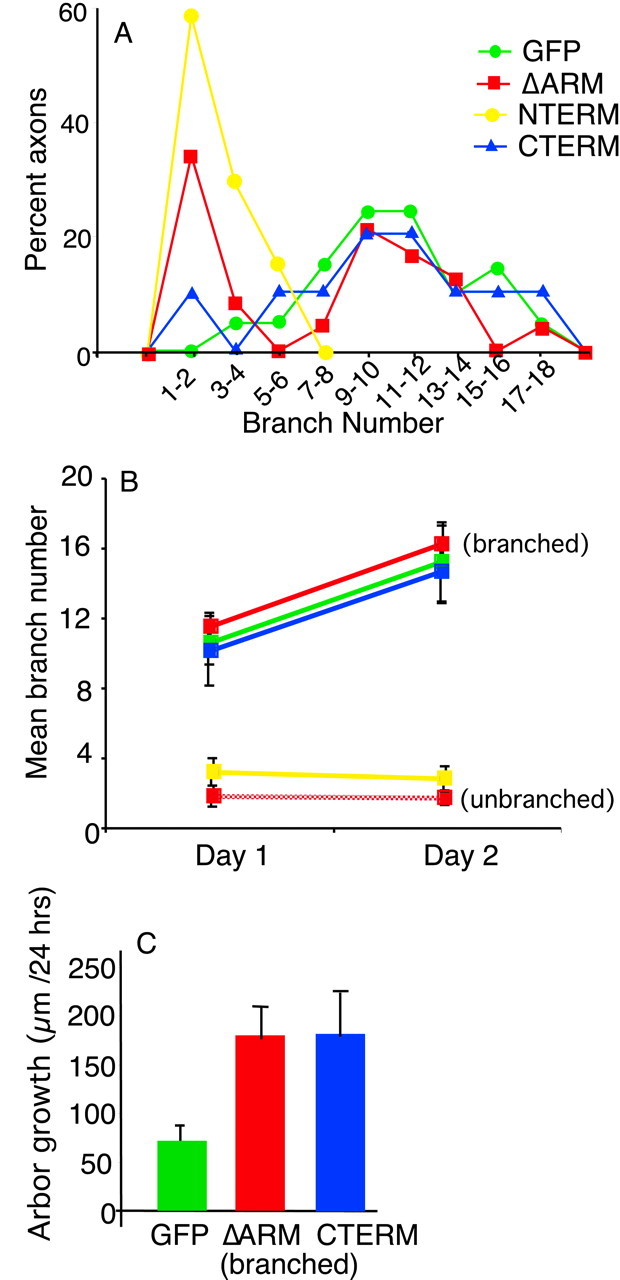
Quantification of morphological parameters for arbors from RGCs expressing β-catenin deletion mutants confirm phenotypes shown in Figures 3 and 4. The distribution of branch numbers in arbors at stage 45 shows that GFP axon arbors are distributed about a mean of ∼11 branches (A). In contrast, ΔARM axons are distributed about two distinct means: ΔARM arbors either have very few (1-2) branches or they have approximately the same number of branches as control GFP arbors (A). In addition, NTERM arbors have no or very few (1-6) branches, whereas CTERM arbors have numbers of branches comparable with GFP arbors (A). Plot of mean numbers of branches in arbors over 2 d shows that GFP arbors acquire more branches over time (B). ΔARM arbors with branches and CTERM arbors behave similarly to GFP arbors (B). In contrast, ΔARM axons without arbors and NTERM axons form few or no additional branches over time (B). Branched ΔARM arbors (with 9 or more branches) and CTERM arbors also grow faster than GFP arbors (C) (p < 0.05 for both, Student's t test). We did not measure the growth rate for unbranched ΔARM and NTERM arbors because many RGCs in these two classes did not form arbors (C). The number of axons analyzed is as follows: A, GFP (21), ΔARM (24), NTERM (7), CTERM (10); B, GFP (10), ΔARM-branched (6), ΔARM-unbranched (6), NTERM (7), CTERM (10), (C) GFP (9), ΔARM-branched (6), CTERM (10).
Table 1.
Total arbor branch length (TABL) in RGCs expressing β-catenin deletion mutants
|
|
TABL (μm) ± SE (n) |
||
|---|---|---|---|
| Construct
|
Day 1
|
Day 2
|
|
| GFP | 319 ± 27 (15) | 385 ± 35 (9) | |
| ΔARM (with branches) | 470 ± 34** | 708 ± 70*(6) | |
| CTERM
|
434 ± 46* (10)
|
615 ± 72*(10)
|
|
RGC axon arbors expressing β-catenin deletion mutants ΔARM and CTERM are longer than controls. Measurements of total arbor length for both branched ΔARM arbors and CTERM arbors over 2 d (day 1 corresponds to stage 45/46) show that they are significantly longer than arbors of control RGCs expressing GFP. Student's t test was used to test for statistical significance of differences. *p < 0.05; **p < 0.005.
Many axons of RGCs that express ΔARM do not form arbors
In contrast to RGC axons expressing GFP alone, approximately half of RGC axons expressing ΔARM failed to form arbors in the tectum (Figs. 3B, 4B, 5A). At stage 45/46, these axons had no or very few (at most three) terminal branches (Figs. 3B, 4B, 5A,B). Moreover, when imaged over several days, we observed that these axons did not form additional branches with time (Figs. 4B, 5B). The first example of an axon shown in Figure 4B shows a representative ΔARM-expressing RGC axon that initially had two branches; during the following 2 d period, this axon did not add additional branches. Thus, in a significant fraction of ΔARM expressing RGCs, axon branching is severely inhibited.
The axons of RGCs expressing ΔARM that formed severely reduced arbors also exhibited other morphological features distinct from those of control axons of RGCs expressing GFP. First, the axons (and a few secondary branches) formed by this subpopulation of ΔARM-expressing RGCs were thicker than the axons and branches formed by control GFP-expressing RGCs (Fig. 3, compare A, B). Second, the tips of the axons and branches formed by these ΔARM-expressing RGCs with reduced arborization often terminated in bulbs or club-like endings (Figs. 3B, arrow, 4B). Both of these features of ΔARM-expressing RGC axons became more pronounced over time. On the second (and third) day of observation, axons of RGCs expressing ΔARM became thicker, and the bulbs at the tips of their branches became larger (Fig. 4B).
Other RGCs overexpressing ΔARM form misshapen arbors
The remaining ΔARM-expressing RGC axons that we imaged in tecta of live tadpoles did form arbors (Figs. 3C, 4C). At stage 45/46, these arbors had comparable numbers of branches to control arbors of RGCs expressing GFP alone (Figs. 3C, 4C, 5A,B). The mean number of branches for ΔARM arbors with branches was ∼12, compared with a mean number of branches of 11 for arbors of controls expressing GFP alone (Fig. 5A,B). Over a 2 d period, these ΔARM arbors increased their total branch number, with an increase comparable with that observed in GFP arbors (Fig. 5B). Thus in this group of arbors, branching per se was not inhibited by expression of ΔARM.
The arbors of axons expressing ΔARM, however, formed branches that were shaped differently than control arbors. First, the total length of axonal branches of arbors expressing ΔARM was greater than the length of control arbors expressing GFP (Figs. 3C, 4C, Table 1). When examined over several days, arbors of ΔARM-expressing RGCs also grew faster than GFP arbors (Figs. 4C, 5C). Second, the branches of these arbors were more tangled than branches of control arbors. In the arbors of RGCs expressing ΔARM, secondary branches crossed back and forth several times across the main axis of the arbor (Fig. 4C, stippling). In contrast, in control arbors, secondary branches extended straight from the main axis of arborization and rarely recrossed this axis (Fig. 4A). Thus, in this group of RGCs, expression of ΔARM resulted in accelerated elongation and more tangled morphology of arbor branches.
The two arborization phenotypes that we observed in GFPΔARM-expressing axons were not caused by the inadvertent expression of GFPΔARM in neurons that were not RGCs. For both phenotypes we followed several bright fluorescent axons to their points of origin in the retinal ganglion cell layer of the contralateral eye. The GFPΔARM arborization phenotypes were also not caused by localization of the fusion protein GFPΔARM to only a portion of the arbor of RGCs. RGCs co-lipofected with GFP and ΔARM in separate plasmids showed the same arborization phenotypes as did GFPΔARM-expressing RGCs [n = 10 RGCs co-lipofected; 4 did not arborize, and 6 arborized but had long, tangled arbors (data not shown)]. Thus, these data implicate β-catenin in axon arborization of RGCs in vivo. They show that β-catenin is required for both the initiation and shaping of RGC arbors.
Overexpression of the N- and C-terminal domain fragments of β-catenin, respectively, inhibit and misshape axon arbors in vivo
ΔARM is a fusion of the N-terminal and C-terminal domains of β-catenin. To determine whether the two distinct arborization phenotypes observed in ΔARM RGCs reflect distinct activities of these two domains, we expressed each domain alone in RGCs (Fig. 2).
RGC axons expressing NTERM had no or severely reduced arborization (Figs. 3D, 4D). At stage 45/46, NTERM RGC axons had very few branches (one to six branches) (Figs. 3D, 4D, 5A,B). Similar to ΔARM axons without branches, NTERM axons did not acquire more branches over time (Figs. 4D, 5B). Instead, the average number of branches formed by these neurons was slightly reduced with time (Fig. 5B). Moreover, NTERM axons were also thicker than control axons and contained bulbs at their terminal ends (Figs. 3D, arrow, 4D). Thus, NTERM axons appeared and behaved very similar to the subpopulation of ΔARM axons without arbors (Fig. 3B,D, compare with Fig. 4B, D).
In contrast, RGCs expressing CTERM formed severely misshapen arbors (Figs. 3E, 4E). At stage 45, CTERM arbors had numbers of branches comparable with those arbors formed by RGCs expressing GFP (Fig. 5A,B). Over 2 d, the mean number of branches formed by RGCs expressing CTERM increased significantly (Fig. 5B). However, similar to the arbors of ΔARM expressing RGCs with extensive branching, the arbors formed by RGCs expressing CTERM were significantly longer and grew faster than arbors formed by control RGCs expressing GFP alone (Figs. 3E, 4E, 5C, Table 1). CTERM arbors were also very tangled with long branches that crossed and overlapped other branches of the arbor (Figs. 3E, stippling, 4E). Thus, expression of CTERM in RGCs generates arbors that resemble those formed by the second class of RGCs expressing ΔARM (Figs. 3, 4, compare C, E).
We confirmed that the presence of overextended, misshapen arbors in RGCs expressing CTERM was not caused by interference of protein interactions by the GFP and myc tags attached to the CTERM construct. Co-lipofection of a modified CTERM without the attached GFP and myc tags (see Materials and Methods) and GFP into RGCs also generated misshapen axon arbors with long, tangled branches (in three of three RGC axons). Consistent with this finding, recent work in our laboratory also indicates that attachment of a myc tag at the C-terminal end of β-catenin does not abolish (although it may weaken) PDZ protein binding to this domain (S. Bamji, N. E. Kimes, and L. F. Reichardt, personal communication).
These results demonstrate that the N- and C-terminal domains of β-catenin have distinct functions in axon arborization. They suggest that interactions mediated through the N-terminal domain of β-catenin are required for formation of arbors by RGC axons in the tectum, whereas interactions mediated by the C-terminal domain are required for normal extension and shaping of arbors.
RGC axons that express the β-catenin deletion mutants Δ ARM and NTERM are frequently mistargeted within the tectum
Previous studies have shown that in Xenopus tadpoles RGC axons do not branch until they reach their topographically appropriate locations in the tectal neuropil (Sakaguchi and Murphey, 1985). This suggests that specific topographic cues are required to promote formation of axonal arbors by RGCs. Thus, one possibility is that ΔARM- and NTERM-expressing RGC axons do not form arbors because they are mistargeted within the tectum. Examination of the tectal projections of the subpopulation of ΔARM-expressing axons without arbors and of NTERM-expressing axons demonstrated that they did not always terminate in a topographically appropriate location (Table 2). Two types of targeting errors were observed. The poorly branched arbors of RGCs expressing ΔARM or NTERM frequently formed abnormally short projections into the tectum, or they had unusually long projections that meandered over the tectum and extended beyond the midline of the tectum, terminating in the ipsilateral tectum (Table 2). In contrast, the well branched arbors formed by RGC neurons expressing GFP, ΔARM, or CTERM appeared to project consistently into and terminate within a relatively central region of the appropriate tectal hemisphere and to thus be correctly targeted (Table 2). In conclusion, some of the poorly branched axons of RGCs expressing ΔARM or NTERM were clearly mistargeted within the tectum.
Table 2.
Mistargeting of RGC axons that express β-catenin deletion mutants
|
|
Types of targeting errors |
|
|
||
|---|---|---|---|---|---|
| Construct
|
Very short axonal projection into tectum (<40 μm along the mediolateral axis)
|
Long axonal projection extending beyond tectal midline
|
Total axons mistargeted within the tectum
|
No. of axons analyzed
|
|
| GFP | 0 | 0 | 0 | 10 | |
| ΔARM (unbranched) | 4 | 2 | 6 | 10 | |
| ΔARM (branched) | 0 | 0 | 0 | 9 | |
| NTERM | 3 | 1 | 4 | 7 | |
| CTERM
|
0
|
0
|
0
|
10
|
|
Discussion
N- and C-terminal domains of β-catenin, respectively, initiate and shape axon arbors in RGCs in vivo
Results presented above show that overexpression of the N- and C-terminal domains of β-catenin, respectively, inhibit and misshape axon arbors in RGCs in the tecta of live Xenopus laevis tadpoles. These results suggest that interactions of the N- and C-terminal domains of β-catenin are required, respectively, to form and shape axon arbors. This evidence implicates two distinct domains of β-catenin in two different processes of axon arborization.
Cadherin-based adhesion and formation of axon arbors in RGCs
Overexpression of β-catΔARM or β-catNTERM in RGCs is expected to interfere with functions mediated by the N-terminal domain of endogenous β-catenin by sequestering proteins that normally bind to this domain. Results in this paper show that interference with interactions mediated by this domain prevents formation of axon arbors by RGCs in the optic tectum. The N-terminal domain of β-catenin has been shown to interact with α-catenin, which in turn links the cadherin-β-catenin complex to the F-actin cytoskeleton, thereby strengthening cadherin-based cell-cell adhesion (Ivanov et al., 2001; Schneider et al., 2003). Thus, the N-terminal domain of endogenous β-catenin may promote axon branching in RGCs by strengthening cadherin-based cell adhesion. How could strong cadherin-mediated cell-cell adhesion promote axon arborization in RGCs? We observed that NTERM- and ΔARM-expressing RGC axons that did not form arbors were also frequently mistargeted within the tectum. Other work has suggested that in Xenopus laevis specific topographic signals in the tectal target promote branching (Sakaguchi and Murphey, 1985). Thus, one possibility is that cadherin-based cell-cell adhesion is required for cell-cell interactions that target RGC axons to their correct location within the tectum. Consistent with this possibility, in two different species, cadherin function has been shown to be required for targeting optic axons within their target tissue (Inoue and Sanes, 1997; Lee et al., 2001).
The N-terminal domain of β-catenin also contains a glycogen synthase kinase-3β (GSK-3β) binding site, and several phosphorylation sites that regulate ubiquitination and degradation of the protein (Schneider et al., 2003). Thus another possibility for the mechanism of action of the overexpressed N-terminal domain of β-catenin is that it sequesters GSK-3β and the ubiquitination machinery away from endogenous, full-length β-catenin. This could conceivably reduce the normal level of phosphorylation and destruction of β-catenin, thereby mimicking the action of activated Wnt-based signaling. However, in a different system, activation of Wnt-based signaling has been shown to promote, not inhibit, axon branching (Hall et al., 2000), the opposite phenotype of what we observe in Xenopus RGCs overexpressing the N-terminal domain of β-catenin.
β-catenin-PDZ protein interactions and shaping of RGC axon arbors
Our results also demonstrate that interactions mediated by the C-terminal domain of β-catenin are required to shape RGC axon arbors. When this domain was overexpressed in RGCs, longer and more tangled axon branches were formed by RGCs. These overgrown, tangled axon arbors of RGCs overexpressing the C-terminal domain of β-catenin may correlate with and perhaps be caused by an inhibition of synaptogenesis. Cadherin function has been shown to be required for synapse formation in hippocampal neurons (Togashi et al., 2002). Introduction of dominant-negative cadherin molecules into cultured hippocampal cells removed endogenous cadherin and β-catenin from synapses, which in turn dispersed synaptic proteins, disrupted synaptic function, and caused morphological changes in dendritic spines. Moreover, the last three amino acids of the C-terminal domain of β-catenin comprise a PDZ-binding motif through which β-catenin interacts with, and thereby links cadherin to, at least two synaptic PDZ proteins (Perego et al., 2001; Nishimura et al., 2002). Indeed, overexpression of the C-terminal domain of β-catenin in hippocampal neurons has been shown to inhibit recruitment of the synaptic PDZ protein S-SCAM (Nishimura et al., 2002). Consistent with the possibility that the tangled arbors are caused by an inhibition of synaptogenesis, perturbation of other proteins involved in synaptogenesis has been shown to also cause excessive growth of arbors. For example, in the Xenopus retinotectal system, inhibition of CaMKII (Ca2+/calmodulin-dependent protein kinase II) function inhibits maturation of synapses and also promotes excessive growth of axonal arbors (Zou and Cline, 1999). In addition, in mice lacking either agrin or MuSK (muscle-specific kinase), the neuromuscular junction fails to differentiate. In these mice, the axons that innervate the neuromuscular junction also form extremely abnormal arbors that extend over the entire surface of the muscle instead of being restricted to a small region, as in normal mice (Lin et al., 2001).
Several transcriptional coactivators have also been shown to bind to the C-terminal region of β-catenin from armadillo repeat 10 through the C-terminal end of the protein (Hecht et al., 1999; Takemaru and Moon, 2000). Thus, another possibility is that the overexpressed C-terminal domain of β-catenin competes with endogenous full-length β-catenin for binding to these transcriptional activators and perturbs the normal pattern of transcription.
β-catΔARM generates two distinct arborization phenotypes in RGCs that appear to be regulated by distinct signaling pathways
The deletion mutant β-catΔARM is a fusion of the N- and C-terminal domains of β-catenin. Expression of ΔARM in RGCs generated two distinct arborization phenotypes that mimicked the separate activities of the N- and C-terminal domains of β-catenin. These results suggest that in ΔARM-expressing RGCs that do not form arbors, binding of proteins to the N-terminal domain of β-catenin is perturbed, whereas in ΔARM-expressing RGCs that form extended, misshapen arbors, perturbation of C-terminal domain interactions is dominant. How does β-catΔARM result in two very distinctive phenotypes that appear to be caused by perturbation of two distinct signaling pathways? A likely possibility is that in the two classes of RGCs, different levels of ΔARM are present that selectively perturb distinct protein-protein interactions. In RGCs with lower levels of ΔARM, binding of proteins to the C-terminal domain could be perturbed, resulting in misshapen arbors. In RGCs with higher levels of ΔARM, N-terminal (and C-terminal) domain interactions could be perturbed, thereby inhibiting formation of arbors. Consistent with this hypothesis, we observed that GFPΔARM-expressing RGCs that did not form arbors tended to exhibit brighter GFP fluorescence than GFPΔARM-expressing RGCs that formed overextended, misshapen arbors (data not shown). This suggests that those RGCs that did not form arbors may have contained higher levels of GFPΔARM.
Regulation of β-catenin-cadherin interactions during axon outgrowth and arborization in RGCs
Results in this study show that RGC axons expressing β-catenin deletion mutants grow normally from the retina to the tectum (Loureiro and Peifer, 1998; Loureiro et al., 2001). In particular, our data indicate that interactions of the N-terminal domain of β-catenin with proteins such as α-catenin are not required for growth of RGC axons from the retina to the tectum. A previous study showed that N-cadherin function is required for RGC axon outgrowth to the tectum (Riehl et al., 1996). However, this study showed that overexpression of the juxtamembrane domain of N-cadherin (required for p120 catenin binding) and not the more C-terminal β-catenin binding domain interfered with RGC axon outgrowth (Riehl et al., 1996). Consistent with this, our data also indicate that normal RGC axon growth does not require cadherin interaction with β-catenin (and α-catenin).
We also show here that interactions of the N-terminal domain of β-catenin with proteins such as α-catenin are required within the tectum for targeting and arbor formation by RGC axons (see Discussion above). Cadherin function is also required for targeting and formation of arbors by RGC axons in the tectum (Inoue and Sanes, 1997). Thus, the cadherin-β-catenin-α-catenin adhesion complex may function together at a later stage to promote targeting and formation of arbors by RGC axons in the tectum. One mechanism to regulate the associations and function of the cadherin-β-catenin-α-catenin adhesion complex is through tyrosine phosphorylation of β-catenin (Fujita et al., 2002, Lilien et al., 2002). Previous work has suggested that tyrosine phosphorylation of β-catenin may mediate axon outgrowth by RGCs, likely by dissociation of the cadherin-β-catenin-α-catenin adhesion complex (Kypta et al., 1996; Lilien et al., 2002). It will be interesting to test whether dephosphorylation of tyrosine residues on β-catenin is subsequently required during the later developmental events of targeting and formation of arbors by RGC axons in the tectum.
Footnotes
This work was supported by National Research Service Award F32 MH 12613 to T.M.E. and by the Howard Hughes Medical Institute (HHMI). L.F.R. is an investigator of the HHMI. We thank members of our laboratory, especially S. Bamji, L. Elia, and B. Rico, for comments on this manuscript.
Correspondence should be addressed to Tamira Elul, Physiology Department, University of California San Francisco, 533 Parnassus Avenue, San Francisco, CA 94143. E-mail: tamira@itsa.ucsf.edu.
Copyright © 2003 Society for Neuroscience 0270-6474/03/236567-09$15.00/0
References
- Alsins B, Vu T, Cohen-Cory S ( 2001) Visualizing synapse formation in arborizing optic axons in vivo: dynamics and modulation by BDNF. Nat Neurosci 4: 1093-1101. [DOI] [PubMed] [Google Scholar]
- Benson DL, Tanaka H ( 1998) N-cadherin redistribution during synaptogenesis in hippocampal neurons. J Neurosci 18: 6892-6904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantallops I, Haas K, Cline HT ( 2000) Postsynaptic CPG15 promotes synaptic maturation and presynaptic axon arbor elaboration in vivo. Nat Neurosci 3: 1004-1011. [DOI] [PubMed] [Google Scholar]
- Cohen-Cory S, Fraser S ( 1995) Effects of brain-derived neurotrophic factor on optic axon branching and remodeling in vivo. Nature 378: 192-196. [DOI] [PubMed] [Google Scholar]
- Fujita Y, Krause G, Scheffner M, Zechner D, Leddy HE, Behrens J, Sommer T, Birchmeier W ( 2002) Hakai, a c-Cbl-like protein, ubiquitinates and induces endocytosis of the E-cadherin complex. Nat Cell Biol 4: 222-231. [DOI] [PubMed] [Google Scholar]
- Funayama N, Fagotto F, McCrea P, Gumbiner BM ( 1995) Embryonic axis induction by the Armadillo repeat domain of β-catenin: evidence for intracellular signaling. J Cell Biol 128: 959-968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottardi CJ, Gumbiner BM ( 2001) Adhesion signaling: how β-catenin interacts with its partners. Curr Biol 11: R792-794. [DOI] [PubMed] [Google Scholar]
- Hall AC, Lucas FR, Salinas PC ( 2000) Axonal remodeling and synaptic differentiation in the cerebellum is regulated by WNT-7a signaling. Cell 100: 525-535. [DOI] [PubMed] [Google Scholar]
- Hecht A, Litterst CM, Huber O, Kemler R ( 1999) Functional characterization of multiple transactivating elements of β-catenin, some of which interact with the TATA-binding protein in vitro. J Biol Chem 274: 18017-18025. [DOI] [PubMed] [Google Scholar]
- Holt CE ( 1984) Does timing of axon outgrowth influence initial retinotectal topography in Xenopus? J Neurosci 4: 1130-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt CE, Garlick N, Cornel E ( 1990) Lipofection of cDNAs in the embryonic vertebrate central nervous system. Neuron 4: 203-214. [DOI] [PubMed] [Google Scholar]
- Hulsken J, Birchmeier W, Behrens J ( 1994) E-cadherin and APC compete for the interaction with β-catenin and the cytoskeleton. J Cell Biol 127: 2061-2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue A, Sanes JR ( 1997) Lamina-specific connectivity in the brain: regulation by N-cadherin, neurotrophins, and glycoconjugates. Science 276: 1428-1431. [DOI] [PubMed] [Google Scholar]
- Ivanov DE, Philippova MP, Tkachuck VA ( 2001) Structure and function of classical cadherins. Biochemistry 66: 1174-1186. [DOI] [PubMed] [Google Scholar]
- Kypta RM, Su H, Reichardt LF ( 1996) Association between a transmembrane protein tyrosine phosphatase and the cadherin-catenin complex. J Cell Biol 134: 1519-1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Herman T, Clandinin TR, Lee R, Zipursky SL ( 2001) N-cadherin regulates target specificity in the Drosophila visual system. Neuron 30: 437-450. [DOI] [PubMed] [Google Scholar]
- Lilien J, Balsamo J, Arregui C, Xu G ( 2002) Turn-off, drop-out: functional state switching of cadherins. Dev Dyn 224: 18-29. [DOI] [PubMed] [Google Scholar]
- Lin W, Burgess RW, Dominguez B, Pfaff SL, Sanes JR, Lee KF ( 2001) Distinct roles of nerve and muscle in postsynaptic differentiation of the neuromuscular synapse. Nature 410: 1057-1064. [DOI] [PubMed] [Google Scholar]
- Loureiro J, Peifer M ( 1998) Roles of Armadillo, a Drosophila catenin, during central nervous system development. Curr Biol 8: 622-632. [DOI] [PubMed] [Google Scholar]
- Loureiro JJ, Akong K, Cayirlioglu P, Baltus AE, DiAntonio A, Peifer M ( 2001) Activated armadillo/β-catenin does not play a general role in cell migration and process extension in Drosophila Dev Biol 235: 33-44. [DOI] [PubMed] [Google Scholar]
- Miskevich F, Zhu Y, Ranscht B, Sanes JR ( 1998) Expression of multiple cadherins and catenins in the chick optic tectum. Mol Cell Neurosci 12: 240-255. [DOI] [PubMed] [Google Scholar]
- Murphy-Erdosh C ( 1994) The molecular function and embryonic expression of B-cadherin and cadherin associated proteins. PhD thesis, University of California San Francisco.
- Nieuwkoop PD, Faber J ( 1956) Normal table of Xenopus laevis. Amsterdam: Daudin.
- Nishimura W, Yao I, Iida J, Tanak N, Hata Y ( 2002) Interaction of synaptic-scaffolding molecule and β-catenin. J Neurosci 22: 757-765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Rourke NA, Fraser SE ( 1990) Dynamic changes in optic fibres terminal arbors lead to retinotectal map formation: an in vivo confocal microscopic study. Neuron 5: 159-171. [DOI] [PubMed] [Google Scholar]
- Perego C, Vanoni C, Massari S, Longhi R, Pietrini G ( 2001) Mammalian Lin-7 PDZ proteins associate with β-catenin at cell-cell junctions of epithelia and neurons. EMBO J 19: 3978-3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinches E, Cline HT ( 1998) Distribution of synaptic vesicle proteins within single retinotectal axons of Xenopus tadpoles. J Neurobiol 426-434. [PubMed]
- Pokutta S, Weis WI ( 2002) The cytoplasmic face of cell contact sites. Curr Opin Struct Biol 12: 255-262. [DOI] [PubMed] [Google Scholar]
- Riehl R, Johnson K, Bradley R, Grunwald GB, Cornel E, Lillienbaum A, Holt CE ( 1996) Cadherin function is required for axon outgrowth in retinal ganglion cells in vivo. Neuron 17: 837-848. [DOI] [PubMed] [Google Scholar]
- Sakaguchi DS, Murphey RK ( 1985) Map formation in the developing Xenopus retinotectal system: an examination of ganglion cell terminal arborizations. J Neurosci 5: 3228-3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider SQ, Finnerty JR, Martindale MQ ( 2003) Protein evolution: structure-function relationships of the oncogene β-catenin in the evolution of multicellular animals. J Exp Zool 295B: 25-44. [DOI] [PubMed] [Google Scholar]
- Sehgal RN, Gumbiner BM, Reichardt LF ( 1997) Antagonism of cell adhesion by an α-catenin mutant, and of the Wnt-signaling pathway by α-catenin in Xenopus embryos. J Cell Biol 139: 1033-1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemaru KI, Moon RT ( 2000) The transcriptional coactivator CBP interacts with β-catenin to activate gene expression. J Cell Biol 149: 249-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang L, Huang CP, Schuman EM ( 1998) A role for the cadherin family of cell adhesion molecules in hippocampal long-term potentiation. Neuron 20: 1165-1175. [DOI] [PubMed] [Google Scholar]
- Togashi H, Abe K, Mizoguchi A, Takaoka K, Chisaka O, Takeichi M ( 2002) Cadherin regulates dendritic spine morphogenesis. Neuron 35: 77-89. [DOI] [PubMed] [Google Scholar]
- Uchida N, Honjo Y, Johnson KR, Wheelock MJ, Takeichi M ( 1996) The cadherin/catenin adhesion system is localized in synaptic junctions bordering transmitter release zones. J Cell Biol 135: 767-779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KH, Brose K, Arnott D, Kidd T, Goodman CS, Henzel W, Tessier-Lavigne M ( 1999) Biochemical purification of a mammalian slit protein as a positive regulator of sensory axon elongation and branching. Cell 96: 771-784. [DOI] [PubMed] [Google Scholar]
- Yates PA, Roskies AL, McLaughlin T, O'Leary DDM ( 2001) Topographic-specific axon branching controlled by Ephrin-A is the critical event in retinotectal map development. J Neurosci 21: 8548-8563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou DJ, Cline HT ( 1996) Expression of constitutively active CaMKII in target tissue modifies presynaptic axon arbor growth. Neuron 16: 529-539. [DOI] [PubMed] [Google Scholar]
- Zou DJ, Cline HT ( 1999) Postsynaptic calcium/calmodulin-dependent protein kinase II is required to limit elaboration of presynaptic and postsynaptic neuronal arbors. J Neurosci 19: 8909-8918. [DOI] [PMC free article] [PubMed] [Google Scholar]