Summary
The neuronal protein FE65 functions in brain development and Amyloid Precursor Protein (APP) signaling through its interaction with the mammalian enabled (Mena) protein and APP, respectively. The recognition of short polyproline sequences in Mena by the FE65 WW domain plays a central role in axon guidance and neuronal positioning in the developing brain. We have determined the crystal structures of the human FE65 WW domain (residues 253–289) in the apo form and bound to the peptides PPPPPPLPP and PPPPPPPPPL, which correspond to human Mena residues 313–321 and 347–356, respectively. The FE65 WW domain contains two parallel ligand-binding grooves, XP (formed by residues Y269 and W280) and XP2 (formed by Y269 and W271). Both Mena peptides adopt a polyproline helical II conformation and bind to the WW domain in a forward (N–C) orientation through selection of the PPPPP motif by the XP and XP2 grooves. This mode of ligand recognition is strikingly similar to polyproline interaction with SH3 domains. Importantly, comparison of the FE65 WW structures in the apo and liganded forms shows that the XP2 groove is formed by an induced-fit mechanism that involves movements of the W271 and Y269 side chains upon ligand binding. These structures elucidate the molecular determinants underlying polyproline ligand selection by the FE65 WW domain and provide a framework for the design of small molecules that would interfere with FE65 WW–ligand interaction and modulate neuronal development and APP signaling.
Keywords: FE65, WW domain, Mena, polyproline helix II, crystal structure
Introduction
FE65 is expressed predominantly in the brain, where it plays critical roles in cortical development and APP signaling.1 FE65 possesses a WW domain (FE65 WW hereafter) and two phosphotyrosine-binding (PTB) domains with distinct binding specificities. The FE65 WW recognizes polyproline sequences present in several proteins, including the c-Abl tyrosine kinase,2 the mammalian homolog of the Drosophila protein enabled (Mena),3 and the neuronal P2X2 receptor.4 The FE65 PTB1 binds to the histone acetyltransferase TIP60,5 the transcription factor CP2,6 and the cytoplasmic domains of the low density lipoprotein receptor-related protein7 and ApoER2 receptor,8 whereas PTB2 interacts with the APP intracellular domain.9 The multi-protein complexes resulting from FE65-mediated interactions underlie the function of this protein in regulation of neuronal growth cone motility,10,11 ATP-mediated synaptic transmission,4 learning and memory,12 APP processing,13–15 and transcriptional regulation of gene expression.5,16–22 The FE65 WW is essential for many of these functions. In particular, the FE65 WW-mediated interaction with Mena plays a central role in axonal growth cone dynamics, axon guidance, and neuronal positioning in the developing brain.23–27
WW domains, named for two conserved tryptophan residues (Figure 1(a)), are highly compact protein-protein interaction modules that fold into stable, slightly curved, three-stranded antiparallel β-sheet structures.28–31 These domains interact with short proline-rich sequences and are found in proteins of diverse functions, including regulation of transcription, RNA processing, ubiquitin ligation, protein trafficking, receptor signaling, and control of the cytoskeleton.28,32 WW domains were initially categorized into four groups based on their ligand-binding specificity. Group I binds polypeptides with the core consensus PPXY (PY motif), where X can be any residue; group II recognizes the sequence PPLP (PL motif); group III selects for proline-rich sequences with arginine residues (PR motif); and group IV interacts with phospho(serine/threonine)-proline containing peptides (pS/pTP motif).28 The concave surface of the WW domain contains a series of exposed, nearly parallel aromatic residues, forming a hydrophobic cavity that receives the polyproline peptide. Group I and IV domains have a single groove formed by two highly conserved aromatic residues (typically a tyrosine in β2 strand and a tryptophan in β3 strand), which receives the XP residues of the peptide (XP groove).28,29 By contrast, group II and III WW domains have an additional groove, XP2, which is formed by the conserved tyrosine and an aromatic residue in β2 strand.33,34 In this respect, groups II and III are more similar to SH3 domains, which have two successive XP grooves, suggesting that these structurally and evolutionary unrelated protein modules have converged on a similar mechanism for polyproline recognition. Based on the distinct structural features and similar binding specificities of group II and III WW domains, including recognition of uninterrupted polyproline sequences (PPPPP) in addition to PL and PR motifs, it was recently proposed that these groups be merged into a larger class II/III.35
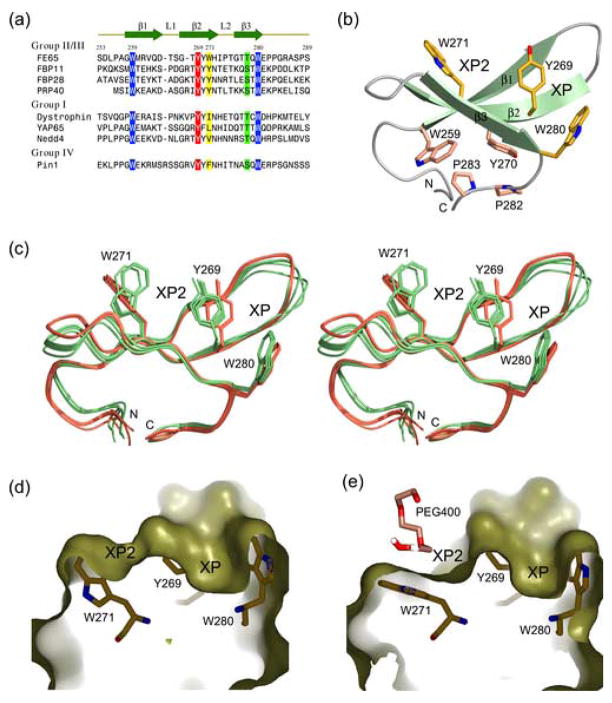
Figure 1.
(a) Sequence comparison of representative WW domains. The sequences of the single WW of FE65, dystrophin, and Pin1, the first WW of FBP11, PRP40, YAP65, and Nedd4, and the second WW of FBP28, were aligned using the program CLUSTAL W.56 Hyphens represent gaps inserted for optimum alignment. The secondary structure elements of FE65 WW are indicated at the top. Residues of human FE65 are numbered. Two conserved tryptophan residues (after which the domain is named) are shown in white on blue background. Invariant tyrosine residues (shown in white on red background) and bulky hydrophobic residues (highlighted in yellow) form the XP2 groove in group II/III domains. Conserved threonine/serine residues that hydrogen bond with the ligand are highlighted in green. (b) Ribbon diagram of the FE65 WW domain. Residues that form the XP and XP2 groves are shown as yellow stick models. The side chains of residues forming a hydrophobic core that stabilizes the fold are shown in pink. (c) Stereo view of eight FE65 WW molecules present in the asymmetric unit. Superposition of these independent structures shows that they fall into two distinct conformations highlighted in shades of green (apo form) and pink (bound to a PEG400 molecule). Note the conformational changes of the W271 and Y269 side chains in the bound form. (d) In the apo form, the XP2 groove is shallow. (e) Binding of a PEG400 molecule induces the formation of a deep XP2 groove, primarily through conformational changes of the indole side chain of W271 and to a lesser degree of the phenyl ring of Y269. The figure was made using PYMOL [www.pymol.org].
Structural studies of WW domains bound to their cognate ligands, including the crystal structures of dystrophin36 (group I) and Pin137 (group IV), and the solution structures of YAP65,38 Nedd4,39,40 and Smurf241 (group I), and FBP1133,34 (group II/III) WW–peptide complexes, provided insights into the molecular mechanisms underlying ligand selectivity by these modules. The peptides adopt a polyproline II (PPII) helical conformation with core proline residues packing against the XP and XP2 grooves and carbonyl oxygen atoms hydrogen bonding with WW residues. Interactions between residues outside the ligand core motif and variable loops of the WW domain, as well as adjacent domains, contribute to the specificity and affinity.29,30,41 Importantly, WW domains can bind to ligands in opposite orientations, as was first observed with SH3 domains.42,43 For example, the Pin1 WW binds to the ligand in an N–C or “+” orientation,37 whereas the dystrophin WW interacts with the peptide in a C–N or “−” orientation,36 because of a twofold rotational pseudosymmetry in the shapes and hydrogen bonding networks of the WW grooves and PPII helices.29
Insights into the architecture of the XP and XP2 grooves of group II/III domains were obtained from the solution structure of FBP11 WW1 bound to a peptide having the core sequence PPLP.33,34 A prominent feature of this structure is the non-parallel arrangement of the XP2 and XP grooves, with the former being occupied by the pair PP and the latter receiving the residues LP. However, because this is currently the only structure of a group II/III WW domain, it is not clear whether the observed orientation of the XP and XP2 grooves is a common feature of all members of this group. The FE65 WW in particular, is distinct from other group II/III domains because it has a tryptophan (W271) instead of a tyrosine in the β2 strand that forms the XP2 groove (Figure 1(a)), suggesting that the size and orientation of this groove might be different. Therefore, structures of the FE65 WW and other group II/III WW domains bound to their ligands will advance our understanding of the molecular basis underlying ligand recognition by these protein modules.
Here we report the atomic structures of FE65 WW in the apo form and bound to two polyproline peptides from human Mena, which represent the first high-resolution crystallographic analysis of a group II/III WW domain. The FE65 WW contains two parallel ligand-binding grooves, XP and XP2, the latter being formed by an induced-fit mechanism. These grooves select for the PPPPP motif of the Mena peptides using a mechanism strikingly similar to that of polyproline recognition by SH3 domains. These structures will facilitate the design of small molecules that could modulate the function of FE65, with possible clinical applications.
Results and Discussion
Structure determination
We expressed a protein fragment of human FE65 (residues 253–289), which encompasses the WW domain, as a glutathione S-transferase (GST) fusion in Escherichia coli cells, purified it by affinity chromatography, and released the FE65 WW from the GST moiety by digestion with the tobacco etch virus (TEV) protease. The FE65 WW protein was further purified by size exclusion chromatography. The purified protein formed multimeric complexes in solution, as judged by size exclusion chromatography and dynamic light scattering experiments (data not shown), and it was crystallized by the sitting-drop vapor-diffusion method. Initial attempts to determine the crystal structure of the apo FE65 WW by molecular replacement using other WW structures as search models failed (data not shown). The structure was determined by single-wavelength anomalous dispersion (SAD) methods of selenomethionine (SeMet)-substituted crystals. The structure was refined to 2.28 Å resolution with a crystallographic factor Rcryst of 21.6% and an Rfree of 28.2% (Table 1). Interestingly, there are eight molecules (A–H) in the asymmetric unit that adopt two distinct conformations (see below). We also crystallized the FE65 WW bound to the peptide Mn10 (PPPPPPPPPL), corresponding to residues 347–356 of human Mena, and determined the structure by multiwavelength anomalous dispersion (MAD) methods, using crystals of Mn10 bound to SeMet-substituted FE65 WW. The crystal structure was refined to 1.33 Å resolution with an Rcryst of 17.2% and an Rfree of 21.4%. Using this structure as a search model, we also solved by molecular replacement the crystal structure of the FE65 WW bound to the peptide Mn9 (PPPPPPLPP) spanning Mena residues 313–321, and refined it to 1.35 Å resolution with an Rcryst of 19.2% and an Rfree of 22.4%. Both FE65 WW–Mn9 and FE65 WW–Mn10 crystals have one molecule in the asymmetric unit.
Table 1.
Statistics of structure determination and refinement
| WW Apoa | WW–Mn10b | WW–Mn9b | |||||
|---|---|---|---|---|---|---|---|
| Data collection and phasing | |||||||
| Data set | Native | SAD λ1 | Native | MAD λ1 | MAD λ2 | MAD λ3 | Native |
| Wavelength (Å) | 0.975 | 0.9789 | 0.985 | 0.9792 | 0.9793 | 0.960 | 1.000 |
| Resolution (Å) | 50–2.19 | 50–2.53 | 50–1.33 | 50–1.58 | 50–1.58 | 50–1.50 | 50–1.27 |
| Unique reflections | 20584 | 13311 | 9129 | 5591 | 5595 | 6494 | 14379 |
| Completeness (%)c | 99.4(98.6) | 98.9(89.2) | 98.6(90.3) | 99.9(100) | 99.9(100) | 99.3(93.2) | 97.6(93.5) |
| Redundancy (%) | 9.2(8.2) | 36.0(27.1) | 6.5(5.4) | 20.5(19.6) | 19.9(19.1) | 12.8(9.6) | 7.5(5.4) |
| Rsym (%)d | 4.5(32.3) | 8.5(42.7) | 5.7(44.0) | 7.1(31.3) | 6.3(32.8) | 6.5(43.8) | 4.4(38.6) |
| <I> / <σ(I)> | 43.3(5.3) | 60.7(7.8) | 30.1(3.8) | 53.3(12.9) | 52.6(12.1) | 41.3(5.3) | 27.5(3.7) |
| Refinement | |||||||
| Resolution range (Å) | 37.3–2.28 | 17.03–1.33 | 25.8–1.35 | ||||
| Reflections in working/test | 17415/924 | 8654/431 | 11423/586 | ||||
| Rcryst (%)e | 21.6 | 17.2 | 19.2 | ||||
| Rfree (%)f | 28.2 | 21.4 | 22.4 | ||||
| Average isotropic B factors (Å2) | |||||||
| Protein | 47.2 | 18.9 | 23.6 | ||||
| Peptide | 20.3 | 29.6 | |||||
| Ramachandran plot (%) | |||||||
| Allowed | 87.8 | 91.3 | 92.9 | ||||
| Additional | 12.2 | 8.7 | 3.6 | ||||
| Generous | 0.0 | 0.0 | 3.6 | ||||
| Disallowed | 0.0 | 0.0 | 0.0 | ||||
| Number of water molecules | 119 | 34 | 36 | ||||
Crystals of Fe65 WW in the apo form were analyzed at NSLS beamline X12C.
Crystals of the Fe65 WW–Mn10 and Fe65 WW–Mn9 complexes were analyzed at NSLS beamline X12B.
The numbers in parentheses correspond to the highest resolution shell (2.27–2.19 Å for WW Apo Native, 2.62–2.53 Å for SAD, 1.38–1.33 Å for WW–Mn10 Native, 1.64–1.58 Å for MAD λ1 and λ2, 1.55–1.50 Å for MAD λ3, and 1.32–1.27 Å for WW–Mn9 Native).
Rsym = Σ|(I − <I>)|/Σ(I), where I is the observed integrated intensity, <I> is the average integrated intensity obtained from multiple measurements, and the summation is over all observed reflections.
Rcryst = Σ||Fobs| − k|Fcalc||/Σ|Fobs|, where Fobs and Fcalc are the observed and calculated structure factors, respectively.
Rfree is calculated as Rcryst using 5% of the reflection data chosen randomly and omitted from the refinement calculations.
Crystal structure of FE65 WW in the apo form
The FE65 WW adopts the typical WW fold, comprising an antiparallel β-sheet formed by three β-strands (β1–β3) and two short loops (L1 and L2) (Figure 1(a) and (b)). At the convex surface of the β-sheet the conserved aromatic residues Y270 and W259 stack against P282 and the invariant P283, respectively, forming a hydrophobic cluster that stabilizes the fold (Figure 1(b)). This hydrophobic core is shielded from the solvent by L255 and P256 and it brings together the N and C termini, generating a domain that can be transferred as a functional unit between proteins. The concave ligand-binding surface contains two distinct grooves, XP and XP2. The canonical XP groove is formed by Y269 and W280 and its size is determined by the L1 loop, which is essentially a short β-turn, and T278 in β3 strand. The XP2 groove is formed primarily by Y269, W271, and V262. In loop L2, H272 stacks against P274 exposing the hydrophobic side chain of I273, which in turn restricts the movement of the W271 indole ring and indirectly influences the XP2 groove. Because the FE65 WW has only twenty residues between the conserved W259 and W280 (Figure 1(a)), its H272 has shifted towards the N terminus by one residue compared to other WW domains and as a result, its imidazole ring lies below the β-sheet. By contrast, the corresponding histidine in group I WW domains lies above the β-sheet and participates in ligand recognition by hydrogen bonding with the tyrosine of the PY motif.36,38–41
Formation of the FE65 WW XP2 groove by an induced-fit mechanism
Superimposition of the eight molecules present in the asymmetric unit shows that they are grouped into two distinct conformations (Figure 1(c)). Molecules B, C, E, and H adopt a conformation in which the XP2 groove is unoccupied and the side chains of W271 and Y269 are approaching each other, effectively closing this groove (Figure 1(c) and (d)). By contrast, molecules A, D, F and G have a PEG400 molecule (from the crystallization solution) bound to the XP2 groove, resulting in a rotation of the W271 indole ring plane by an average of 60° relatively to the unbound one (Figure 1(c) and (e)). The phenyl ring of Y269 is also displaced by ~1.5 Å, effectively increasing the width of the XP2 groove to accommodate the ligand. Sequence alignment shows that W271 corresponds to smaller aromatic residues (Tyr or Phe) in other group II/III and IV domains and hydrophobic residues in group I domains (Figure 1(a)). Notably, mutation of the corresponding leucine (L190) to tryptophan in the YAP65 WW1 domain shifts the specificity of this domain from group I to that of group II.44 The present crystal structure provides an explanation for this switch and elucidates the molecular basis for the important role of the W271 indole ring in the formation of the XP2 groove by an induced-fit mechanism.
Structural determinants of polyproline recognition by FE65 WW
It was previously shown that the human Mena polyproline sequences spanning residues 313–321 and 347–356 are ligands for the FE65 WW.3 To evaluate the contribution of these motifs to the Mena interaction with FE65 WW, we synthesized the peptides Mn9 and Mn10 that correspond to these regions and determined their binding affinities for FE65 WW using isothermal titration calorimetry. FE65 WW binds to Mn9 and Mn10 peptides with dissociation constants 124 ± 9.5 μM and 116 ± 6.5 μM, respectively (Figure 2). The obtained micromolar affinities are in agreement with previously measured affinities for other WW–ligand complexes,45 and are thought to underlie the formation of transient complexes by these modules during dynamic cellular processes.
Figure 2.
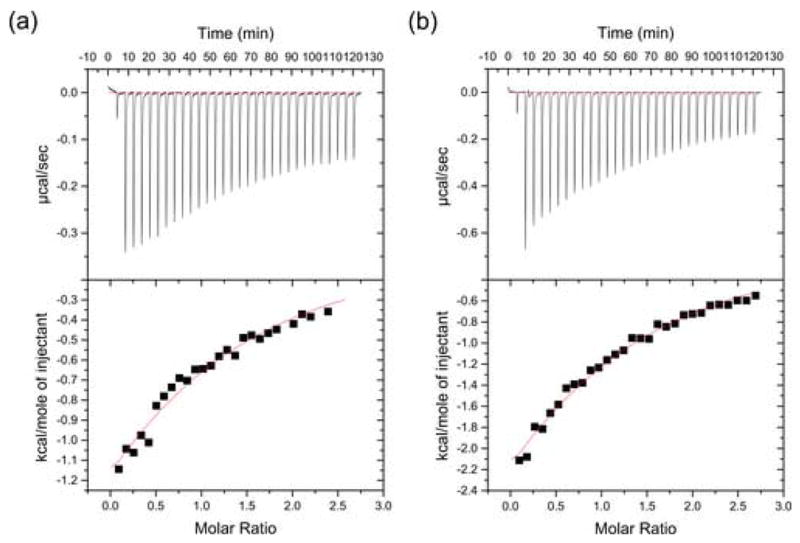
Isothermal titration calorimetry results obtained for the FE65 WW interaction with (a) peptide Mn10 and (b) peptide Mn9, as described in Experimental Procedures.
The structural basis for the specificity and affinity of the FE65 WW–Mn10 interaction was elucidated by the crystal structure of this complex (Figure 3(a) and (b)). The peptide adopts a PPII helix and each of the XP and XP2 grooves is occupied by two proline residues: P5′P6′ insert into XP2 and P8′P9′ occupy XP (peptide residues denoted with primes hereafter). The residues P1′–P4′ extend beyond the WW molecule (Figure 3(a)), with P1′P2′ contacting the loop L2 of an adjacent crystallographic molecule. Notably, the side chain of L10′ faces toward the solvent and does not make any contacts with the protein (Figure 3(a)). The XP2 groove is in the open conformation, as observed in the PEG400-bound FE65 WW. Indeed, the W271 and Y269 side chains of FE65 WW bound to Mn10 superimpose almost exactly with the corresponding side chains of the PEG400-bound FE65 WW, indicating that an induced fit mechanism underlies polyproline ligand recognition by FE65 WW. Importantly, the separation of the two grooves by Y269 and T278 necessitates the presence of a spacer residue between the two pairs of prolines that occupy the grooves. Although in the FE65 WW–Mn10 complex the spacer is proline (P7′), this position could be occupied by any amino acid (X) without breaking the PPII conformation, indicating that the sequence PPXPP is the general recognition motif of FE65 WW.
Figure 3.
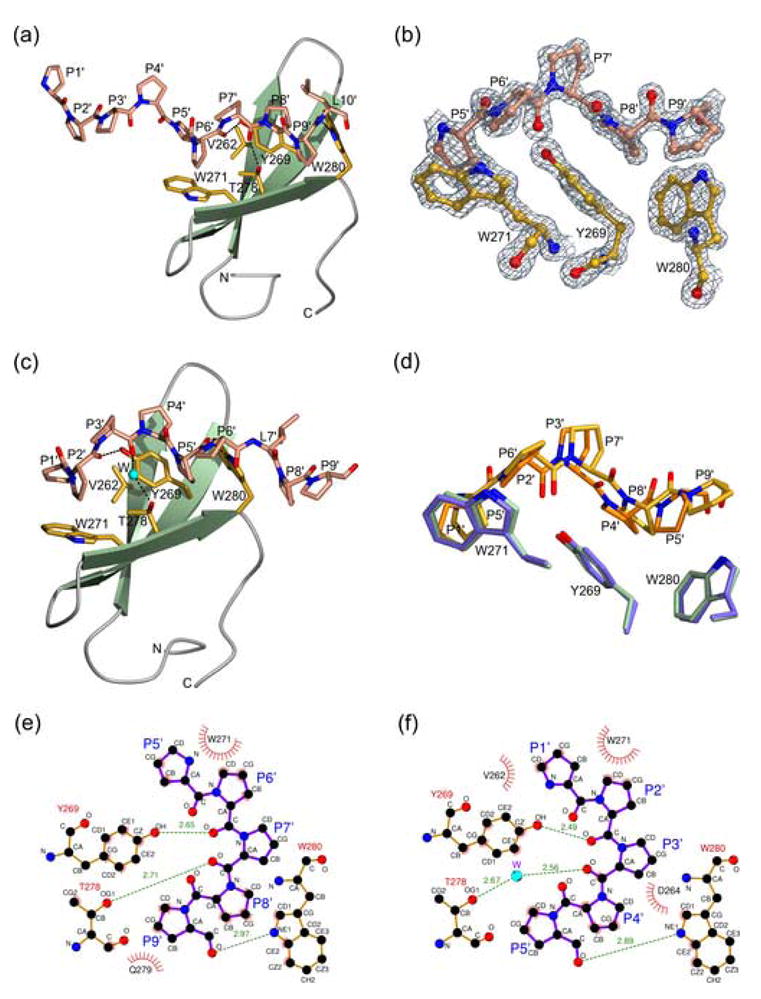
(a) Ribbon diagram of the FE65 WW bound to peptide Mn10, shown as a stick model. WW residues critical for interaction with the peptide are shown as yellow stick models. N and O atoms are colored blue and red, respectively. Hydrogen bonds are denoted by dashed lines. (b) A weighted 2Fobs–Fcalc electron density map of the FE65 WW–Mn10 structure calculated at 1.33 Å and contoured at 2σ. (c) Ribbon diagram of the FE65 WW bound to peptide Mn9. Atoms are colored as in Figure 3(a). A water molecule is indicated by a cyan sphere. (d) Superposition of the FE65 WW (green) bound to Mn10 (yellow) with the FE65 WW (cyan) bund to Mn9 (orange) structures. For clarity, only the parts of the ligands that interact with the XP- and XP2-forming aromatic residues of FE65 WW are shown. The figure was made using BOBSCRIPT57 and POV-Ray [www.povray.org]. (e) and (f) Two-dimensional representation of the FE65 WW interactions with the Mn10 and Mn9 peptides, respectively (made using LIGPLOT58). FE65 and Mena residues are colored orange and purple, respectively. Hydrogen bonds are shown as dashed lines, hydrophobic interactions as arcs with radial spokes, and a water molecule as a cyan sphere.
Because the Mn10 peptide does not contain an intact PPLP motif, previously reported to be recognized by the FE65 WW,3 we also determined the crystal structure of the FE65 WW bound to the peptide Mn9 that contains this sequence. Surprisingly, the XP and XP2 grooves of FE65 WW also bind to the PPPPP and not to the PPLP motif (Figure 3(c)). Like Mn10, the Mn9 peptide also forms a PPII helix, with P1′P2′ residues inserting into XP2 and P4′P5′ occupying the XP groove, further corroborating the notion that the FE65 grooves have a stronger preference for proline rather than leucine residues. It appears that the small size of the FE65 XP groove, which is determined by the short loop L1, plays an important role in the selection of the pyrrolidine ring of proline over the bulkier isobutyl group of a leucine residue. By contrast, the longer L1 loop of FBP11 WW1 generates a larger XP groove that can accommodate a leucine residue. Nevertheless, in the FE65 WW–Mn9 structure the side chain of L7′ clamps the indole group of W280 outside the XP groove, providing additional stabilization to the complex (Figure 3(c)).
Superposition of the FE65 WW–Mn10 and FE65 WW–Mn9 complexes shows that the overall structures are very similar (root-mean-square deviation 0.61 Å for all Cα atoms of the WW region spanning L255–P283 and the atoms of the five proline residues that occupy the two grooves). However, it is noted that the Mn10 and Mn9 proline residues occupying the XP groove are not as well superimposed as those inserting into the XP2 pocket (Figure 3(d)), likely reflecting the different crystal packing arrangements of the two complexes due to differences in the crystallization conditions and/or peptide composition. Notably, the C-terminal region of the Mn9 peptide extends beyond the WW domain and the side chain of L7′ makes hydrophobic contacts with adjacent crystallographic molecules, resulting in a more tightly coiled PPII helix. Notwithstanding the possible crystal lattice effects that may influence the position of the peptide inside the XP and XP2 grooves, these structures demonstrate the flexibility of the FE65 WW grooves to accommodate slightly different positions of the ligand (Figure 3(d)). Both the FE65 WW complexes with the Mn10 and Mn9 peptides are stabilized by similar interactions, although subtle differences are observed in the hydrophobic contacts and hydrogen bonding networks of the two structures (Figure 3(e) and (f)). Most notably, the carbonyl oxygen atoms of P6′ and P9′ in Mn10 (P2′ and P5′ in Mn9) form direct hydrogen bonds with the hydroxyl group of Y269 and the Nε1 of W280, respectively. By contrast, the Oγ1 atom of T278 hydrogen bonds directly with the carbonyl oxygen of the spacer residue P7′ in the FE65 WW–Mn10 complex but indirectly through a water molecule with the carbonyl oxygen of P3′ in the FE65 WW–Mn9 structure.
Similarities between the FE65 WW and SH3 ligand-binding surfaces
A prominent feature of the FE65 WW–Mn10 and FE65 WW–Mn9 structures is the parallel orientation of the XP and XP2 grooves, which is in large part due to the W271 indole ring that forms the external wall of XP2 (Figure 4(a) and (b)). By comparison, two solution structures of the FBP11 WW1 bound to the peptide PPLP revealed that the XP and XP2 grooves are not parallel33,34 (Figure 4(c)). The FBP11 WW1 XP2 groove is formed by two tyrosine residues (Y23 and Y25) instead of a tyrosine and a tryptophan in FE65 (Y269 and W271), and the FBP11 grooves are nearly continuous because of the smaller side chain of S32 compared to the T278 in FE65 WW. As a result, the FBP11 WW1 grooves are occupied by successive pairs of residues, without a spacer amino acid. In contrast, the parallel and non-continuous nature of the FE65 WW grooves underlies the recognition of a ligand having two pairs of proline residues separated by one spacer amino acid, the side chain of which makes no contributions to the interaction.
Figure 4.
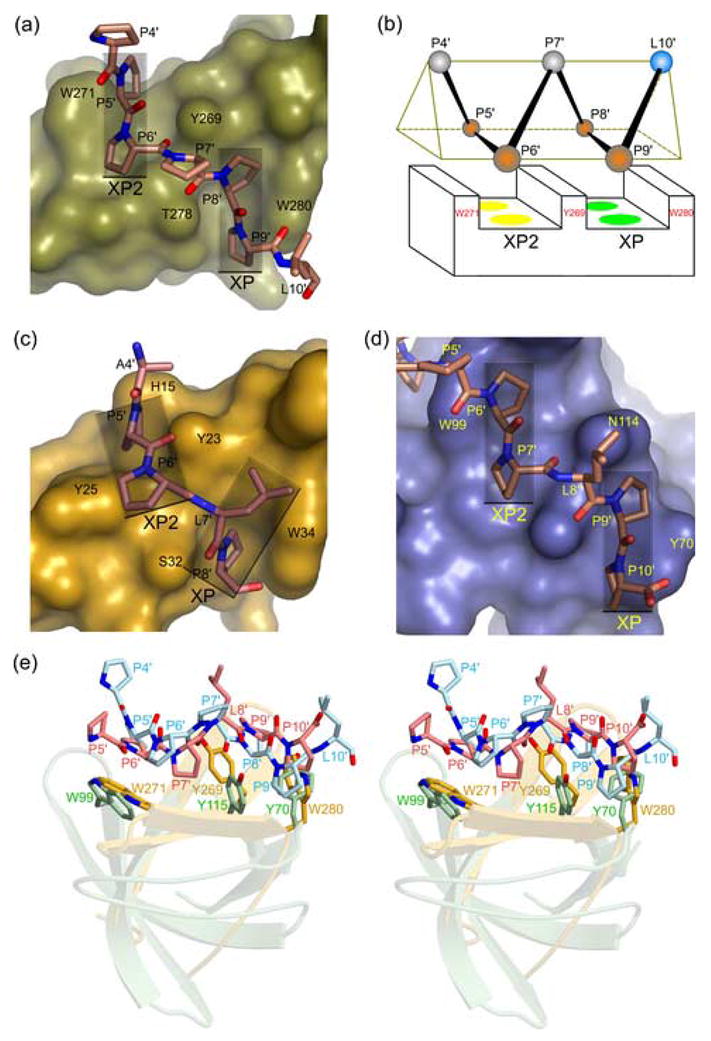
(a) Surface representation of the FE65 WW bound to the Mn10 peptide (shown as a stick model). Semitransparent rectangles denote the parallel orientation of the XP and XP2 grooves. (b) Schematic diagram of the FE65 WW bound to Mn10 PPII helix (shown as a triangular prism), illustrating the parallel arrangement of the XP and XP2 grooves, the three aromatic residues that form them, and the proline residues of the ligand that occupy them (orange spheres). Unbound proline and leucine residues are shown as gray and cyan spheres, respectively. (c) Surface representation of the FBP11 WW1 bound to PPLP sequence (PDB code 2DYF), showing the non-parallel arrangement of the XP and XP2 grooves (rectangles). (d) Surface representation of the Abl tyrosine kinase SH3 domain bound to the peptide 3BP1 containing the sequence PPLPP (PDB code 1ABO), showing the parallel XP and XP2 grooves (rectangles). (e) Stereo view of the FE65 WW–Mn10 structure superimposed to the Abl SH3–3BP1 complex, showing the similar mode of PPII recognition by the aromatic triad of WW (yellow) and SH3 (light green) domains. The Mn10 and 3BP1 peptides are shown in cyan and pink, respectively.
The parallel orientation of the FE65 WW grooves is a novel structural feature of group II/III domains and is remarkably similar to the arrangement of the SH3 proline-binding grooves (Figure 4(a) and (d)), despite the different folds of these protein modules. Indeed, superposition of the FE65 WW–Mn10 and the Abl SH3–3BP1 crystal structures shows strikingly similar positions of the three aromatic residues, which emanate from different secondary structure elements in the WW and SH3 domains to form the two polyproline-binding grooves (Figure 4(e)). The almost identical spatial position of these critical aromatic triads provides strong evidence that these evolutionary unrelated domains have converged upon a similar structural mechanism for PPII recognition.29,46
Perspective
This work presents the first crystal structure of a group II/III WW domain and provides mechanistic insights into uninterrupted polyproline ligand recognition by FE65 WW at the atomic level. A prominent feature of this domain is the presence of a second groove, XP2, which is parallel to the conserved groove XP and is formed by an induced-fit mechanism. The parallel and non-continuous arrangement of the XP and XP2 grooves necessitates a spacer residue between the two pairs of proline residues, leading to the recognition of the PPXPP sequence, where X is any residue. Remarkably, the architecture of these grooves is more similar to that of SH3 domains than FBP11 WW1, the only other group II/III WW domain with known structure, providing evidence for evolution convergence. Thus, the high-resolution crystal structures of FE65 WW bound to polyproline ligands advances our understanding of the principles underlying ligand selection by this versatile domain. This information will facilitate the design of novel WW domains with desired ligand specificities and affinities, as described previously.47,48 Importantly, the specific and low-affinity interactions of WW domains with their polyproline ligands make them excellent targets for drug development.45 Given the significance of the FE65 WW-mediated protein interactions in brain development and pathogenesis of human diseases, including Alzheimer’s disease, the present work will provide a framework for the development of small molecules that could modulate these interactions and regulate the function of FE65, with possible clinical applications.
Experimental Procedures
Protein expression and crystallization
A DNA fragment encoding the human FE65 WW (residues 253–289) with a recognition site for the TEV protease (ENLYFQ↓G) at its N terminus was amplified by the polymerase chain reaction method and was cloned into a modified pGEX-2T vector (GE Healthcare). The FE65 WW protein was expressed in E. coli BL21(DE3) cells as a GST fusion, purified on glutathione Sepharose, released from GST with digestion with TEV, and was further purified by size exclusion chromatography on a Superdex 30 column (GE Healthcare). SeMet-substituted FE65 WW protein was produced in E. coli B834 cells (Novagen) grown in M9 medium supplemented with 40 mg/l SeMet (Sigma), and the protein was purified in the same manner as the unlabelled protein in the presence of 5 mM DTT throughout the entire purification process.
The FE65 WW protein was concentrated by ultracentrifugation to 40 mg/ml in 20 mM Tris–HCl (pH 7.6), 150 mM NaCl, and was used for crystallization experiments by the sitting-drop vapor-diffusion method. Crystals of FE65 WW in the apo form were obtained in 2.2 M ammonium sulfate, 0.1 M HEPES (pH 7.5), 2% PEG400 at 20 °C. Crystals were cryoprotected by stepwise transfer in 2.3 M ammonium sulfate, 0.1 M HEPES (pH 7.5), 10% glycerol, 10% xylitol. The crystals belong to space group P6322 with unit cell dimensions a = b = 75.61 Å, c = 226.49 Å, α = β = 90°, γ = 120°. There are eight molecules in the asymmetric unit. The crystallized FE65 WW protein includes the vector-derived residue G at its N terminus.
For crystallization of FE65 WW–Mena complexes, the protein concentration was adjusted to 18 mg/ml in 20 mM Tris–HCl (pH 7.6), 150 mM NaCl, mixed with the peptides Mn10 or Mn9 at ~1:3 (protein/peptide) molar ratios, incubated at 4 °C for 1–2 h, and used for crystallization by the sitting-drop vapor-diffusion method. Crystals of the FE65 WW–Mn10 complex grew in 1.7–1.9 M ammonium sulfate, 0.1 M HEPES (pH 7.4), at 20 °C, and they were cryoprotected in 1.8 M ammonium sulfate, 0.1 M HEPES (pH 7.4), 20% glycerol. They belong to space group P3121 with unit cell dimensions a = b = 41.66 Å, c = 38.63 Å, α = β = 90°, γ = 120°, and have one molecule in the asymmetric unit. Crystals of the FE65 WW–Mn9 complex grew in 1.3 M sodium citrate, 0.1 M HEPES (pH 7.6), and were cryoprotected in 1.52 M sodium citrate, 0.05 M HEPES (pH 7.6). They belong to space group P41212 with unit cell dimensions a = b = 37.50 Å, c = 78.46 Å, α = β = γ = 90°, and there is one molecule in the asymmetric unit. The data were processed using the HKL2000 package.49 Data collection and processing statistics are given in Table 1.
Structure determination and refinement
The crystal structure of FE65 WW in the apo form was determined by SAD methods, using SeMet-substituted protein and data collected on beamline X12C at the National Synchrotron Light Source (NSLS), Brookhaven National Laboratory, Long Island, New York. The structure of the FE65 WW–Mn10 complex was solved by MAD methods, using SeMet-substituted FE65 WW protein and data collected at NSLS, beamline X12B. Initial SAD and MAD phases were calculated using SHELX,50 followed by several cycles of model building and refinement using COOT,51 REFMAC5,52 and ARP/wARP.53 The FE65 WW–Mn9 structure was determined by molecular replacement using MOLREP54 and the FE65 WW–Mn10 complex as a search model. PROCHECK55 was used for analysis and validation of the refined structures (Table 1).
Isothermal titration calorimetry
Binding constants of FE65 WW for the synthetic peptides Mn9 and Mn10 were measured using a VP-ITC microcalorimeter (MicroCal). Briefly, 0.71 mM Mn9 and 0.74 mM Mn10 were titrated against 63.9 μM FE65 WW protein in 20 mM Tris–HCl (pH 7.6), 150 mM NaCl at 23 °C. Titration curves were analyzed using the program ORIGIN 5.0 (OriginLab). Protein and peptide concentrations were determined by quantitative amino acid analysis on an ABI 420A derivatizer/analyzer and an ABI 103A separation system (Applied Biosystems).
Acknowledgments
We thank the staff at NSLS for assistance with diffraction data collection. This work was supported by grants AG021964, DK062162, and GM065520 from the National Institutes of Health, DAMD170210300, DAMD170310563, W81XWH0510622, and W81XWH0710178 from the US Department of Defense, and the Temple Discovery Award TLL035927 from the Alzheimer’s Association to J.A.A.L.
Abbreviations
- APP
Amyloid Precursor Protein
- GST
glutathione S-transferase
- Mena
mammalian enabled
- PPII
polyproline helix II
- SH3
Src homology 3
Footnotes
Coordinates
The atomic coordinates and structure factors of FE65 WW in the apo form and bound to Mena peptides Mn9 and Mn10 have been deposited in the Protein Data Bank with accession codes 2IDH, 2OEI, and 2HO2, respectively.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.King GD, Turner RS. Adaptor protein interactions: modulators of amyloid precursor protein metabolism and Alzheimer’s disease risk? Exp Neurol. 2004;185:208–219. doi: 10.1016/j.expneurol.2003.10.011. [DOI] [PubMed] [Google Scholar]
- 2.Zambrano N, Bruni P, Minopoli G, Mosca R, Molino D, Russo C, Schettini G, Sudol M, Russo T. The beta-amyloid precursor protein APP is tyrosine-phosphorylated in cells expressing a constitutively active form of the Abl protoncogene. J Biol Chem. 2001;276:19787–19792. doi: 10.1074/jbc.M100792200. [DOI] [PubMed] [Google Scholar]
- 3.Ermekova KS, Zambrano N, Linn H, Minopoli G, Gertler F, Russo T, Sudol M. The WW domain of neural protein FE65 interacts with proline-rich motifs in Mena, the mammalian homolog of Drosophila enabled. J Biol Chem. 1997;272:32869–32877. doi: 10.1074/jbc.272.52.32869. [DOI] [PubMed] [Google Scholar]
- 4.Masin M, Kerschensteiner D, Dumke K, Rubio ME, Soto F. Fe65 interacts with P2X2 subunits at excitatory synapses and modulates receptor function. J Biol Chem. 2006;281:4100–4108. doi: 10.1074/jbc.M507735200. [DOI] [PubMed] [Google Scholar]
- 5.Cao X, Sudhof TC. A transcriptionally active complex of APP with Fe65 and histone acetyltransferase Tip60. Science. 2001;293:115–120. doi: 10.1126/science.1058783. [DOI] [PubMed] [Google Scholar]
- 6.Zambrano N, Minopoli G, de Candia P, Russo T. The Fe65 adaptor protein interacts through its PID1 domain with the transcription factor CP2/LSF/LBP1. J Biol Chem. 1998;273:20128–20133. doi: 10.1074/jbc.273.32.20128. [DOI] [PubMed] [Google Scholar]
- 7.Trommsdorff M, Borg JP, Margolis B, Herz J. Interaction of cytosolic adaptor proteins with neuronal apolipoprotein E receptors and the amyloid precursor protein. J Biol Chem. 1998;273:33556–33560. doi: 10.1074/jbc.273.50.33556. [DOI] [PubMed] [Google Scholar]
- 8.Hoe HS, Magill LA, Guenette S, Fu Z, Vicini S, Rebeck GW. Fe65 interaction with the ApoE receptor ApoEr2. J Biol Chem. 2006;281:24521–24530. doi: 10.1074/jbc.M600728200. [DOI] [PubMed] [Google Scholar]
- 9.Borg JP, Ooi J, Levy E, Margolis B. The phosphotyrosine interaction domains of X11 and FE65 bind to distinct sites on the YENPTY motif of amyloid precursor protein. Mol Cell Biol. 1996;16:6229–6241. doi: 10.1128/mcb.16.11.6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sabo SL, Ikin AF, Buxbaum JD, Greengard P. The Alzheimer amyloid precursor protein (APP) and FE65, an APP-binding protein, regulate cell movement. J Cell Biol. 2001;153:1403–1414. doi: 10.1083/jcb.153.7.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sabo SL, Ikin AF, Buxbaum JD, Greengard P. The amyloid precursor protein and its regulatory protein, FE65, in growth cones and synapses in vitro and in vivo. J Neurosci. 2003;23:5407–5415. doi: 10.1523/JNEUROSCI.23-13-05407.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang B, Hu Q, Hearn MG, Shimizu K, Ware CB, Liggitt DH, Jin LW, Cool BH, Storm DR, Martin GM. Isoform-specific knockout of FE65 leads to impaired learning and memory. J Neurosci Res. 2004;75:12–24. doi: 10.1002/jnr.10834. [DOI] [PubMed] [Google Scholar]
- 13.Sabo SL, Lanier LM, Ikin AF, Khorkova O, Sahasrabudhe S, Greengard P, Buxbaum JD. Regulation of beta-amyloid secretion by FE65, an amyloid protein precursor-binding protein. J Biol Chem. 1999;274:7952–7957. doi: 10.1074/jbc.274.12.7952. [DOI] [PubMed] [Google Scholar]
- 14.Santiard-Baron D, Langui D, Delehedde M, Delatour B, Schombert B, Touchet N, Tremp G, Paul MF, Blanchard V, Sergeant N, Delacourte A, Duyckaerts C, Pradier L, Mercken L. Expression of human FE65 in amyloid precursor protein transgenic mice is associated with a reduction in beta-amyloid load. J Neurochem. 2005;93:330–338. doi: 10.1111/j.1471-4159.2005.03026.x. [DOI] [PubMed] [Google Scholar]
- 15.Pietrzik CU, Yoon IS, Jaeger S, Busse T, Weggen S, Koo EH. FE65 constitutes the functional link between the low-density lipoprotein receptor-related protein and the amyloid precursor protein. J Neurosci. 2004;24:4259–4265. doi: 10.1523/JNEUROSCI.5451-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kimberly WT, Zheng JB, Guenette SY, Selkoe DJ. The intracellular domain of the beta-amyloid precursor protein is stabilized by Fe65 and translocates to the nucleus in a Notch-like manner. J Biol Chem. 2001;276:40288–40292. doi: 10.1074/jbc.C100447200. [DOI] [PubMed] [Google Scholar]
- 17.Cao X, Sudhof TC. Dissection of amyloid-beta precursor protein-dependent transcriptional transactivation. J Biol Chem. 2004;279:24601–24611. doi: 10.1074/jbc.M402248200. [DOI] [PubMed] [Google Scholar]
- 18.Telese F, Bruni P, Donizetti A, Gianni D, D’Ambrosio C, Scaloni A, Zambrano N, Rosenfeld MG, Russo T. Transcription regulation by the adaptor protein Fe65 and the nucleosome assembly factor SET. EMBO Rep. 2005;6:77–82. doi: 10.1038/sj.embor.7400309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hass MR, Yankner BA. A gamma-secretase-independent mechanism of signal transduction by the amyloid precursor protein. J Biol Chem. 2005;280:36895–36904. doi: 10.1074/jbc.M502861200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sumioka A, Nagaishi S, Yoshida T, Lin A, Miura M, Suzuki T. Role of 14-3-3gamma in FE65-dependent gene transactivation mediated by the amyloid beta-protein precursor cytoplasmic fragment. J Biol Chem. 2005;280:42364–42374. doi: 10.1074/jbc.M504278200. [DOI] [PubMed] [Google Scholar]
- 21.Nakaya T, Suzuki T. Role of APP phosphorylation in FE65-dependent gene transactivation mediated by AICD. Genes Cells. 2006;11:633–645. doi: 10.1111/j.1365-2443.2006.00968.x. [DOI] [PubMed] [Google Scholar]
- 22.Yang Z, Cool BH, Martin GM, Hu Q. A dominant role for FE65 (APBB1) in nuclear signaling. J Biol Chem. 2006;281:4207–4214. doi: 10.1074/jbc.M508445200. [DOI] [PubMed] [Google Scholar]
- 23.Guenette S, Chang Y, Hiesberger T, Richardson JA, Eckman CB, Eckman EA, Hammer RE, Herz J. Essential roles for the FE65 amyloid precursor protein-interacting proteins in brain development. EMBO J. 2006;25:420–431. doi: 10.1038/sj.emboj.7600926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gertler FB, Niebuhr K, Reinhard M, Wehland J, Soriano P. Mena, a relative of VASP and Drosophila Enabled, is implicated in the control of microfilament dynamics. Cell. 1996;87:227–239. doi: 10.1016/s0092-8674(00)81341-0. [DOI] [PubMed] [Google Scholar]
- 25.Lanier LM, Gates MA, Witke W, Menzies AS, Wehman AM, Macklis JD, Kwiatkowski D, Soriano P, Gertler FB. Mena is required for neurulation and commissure formation. Neuron. 1999;22:313–325. doi: 10.1016/s0896-6273(00)81092-2. [DOI] [PubMed] [Google Scholar]
- 26.Goh KL, Cai L, Cepko CL, Gertler FB. Ena/VASP proteins regulate cortical neuronal positioning. Curr Biol. 2002;12:565–569. doi: 10.1016/s0960-9822(02)00725-x. [DOI] [PubMed] [Google Scholar]
- 27.Lebrand C, Dent EW, Strasser GA, Lanier LM, Krause M, Svitkina TM, Borisy GG, Gertler FB. Critical role of Ena/VASP proteins for filopodia formation in neurons and in function downstream of Netrin-1. Neuron. 2004;42:37–49. doi: 10.1016/s0896-6273(04)00108-4. [DOI] [PubMed] [Google Scholar]
- 28.Sudol M, Hunter T. NeW wrinkles for an old domain. Cell. 2000;103:1001–1004. doi: 10.1016/s0092-8674(00)00203-8. [DOI] [PubMed] [Google Scholar]
- 29.Zarrinpar A, Lim WA. Converging on proline: the mechanism of WW domain peptide recognition. Nature Struct Biol. 2000;7:611–613. doi: 10.1038/77891. [DOI] [PubMed] [Google Scholar]
- 30.Macias MJ, Wiesner S, Sudol M. WW and SH3 domains, two different scaffolds to recognize proline-rich ligands. FEBS Lett. 2002;513:30–37. doi: 10.1016/s0014-5793(01)03290-2. [DOI] [PubMed] [Google Scholar]
- 31.Ball LJ, Kuhne R, Schneider-Mergener J, Oschkinat H. Recognition of proline-rich motifs by protein-protein-interaction domains. Angew Chem Int Ed. 2005;44:2852–2869. doi: 10.1002/anie.200400618. [DOI] [PubMed] [Google Scholar]
- 32.Ingham RJ, Colwill K, Howard C, Dettwiler S, Lim CS, Yu J, Hersi K, Raaijmakers J, Gish G, Mbamalu G, Taylor L, Yeung B, Vassilovski G, Amin M, Chen F, Matskova L, Winberg G, Ernberg I, Linding R, O’Donnell P, Starostine A, Keller W, Metalnikov P, Stark C, Pawson T. WW domains provide a platform for the assembly of multiprotein networks. Mol Cell Biol. 2005;25:7092–7106. doi: 10.1128/MCB.25.16.7092-7106.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pires JR, Parthier C, Aido-Machado R, Wiedemann U, Otte L, Bohm G, Rudolph R, Oschkinat H. Structural basis for APPTPPPLPP peptide recognition by the FBP11WW1 domain. J Mol Biol. 2005;348:399–408. doi: 10.1016/j.jmb.2005.02.056. [DOI] [PubMed] [Google Scholar]
- 34.Kato Y, Miyakawa T, Kurita J, Tanokura M. Structure of FBP11 WW1-PL ligand complex reveals the mechanism of proline-rich ligand recognition by group II/III WW domains. J Biol Chem. 2006;281:40321–40329. doi: 10.1074/jbc.M609321200. [DOI] [PubMed] [Google Scholar]
- 35.Kato Y, Nagata K, Takahashi M, Lian L, Herrero JJ, Sudol M, Tanokura M. Common mechanism of ligand recognition by group II/III WW domains: redefining their functional classification. J Biol Chem. 2004;279:31833–31841. doi: 10.1074/jbc.M404719200. [DOI] [PubMed] [Google Scholar]
- 36.Huang X, Poy F, Zhang R, Joachimiak A, Sudol M, Eck MJ. Structure of a WW domain containing fragment of dystrophin in complex with beta-dystroglycan. Nature Struct Biol. 2000;7:634–638. doi: 10.1038/77923. [DOI] [PubMed] [Google Scholar]
- 37.Verdecia MA, Bowman ME, Lu KP, Hunter T, Noel JP. Structural basis for phosphoserine-proline recognition by group IV WW domains. Nature Struct Biol. 2000;7:639–643. doi: 10.1038/77929. [DOI] [PubMed] [Google Scholar]
- 38.Macias MJ, Hyvonen M, Baraldi E, Schultz J, Sudol M, Saraste M, Oschkinat H. Structure of the WW domain of a kinase-associated protein complexed with a proline-rich peptide. Nature. 1996;382:646–649. doi: 10.1038/382646a0. [DOI] [PubMed] [Google Scholar]
- 39.Kanelis V, Rotin D, Forman-Kay JD. Solution structure of a Nedd4 WW domain-ENaC peptide complex. Nature Struct Biol. 2001;8:407–412. doi: 10.1038/87562. [DOI] [PubMed] [Google Scholar]
- 40.Kanelis V, Bruce MC, Skrynnikov NR, Rotin D, Forman-Kay JD. Structural determinants for high-affinity binding in a Nedd4 WW3* domain-Comm PY motif complex. Structure. 2006;14:543–553. doi: 10.1016/j.str.2005.11.018. [DOI] [PubMed] [Google Scholar]
- 41.Chong PA, Lin H, Wrana JL, Forman-Kay JD. An expanded WW domain recognition motif revealed by the interaction between Smad7 and the E3 ubiquitin ligase Smurf2. J Biol Chem. 2006;281:17069–17075. doi: 10.1074/jbc.M601493200. [DOI] [PubMed] [Google Scholar]
- 42.Feng S, Chen JK, Yu H, Simon JA, Schreiber SL. Two binding orientations for peptides to the Src SH3 domain: development of a general model for SH3-ligand interactions. Science. 1994;266:1241–1247. doi: 10.1126/science.7526465. [DOI] [PubMed] [Google Scholar]
- 43.Lim WA, Richards FM, Fox RO. Structural determinants of peptide-binding orientation and of sequence specificity in SH3 domains. Nature. 1994;372:375–379. doi: 10.1038/372375a0. [DOI] [PubMed] [Google Scholar]
- 44.Espanel X, Sudol M. A single point mutation in a group I WW domain shifts its specificity to that of group II WW domains. J Biol Chem. 1999;274:17284–17289. doi: 10.1074/jbc.274.24.17284. [DOI] [PubMed] [Google Scholar]
- 45.Nguyen JT, Turck CW, Cohen FE, Zuckermann RN, Lim WA. Exploiting the basis of proline recognition by SH3 and WW domains: design of N-substituted inhibitors. Science. 1998;282:2088–2092. doi: 10.1126/science.282.5396.2088. [DOI] [PubMed] [Google Scholar]
- 46.Zarrinpar A, Bhattacharyya RP, Lim WA. The structure and function of proline recognition domains. Sci STKE. 2003;179:RE8. doi: 10.1126/stke.2003.179.re8. [DOI] [PubMed] [Google Scholar]
- 47.Socolich M, Lockless SW, Russ WP, Lee H, Gardner KH, Ranganathan R. Evolutionary information for specifying a protein fold. Nature. 2005;437:512–518. doi: 10.1038/nature03991. [DOI] [PubMed] [Google Scholar]
- 48.Russ WP, Lowery DM, Mishra P, Yaffe MB, Ranganathan R. Natural-like function in artificial WW domains. Nature. 2005;437:579–583. doi: 10.1038/nature03990. [DOI] [PubMed] [Google Scholar]
- 49.Otwinowski Z, Minor W. Processing of X-ray crystallographic data in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 50.Sheldrick G, Schneider T. SHELXL: High-resolution refinement. Methods Enzymol. 1997;277:319–343. [PubMed] [Google Scholar]
- 51.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallog sect D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 52.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallog sect D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 53.Perrakis A, Morris R, Lamzin VS. Automated protein model building combined with iterative structure refinement. Nature Struct Biol. 1999;6:458–463. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- 54.Vagin AA, Teplyakov A. MOLREP: an automated program for molecular replacement. J Appl Crystallog. 1997;30:1022–1025. [Google Scholar]
- 55.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallog. 1993;26:283–291. [Google Scholar]
- 56.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Esnouf RM. An extensively modified version of MolScript that includes greatly enhanced coloring capabilities. J Mol Graph Model. 1997;15:132–134. doi: 10.1016/S1093-3263(97)00021-1. [DOI] [PubMed] [Google Scholar]
- 58.Wallace AC, Laskowski RA, Thornton JM. Derivation of 3D coordinate templates for searching structural databases: application to Ser-His-Asp catalytic triads in the serine proteinases and lipases. Protein Eng. 1995;8:127–134. doi: 10.1002/pro.5560050603. [DOI] [PMC free article] [PubMed] [Google Scholar]