Abstract
We describe an automated method to isolate mutant C. elegans animals, which fail to appropriately execute a cellular differentiation program that we monitor with gfp-based reporter gene technology. A fluorescence activated sorting mechanism implemented in the COPAS Biosort machine is able to isolate mutants with subtle alterations in the cellular specificity of gfp expression. This methodology is significantly more efficient than comparable manual screens and enabled us to isolate mutants in which dopamine neurons fail to differentiate appropriately.
Text
Forward genetic screens for mutant organisms provide a powerful tool for understanding the molecular basis underlying a plethora of biological processes. The specific advantages of the model organism Caenorhabditis elegans, such as self-fertilization, short generation time, transparency and ease of handling, prompted its extensive use in forward genetic screens that have yielded valuable insights into animal development and physiology 1,2. Depending on the phenotype screened for, the actual isolation of mutants can be a major limitation in a forward genetic analysis. For example, the screening for mutants in which individual cellular fates, visualized by gfp-based reporter gene technology 3, are not appropriately executed, can be a tedious undertaking. A technology that bears promise to automate the phenotypic assessment step in cell fate screens is the COPAS Biosort system (Union Biometrica; “worm sorter”). The worm sorter is a flow cytometry instrument adjusted to analyze and sort small living organisms the size of C. elegans on the basis of their optical density, size, and fluorescence 4. The worm sorter has not been previously tested for isolating mutants that fail to execute specific cellular fates visualized by gfp reporters. The very nature of the conventional fluorescence-based screen for cell-fate mutants poses specific challenges for this technology as such screens often require the detection of animals in which only a small number of cells is labeled with a cell type-specific fluorescent marker. Moreover, desirable mutants may only lose marker gene expression in a subset of cells, and the overall changes in fluorescent intensity may therefore be subtle. Testing the performance of the COPAS Biosort system in such specific conditions is necessary for assessing its applicability in screens for cell fate mutants.
We describe here the use of the COPAS Biosort machine in a forward genetic screen for mutants defective in executing the dopaminergic cell fate. Dopaminergic neurons fulfill important functions across phylogeny, yet the genetic pathways that control dopaminergic cell fate are poorly understood to date 5. A gfp reporter fusion to the promoter of the dopamine neuron-specific dopamine re-uptake transporter dat-1 exclusively labels all dopaminergic neurons in vtIs1 [dat-1∷gfp], a transgenic strain that expresses gfp exclusively in all 8 dopaminergic neurons of the worm 14 6 (Fig. 1a). As there are only eight dopaminergic neurons in the ~1000 cell-containing C.elegans hermaphrodite, the use of dat-1∷gfp provides a challenging test system for the sensitivity of the worm sorter. We performed test runs to determine whether these eight cells provide enough signal strength for the worm sorter. We first ran a sorting test in which the transgenic strain carrying dat-1∷gfp was mixed with non-transgenic, i.e. non-gfp expressing worms. Non-transgenic animals could indeed be sorted out efficiently (Supplementary Table 1). To control for the variability in fluorescent intensities among transgenic individuals, we introduced a chromosomally integrated, broadly expressed rfp transgene (vsIs33 [dop-3∷rfp])7 in the background of our screening strain as an internal reference of fluorescence intensity. This strain expresses rfp in some body-wall muscles and in various head neurons, ventral nerve cord neurons, tail neurons, and two mechanosensory neurons -the PVDs- but not in dopaminergic neurons (Fig. 1b). The sorter was set to compare fluorescence between green and red channels and plot their ratio. The sorting region was set to isolate individuals with reduced green to red ratio of fluorescence. This substantially increased the efficiency of sorting gfp negative animals (Supplementary Table 1).
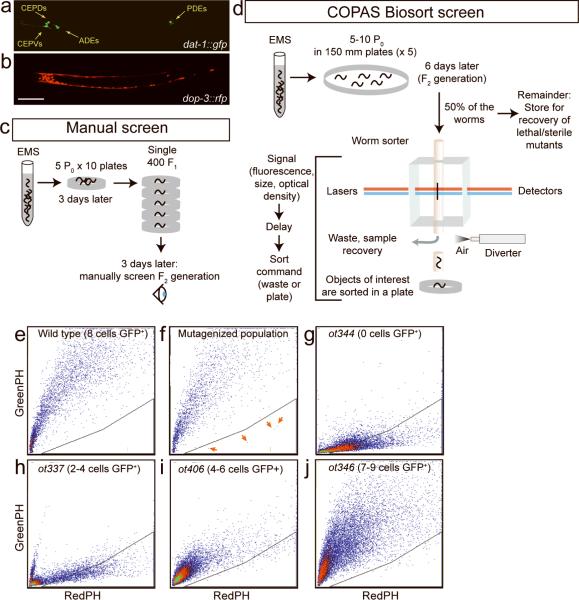
Figure 1. Screening for dopaminergic cell fate mutants.
(a, b) Transgenic strains used for the worm sorter screen. Scale bar represents 100 μm. (c) Experimental procedure of the manual screen. (d) Experimental procedure of the COPAS worm sorter screen. (e)-(j) Relative fluorescence intensity plots between red and green channels of sorted worms. Scale is defined by the sorting parameters (see Supplementary Methods). Each dot corresponds to a single worm. (e) Profile of a non-mutagenized population of the sorting strain. (f) Example profile of a mutagenized population of worms. Red arrows indicate individual animals that fall into the manually-set sorting window. (g-j) Profiles of homogenous populations of retrieved mutants: (g) ot344 (0 out of 8 cells expressing gfp), (h) ot337 (2-4 out of 8 cells expressing gfp), (i) ot406 (4-6 out of 8 cells expressing gfp), and (j) ot346 (7-9 out of 8 cells expressing gfp). The triangle area at the bottom right of each panel is an example of a sorting region chosen to demonstrate the profile differences between various populations. The actual sorting region used in the screening process was usually larger, to minimize the possibility of loosing mutants.
To test the practicability of the worm sorter-based genetic screens for dopaminergic fate mutants, we conducted two different genetic screens for EMS-induced mutations, a manual one and an automated worm sorter screen, and compared the outcomes. For the manual F1 clonal screen we mutagenized animals with EMS, singled F1 progeny onto individual plates and screened through their progeny with the help of a stereo dissecting microscope with a fluorescent light source (Fig. 1c). We screened through 11,000 EMS-mutagenized haploid genomes in the course of several months and identified 10 mutants that express dat-1∷gfp in fewer cells than normal (Table 2).
Table 2.
Mutants retrieved from genetic screens
| Gene name | Molecular identity | Manual screen | Sorter screen | Allele names | Dom/Rec | Pleiotropies |
|---|---|---|---|---|---|---|
| dopy-2 | unknown | 0 alleles | 3 alleles | ot340, ot345 ot406 | R | - |
| dopy-3 | unknown | 0 alleles | 1 allele | ot337 | D | - |
| dopy-4 | unknown | 1 allele * | 0 alleles | ot260 | R | - |
| dopy-5 | unknown | 4 alleles ** | 0 alleles | ot283, ot284, ot296, ot298 | R | sterile |
| dopy-6 | unknown | 1 allele * | 0 alleles | ot263 | R | - |
| dopy-7 | unknown | 0 alleles | 2 alleles | ot399, ot347 | R | sick |
| lin-32 | bHLH | 2 alleles | 4 alleles | ot259, ot297, ot341, ot343, ot338, ot366 | R | - |
| ham-1 | no homologies | 2 alleles | 6 alleles | ot253, ot257, ot342, ot361, ot367, ot339, ot371, ot364 | R | - |
| vab-3 | paired + homeodomain | 0 alleles | 1 allele | ot346 | R | notched head |
| Total allele number | 10 | 17 | ||||
| Genomes screened | 11,000 | 80,000 | ||||
| Allele frequency per genomes screen | 1/1,100 | 1/4,700 | ||||
| Time investment | 100 days § | 25 days § | ||||
| Allele frequency per time | 1 allele / 10 days | 1 allele / 1.5 days | ||||
Dom/Rec indicates “dominant/recessive”.
These days are differentially spent. 100 days dissecting scope work mean full time work at the microscope while 25 days of worm sorting involves mainly machine running and casual observation of functioning of sorter.
As these two mutants only affect gfp expression in the 2 PDE neurons, it is possible that these two mutants were only retrieved by the manual screen because gfp expression in the PDEs is dimmer than in the other dopaminergic neurons and a loss of gfp expression only in these neurons is the most challenging phenotype to detect. In fact, one of the two missing PDE mutants was isolated in the manual screen only for its concurrent `extra PDEs' phenotype (see quantification of phenotypic data in Fig. 2).
This sterile mutant was not isolated by the worm sorter screen because we did not pursue sorted mutants that could not easily be maintained as homozygotes. In contrast to semi-clonal manual screens, in which it is easy to maintain a sterile mutant strain by identifying and maintaining heterozygous adult animals from the parental plate, plates that were analyzed by the worm sorter contained significantly more complex population of mutants, which makes the re-isolation of the parents labor intensive, though still possible in principle.
A more comprehensive version of this table is presented in Supplementary Table 6.
In the automated screen we screened through approximately 80,000 EMS-mutagenized haploid genomes with the COPAS Biosort instrument (Fig. 1d), focusing on isolating mutants that show reduced gfp expression. Figure 1e,f shows an example of a fluorescence distribution graph of a mutagenized population, in which some animals show reduced green to red ratio compared to wild-type animals. The animals falling into the sorting region (arrows in Fig. 1f) were sorted out by the COPAS instrument onto a plate and were then inspected under a fluorescence stereoscope to confirm the presence of a phenotype.
We identified 22 mutants with reduced gfp expression. 3 mutants were isolated in which the overall gfp expression level was merely reduced in all 8 dopaminergic neurons (data not shown) and 2 mutants showed complete loss of gfp expression, but proved to be mutations in the transgenic array (see Supplementary Methods); both mutant classes were not further considered. The remaining 17 show a reduced number of cells expressing gfp in at least one class of dopaminergic neurons. The mutants show striking cellular specificity, with individual mutants affecting distinct subsets of dopaminergic neurons; one mutant affects as little as one subclass of the eight dopaminergic neurons (Table 1). The sorting profiles of homozygous population of some of these mutants are presented in Fig. 1g-j.
Table 1.
Types of phenotypes retrieved using the worm sorter
| Number of cells expressing gfp | Number of alleles | Allele name |
|---|---|---|
| 0 out of 8 | 2 | ot344, ot373 (array mutations) |
| 2 out of 8 | 3 | ot399, ot347, ot366 |
| 3 out of 8 | 1 | ot337 |
| 4 out of 8 | 5 | ot340, ot406, ot341, ot343, ot338 |
| 6 out of 8 | 7 | ot342, ot361, ot367, ot339, ot371, ot363, ot346 |
| 7 out of 8 | 1 | ot345 |
For mutants displaying multiple phenotypes (see Supplementary Fig. 2), we consider here only the phenotype on the basis of which the mutant was isolated.
We mapped the mutants retrieved from both screens using high throughput SNP mapping 8, performed complementation tests (data not shown) and sequenced candidate genes in the regions in which we mapped individual mutants. In total, we recovered 6 alleles of the Math/atonal-like bHLH gene lin-32, previously known to play a role in specifying one subtype of dopaminergic neurons 9, but not previously known to also affect the specification of other dopaminergic neurons (Table 2, Supplementary Fig.1 and Supplementary Table 2). We recovered one allele of the Pax6/Eyeless-like gene vab-3 10, which had not previously been implicated in dopamine neuron development (Table 2, Supplementary Fig. 2 and Supplementary Table 3) and eight alleles of the ham-1 gene, previously known to be involved in controlling asymmetric cell divisions 11 (Table 2, Supplementary Fig. 2 and Supplementary Table 4). In addition to these known genes, we isolated a number of additional, apparently novel mutants that fall into six complementation groups that we mapped to different chromosomal locations (Table 2, Fig. 2 and Supplementary Fig. 1). We termed these genes “dopy” for “dop aminergic neuron atypical”. dopy mutants appear to affect dat-1∷gfp expression in a cell-type specific manner, suggesting that on some level, there is differential regulation of the dopaminergic fate among the various classes of dopaminergic neurons (Fig. 2h). Based on the allele recovery rate, we estimate that our worm sorter screen has reached at least 78% saturation (Supplementary Table 5).
Figure 2. Phenotypes of isolated dopy mutants.
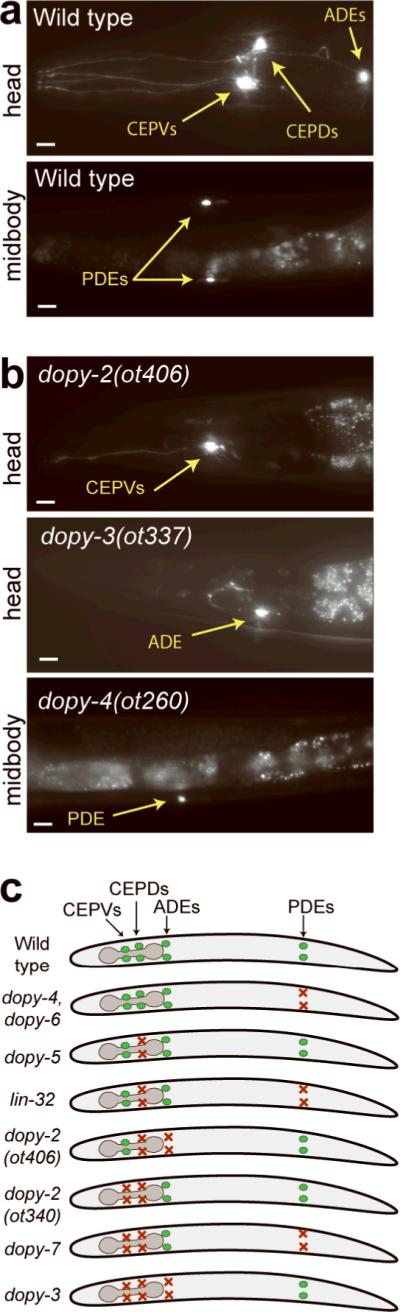
Micrographs of representative wild-type (a) and mutant animals (b) expressing dat-1∷gfp. Scale bars represent 10 μm. See Supplementary Fig. 1 for additional mutants and quantification of data. (c) Schematic summary of cell-type specificity of dopaminergic mutant phenotypes.
Comparing the manual and automated screening approach, we find that the worm sorter screen is more efficient in several regards. First, as less worm picking is involved, the preparation of mutagenized worms for screening with the sorter requires much less operating time than in the manual screen (compare procedures in Fig. 1c and 1d). Moreover, in the actual screening process, it takes about one hour of intense effort at the stereomicroscope to screen through 60 mutagenized genomes (for which ~1,000 individual worms need to be screened in our screening design) while the worm sorter can screen up to 2,000 mutagenized genomes (~50,000 individual worms) in the same amount of time (Table 2). Considering the total amount of time we invested in each screening strategy, the work load was on average ten laborious days of manual screen at the stereomicroscope per isolated mutant as opposed to one and a half part-time days that involve mostly casual supervision of the proper function of the COPAS Biosort machine (Table 2; see Supplementary Methods for details). This high-throughput screening rate of the COPAS method is unprecedented as the only other technology available for automated mutant isolation, the use of microfluids 12, offers a screening speed that is orders of magnitude lower (few hundred animals per hour as opposed to several thousands).
Comparison of the spectrum of mutants retrieved by manual and automated screening further illustrates the effectiveness and sensitivity of the worm sorter approach. While we retrieved alleles of two loci (lin-32, ham-1) by both the manual and automated screen (with more alleles retrieved by the latter), four loci (dopy-2, dopy-3, dopy-7, vab-3) were retrieved exclusively by the automated screen. This is likely because we were able to screen through a larger number of mutagenized genomes. These mutant alleles would have likely surfaced in manual screens as well, but those mutants would have taken much more time to isolate. The sensitivity of the worm sorter is further illustrated by its ability to recognize mutants with as little as one cell having lost gfp expression (Table 1). Three loci were only retrieved by the manual screen for reasons explained in the legend to Table 2.
In conclusion, we have demonstrated that the worm sorter can be used to automate the phenotypic selection step in cell fate screens. This approach can be used for a variety of different cell fate markers; for example, we have also used the worm sorter to isolate mutants with aberrant expression of neuronal fate markers normally only expressed in two (AIYL and AIYR) or as little as one (ASEL) neuronal subtypes (V. Bertrand, F. Zhang, M.D. and O.H., unpub. data). Use of the worm sorter dramatically speeds up the screening process and, in the case described here in detail, identified a variety of mutants that displayed abnormal dopaminergic neuron differentiation. An implicit feature of automation is scalability. Given the minimal effort involved in growing and sorting worms, one can interrogate the genome to an as saturating degree as any mutagenesis approach may allow. In combination with the recently developed strategies which allow for easy identification of the molecular lesion such as transposon tagging13, or whole genome sequencing (Sarin et al., accompanying paper), worm sorter based screens will enable an exhaustive analysis of genetic pathways involved in development.
Supplementary Material
ACKNOWLEDGEMENTS
We thank A. Kruyer, A. Pretorian, J. Recio and Q. Chen for technical assistance. S. Sarin for calculating the degree of saturation, V. Bertrand and F. Zhang for communicating unpublished results and S. Mitani for providing lin-32 deletion alleles. This work was funded by the National Institutes of Health (R01NS039996-05; R01NS050266-03), the Howard Hughes Medical Institute, an EMBO long term fellowship to M.D., an EMBO long term fellowship and a Marie Curie Outgoing International fellowship to N.F.
Footnotes
The authors declare no competing financial interest.
REFERENCES
- 1.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jorgensen EM, Mango SE. The art and design of genetic screens: Caenorhabditis elegans. Nat Rev Genet. 2002;3:356–69. doi: 10.1038/nrg794. [DOI] [PubMed] [Google Scholar]
- 3.Chalfie M, Tu Y, Euskirchen G, Ward WW, Prasher DC. Green fluorescent protein as a marker for gene expression. Science. 1994;263:802–5. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- 4.Pulak R. Techniques for analysis, sorting, and dispensing of C. elegans on the COPAS flow-sorting system. Methods Mol Biol. 2006;351:275–86. doi: 10.1385/1-59745-151-7:275. [DOI] [PubMed] [Google Scholar]
- 5.Abeliovich A, Hammond R. Midbrain dopamine neuron differentiation: factors and fates. Dev Biol. 2007;304:447–54. doi: 10.1016/j.ydbio.2007.01.032. [DOI] [PubMed] [Google Scholar]
- 6.Nass R, Hall DH, Miller DM, 3rd, Blakely RD. Neurotoxin-induced degeneration of dopamine neurons in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2002;99:3264–9. doi: 10.1073/pnas.042497999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chase DL, Pepper JS, Koelle MR. Mechanism of extrasynaptic dopamine signaling in Caenorhabditis elegans. Nat Neurosci. 2004;7:1096–103. doi: 10.1038/nn1316. [DOI] [PubMed] [Google Scholar]
- 8.Davis MW, et al. Rapid single nucleotide polymorphism mapping in C. elegans. BMC Genomics. 2005;6:118. doi: 10.1186/1471-2164-6-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao C, Emmons SW. A transcription factor controlling development of peripheral sense organs in C. elegans. Nature. 1995;373:74–8. doi: 10.1038/373074a0. [DOI] [PubMed] [Google Scholar]
- 10.Hack MA, et al. Neuronal fate determinants of adult olfactory bulb neurogenesis. Nat Neurosci. 2005;8:865–72. doi: 10.1038/nn1479. [DOI] [PubMed] [Google Scholar]
- 11.Guenther C, Garriga G. Asymmetric distribution of the C. elegans HAM-1 protein in neuroblasts enables daughter cells to adopt distinct fates. Development. 1996;122:3509–18. doi: 10.1242/dev.122.11.3509. [DOI] [PubMed] [Google Scholar]
- 12.Chung K, Crane MM, Lu H. Automated on-chip rapid microscopy, phenotyping and sorting of C. elegans. Nat Methods. 2008;5:637–43. doi: 10.1038/nmeth.1227. [DOI] [PubMed] [Google Scholar]
- 13.Boulin T, Bessereau JL. Mos1-mediated insertional mutagenesis in Caenorhabditis elegans. Nat Protoc. 2007;2:1276–87. doi: 10.1038/nprot.2007.192. [DOI] [PubMed] [Google Scholar]
- 14.Nass R, et al. A genetic screen in Caenorhabditis elegans for dopamine neuron insensitivity to 6-hydroxydopamine identifies dopamine transporter mutants impacting transporter biosynthesis and trafficking. J Neurochem. 2005;94:774–85. doi: 10.1111/j.1471-4159.2005.03205.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.