Abstract
Double-stranded (ds) RNA virus particles are organized around a central icosahedral core capsid made of 120 identical subunits. This core capsid is unable to invade cells from outside, and animal dsRNA viruses have acquired surrounding capsid layers that are used to deliver a transcriptionally active core particle across the membrane during cell entry. In contrast, dsRNA viruses infecting primitive eukaryotes have only a simple core capsid, and as a consequence are transmitted only vertically. Here, we report the 3.4 Å X-ray structure of a picobirnavirus—an animal dsRNA virus associated with diarrhoea and gastroenteritis in humans. The structure shows a simple core capsid with a distinctive icosahedral arrangement, displaying 60 two-fold symmetric dimers of a coat protein (CP) with a new 3D-fold. We show that, as many non-enveloped animal viruses, CP undergoes an autoproteolytic cleavage, releasing a post-translationally modified peptide that remains associated with nucleic acid within the capsid. Our data also show that picobirnavirus particles are capable of disrupting biological membranes in vitro, indicating that its simple 120-subunits capsid has evolved animal cell invasion properties.
Keywords: double-stranded RNA viruses, gastroenteritis viruses, structural virology, virus crystallography
Introduction
Double-stranded (ds) RNA viruses constitute a broad category of icosahedral viruses infecting a large spectrum of living organisms. The hosts range from prokaryotes—exemplified by the Pseudomonas phage φ6 in the Cystoviridae family of viruses—to humans, like rotaviruses (Reoviridae family (Estes and Kapikian, 2007), see http://www.ncbi.nlm.nih.gov/ICTVdb/Ictv/index.htm for virus taxonomy) or the more recently discovered picobirnaviruses (PBVs), which are associated with pathogenic symptoms like diarrhoea and gastroenteritis (Grohmann et al, 1993). Moreover, many of the primitive unicellular eukaryotes—yeast (McCabe et al, 1999) and protozoa (Wang and Wang, 1991)—contain cytoplasmic genetic elements that have been identified as simple capsid dsRNA viruses (as opposed to the complex capsid organization of viruses of the Reoviridae family (Estes and Kapikian, 2007; Roy, 2007; Schiff et al, 2007)). These simple-capsid dsRNA viruses never leave the cytoplasmic environment and are unable of invading a cell from outside. They are transmitted only during cell division or through cytoplasmic bridges that open during cell mating (McCabe et al, 1999). The best-studied example is L-A virus of yeast (Naitow et al, 2002), which belongs to the Totiviridae family. A characteristic feature of dsRNA virus particles in general is that their genome is always confined and transcribed within the particle, hidden from detection by the innate immunity sensors of the infected cell. On transcription, viral mRNAs are extruded through specific portals at the five-fold axes of the particle. This was experimentally shown for the yeast L-A virus (Fujimura et al, 1986) and also for the core particles of viruses in the Reoviridae family (Lawton et al, 2000) and the Cystoviridae family (Qiao et al, 2008).
A number of elegant 3D structural studies have shown that the complex, multilayered capsid dsRNA viruses in the Reoviridae family (Grimes et al, 1998; Reinisch et al, 2000; Nakagawa et al, 2003; Yu et al, 2008) and also the φ6 bacteriophage (Huiskonen et al, 2006) share with the simple capsid totiviruses the quaternary organization of their inner capsid layer (Bamford et al, 2005), which has an icosahedral architecture made of 120 subunits. The two independent coat protein (CP) polypeptides in the icosahedral asymmetric unit of this core capsid layer make non-equivalent contacts with each other. This is in contrast to the packing symmetry observed in the majority of icosahedral virus particles analysed, which display a surface lattice triangulation originating from quasi-equivalent contacts between capsid proteins (Caspar and Klug, 1962). The 120-subunits icosahedral architecture is a hallmark of dsRNA viruses, and has not been observed in infectious virions of any other category of viruses. In the Reoviridae and Cystoviridae, this inner capsid is enclosed within a second icosahedral layer, which is organized with triangulation T=13. This second layer is in turn enclosed within a third layer in the case of rotaviruses. In this particular case, the third layer is used for membrane translocation when invading a new cell, delivering a double-layered ‘core' particle, which becomes transcriptionally active in the cytoplasm of the target cell (Pesavento et al, 2006). Orthoreoviruses (the prototype members of the Reoviridae) use, instead, the capsid protein of the second layer (termed mu1) for entry, delivering a transcriptionally active single-layered core particle into the cytoplasm (Reinisch et al, 2000). Mu1 undergoes an activating autoproteolytic cleavage generating a myristoylated N-terminal peptide, which was shown to be critical for entry into cells (Odegard et al, 2004; Ivanovic et al, 2008). Notably different are the Birnaviruses (Coulibaly et al, 2005), which are dsRNA viruses having only a single-layered capsid of icosahedral surface symmetry of triangulation T=13. In this case, the capsid protein (CP) is also matured to release peptides from its C-terminal end that are used for cell invasion (Chevalier et al, 2005; Galloux et al, 2007). Birnaviruses are the exception among dsRNA viruses because they lack the inner 120-subunits layer, and the CP has a shell domain that is homologous to the CP of simple +sRNA insect viruses like the T=3 nodaviruses; yet its projecting domain is homologous to that of the counterpart in the T=13 second layer CP in the members of the Reoviridae.
Here, we report the X-ray structure of a PBV particle, using recombinant virion-like particles (VLPs) obtained by expressing the rabbit PBV capsid protein gene in insect cells. PBVs are frequently detected in stool samples from children with diarrhoea (Bhattacharya et al, 2007; Finkbeiner et al, 2008; Giordano et al, 2008) as well as in samples from immunocompromised patients. Although PBVs are widespread in humans and mammals in general—as well as birds (Chandra, 1997; Masachessi et al, 2007)—our current knowledge of their biology is very limited, mainly because of their non-cultivable status, that is, the absence of a cell culture system for propagating the virus. The first complete nucleotide sequence of the two genomic segments of PBV isolated from humans was published in 2005 (Wakuda et al, 2005). Genomic segment 1 (which is between 2.3 and 2.6 kilobase-pairs long, depending on the genotype) has two open reading frames (ORFs). Although the first one codes for a protein of unknown function, the second one is shown here to encode the CP. The smaller segment 2 has a single-ORF coding for the viral polymerase. The crystal structure shows that the PBV particles have a single-layered icosahedral architecture made of 120 subunits, with the notable difference that the particles are formed by 60 two-fold symmetric dimers of a CP with a new fold—an organization not observed in any icosahedral virus crystallized so far. The architecture is clearly different to that of totiviruses and the inner layer of the Reoviridae, but appears related to that of the simple capsid dsRNA viruses of the Partitiviridae family (Ochoa et al, 2008), which infect yeast and plants. We show that a specific feature of PBV is that the CP undergoes an autocatalytic maturation releasing a positively charged, post-translationally modified peptide that remains associated within the virion, reminiscent of the process involved in the maturation of other non-enveloped animal viruses.
Results
PBV CP spontaneously assembles into VLPs
The identity of the capsid protein coding gene in the PBVs had not been shown earlier. We cloned the second ORF present in segment 1 of rabbit PBV (Green et al, 1999) into a baculovirus vector for production in insect Sf9 cells. The protein encoded displays 25% amino-acid sequence identity with its human counterpart (see Figure 1A). CsCl gradient analysis of recombinant baculovirus infected cell extracts showed the presence of particles that segregate into two discrete bands corresponding to densities of 1.320 and 1.345 g/ml, termed low- (LD) and high-density (HD) bands, respectively. Electron microscopy observations of the material present in the HD band, which contained about 80% of the total material, showed homogeneous 35 nm diameter spherical particles. The diameter of the particles—as well as the overall morphology—is the same as those reported for intact PBV virions isolated from pigs (Ludert et al, 1991). This study thus experimentally identifies the second ORF of PBV genomic segment 1 as coding for the CP.
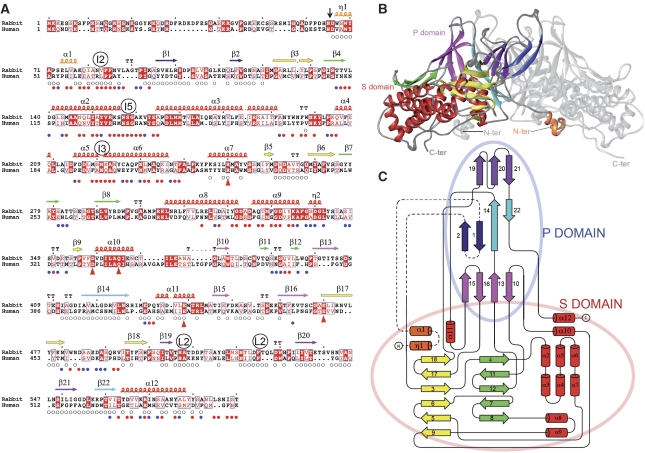
Figure 1.
Structure of the PBV CP. (A) Amino-acid sequence alignment of rabbit and human PBV CP (accession Q9Q1V2 and Q50LE5, respectively) highlighting conserved amino acids. A vertical arrow points to the N-terminal residue of the 55 kD CP found in the particle. Secondary structure elements (SSE) are labelled as α, η and β, for α-helices, 3/10-helices and β-strands, respectively, numbered sequentially from the N terminus and coloured to match the diagrams of the other panels. A labeled circle above the sequence marks residues approaching the icosahedral (I2, I3 and I5) and local (L2) symmetry axes. Small circles below the sequence flag residues involved in L2-related intra-dimer contacts (open circles) and between dimers in the capsid (full coloured circles: blue and red denote single and double (meaning two non-equivalent) contacts in the capsid, respectively. Red triangles below the sequence flag residues involved in the hydrogen-bonding network surrounding the transproteolytic cleavage site. (B) Ribbon diagram of the CP dimer. Shell and Projecting domains are labelled S and P, and the N- and C-termini are indicated. One subunit is coloured by SSE, with all helices in red except for the swapped region (η1-α1, orange) and β-sheets in yellow and green in the S domain, and purple, blue and pink in the P domain. In the blue sheet, dark blue indicates the swapped, N-terminal β1–β2 hairpin. For clarity, the second subunit is shown in grey. (C) Topology diagram. β-strands and α or η helices are indicated by arrows and cylinders, respectively, coloured and numbered as in A, and connected by a black line. A broken line connects elements that are swapped between the two subunits in the dimer. Pale red and blue ovals encompass secondary structure elements of the S and P domains, respectively.
Particles in the HD band contained nucleic acid molecules of cellular origin ranging between 0.1 and 2 kb. In contrast, the material recovered from the LD band was a heterogeneous mix of different sizes and also broken particles containing essentially no nucleic acid. SDS–PAGE analysis revealed a protein of 55 kD present in both bands, as well as a small 7 kD protein present only in the HD band (Supplementary Figure S1). Mass spectrometry (MS) of the 7 kD polypeptide indicated that it consists of a post-translationally modified segment spanning the 65 N-terminal amino acids of CP (Supplementary Figure S2). N-terminal sequencing of the 55 kD protein showed that it corresponds to the remaining of the precursor molecule, amino acids 66–590, which will be referred to as the capsid protein (CP) in the rest of the manuscript. The presence of the 7-kD peptide was observed only when nucleic acid was packaged in the capsid, suggesting a role in genome packing—consistent with the numerous positively charged residues present in its amino-acid sequence, as can be seen in Figure 1A—in addition to a possible role in cell entry (see below ‘Maturation of the CP'). Indeed, preliminary experiments showed that the purified PBV particles have a membrane disrupting activity, inducing leakage of liposome-encapsulated fluorescent dyes (Supplementary Figure S3). Moreover, as had been observed for rotaviruses (Nandi et al, 1992), trypsin treatment of the PBV particles augmented liposome leakage (Supplementary Figure S3). We were however unable to identify the trypsin cleavage site in the PBV particles.
Structural studies
VLPs recovered from the HD band were used for crystallization trials and for electron cryomicroscopy (cEM) analyses. Diffraction quality crystals were obtained as described in Materials and methods. An initial cEM reconstruction to 20 Å resolution (Figure 2B) was combined with X-ray diffraction data collected at synchrotron sources to extend phases using 30-fold averaging to a resolution of 3.4 Å. This procedure resulted in a very clear electron density map of the VLP (Supplementary Figure S4), allowing the trace of the CP polypeptide chain. No density corresponding to the 7-kD peptide was observed. The model was refined against structure factors measured between 50 and 3.4 Å resolution, the final statistics are shown in Table I. The final model contains two independent polypeptide chains, labelled A and B, which are not related by the icosahedral symmetry of the particle, with residues A66 to A590 and B66 to B586 (i.e. the last four residues of chain B are disordered).
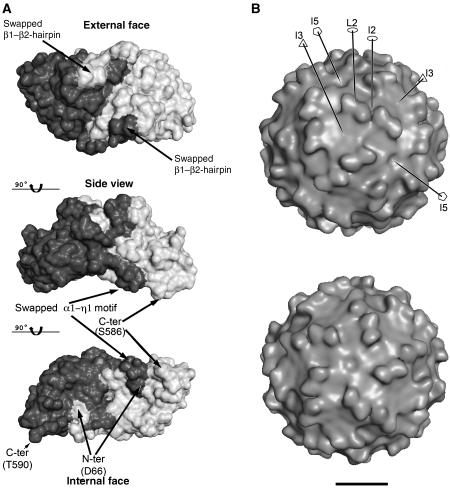
Figure 2.
Surface features of the PBV capsid. (A) CP dimer with the subunits in different shades of grey. Top and bottom are views down the L2 axis, from outside and inside the VLP, respectively, with a side view in the middle. The internal curvature of the CP dimer matches roughly the internal radius of the particle. Note the intricate interface between subunits, generated by the N-terminal β1β2 and α1-η1 swapping, as indicated. The location of the N-terminus marks the site of the transproteolytic cleavage. (B) Surface of the VLP. cEM 3D-reconstruction showing the presence of 60 dimeric protrusions formed by domain P. The contour level was chosen to accommodate the volume of the CP dimer in the icosahedral asymmetric unit. A few symmetry axes are labelled for orientation. Top and bottom panels are seen down the I3 and I5 axes, respectively, slightly miss-oriented in order to help visualize certain features, like the cleft at the I2 axes, the I5 depression and the flat I3 region. Some of the symmetry axes are drawn, with standard symbols for 2-, 3- and 5-fod axes (empty ellipse, triangle and pentagon, respectively) and labelled. Bar: 100 Å.
Table 1.
Data collection and refinement statistics
| Data collection | |
| Space group | P3221 |
| Cell dimensions | |
| a, b, c (Å) | 407.42, 407.42, 808.63 |
| α, β, γ (deg) | 90.0, 90.0, 120.0 |
| Resolution (Å) | 80.0–3.4 (3.6–3.4)a |
| Rsym or Rmerge | 24.0 (52.6)a |
| I/σI | 3.3 (1.0)a |
| Completeness (%) | 69.9 (41.6)a |
| Redundancy | 1.5 (1.3)a |
| Refinement | |
| Resolution (Å) | 50.0–3.4 |
| No. of reflections | 736678/3758 |
| Rwork/Rfree b | 27.2/27.3 |
| No. atoms | |
| Protein | 8234 |
| Water | 52 |
| B-factors | |
| Protein | 34.1 |
| Water | 5.0 |
| RMS deviations | |
| Bond lengths (Å) | 0.010 |
| Bond angles (deg) | 1.5 |
aValues in parentheses are for highest-resolution shell.
bRfree value calculated with 0.5% of the relflections data set, selected by resolution shells. Because of the high redundancy in the data set due to a 30-fold non-crystallographic symmetry, the Rfree does not represent a ‘free' set, hence the very small difference with the working R. To avoid over-fitting, refinement was therefore carried out with a high weight to the geometry of the model, and the NCS were maintained as constraints throughout, to reduce the number of parameters to be fitted.
The PBV capsid is built of 60 symmetric dimers
The 3D structure revealed a particle made of 60 symmetric CP dimers displaying an intricate interface, with the N-terminal 55 residues (amino acids 66–120, see Figures 1 and 2A) exchanged between the two subunits. The trace of the polypeptide chain displays a complex topology (Figure 1B and C); a search in the protein database identified no entry with significant structural homology, indicating that the 3D-fold of PBV CP has not been observed earlier. The tertiary organization gives rise to a compact structure in which two domains can be recognized, a shell (S) domain that contains all of the α-helices and two β-sheets (yellow and green in Figure 1B). The remaining β-sheets are part of the projecting (P) domain, which forms the notable protrusions that stand out in the low resolution cEM 3D reconstruction (Figure 2B). The CP dimer is such that each domain is shared by the two subunits, with the N-terminal β-hairpin (β1–β2, dark blue in Figure 1) inserted in the P domain and the N-terminal helix-turn-helix motif (η1–α1, orange) in the S domain of the partner subunit in the dimer. This intricate interface, highlighted in Figure 2A, indicates that dimerization takes place during folding of the polypeptide chains, giving rise to an extensive buried surface of about 12400 Å2 (6200 Å2 per subunit), implying that the CP dimer is the building block for particle assembly.
The inner surface of the dimer has a smooth curvature (Figure 2B, middle row) roughly matching that of the inner radius of the particle (130 Å, see also Figure 3). Although each subunit of the CP dimer makes non-equivalent contacts in the particle, there are only minor distortions of the local two-fold symmetry of the dimer, affecting mainly side-chains and loops in the contact regions (Supplementary Figure S5). The overall root mean square deviation after superposition of the refined models of the two subunits of the dimer is 0.95 Å for 521 α carbons; the rotation relating the two subunits is very close to 180.0 degrees (Supplementary Table S1). This axis is indicated by L2 (for ‘local' two-fold axis, Figure 2B) to differentiate from the icosahedral symmetry axes, which are denoted by I2, I3 and I5 (for two-fold, three-fold and five-fold icosahedral axes, respectively) in the descriptions below. Figure 2B shows that the L2-related dimeric protrusions of CP are in turn related by the I2 axes, which lie in a deep cleft between adjacent CP dimers. The reconstruction also shows that there is a small depression at each of the I5 and a flat region at the I3 axes.
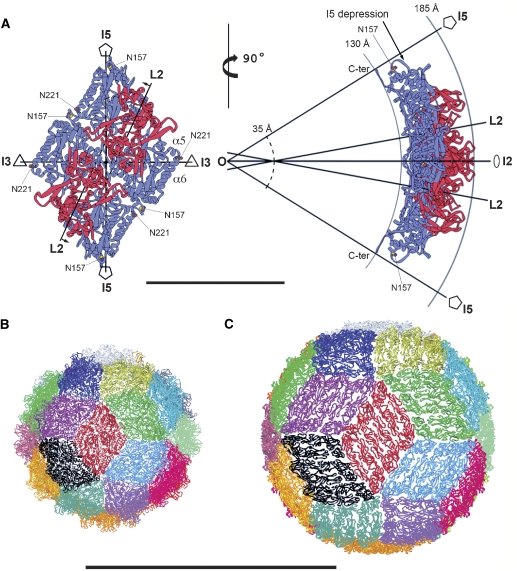
Figure 3.
Parallel dimers define the outline of a rhombus-shaped tile on the icosahedral particle. (A) Two CP dimers related by an I2 axis of the particle are displayed as ribbons, with the S and P domains coloured blue and red, respectively. The nearest icosahedral and local symmetry axes are drawn in projection and labelled. The left panel is projected down the I2 axis. The L2 axes make an angle of about 11.5 degrees with the I2 axis and do not intersect each other, but lie in parallel planes perpendicular to the paper; small arrows by the L2 labels indicate the direction and length (6.2 Å) of the translation needed to bring these planes into coincidence with a parallel vertical plane containing the I2 axis. The right panel shows an orthogonal view rotated about a vertical axis. Full-line arcs indicate the inner and outer radii of the particle (labelled), highlighting the curvature of the diamond tile. The L2 axes cross the vertical plane containing the I2 axis (which is in the plane of the paper) along a vertical line, at a distance of 14.5 Å from each other, above and below the I2 axis. This vertical line intersects the I2 axis at a radius of about 35 Å (broken line arc), indicating a sharper local curvature at the I2 contact than that of the overall particle. The particle centre is indicated by O, where I5, I3 and I2 axes intersect. Residues Asn 157 and Asn 221 are displayed as yellow and red spheres and labelled, as well as helices α5 and α6 mentioned in the text. C-ter labels the last four amino acids of chain A (587–590, disordered in chain B) running along the I5 axis. Bar: 100 Å. (B). Triacontahedral design of the PBV particle, each of the 30 rhombic tiles is coloured differently. The red tile is oriented as the one displayed in the top panel. (C) Convergence with the icosahedral organization with the flavivirus particle, where each of the 30 tiles (coloured as in B) is composed of 3 parallel dimers of the envelope protein. Coordinates obtained from PDB entry 1THD. Panels B and C are at the same scale. Bar: 500 Å.
Conserved segments are involved in capsid contacts
The alignment of the two available sequences of PBV CP (rabbit and human) displayed in Figure 1A shows blocks of amino-acid conservation spread apparently at random throughout the primary structure of the protein. In contrast, in the 3D structure these patches display a consistent pattern, mapping to regions that are mainly involved in dimer–dimer contacts in the icosahedral capsid (indicated with small full circles underneath the sequence), whereas the intra-dimer contacts (small open circles) appear less conserved. One exception is the conserved block at the centre of helix α10, buried within the CP subunit, which includes residues participating in the hydrogen-bonding network surrounding the transproteolytic cleavage site used to generate the CP N terminus (see below and Supplementary Figure S6). The conservation pattern also shows that the P domain spans the least conserved regions of the polypeptide chain, consistent with being exposed and likely to undergo antigenic drift, as well as being potentially used for receptor binding with different specificities in viruses infecting different hosts.
Important conserved motifs are located in regions approaching the icosahedral axes of the particle. For instance, the α2–α3 loop has the conserved sequence 157-NSG-159 lying right at the I5 axis (Figures 1A and 3A). The structure shows that Gly 159 is conserved because side chain atoms would collide with their five-fold-related counterparts. The side chain of the preceding Ser 158 contributes to a ring of interactions about the I5 axis, whereas its main chain carbonyl hydrogen bonds the main chain amide of Gly 159 of the neighbouring subunit, in a β interaction that results in a thin five-fold β-annulus sealing the I5 depression of the particle. Finally, the Asn 157 side chain hydrogen bonds its main chain carbonyl, stabilizing the polypeptide chain in the conformation required to allow the described interactions formed by the immediately downstream residues. The five α2–α3 loops appear to constitute a sort of diaphragm—as seen down the I5 axis (Figure 4). The I5 depression leaves room for a concerted movement of the five loops to open the diaphragm by breaking open the five-fold hydrogen bonds of the β-annulus, triggered by a putative conformational change during transcription, as is the case in other dsRNA viruses (see Discussion).
Figure 4.
View of the PBV particle down an I5 axis. The individual subunits are coloured according to secondary structure elements: α-helices blue, β-sheets red and random coil orange/light brown. Note the I5 loop with the conserved 157-NSG-159 segment at the centre, forming a diaphragm-like structure at the I5 axis. Bar: 100 Å
At the I3 axes, the side chain of conserved Asn 221 breaks the short helix α5 by capping its C-terminal end, resulting in a 120 degrees kink of the polypeptide chain, stabilizing this corner of the CP dimer and contributing to the compact packing of dimers at the particle surface (Figure 3). The conserved residues at both I5 and I3 axes display a nearly identical conformation at the L2-related site (see Supplementary Figure S3). Moreover, the conserved Asn 159 and Asn 221 (at the I5 and I3 axes, respectively) come together to contact each other at the L2-related site (highlighted in Figure 3A). In summary, considering the limited sequence identity between human and rabbit PBVs (25%), the conservation pattern helps in pointing to residues with likely functional properties.
Maturation of the CP
Many non-enveloped animal viruses undergo an autoproteolytic cleavage reaction, which activates the virion for entry usually by priming the particle for the release of a membrane-active peptide on interactions with the host. This maturation step often takes place on assembly of the particle. The PBV CP structure provides evidence for intra-dimer, transproteolytic cleavage between residues 65 and 66 (arrow in Figure 1A). The N-terminal residue Asp 66 of one subunit remains in the presumed catalytic site of the opposite subunit in the dimer. The side chain of Asp 66 points to solvent at the interior of the particle. The very clear experimental electron density map shows that the charged amino-terminal group of the polypeptide chain is positioned to make a bidentate hydrogen bonding interaction (and salt bridge) with a conserved buried glutamic acid, Glu 439 of the partner subunit (Supplementary Figure S6). This interaction stabilizes the post-cleavage structure. Proteolysis takes place C-terminal to conserved Asn 65 (Figure 1A), similar to the described autocatalytic nodavirus and tetravirus endopeptidase activity (Reddy et al, 2003), in which cleavage occurs after a conserved asparagine residue (Hosur et al, 1987; Agrawal and Johnson, 1992). The proposed catalytic mechanism involves a carboxylate group (the side chain of an aspartic or glutamic acid) of CP that gets positioned during assembly such that it activates the catalytic asparagine, resulting in nucleophilic attack of its side chain amide nitrogen onto its own carbonyl carbon, cleaving the peptide bond. A candidate residue for the activation in PBV is the conserved Asp 359 in the adjacent subunit. This residue was not mutated in this work to assess its role, but mutating Asn 65 did abolish the proteolytic reaction, giving rise to aberrant particle morphogenesis (Supplementary Figure S1).
The proteolytic maturation of PBV CP is also reminiscent of the autocatalytic cleavage of the orthoreovirus capsid protein mu1, which takes place after a conserved asparagine residue and is critical for cell entry (Odegard et al, 2004). This process generates an N-terminal peptide (mu1N) 41 amino acids long, myristoylated at its N-terminal glycine residue, which remains attached to the capsid. In PBV, there is no N-terminal glycine that could undergo myristoylation. Nevertheless, our MS analysis of the peptide shows a complex profile (Supplementary Figure S2), indicating the presence of multiple post-translational modifications. First, this analysis shows that the initiating methionine is retained in the mature protein. Second, the MS plot shows peaks corresponding to acetylation at the N-terminus and at multiple other sites, most likely the side chains of the many lysine residues present. Indeed, the spectrum has peaks with a periodicity of 42 daltons—the mass difference between modified peptides differing by one acetyl group—that extend up to the mass corresponding to acetylation at all 11 lysine side chains present in the peptide, in addition to the acetyl group at the N terminus. Third, the plot displays peaks corresponding to up to two oxidations of the multiply acetylated peptide, consistent with the presence of two methionines in the sequence (Figure 1A), which are susceptible to oxidation. The PBV CP maturation cleavage thus generates a charged peptide that is likely to interact electrostatically with nucleic acid, with the total charge apparently controlled by acetylation (reminiscent of histone tails, An, 2007). The released peptide does not display an obvious hydrophobic region or amphipathic pattern that could be proposed to interact with membranes, but it is possible that the acetylated lysine side chains contribute a sufficiently hydrophobic surface to play this role. Whatever the case, the mechanism of the observed liposome leakage effect documented in Supplementary Figure S3 remains to be further investigated to understand the possible roles of the modified 7-kD peptide during cell entry.
Triacontahedral design of the PBV particle
The outline of the CP dimer is a parallelogram when looking down its local two-fold axis (Figure 2A). The I2 axes of the particle relate two CP dimers such that they form a rhombus, or diamond tile (Figure 3A). The side view shows that the tile is concave in shape, resulting from the dimer curvature (Figure 2A, middle panel) and the angle between the L2 axes of the tile (Figure 3A, right panel). Together, the 30 tiles give rise to a spherical particle with triacontahedral design (Figure 3B). None of the icosahedral non-enveloped viruses crystallized so far displays a spherical triacontahedral organization made of parallel dimers. Only the enveloped flaviviruses have this type of arrangement of the envelope proteins at the virus surface (Figure 3C). The mature flavivirus particles display 30 concave diamond tiles made of three parallel dimers of the envelope (E) protein, arranged with icosahedral symmetry in a so-called ‘herringbone' pattern (Kuhn et al, 2002). As indicated in Figure 3B, in both particles the dimers are shifted laterally along their long axes to match the edges of the diamond tile. The structures of the PBV and flavivirus particles, thus, provide an example of convergence of two totally unrelated proteins towards a common motif, giving rise to a very compact spherical shell with icosahedral symmetry.
Comparison with dsRNA viruses of known structure
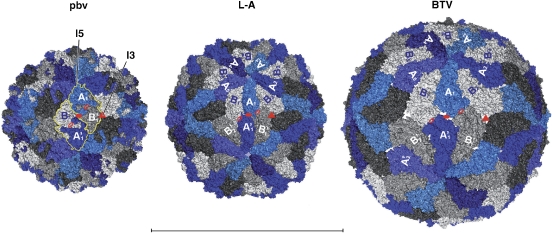
Icosahedral capsids with 120 subunits are found exclusively in dsRNA viruses (reviewed in Bamford et al, 2005). Under special in vitro conditions, the capsid protein of the T=3 +sRNA bromoviruses can also form subviral particles made of 60 dimers of the CP (Krol et al, 1999; Zlotnick et al, 2000), but the infectious virions display a T=3 quasi-equivalent icosahedral architecture made of 90 CP dimers. In contrast to PBV, the 120 CP subunits of dsRNA virions crystallized so far are not arranged as 60 two-fold symmetric dimers. In this respect, the PBV particle exhibits an overall icosahedral architecture closer to that of the bromovirus subviral particles. Figure 5 shows a comparison of the PBV capsid with its counterparts from L-A virus (Naitow et al, 2002) and the blue tongue virus (BTV, a member of the Reoviridae family (Grimes et al, 1998)) inner capsid layer. In the latter two viruses, the two independent subunits in the icosahedral asymmetric unit—one by the I5 (chain A, in blue) and the other by the I3 axes (chain B, in grey)—adopt different conformations and are not two-fold related. Particle assembly has been proposed to take place through decameric intermediates (Grimes et al, 1998)—indicated with different shades of grey in Figure 5. It is clear that the dimeric building blocks are assembled differently in PBV, with each CP dimer combining subunits that would be across decameric caps in the others. However, a distorted quasi-two-fold local axis relating decameric caps is present in the latter, as indicated in Figure 5. The best superposition of the subunits about this axis involves a rotation of close to 160 instead of 180 degrees (Supplementary Table S1).
Figure 5.
Comparison with L-A virus (PDB entry 1M1C) and the BTV inner layer (PDB entry 2 BTV). The particles are displayed in molecular surface representations viewed down an I2 axis, with the two independent subunits coloured blue (chain A, by the I5 axes) and grey (chain B, by the I3 axes). For clarity, a yellow contour outlines two I2-related CP dimers in the PBV particle (left panel). Red full symbols indicate icosahedral axes (triangle for I3 and ellipse for I2). The L2 axes (or distorted L2 axes in the case of L-A virus and BTV, see Supplementary Table S1) are shown by open red ellipses. One of the 12 decameric CP caps is labelled An and Bn, n=1 to 5, with the 5 B subunits in light grey. Note the very large interface of each B subunit with its two nearest A neighbors (A5 and A1 for B1, etc). Subunits labelled A'n and B'n belong to an I2-related decameric cap with the B subunits coloured intermediate grey; a third cap to the right, related by the labelled I3 axis, is drawn with the B subunits coloured dark grey (not labelled). In contrast to L-A virus and BTV, the PBV dimer mixes subunits from two adjacent decameric caps (AB' and BA'), indicating that the assembly pathway is necessarily different. All three panels are drawn at the same scale. Bar: 500 Å.
Similarities with partitiviruses
A 7.3 Å 3D cEM reconstruction of PsV-S—a partitivirus infecting the fungus Penicillum stolloniferum—was very recently reported (Ochoa et al, 2008). This structure shows that the capsid of PsV-S also features 60 symmetric dimers, with an overall organization similar to the PBV particle described here. The PsV-S particles appear slightly smaller than the PBV counterparts, and instead of the projections observed in PBV, display an ‘arch' that protrudes at the centre of each dimer. Inspection of an alignment of the amino-acid sequences of the CP of 12 partitiviruses available in the database does not show a conserved asparagine that would be responsible for an autoproteolytic cleavage as in PBV, in line with the fact that PsV-S, infecting a primitive eukaryote, is not likely to have the ability to infect cells from outside. The crystal structure of a partitivirus is in progress (Yizhi Tao, personal communication) and will provide a way to compare the 3D-folds of the CPs in the two viruses, and their precise interactions to make the particle. It will be very interesting at that point to carry out a close comparative analysis of the corresponding architectures.
Discussion
One important point that is clarified by the structure reported here is the filiation of PBVs, which had been widely believed to be a small version of birnaviruses. PBV virions had actually been reported as having a similar diameter to noda- and tetravirus particles—and were tentatively assigned an icosahedral surface lattice of triangulation T=3 (Ludert et al, 1991). Because of our earlier discovery that the birnavirus capsid protein is homologous to that of small +sRNA viruses—like the T=3 nodaviruses and the T=4 tetraviruses (Coulibaly et al, 2005), we set out to explore in more detail the PBV particle structure to see whether they are intermediate between small, non-enveloped +sRNA viruses and the dsRNA birnaviruses. As PBVs are non-cultivable, we used an approach that proved successful to obtain the X-ray structure of the Norwalk virus capsid (Prasad et al, 1999)—also a non-cultivable virus that is responsible for gastroenteritis in humans—which made use of recombinant VLPs produced with the baculovirus system. The only electron micrographs of authentic PBV virions are those reported from pig samples (Ludert et al, 1991). The size and features observed in those micrographs are compatible with the structure reported here, indicating that the recombinant VLPs have the same structure as authentic PBV virions. The latter are expected to contain at least one of the two genomic segments as well as the viral polymerase, in contrast to the VLPs, which contain random cellular nucleic acid. The internal molecules are, however, not organized with the symmetry of the particle, and neither the polymerase nor the genomic dsRNA would have been visible if the crystallized particles had been authentic virions. A possible difference could be the conformation of one or two of the five-fold portals for extruding mRNAs during transcription, if the RNA polymerase is located directly underneath, as has been observed in the case of viruses in the Reoviridae family. Notably, there is a conserved sequence element at the five-fold axes, and a diaphragm-like organization of the five loops that seal this pore (Figure 4), with dimensions compatible with a putative ‘portal' function during translocation of a single-stranded RNA transcript.
The structure of the PBV VLPs thus shows that they are not related to birnaviruses, and display a different capsid architecture made of 60 symmetric dimers, with the closest relatives appearing to be viruses infecting unicellular eukaryotes and plants, like the partitiviruses (Ghabrial et al, 2005). The comparison with totiviruses and the inner layer of the members of the Reoviridae shows that the PBV particles are organized differently (Figure 5). The structural organization of the former, however, has a remnant of a dyad relating the two independent subunits in the particle. Our data thus provide new insights into the organization of dsRNA viruses in general, pointing to a simple common organization of the simplest—perhaps the most ‘primitive'—members of this category of viruses. Bacteriophages of the Cystoviridae family also have an icosahedral inner core made of 120 protein subunits within a complex icosahedral capsid, with an organization of the core that appears different to both, totiviruses and PBVs, at least to the resolution to which these particles have been analysed (Huiskonen et al, 2006). Higher resolution structural data on cystoviruses are necessary to assess the extent of their possible relationship to dsRNA viruses infecting eukaryotic cells.
The discovery of the strong similarity in the organization of PBVs and partitiviruses—which are known to infect fungi and also plants—is in line with reports of a correlation between the detection of PBV-like viruses with the presence of oocysts of Cryptosporidium typical of Cryptosporidium parvum (Gallimore et al, 1995) in human stools. C. Parvum is a protozoan parasite that is responsible for gastroenteritis in humans (Caccio, 2005). Incidentally, the only characterized virus of C. Parvum has been identified as a partitivirus (Khramtsov and Upton, 2000), and it would be interesting to investigate more closely the structural organization of these PBV-like viruses (reported to have a considerably smaller genome than standard PBVs) and to compare them to the partitiviruses. A specific feature of partitiviruses is that each individual particle carries only one genomic segment, thereby prompting the question of whether each PBV particle carries the full genomic information.
The autoproteolytic processing of the PBV capsid protein, together with the liposome-perforating capacity of the particles provide strong weight to the interpretation that PBVs are indeed animal viruses, and not viruses of some eukaryotic parasite present in the intestinal flora of vertebrates. In contrast to viruses infecting plants or unicellular eukaryotes, animal viruses need a means of translocating across the cell membrane for entry. Several non-enveloped animal viruses have been shown to use a similar autocatalytic cleavage of the capsid protein precursor for this purpose, generating a peptide that is retained in their capsid and which is used for entry, as discussed above for the reovirus Mu1 protein. Similar cleavage events have been observed in the capsid protein of many other non-enveloped animal viruses, the picornaviruses for instance, but not in plant viruses nor in the simple capsid dsRNA viruses characterized to date. The mechanism of membrane destabilization by the PBV particles remains, however, to be elucidated, in particular the role—if any—of the post-translational modifications of the otherwise hydrophilic and charged 7-kD peptide, which is unlike viral membrane-active peptides characterized this far. Another feature presented by PBVs in common with other animal viruses is the variability of the projecting domain, which despite the limited data, can be inferred from the comparison between the rabbit and human PBV sequences of Figure 1A. These domains are the most exposed, and would be targeted by the adaptive immune system of vertebrates, as well as being responsible for interactions with an entry receptor that may vary from species to species. Serological studies of patients in which PBV has been detected are needed to understand the antigenicity of the virus.
Concluding remarks
The structural data indicate that the complex-capsid dsRNA animal viruses of the large Reoviridae family appear to have their roots in the simple capsid totiviruses. PBVs, in contrast, appear related to the simple partitiviruses, and evolved a different solution to undergo an extra-cytoplasmic cycle. Moreover, the structural relation of PBVs with partitiviruses suggests the possibility that PBVs may have crossed the species barrier from putative unicellular eukaryotic organism to infect vertebrates. Given the large number of microorganisms present in the intestinal flora of vertebrates, such species-crossing events are likely to be more common than anticipated earlier, and as long as the resulting viruses remain apathogenic for their new host, they are essentially unnoticed. With more and more outbreaks of new and emerging viral infections—and with indirect signals indicating that PBVs maybe involved in pathogenic symptoms, at least in immunocompromised patients—it becomes important to gather as much knowledge as possible on all the different viruses dwelling in the human environment, and understand the adaptation mechanisms at play.
Materials and methods
Plasmids and recombinant baculovirus construct
A DNA plasmid containing the PBV CP sequence was used to generate the BacPBV recombinant baculovirus. Briefly, competent DH10Bac cells containing the bacmid were transformed with the pFastBac plasmid derivative. Colonies containing recombinant bacmids were identified by disruption of the lacZ gene. High-molecular-weight mini-prep DNA was prepared from selected colonies, and this DNA was used to transfect Sf9 cells with Lipofectin. Recombinant baculovirus were obtained by standard procedures. High-titer viral stocks of the recombinant baculoviruses (108 PFU/ml) were then prepared for subsequent use.
VLP Preparation
Sf9 cells were infected with the recombinant BAcPBV at a multiplicity of infection higher than 5 PFU/ml in the presence of the protease inhibitors leupeptin (0.5 μg/ml) and aprotinin (1 μg/ml), collected 100 h after infection (after addition of the same protease inhibitors at identical concentration), and then treated with Freon 113. Purification was carried out by density gradient centrifugation in a CsCl solution at a density of 1.32.
Electron microscopy
Adequate VLP samples were studied by cEM using holey carbon film coated grid vitrified in liquid nitrogen cooled ethane. The grids were transferred with a Gatan cryo-holder 626 in a Philips CM 12 microscope operated at 80 kV. Images were recorded at a magnification of 35 000 on Kodak SO-63 developed 12 min in D-19 developer at room temperature.
For image analysis, the quality of micrographs was checked by optical diffraction. Suitable micrographs were scanned with a Nikon Coolscan 8000 with a pixel size of 12.7 μm, corresponding to 0.363 nm at the specimen level. 520 particles were manually selected with the × 3d program (Conway and Steven, 1999). Individual images were corrected for the phase contrast effects using the ctfit program (Ludtke et al, 1999).
All further image analysis was performed using the Imagic-5 software package (van Heel et al, 2000). The images were band-pass filtered between 700 and 5 Å, and translationally centred and classified using multivariate statistical analysis. This analysis revealed the existence of classes of views presenting two, three and five rotational axes, compatible with icosahedral symmetry. A preliminary 3D map was calculated by angular reconstitution imposing icosahedral symmetry. Images of irregular particles were discarded during this procedure. The view orientation was further refined by projection matching using an angular search step of 2 degrees. A final map was calculated using 416 particle images, at a resolution of about 20 Å estimated by Fourier shell correlation with a 0.5 correlation cutoff.
Crystallization and diffraction data collection
The VLP preparation was concentrated to 8 mg/ml in Tris 25 mM pH 7.5 and 50 mM NaCl. Crystals were grown by vapour diffusion at 18 °C in sitting drops by mixing 1 μl of a reservoir solution consisting of 1.2 M ammonium sulfate and 100 mM Tris pH 7.5. Crystal growth was slow, with the best crystals reaching the maximum size (200 × 150 × 100 μm) over a period of 3 to 4 months. For data collection, the crystals were transferred to a cryoprotectant consisting of 20% (w/w) glycerol, 10% PEG-400, 1.5 M ammonium sulphate and 100 mM Tris pH 7.5 and then cryo-cooled in liquid ethane and stored in solid ethane in a liquid nitrogen dewar until defrosted in the cryostream at beamline PX06SA at the Swiss Light Source, where diffraction data were collected at a wavelength of 1.0 Å, using a mar225 CCD detector (distance 350 mm and oscillations 0.25° per frame at 100°K). Programs MOSFLM (Leslie, 2006) and SCALA (Evans, 2006) were used for data processing. Crystals belonged to the P3221 space group, with unit cell dimensions a=b=408.0 Å and c=811.3 Å and contained half a particle in the asymmetric unit, allowing 30-fold non-crystallographic symmetry (NCS) averaging for phase extension.
Structure determination
The orientation and position of the particle was determined by molecular replacement using AMoRe (Navaza, 1994) and the cryo-EM map at 20 Å resolution. 30-fold NCS averaging with RAVE, MAMA and other programs of the Uppsala suite (Jones, 1992), in combination with the CCP4 suite (CCP4, 1994) were used for phase extension to 3.4 Å resolution. This procedure yielded a very clear electron density map that allowed tracing amino acids 66 to 590 for subunit A and 66 to 586 for subunit B with program O (Jones et al, 1991) and subsequent refinement of the model using CNS (Brunger et al, 1998) to final R-factor 27.2% for data between 50 and 3.4 Å. The Ramachandran plot indicates that 74.9%, 23.9% and 1.1% of the residues are in most favoured, additional allowed, and generously allowed regions, respectively, according to PROCHECK (Laskowski et al, 1993). Data collection and refinement statistics are summarized in Table I.
Illustrations
Figures were prepared using Program ESPript (Gouet, 1999), Ribbons (Carson, 1997) and Pymol (http://pymol.sourceforge.net).
Accession numbers
Coordinates and structure factor amplitudes have been deposited in the Protein Data Bank under accession number 2VF1.
Supplementary Material
Supplementary Material
Acknowledgments
We thank F Ternois for help in the initial stages of the project, as well as B Valot and J Chamot-Rooke who carried out the ETD and FTICR mass spectrometry analyses, respectively. The cDNA coding for the PBV CP ORF was kindly provided by CI Gallimore, CPHL, London, UK. Diffraction data were collected at the PX06SA beamline at SLS, Villigen, Switzerland. This work was supported by CNRS, INRA, I Pasteur, Unversité Paris XI and Merck-Serono.
References
- Agrawal DK, Johnson JE (1992) Sequence and analysis of the capsid protein of Nudaurelia capensis omega virus, an insect virus with T=4 icosahedral symmetry. Virology 190: 806–814 [DOI] [PubMed] [Google Scholar]
- An W (2007) Histone acetylation and methylation: combinatorial players for transcriptional regulation. Subcell Biochem 41: 351–369 [PubMed] [Google Scholar]
- Bamford DH, Grimes JM, Stuart DI (2005) What does structure tell us about virus evolution? Curr Opin Struct Biol 15: 655–663 [DOI] [PubMed] [Google Scholar]
- Bhattacharya R, Sahoo GC, Nayak MK, Rajendran K, Dutta P, Mitra U, Bhattacharya MK, Naik TN, Bhattacharya SK, Krishnana T (2007) Detection of Genogroup I and II human picobirnaviruses showing small genomic RNA profile causing acute watery diarrhoea among children in Kolkata, India. Infect Genet Evol 7: 229–238 [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr 54: 905–921 [DOI] [PubMed] [Google Scholar]
- Caccio SM (2005) Molecular epidemiology of human cryptosporidiosis. Parassitologia 47: 185–192 [PubMed] [Google Scholar]
- Carson M (1997) Ribbons. Methods Enzymol 277: 493–505 [PubMed] [Google Scholar]
- Caspar DL, Klug A (1962) Physical principles in the construction of regular viruses. Cold Spring Harb Symp Quant Biol 27: 1–24 [DOI] [PubMed] [Google Scholar]
- CCP4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr 50: 760–763 [DOI] [PubMed] [Google Scholar]
- Chandra R (1997) Picobirnavirus, a novel group of undescribed viruses of mammals and birds: a minireview. Acta Virol 41: 59–62 [PubMed] [Google Scholar]
- Chevalier C, Galloux M, Pous J, Henry C, Denis J, Da Costa B, Navaza J, Lepault J, Delmas B (2005) Structural peptides of a nonenveloped virus are involved in assembly and membrane translocation. J Virol 79: 12253–12263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway JF, Steven AC (1999) Methods for reconstructing density maps of ‘single' particles from cryoelectron micrographs to subnanometer resolution. J Struct Biol 128: 106–118 [DOI] [PubMed] [Google Scholar]
- Coulibaly F, Chevalier C, Gutsche I, Pous J, Navaza J, Bressanelli S, Delmas B, Rey FA (2005) The birnavirus crystal structure reveals structural relationships among icosahedral viruses. Cell 120: 761–772 [DOI] [PubMed] [Google Scholar]
- Estes M, Kapikian A (2007) Rotaviruses. In Fields Virology, Knipe DM, Howley PM (eds), Vol. 2, 5 edn, pp 1917–1974. Philadelphia: Lippincott Williams and Wilkins [Google Scholar]
- Evans P (2006) Scaling and assessment of data quality. Acta Crystallogr 62: 72–82 [DOI] [PubMed] [Google Scholar]
- Finkbeiner SR, Allred AF, Tarr PI, Klein EJ, Kirkwood CD, Wang D (2008) Metagenomic analysis of human diarrhea: viral detection and discovery. PLoS Pathog 4: e1000011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimura T, Esteban R, Wickner RB (1986) In vitro L-A double-stranded RNA synthesis in virus-like particles from Saccharomyces cerevisiae. Proc Natl Acad Sci USA 83: 4433–4437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallimore CI, Green J, Casemore DP, Brown DW (1995) Detection of a picobirnavirus associated with Cryptosporidium positive stools from humans. Arch Virol 140: 1275–1278 [DOI] [PubMed] [Google Scholar]
- Galloux M, Libersou S, Morellet N, Bouaziz S, Da Costa B, Ouldali M, Lepault J, Delmas B (2007) Infectious bursal disease virus, a non-enveloped virus, possesses a capsid-associated peptide that deforms and perforates biological membranes. J Biol Chem 282: 20774–20784 [DOI] [PubMed] [Google Scholar]
- Ghabrial S, Buck K, Hillman B, Milne R (2005) Partitiviridae. London: Elsevier/Academic Press [Google Scholar]
- Giordano MO, Masachessi G, Martinez LC, Barril PA, Ferreyra LJ, Isa MB, Nates SV (2008) Two instances of large genome profile picobirnavirus occurrence in Argentinian infants with diarrhea over a 26-year period (1977-2002). J Infect 56: 371–375 [DOI] [PubMed] [Google Scholar]
- Gouet P, Courcelle E, Stuart DI, Metoz F (1999) ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics 15: 305–308 [DOI] [PubMed] [Google Scholar]
- Green J, Gallimore CI, Clewley JP, Brown DW (1999) Genomic characterisation of the large segment of a rabbit picobirnavirus and comparison with the atypical picobirnavirus of Cryptosporidium parvum. Arch Virol 144: 2457–2465 [DOI] [PubMed] [Google Scholar]
- Grimes JM, Burroughs JN, Gouet P, Diprose JM, Malby R, Zientara S, Mertens PP, Stuart DI (1998) The atomic structure of the bluetongue virus core. Nature 395: 470–478 [DOI] [PubMed] [Google Scholar]
- Grohmann GS, Glass RI, Pereira HG, Monroe SS, Hightower AW, Weber R, Bryan RT (1993) Enteric viruses and diarrhea in HIV-infected patients. Enteric Opportunistic Infections Working Group. N Engl J Med 329: 14–20 [DOI] [PubMed] [Google Scholar]
- Hosur MV, Schmidt T, Tucker RC, Johnson JE, Gallagher TM, Selling BH, Rueckert RR (1987) Structure of an insect virus at 3.0 A resolution. Proteins 2: 167–176 [DOI] [PubMed] [Google Scholar]
- Huiskonen JT, de Haas F, Bubeck D, Bamford DH, Fuller SD, Butcher SJ (2006) Structure of the bacteriophage phi6 nucleocapsid suggests a mechanism for sequential RNA packaging. Structure 14: 1039–1048 [DOI] [PubMed] [Google Scholar]
- Ivanovic T, Agosto MA, Zhang L, Chandran K, Harrison SC, Nibert ML (2008) Peptides released from reovirus outer capsid form membrane pores that recruit virus particles. EMBO J 27: 1289–1298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones TA (1992) A, yaap, asap, @*? A set of averaging programs. In Molecular Replacement, EJ Dodson, S Gover and W Wolf (eds) pp 91–105. Warrington, England: SERC Daresbury Laboratory [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard M (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr 47 (Pt 2): 110–119 [DOI] [PubMed] [Google Scholar]
- Khramtsov NV, Upton SJ (2000) Association of RNA polymerase complexes of the parasitic protozoan Cryptosporidium parvum with virus-like particles: heterogeneous system. J Virol 74: 5788–5795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krol MA, Olson NH, Tate J, Johnson JE, Baker TS, Ahlquist P (1999) RNA-controlled polymorphism in the in vivo assembly of 180-subunit and 120-subunit virions from a single capsid protein. Proc Natl Acad Sci USA 96: 13650–13655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn RJ, Zhang W, Rossmann MG, Pletnev SV, Corver J, Lenches E, Jones CT, Mukhopadhyay S, Chipman PR, Strauss EG, Baker TS, Strauss JH (2002) Structure of dengue virus: implications for flavivirus organization, maturation, and fusion. Cell 108: 717–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski RA, Moss DS, Thornton JM (1993) Main-chain bond lengths and bond angles in protein structures. J Mol Biol 231: 1049–1067 [DOI] [PubMed] [Google Scholar]
- Lawton JA, Estes MK, Prasad BV (2000) Mechanism of genome transcription in segmented dsRNA viruses. Adv Virus Res 55: 185–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie AG (2006) The integration of macromolecular diffraction data. Acta Crystallogr 62: 48–57 [DOI] [PubMed] [Google Scholar]
- Ludert JE, Hidalgo M, Gil F, Liprandi F (1991) Identification in porcine faeces of a novel virus with a bisegmented double stranded RNA genome. Arch Virol 117: 97–107 [DOI] [PubMed] [Google Scholar]
- Ludtke SJ, Baldwin PR, Chiu W (1999) EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol 128: 82–97 [DOI] [PubMed] [Google Scholar]
- Masachessi G, Martinez LC, Giordano MO, Barril PA, Isa BM, Ferreyra L, Villareal D, Carello M, Asis C, Nates SV (2007) Picobirnavirus (PBV) natural hosts in captivity and virus excretion pattern in infected animals. Arch Virol 152: 989–998 [DOI] [PubMed] [Google Scholar]
- McCabe PM, Pfeiffer P, Van Alfen NK (1999) The influence of dsRNA viruses on the biology of plant pathogenic fungi. Trends Microbiol 7: 377–381 [DOI] [PubMed] [Google Scholar]
- Naitow H, Tang J, Canady M, Wickner RB, Johnson JE (2002) L-A virus at 3.4 A resolution reveals particle architecture and mRNA decapping mechanism. Nat Struct Biol 9: 725–728 [DOI] [PubMed] [Google Scholar]
- Nakagawa A, Miyazaki N, Taka J, Naitow H, Ogawa A, Fujimoto Z, Mizuno H, Higashi T, Watanabe Y, Omura T, Cheng RH, Tsukihara T (2003) The atomic structure of rice dwarf virus reveals the self-assembly mechanism of component proteins. Structure 11: 1227–1238 [DOI] [PubMed] [Google Scholar]
- Nandi P, Charpilienne A, Cohen J (1992) Interaction of rotavirus particles with liposomes. J Virol 66: 3363–3367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navaza J (1994) AMoRe: an automated package for molecular replacement. Acta Crystallogr 50: 157–163 [Google Scholar]
- Ochoa WF, Havens WM, Sinkovits RS, Nibert ML, Ghabrial SA, Baker TS (2008) Partitivirus structure reveals a 120-subunit, helix-rich capsid with distinctive surface arches formed by quasisymmetric coat-protein dimers. Structure 16: 776–786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odegard AL, Chandran K, Zhang X, Parker JS, Baker TS, Nibert ML (2004) Putative autocleavage of outer capsid protein μ1, allowing release of myristoylated peptide μ1N during particle uncoating, is critical for cell entry by reovirus. J Virol 78: 8732–8745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesavento JB, Crawford SE, Estes MK, Prasad BV (2006) Rotavirus proteins: structure and assembly. Curr Top Microbiol Immunol 309: 189–219 [DOI] [PubMed] [Google Scholar]
- Prasad BV, Hardy ME, Dokland T, Bella J, Rossmann MG, Estes MK (1999) X-ray crystallographic structure of the Norwalk virus capsid. Science (New York, NY) 286: 287–290 [DOI] [PubMed] [Google Scholar]
- Qiao X, Sun Y, Qiao J, Mindich L (2008) Temporal control of message stability in the life cycle of ds. J Virol 83: 633–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy V, Schneemann A, Johnson JE (2003) Nodavirus endopeptidase. Handbook of Proteolytic Enzymes, 2nd edn 1–5 [Google Scholar]
- Reinisch KM, Nibert ML, Harrison SC (2000) Structure of the reovirus core at 3.6 A resolution. Nature 404: 960–967 [DOI] [PubMed] [Google Scholar]
- Roy P (2007) Orbiviruses. In Fields Virology, Knipe DM, Howley PM (eds), Vol. 1, 5 edn, 54, pp 1975–1998. Philadelphia: Lippincott Williams and Wilkins [Google Scholar]
- Schiff LA, Nibert M, Tyler K (2007) Orthoreoviruses and their replication. In Fields Virology, Knipe DM, Howley PM (eds), Vol. 1, 5 edn, pp 1853–1916. Philadelphia: Lippincott Williams and Wilkins [Google Scholar]
- van Heel M, Gowen B, Matadeen R, Orlova EV, Finn R, Pape T, Cohen D, Stark H, Schmidt R, Schatz M, Patwardhan A (2000) Single-particle electron cryo-microscopy: towards atomic resolution. Q Rev Biophys 33: 307–369 [DOI] [PubMed] [Google Scholar]
- Wakuda M, Pongsuwanna Y, Taniguchi K (2005) Complete nucleotide sequences of two RNA segments of human picobirnavirus. J Virol Methods 126: 165–169 [DOI] [PubMed] [Google Scholar]
- Wang AL, Wang CC (1991) Viruses of the protozoa. Annu Rev Microbiol 45: 251–263 [DOI] [PubMed] [Google Scholar]
- Yu X, Jin L, Zhou ZH (2008) 3.88 A structure of cytoplasmic polyhedrosis virus by cryo-electron microscopy. Nature 453: 415–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlotnick A, Aldrich R, Johnson JM, Ceres P, Young MJ (2000) Mechanism of capsid assembly for an icosahedral plant virus. Virology 277: 450–456 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material