Abstract
The mechanism of chromatin compaction in mitosis has been well studied, while little is known about what controls chromatin decompaction in early G1 phase. We have localized the Condensin subunit Brn1 to a compact spiral of rDNA in mitotic budding yeast cells. Brn1 release and the resulting rDNA decompaction in late telophase coincided with mitotic spindle dissociation, and occurred asymmetrically (daughter cells first). We immunoprecipitated the GTP-exchange factor Lte1, which helps activate the mitotic exit network (MEN) in anaphase, with mitotic Brn1. In lteΔ cells Brn1 release was delayed, even at temperatures that do not impair mitotic exit. Mutations in MEN pathway components that act downstream of Lte1 similarly delayed rDNA decompaction. We found that Brn1 release in wild-type cells coincided with the release of Cdc14 phosphatase from the nucleolus and with mitotic CDK inactivation, yet it could be selectively delayed by perturbation of the MEN pathway. This may argue that different levels of Cdk inactivation control spindle disassembly and chromatin decompaction. Mutation of lte1 also impaired rotation of the nucleus in early G1.
Keywords: Brn1, Cdc14, chromatin decondensation, Condensin, mitotic exit network (MEN)
Introduction
Passage through mitosis requires both temporally and spatially coordinated changes in chromatin compaction. The mechanisms that alter the level of chromatin compaction are not fully understood, although major players are the two large and related complexes of Condensin and Cohesin (Guacci et al, 1997; Ciosk et al, 1998; Lavoie et al, 2000). The Condensin complex contains a pair of related structural maintenance of chromosome (SMC) subunits that form a dimeric hinge, as well as three non-SMC subunits: Ycs4/XCAP-D2, Ycs5/XCAP-G and Brn1/Barren (Schleiffer et al, 2003). Brn1 is a member of a conserved family of kleisins, which associate with and bridge between SMC head groups. The genes encoding Condensin subunits are essential for vegetative growth, yet conditional mutations were isolated and shown to impair mitotic chromatin compaction in budding yeast (Strunnikov et al, 1995; Freeman et al, 2000; Lavoie et al, 2000, 2004; Ouspenski et al, 2000; Bhalla et al, 2002). Conditional mutations in Cohesin subunits also have measurable effects on mitotic rDNA compaction (Lavoie et al, 2004).
Mitotic kinases, such as the cyclin-dependent kinase Cdk, control the dynamics of these structural components of chromatin in early mitosis (Stegmeier et al, 2002; D'Amours et al, 2004; Lavoie et al, 2004; Sullivan et al, 2004; reviewed in Toth et al, 2007). For instance, activation of the anaphase promoting complex (APC) by Cdk leads to the destruction of Pds1 and cleavage of the kleisin subunit of Cohesin, Scc1 (Uhlmann et al, 1999). This coincides with a partial release of the Cdc14 phosphatase from its inhibitor Net1 (Queralt et al, 2006), which sequesters Cdc14 in the nucleolus (Shou et al, 1999; Visintin et al, 1999). This early anaphase release of Cdc14 (Stegmeier et al, 2002) correlates with both the accumulation of Condensin in the nucleolus and rDNA compaction, an event that facilitates rDNA segregation (D'Amours et al, 2004; Sullivan et al, 2004). The Ipl1/AuroraB kinase have also been implicated in chromatin compaction at the beginning of mitosis in yeast (Lavoie et al, 2004; Sullivan et al, 2004) and in mammalian cells (Lipp et al, 2007). The vertebrate AuroraB kinase was shown to control the localization of Condensin I to mitotic chromosomes, while the successive phosphorylation of vertebrate Condensin I and II by cyclin–Cdk complexes is thought to promote condensation (reviewed by Hirano, 2005).
After sister chromatids are separated by extension of the mitotic spindle, cells exit mitosis. In budding yeast this is triggered by activation of the mitotic exit network (MEN), which controls the degradation of B-type cyclins and accumulation of the Cdk inhibitor Sic1 (D'Amours et al, 2004; Toth et al, 2007). Both are required to fully inhibit mitotic Cdk. Exit from mitosis requires the breakdown of the mitotic spindle and initiation of G1-phase-specific transcription events, although, unlike mammals, yeast has a closed mitosis that obviates the need to re-assemble the nuclear envelope around daughter nuclei. Nonetheless, in early G1-phase cells, the yeast nucleus rotates to position the nucleolus opposite the spindle pole body (SPB) (Bystricky et al, 2005).
At the top of the MEN signalling cascade is a small GTPase called Tem1. Tem1 is negatively regulated by a GTPase-activating complex composed of Bub2 and Bfa1 (Geymonat et al, 2002) and is positively regulated by Lte1, which contains homology to guanine nucleotide exchange factors (GEF) of the Cdc25 family (Shirayama et al, 1994). Lte1 is not essential for mitosis at 30°C, yet cells that lack Lte1 are cold sensitive for progress through telophase (Shirayama et al, 1994). Indeed, GTP-bound Tem1 is required to activate Cdc15 kinase, which leads in turn to a second wave of Cdc14 phosphatase release. This final release of Cdc14 ensures inactivation of the mitotic Cdk by promoting the degradation of Clb2 by APCCdh1 and the accumulation of the Cdk1 inhibitor Sic1 (Visintin et al, 1998), which jointly signal entry into G1.
In vertebrates, the reformation of the nuclear envelope and reassembly of nuclear lamina precede chromatin decompaction, yet to date no study has examined how chromatin decompaction is coordinated with exit from mitosis. Although chromatin decompaction correlates with breakdown of the long anaphase spindle in wild-type (wt) budding yeast, the temporal coincidence of two events in the cell cycle cannot be taken as evidence of coordinate control. For instance, bud emergence and the initiation of DNA replication coincide temporally in the yeast cell cycle, but are controlled by distinct pathways, which were identified and dissociated by mutagenesis (Hartwell et al, 1974).
Here, we have examined the link between the well-characterized MEN pathway and chromosome decompaction. First, we show that rDNA decompaction correlates with the release of Brn1 from the rDNA. We then found that several spindle-regulatory proteins, among them the Tem1-regulator Lte1, are associated with Brn1 in mitosis. We have used quantitative live microscopy to examine whether Lte1 and the MEN pathway control Brn1 localization. Indeed, lte1 deletion selectively delays Brn1 release and rDNA decompaction with respect to spindle disassembly and formation of the G1 nucleus. The deletion of bub2 rescued the lack of decondensation found in the lte1 mutant at low temperatures, implicating MEN pathway components downstream of Lte1. Decompaction of the rDNA furthermore correlated with the final release of Cdc14 phosphatase from the nucleolus. Moreover, in cells blocked at the cdc15 arrest point in late anaphase, premature inactivation of the cyclin-dependent kinase Cdc28 triggered both Brn1 delocalization and rDNA decompaction. We also found that the absence of Lte1 interferes with the rotation of the nucleus in early G1 phase, which positions the nucleolus opposite to the SPB. Our studies provide the first mechanistic analysis of the coordination of chromatin decompaction with entry into interphase.
Results
Decompaction of the rDNA starts in telophase
To have a molecular marker for the process of chromatin compaction and decompaction in Saccharomyces cerevisiae, we tagged the Condensin subunit Brn1 and monitored its localization both by immunofluorescence (IF) and live microscopy of Brn1-GFP in dividing cells. Both Brn1-13myc and Brn1-GFP were integrated and expressed from the BRN1 promoter and fully complemented the growth arrest phenotype of brn1 deficient cells. High-resolution confocal microscopy of a fixed and immunostained exponential culture revealed a diffuse distribution of Brn1-13myc throughout the nucleus and nucleolus in interphase cells (Figure 1A; Supplementary Figure S1A). In anaphase, however, Brn1 assumed a compact but elongated spiral that extended the length of the nucleus, much like the rDNA (Figure 1D and E). Coincidence of mitotic Brn1 with the rDNA was confirmed by double staining and by colocalization of Brn1-GFP with the nucleolar marker Nop1-CFP (Figure 1D; Supplementary Figure S1A–D; Supplementary Movie 1). Time-lapse microscopy (Supplementary Movie 2) showed similar mitotic spiral structures in strains bearing Brn1-GFP. Finally, we note that in telophase cells Brn1 has a compact, punctate appearance that is lost in G1- and S-phase nuclei (Figure 1A).
Figure 1.
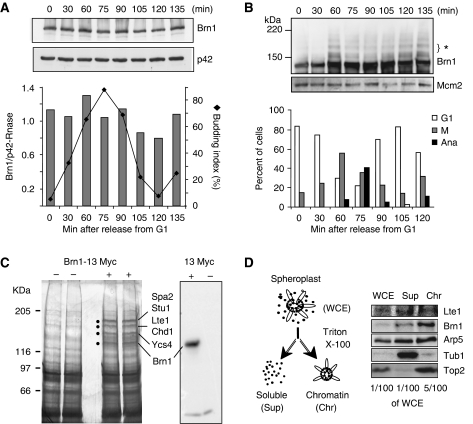
Brn1 localization in interphase and mitosis. (A) IF for Brn1-13myc labelled with anti-Myc (grey) and anti-tubulin (green) antibodies on strain GA-1656. The slightly punctate Brn1-staining in telophase panel can be contrasted to the diffuse staining seen in G1- and S-phase cells. d=daughter cell nucleus. Bar=5 μm. (B) Micrographs show IF for Brn1 and tubulin (as in A) coupled with DAPI for identification of DNA during late mitosis where spindles are either intact (bottom) or starting to disassemble (see arrowheads, upper panel). d=daughter cell nucleus. Bar=5 μm. (C) Selected frames from time-lapse imaging of Brn1-GFP (GA-2663; min in upper left) in which we observe Brn1-GFP segregation to the daughter cell and decompaction occurring initially in the daughter cell nucleus (d). Bar=5 μm. (D) Single confocal section showing Brn1-GFP (green) and Nop1-CFP (red) during chromosome segregation in two adjacent cells by live microscopy. Bar=5 μm. For live imaging of mitosis see Supplementary data. (E) Confocal sections of IF for Brn1-13myc with anti-Myc (green) and for phospho H3 (anti-H3PhosphoS10; red). Two mitotic figures are shown in the larger image, and an interphase cell is in the inset. Schematic figure depicts results from panels D and E. (F) Schematic representation of tetO and lacO array insertion on Chr 12. Underneath is the Perod-Kratky chain equation, where contour length Lc (nm) is the ratio of the genomic distance d (in bp) divided by the linear mass density of the chromatin chain c (in bp/nm) or Lc=d/c. Brn1 is restricted to the nucleolus in the net1-1 mutant as seen in this representative picture showing Brn1-GFP (green) and Nop1-CFP (red) in net1-1 cells (GA-3266; net1-1, Brn1-GFP and Nop1-CFP). Cells were grown at the permissive temperature (25°C), and microscopy was performed after 2 h at nonpermissive temperature (30°C). Bar=5 μm. Below is a schematic of the behavior of two distant points (blue and yellow) on a flexible polymer chain (no Condensin, green) as compared to a less flexible fibre that we propose results from Condensin association. (G) Distances between the centers of gravity of the two spots were measured on strains GA-3779 (wt) and GA-4114 (net1-1) which retains Condensin on the rDNA during interphase. >100 cells were scored for each stage except telophase (n=50). S-phase cells are budded, whereas G2/M have elongated nuclei. End-to-end distance measurements correspond to: 1⩽0.25 μm; 2⩽0.5 μm; 3⩽0.75 μm; 4⩽1 μm; 5⩽1.2 μm; 6⩽1.5 μm; 7⩽1.75 μm; 8⩽2.2 μm.
Careful monitoring of both the spindle (tubulin) and Brn1 staining showed that Brn1 redistributed from a compact to a diffuse staining pattern in the final stages of mitosis (late telophase). This occurred first in daughter cells (73% daughter first, n=50; Figure 1B) when the extended mitotic spindle was still intact (56% intact spindle; Figure 1B). In the remaining 44% of the cells, spindle disassembly was just starting as Brn1 became diffuse (see arrowheads Figure 1B). Time-lapse microscopy further confirmed that Brn1 redistribution occurred first in daughter cell nuclei (see d, Figure 1C), suggesting that the unloading of Condensin in wt cells occurs before or coincident with spindle disassembly, and before establishment of the short G1-phase aster. The lack of coordination between mother and daughter nuclei suggests that decompaction may be controlled by local modifications, and not by a general cell-cycle ‘timer'.
To correlate the Brn1 binding with the compaction status of the rDNA chromatin, we tagged each end of the approximately 200 rDNA repeats with an array of lacO or tetO sites in a strain-expressing fusions of LacI-CFP and TetR-YFP (Figure 1F). Using 3D confocal microscopy (Schober et al, 2008), we monitored the end-to-end distances separating the two fluorescence tags in space (r in nm). Earlier work has shown that the chromatin fibre can be modelled as a flexible polymer chain using parameters described by the Perod–Kratky formula (Figure 1F; Kratky and Porod, 1994; Bystricky et al, 2004). In this equation, the spatial distance r that separates the two points on the polymer is a function of the persistence length (or stiffness, Lp) of the fibre and the linear mass density of the chromatin chain (in bp/nm). These parameters are not separable, as the more compact the chromatin fibre is, the stiffer it becomes, yielding a larger and less variable point-to-point separation for the two sites along the flexible fibre. In brief, the more condensed the local chromatin structure becomes, the less frequently the two distant points along the fibre will come into contact, leading to a larger mean separation in 3D (scheme Figure 1F, bottom).
To correlate changes in compaction (r values) with Brn1 binding, we compared wt with the net1-1 mutant, in which the net1-1 protein fails to inhibit the Cdc14 phosphatase. In these cells, Brn1 staining remains condensed in G1 and is particularly compact in S-phase cells (Figure 1F, see arrowheads; Supplementary Figure S2). We measured the end-to-end distances for markers at the extremities of the rDNA In wt G1- and S-phase cells, which contain dispersed Brn1. Values ranged from 500 to 1500 nm, showing the broad variation typical for a flexible fibre. In net1-1 cells, particularly in S phase, end-to-end distances were concentrated between 1500 and 1750 nm, consistent with the compact Brn1 staining (bar 7, Figure 1G). Similarly, for wt cells in G2/M and early anaphase, the end-to-end distances spanning the rDNA were larger and less variable, suggesting that mitotic rDNA fibre increases its persistence length, that is, becomes stiffer. Thus, coincident with the binding of Brn1, the mitotic rDNA chromatin fibre becomes compact and stiff, reflecting an increase in mass density (more nucleosomes per μm). This resembles mitotic condensation events in mammalian chromosomes (Hirano, 2005).
We can clearly distinguish the staining of Brn1-labelled rDNA from that of genomic DNA in mitosis (see histone H3Ser10 phosphorylation; Figure 1E) and from that of DNA topoisomerase II (TopoII; Supplementary Figure S1E). This is reminiscent of observations made for Condensin and TopoII in mammalian chromosomes (Maeshima and Laemmli, 2003). We note that in yeast H3Ser10-P is not required for chromatin compaction (Lavoie et al, 2002), whereas the role of TopoII in yeast is still unclear (D'Amours et al, 2004; Sullivan et al, 2004; D'Ambrosio et al, 2008). Given that TopoII does not bind the string-like spiral of mitotic rDNA, whereas Brn1 does, we conclude that this Condensin subunit is the better marker for rDNA compaction.
Brn1 is stable throughout the cell cycle
To understand what triggers Brn1 release and rDNA decompaction, we asked whether Brn1 would undergo cleavage like Scc1, the ‘kleisin' counterpart in Cohesin (Schleiffer et al, 2003). Alternatively cell-cycle-dependent modifications of Brn1 might coincide with its relocalization from the rDNA fibre, although Brn1, unlike Barren in higher eukaryotes, does not contain SP/TP consenses for Cdk modification. A western blot for Brn1-13myc on samples taken as cells traverse the cell cycle showed that Brn1 protein levels do not vary significantly through the cell cycle, making it unlikely that its release is mediated by degradation (Figure 2A and B). When low percentage gels were run, we note a ladder of larger bands in mitosis, each one representing a shift of Brn1 by 10–20 kDa (see 60 min, Figure 2B). This modification coincided with the presence of metaphase spindles (1.5–3 μm in length) and disappeared as cells progressed through mitosis to anaphase and telophase (spindle length, 3–10 μm). In a comprehensive analysis of SUMO-conjugated proteins in yeast (Denison et al, 2005), it was shown that Brn1 is sumoylated. We assume, therefore, that the mitosis-specific retardation observed here reflects Brn1 sumoylation, a modification reported for other Condensin subunits as well (D'Amours et al, 2004). However, given that only a fraction of Brn1 is modified during metaphase and that desumoylation does not correlate with rDNA decompaction, SUMO seems unlikely to regulate Brn1 binding and/or release.
Figure 2.
Brn1 is stable during mitosis and precipitates Lte1. (A) Western blot for Brn1-13myc in protein extracts from GA-1656. Samples were taken at the indicated time points after release from α-factor arrest. Blotting for a cytoplasmic p42 RNase serves as a loading control and the graph below shows the budding index and the level of Brn1 relative to the control. (B) As A, except that a 6% polyacrylamide gel was used to resolve larger forms of Brn1. Loading control was Mcm2. The graphs below show cell-cycle phases based on cell morphology at the given time points. Synchrony is lost by 135 min. (C) Silver-stained gel of anti-Myc IP from mitotic extracts of an untagged control strain (GA-180, labelled) or the same strain carrying endogenous Brn1-13myc (GA-1656). Dots indicate specific Brn1-precipitated proteins. This was repeated four times with similar results. The proteins (indicated to right) identified by MALDI ToF mass spectrometry were confirmed by at least five peptides. The specificity of the anti-Myc blot for Brn1-13myc was confirmed (see western blot, right panel). (D) Western blot analysis after chromatin fractionation of nuclear extracts from strain GA-2975. WCE=whole cell extract; Sup=soluble fraction; Chr=chromatin fraction.
Brn1 associates with Lte1
To get a handle on other proteins that might control Condensin's association with chromatin, we next looked for Brn1-interacting partners in mitotic cell extracts. Reciprocal immunoprecipitation (IP) with Smc1, a subunit of Cohesin, and Smc2, a subunit of Condensin, confirmed that Brn1-13myc is indeed part of the Condensin and not the Cohesin complex, although we find that Smc2 and Smc4 are generally less soluble than Brn1 in detergent-lysed cell extracts (data not shown). By IP we recovered Brn1-13myc from the soluble fraction of lysed mitotic spheroplasts and we identified the co-precipitating factors by mass spectroscopy. The cells used for this experiment were blocked in mitosis with nocodazole, which generally yielded approximately 70% G2/M (G2/metaphase) and 25% anaphase cells. A similarly arrested mitotic extract from cells lacking the 13-Myc tag were used as a control.
Four high-molecular-weight bands, ranging from 116 to 205 kDa, were selectively recovered from the tagged-Brn1 extract (Figure 2C). Peptide analysis by MALDI-TOF mass spectrometry showed that bands 3 matched the known Condensin subunit Ycs4, and band 4 was Brn1 itself. As expected from fractionation studies, Smc2 and Smc4 were not recovered in the soluble fraction. Novel Brn1-interacting factors included Spa2, a protein involved in polarized growth, Stu1, which stabilizes the mitotic spindle (Higuchi and Uhlmann, 2005), and Chd1, a chromodomain ATPase that is part of SAGA and helps regulate rDNA transcription (http://db.yeastgenome.org). Finally, the fourth protein recovered in significant levels was the MEN-regulatory factor, Lte1 (Low temperature essential 1). Lte1 localizes to the bud cortex for most of the cell cycle, yet in late mitosis and G1 it redistributes throughout the cell (Supplementary Figure S3E; Bardin et al, 2000; Jensen et al, 2002; Seshan et al, 2002). Given that Lte1 was recovered with Brn1-13myc in mitotic extracts, we asked whether Lte1 could also be recovered with the chromatin fraction after spheroplast lysis. Indeed, after chromatin fractionation, we recovered a nuclear subpool of Lte1 that, like Brn1, was chromatin bound (Figure 2D). We nonetheless are unable to tell whether the interaction between Brn1 and Lte1 is direct, as reciprocal IP was unsuccessful. This failure may stem from the instability of Lte1 in cell extracts.
Loss of Lte1 delays decompaction of the rDNA
The possibility of crosstalk between MEN and Condensin prompted us to examine Brn1 localization and rDNA decompaction in a lte1 deletion strain. LTE1 is not an essential gene, but its deletion renders cells cold sensitive for growth and leads to an anaphase arrest at 14°C (Shirayama et al, 1994). Wild-type (wt) and lte1 deletion cells expressing Brn1-GFP and CFP-Tub1 were synchronized in G1 at 30°C and released at either 30°C or at the semipermissive temperature, 16°C. At 16°C, progression through anaphase and telophase is slower in both wt and mutant cells (Figure 3A and B), allowing us to carefully monitor the timing of Brn1 release. We scored cell-cycle stage by the presence of a bud and the length of the spindle, which is extended in telophase. In cells containing only short microtubule staining (the G1 aster), we scored whether Brn1 was compact (compact, Figure 3C) or dispersed in the nucleoplasm (diffuse, Figure 3C).
Figure 3.
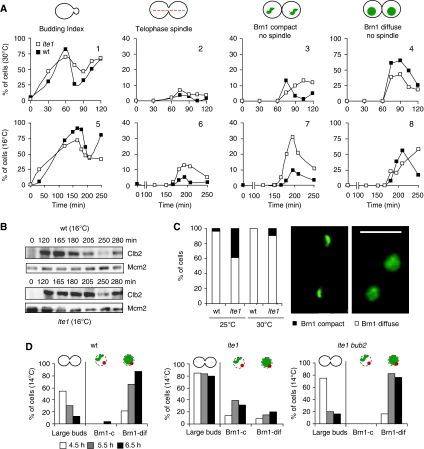
rDNA decompaction is delayed in the lte1 mutant. (A) Cells were arrested in G1 with α-factor and released for strains GA-3263 (BRN1-GFP, CFP-TUB1; wt ▪) and GA-3042 (lte1Δ, BRN1-GFP, CFP-TUB1; lte1Δ □) at the indicated temperatures. Cells were fixed and analysed by confocal microscopy for the indicated phenotypes. 100 cells were scored for each genotype and condition. Panels are labelled 1–8 to facilitate textual reference. (B) Protein extracts were prepared from the same experiment as in A at 16°C. Western blot analysis was performed on all samples for Clb2, and for Mcm2 as a loading control. (C) Brn1-GFP is scored by live microscopy as compact or diffuse in 100 G1-aster containing cells of strains GA-3263 and GA-3042 at the indicated temperatures. Micrographs show representative images of Brn1 fluorescence. (D) Strains GA-3263, GA-3042, and GA-4864 (BRN1-GFP, CFP-TUB1, lte1Δ bub2Δ) were arrested in G1 with α-factor at 30°C and released at 14°C. Samples were taken at indicated times. Mitotic arrest is scored by the number of dumbbell-shaped cells. In cells with G1-phase asters (no matter what the cellular morphology), Brn1 staining was scored as compact or diffuse (c or dif). This frequency is plotted relative to the entire cell population, so that the sum of Brn1-c and Brn1-dif does not equal 100%.
As shown above, in wt cells Brn1 becomes diffuse in daughter nuclei in late telophase just as the spindle disassembles (Figure 1B). At 30°C, the lte1 defect for MEN activation is fully compensated by other pathways or by Tem1 activation (Shirayama et al, 1994), and, therefore, we scored no significant accumulation of telophase spindle structures (Figure 3A, panel 2). Nonetheless, we detected compact Brn1 labelling in approximately 15% of the cells bearing G1-phase asters, indicating inefficient Brn1 unloading, a state that persisted for 2 h (Figure 3A, panel 3). In contrast, the coincidence of compact Brn1 with G1 asters was highly transient in wt cells at 30°C, being lost by 100 min (Figure 3A, panel 3, no spindle). This suggested that there might be a loss of coordination between Brn1 release and spindle disassembly in lte1Δ cells.
At semipermissive temperature (16°C), the loss of coordination between Brn1 unloading and spindle disassembly is sharply aggravated in lte1Δ cells. Although disassembly of the telophase spindle was complete by 250 min, disappearing with almost wt kinetics, compact Brn1 staining was seen to coincide with G1 asters in over 30% of the lte1 cells (Figure 3C, panel 7). This value was 8% in wt cells at this temperature. Given that 16°C is semipermissive for the lte1 strain, Brn1 does eventually become dispersed, although even at 250 min, 10% of the lte1 cells retained compact Brn1 staining (Figure 3A, panel 7). We conclude that the release of Brn1 is inefficient in the lte1 mutant, being delayed by 30–45 min relative to spindle disassembly. A delay can also be detected for the complete degradation of Clb2, which occurs in wt cells by 205 min, but at 250 min in lte1 cells (Figure 3B).
To eliminate possible artefacts due to low temperature, we monitored Brn1 release in wt and lte1 mutant cells at a higher semipermissive temperature. At 25°C, we also found that Brn1-GFP was present in a compact structure together with G1 asters in 38% of the mutant cells, which compares with 3.6% in a wt culture under identical conditions (Figure 3C). Again the absence of Lte1 delayed Brn1 release from the rDNA, even though the switch from an anaphase spindle to G1 aster was affected only slightly (16°C) or not at all (30°C). We conclude that Lte1-controlled events either help coordinate rDNA decompaction with spindle disassembly or directly trigger decompaction by facilitating Brn1 release.
MEN pathway is upstream of Brn1 release
At the top of the MEN cascade, the GTPase Tem1 is negatively regulated by an inhibitory GAP, Bub2, whereas it is positively regulated by Lte1. To see whether the lte1 defect illustrated in Figure 3 correlates with impaired activation of the MEN pathway by Tem1, we tested whether we can suppress the delay in Brn1 release by restoring Tem1 activation indirectly through bub2 deletion (Bardin et al, 2000; Pereira et al, 2000; Wang et al, 2000; Adames et al, 2001; Lee et al, 2001; Geymonat et al, 2002). The effects were monitored by comparing the frequency with which compact Brn1 and small G1 asters coincide in wt, lte1 and double lte1 bub2 mutants, at the restrictive temperature for lte1Δ (14°C).
Cultures were grown at 30°C, blocked in G1 with α-factor and released into precooled media at 14°C. Samples were collected hourly from 4.5 to 6.5 h after release, as within this window full Brn1 release is observed in wt cells (Figure 3D). We scored the abundance of dumbbell (large budded) cells, the disappearance of which signals progression into G1 phase (Figure 3D). As expected, at fully restrictive temperature nearly 80% of the lte1 mutant cells remain dumbbell shaped at 6.5 h, whereas both wt and lte1 bub2 double mutant strains progress into the next cell cycle and show unbudded or budded single cells. The coincidence of compact Brn1 with G1 asters is seen rarely in both wt and lte1 bub2 cells, confirming that Brn1 is usually released by the time that cells bear G1 asters (right hand columns, Figure 3D). However, as seen at semipermissive temperatures, some lte1 cells broke through the late telophase block, and these retained compact Brn1 staining despite the disassembly of the anaphase spindle morphology (Figure 3D). This argues that bub2 deletion largely suppresses the lte1 defect for Brn1 relocalization, as for other events of mitotic exit. We conclude that the MEN pathway has a role in controlling rDNA decompaction.
Mutants of the MEN pathway delay chromatin decompaction
To determine which components of the MEN pathway affect decondensation, we tested mutants downstream of Lte1 for similar defects. We first compared the kinetics of rDNA decompaction between wt and the temperature-sensitive tem1-1 cells. Cells expressing Brn1-GFP and Tub1-CFP from their endogenous promoters were grown at permissive temperature (23°C), blocked in G1 and released at restrictive temperature (37°C). By 120 min 75% of the tem1-1 cells are blocked with long telophase spindles (Figure 4A). In these cells, Brn1 was fully compact (data not shown, see Figure 4C). During the subsequent hour, the tem1-1 mutant progressed through the arrest allowing spindle disassembly, yet Brn1 remained fully compact (Figure 4A, Ca). Indeed, by 180 min over 40% of tem1-1 cells had short G1-phase asters and condensed Brn1 staining (see indicated cells, (a) Figure 4C). At the analogous point in the cell cycle, wt cells had reinitiated budding and Brn1 staining was diffuse (Figure 4A, Cb). Thus, tem1-1 led to a very similar pathology at 37°C as lte1 at low temperatures.
Figure 4.
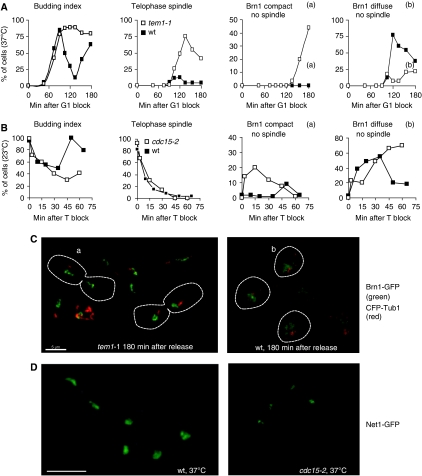
Mutants of the MEN pathway delay unloading of Brn1 from the rDNA. (A) Strains GA-3716 (BRN1-GFP, CFP-TUB1) and GA-3717 (tem1-1, BRN1-GFP CFP-TUB1) were synchronized in G1 with α-factor and released into fresh medium at 37°C. Cells were processed and scored as in Figure 3A. ▪ wt and □ tem1-1. Examples of Brn1 compact (a) and diffuse (b) are circled in C. (B) Strains GA-3263 (BRN1-GFP, CFP-TUB1) and GA-3265 (cdc15-2 BRN1-GFP, CFP-TUB1) were arrested in nocodazole at 23°C, cdc15-2 was inactivated by temperature shift, and then cultures were released into fresh media at 37°C to accumulate telophase cells (T block). Cultures were then released from the block at 23°C and sampled at the indicated times. For the wt control, cells were released from the nocodazole block at 23°C and time points were taken. ▪ wt and □ cdc15-2. (C) Representative micrographs of wt and tem1-1 mutant cells at 180 min after release; a, Brn1 compact with disassembled spindle; b, Brn1-GFP (green) is diffuse with the G1 aster visible by CFP-Tub1 (red). (D) Fields of cells from exponentially growing cultures of wt (GA-5227) and cdc15-2 (GA-5228) strains expressing Net1-GFP fusions after 1.5 h of incubation at 37°C. In wt cells Net1-GFP reveals a compact rDNA structure in telophase, and more open staining in early G1. Bar=5 μm.
Downstream of Tem1 is the kinase Cdc15, whose activation allows progression through late anaphase. The efficiently reversible cdc15-2 temperature-sensitive mutant allowed us to correlate Brn1 release more precisely with telophase progression. Both the cdc15-2 mutant and wt cells were blocked in G2/M with nocodazole at the permissive temperature (23°C), and then shifted to 37°C for 30 min in nocodazole to inactivate the cdc15-2 kinase. Cells were then released from nocodazole into fresh YPAD at 37°C, where telophase cells accumulated with high efficiency (Supplementary Figure S4). When the number of telophase spindles reached its maximum, cells were shifted back to permissive temperature (23°C) and the status of Brn1 compaction was monitored as cells completed mitosis. We scored Brn1 status in similarly treated wt cells as the culture traversed telophase and entered G1 at 23°C, as a control.
At the initial time point, 83% of wt cells and 93% of the cdc15-2 cells were large budded and contained telophase spindles (Figure 4B). After release, the kinetics of spindle disassembly were very similar, although only wt and not cdc15-2 cells rebudded at 45 min after release (Figure 4B). As telophase spindles disassembled, Brn1 rapidly redistributed throughout the nucleoplasm in wt cells (Figure 4Bb). In contrast, in the cdc15-2 mutant Brn1 remained compact and associated with rDNA for 45 min at permissive temperature despite the fact that spindle disassembly and the appearance of the short G1 asters occurred with wt kinetics (Figure 4Ba). The compact nature of the rDNA in arrested cdc15-2 cells at 37°C was confirmed through visualization of Net1-GFP (Figure 4D). These results implicate that the MEN pathway in coordinating Brn1 release with spindle disassembly, and argue that rDNA decondensation is downstream of the Cdc15 arrest point.
Release of Cdc14 during late mitosis coincides with decompaction
The activation of Cdc15 kinase is known to trigger a second release of Cdc14 phosphatase from the rDNA and nucleus, which in turn dephosphorylates Cdk targets (Visintin et al, 1998). To examine the relationship of Cdc14 release and Brn1 relocalization, we triple-tagged wt and lte1 strains with Cdc14-3HA, Brn1-13myc and Tub1-GFP. Cells were grown at 30°C and then shifted to 25°C for 2 h before fixation and IF. We scored telophase cells in which spindles were fully elongated or partially disassembled (see arrowheads, Figure 5A).
Figure 5.
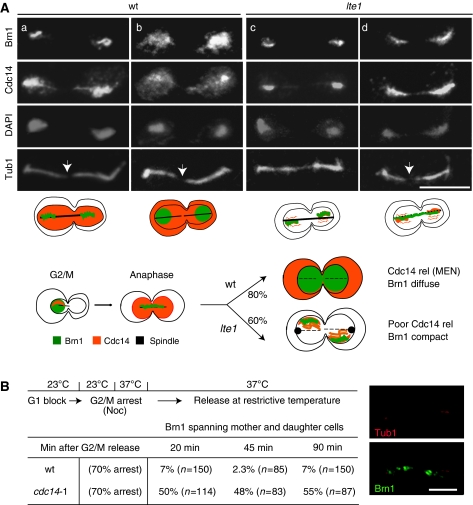
Late mitotic release of Cdc14 coincides with Brn1 relocalization. (A) Micrographs showing IF for tagged Brn1 and Cdc14 performed on exponentially growing cultures of strains GA-3600 (CDC14-3HA, BRN1-13MYC, TUB1-GFP) and GA-3601 (lte1Δ, CDC14-3HA, BRN1-13MYC, TUB1-GFP). Tubulin-GFP and DAPI are visualized by direct epifluorescence. Cultures were grown at 30°C and fixed for IF or incubated for 2 h at 25°C before fixation. Phenotypes shown in panels a, b represent 80% of the wt phenotypes, whereas panels c, d represent 60% of the lte1 cells when grown at 25°C. A scheme shows dynamics of Cdc14, spindle and Brn1 from G2/M to telophase in wt or lte1 mutants. (B) Strain GA-3819 (cdc14-1 BRN1-GFP, CFP-TUB1) was grown at 23°C. On top, a scheme of the experiment, and below, the frequency of compact Brn1 spirals in wt and mutant strains after nocodazole block and cdc14-1 inactivation. A representative micrograph shows cdc14-1 mutant cells after 90 min release at 37°C, with Brn1-GFP (green) and CFP-Tub1 (red). Bar=5 μm.
In wt cells, we see a diffuse distribution of Cdc14 phosphatase whenever Brn1 diffused (Figure 5Ab). On the other hand, in the lte1 mutant at semipermissive temperature, we consistently detected a condensed Brn1 pattern with little or no Cdc14 release (Figure 5Ac, d). This suggests that Lte1-controlled events coordinately promote both Cdc14 release and Brn1 relocalization. This was initially surprising, as the release of Cdc14 from its inhibitor Net1 was reported to promote compaction at the onset of anaphase (D'Amours et al, 2004; Sullivan et al, 2004). In telophase, on the other hand, rDNA decompaction (i.e. Brn1 release) coincided with the second wave of Cdc14 release, which presumably allowed Cdc14 to dephosphorylate other targets. Importantly, this correlation did not occur in an lte1 mutant (Figure 5Ac and d). In late-mitotic wt cells, we could also detect transiently two intermediate states: an elongated spindle, nuclear Cdc14 and condensed Brn1 staining (Figure 5Aa), as well as an elongated spindle, dispersion of Cdc14 into both cytoplasmic and nuclear compartments, and diffuse Brn1 staining (Figure 5Ab). This then converts to a G1 cell with a G1 aster, diffuse Brn1 and dispersed Cdc14 staining. Thus, the complete release of Cdc14 from the nucleolus into the nucleus and cytoplasm in late anaphase correlates with the release of Brn1.
Loss of Cdc14 phosphatase inhibits, whereas Cdk inactivation promotes, Brn1 release
To examine whether Cdc14 phosphatase activity itself affected Brn1 release, we introduced fusions of Brn1-GFP and CFP-Tub1 into wt and temperature-sensitive cdc14-1 cells. Cells were synchronized in G1 at the permissive temperature and then arrested with nocodazole to allow rDNA condensation (Lavoie et al, 2002). Cdc14 function was inactivated by shifting the temperature to 37°C for 30 min, after which cells were released from nocodazole. Initially, 70% of both wt and mutant cells showed a G2/M pattern of DAPI staining (data not shown). As cells progressed from the nocodazole block, the unloading of Brn1 from chromatin was monitored using microscopy, and the frequency of the Brn1 anaphase spiral was scored (see image, Figure 5B). We scored a compact Brn1 spiral in 7% of the wt cells 20 min after release, whereas in the cdc14-1 strain, 50% had this same spiral staining pattern. The absence of Brn1 release persisted in the cdc14-1 cells for 90 min, whereas wt cells progressed through the cell cycle with only a background level of mitotic figures (Figure 5B). Thus, Cdc14 activity is required for Brn1 unloading, and the two proteins show coordinate release in late anaphase.
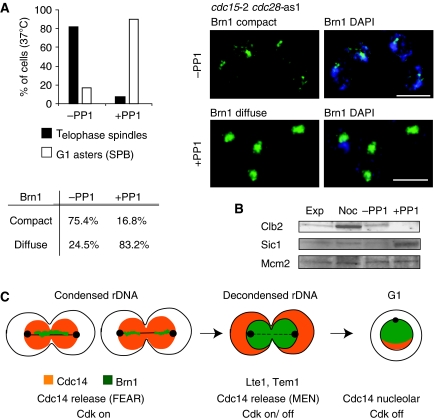
It is well established that Cdc14 activation triggers Clb2 degradation and inactivation of Cdc28 kinase. This latter also results from an accumulation of the Cdk inhibitor, Sic1 (Visintin et al, 1998). To test whether the inactivation of Cdc28 is sufficient to trigger rDNA decompaction in cells arrested in anaphase, we used a double mutant bearing cdc15-2 and cdc28-as1, in which the active site of the Cdc28 kinase is modified to allow inactivation by an ATP analogue, 1NM–PP1 (Bishop et al, 2000). As shown above, the cdc15-2 mutant shows an efficient and synchronous arrest in anaphase (Supplementary Figure S4). We similarly arrested the cdc15-2 cdc28-as1 double mutant in late anaphase using a nocodazole block followed by release into fresh media at nonpermissive temperature. This arrested culture was then split into two parts. To one half we added 1NM-PP1 to inhibit the modified Cdc28 kinase. In cells with an active Cdc28, 80% contain elongated spindles and many retained a compact Brn1 signal (Figure 6A). Sic1, a Cdk inhibitor, was not detectable in these strains, consistent with an anaphase arrest. However, where we inhibited Cdk with the ATP analogue (i.e. cdc28-as1 +PP1), we observed spindle disassembly (Ghiara et al, 1991) and Brn1 assumed a diffuse distribution in over 80% of the cells. Already after nocodazole release, Clb2 was partially degraded, indicating that APCcdc20 had been activated. Further Clb2 degradation was observed on addition of the ATP analogue, which efficiently inactivated Cdc28. Inactivation of Cdc28 was confirmed by monitoring Clb2 degradation and Sic1 accumulation during the cdc15-2 arrest (Figure 6B; Nasmyth et al, 1990; Verma et al, 1997). Under conditions of Cdk1 inhibition, we note that both spindle disassembly and Brn1 release occurred at the same time. Because mutants in the MEN pathway allowed spindle disassembly without coincident Brn1 release, we propose that both Cdc14 release and progressive Cdk inactivation promote Brn1 release and coordinate it with spindle disassembly.
Figure 6.
Cdc28 inactivation in telophase is sufficient for rDNA decompaction. (A) Strain GA-4931 (cdc15-2 cdc28-as1 BRN1-13MYC) was arrested in metaphase, released into a cdc15-2 arrest and divided in two parts. The Cdc28-as1 inhibitor 1NM-PP1 was added to half at 0.5 mM for 45 min. On the left, cells with long telophase spindles or short G1 asters were scored for Brn1 distribution under both conditions. Representative images are on the right. Bar=5 μm. The percentage of compact or diffuse Brn1 patterns were scored for100 telophase-arrested cells (±PP1). (B) Samples were taken from the cultures described in (A) before the block (exp), at the nocodazole block (Noc), and after release 37°C for 45 min with or without the ATP analogue (+PP1 or −PP1). Western blots for the indicated proteins are shown. (C) Model for rDNA decompaction is shown. Brn1 is redistributed from the rDNA to the nucleus coincident with Cdc14 release.
G1 nuclear architecture is altered in the lte1 mutant
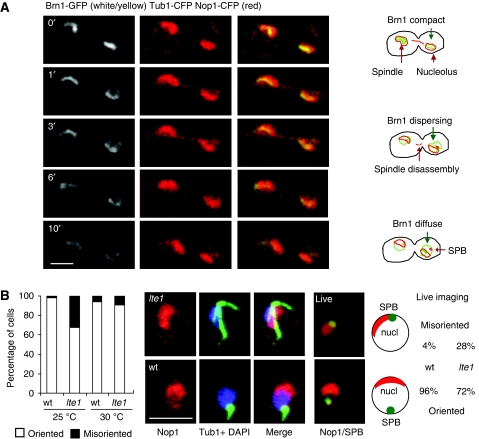
The yeast nucleolus adopts a strikingly reproducible position opposite the SPB when cells are in G1 phase (Yang et al, 1989; Bystricky et al, 2005, see Figure 7A). To examine whether the spatial organization of the G1 nucleus was also linked to the MEN pathway, we examined post-mitotic events in a strain expressing Brn1-GFP, CFP-Tub1 and Nop1-CFP. rDNA decondensation started in cells with elongated spindles and preceded the re-positioning of the nucleolus and the SPB. We asked whether lte1 deletion would also influence the establishment of interphase nuclear architecture by analysing the distribution of Nop1 and tubulin in wt and lte1 strains that were shifted to the semipermissive 25°C after growth at 30°C. Most cells bearing G1 asters had their nucleoli located opposite to the SPB both in wt and mutant cells at permissive temperature (Figure 7B, wt). However, at 25°C over 30% of lte1 cells with G1 asters had a misoriented SPB, such that the SPB and the nucleolus remained in contact on one side of the nucleus (Figure 7B, lte1). The frequency of SPB misorientation was 10-fold higher in the lte1 mutant as compared with wt cells. Live microscopy of strains expressing Tub1-GFP and Nop1-CFP yielded similar results: 28% of lte1 G1-phase cells had abnormal nuclear organization versus 4% in wt cells. Since these cells had already entered G1 phase and disassembled their spindles, our results suggest that Lte1 guides the re-establishment of a polarized interphase nucleus. Intriguingly, decompaction was not a requirement for nuclear reorganization, since nucleoli repositioned correctly in the net1-1 mutant, even though Brn1 staining remained compact (data not shown). This argues that nuclear orientation is independent of Brn1 release, and highlights a second aspect of nuclear and chromatin dynamics coordinated by Lte1 and/or the MEN pathway.
Figure 7.
Nuclear organization is altered in the lte1 mutant. (A) The wt strain GA-3330 (CFP-TUB1, BRN1-GFP, pNOP1-CFP) was grown at 30°C and analysed by time-lapse microscopy. Cells were recorded from late mitosis to G1 and merged images are on the right. Schemes illustrate Brn1 unloading, and the repositioning of the nucleolus and SPB. Bar=2 μm. (B) GA-180 (wt) and GA-3327 (lte1Δ) cells were grown at 30°C and incubated at 25°C before fixation for IF for Nop1 (red), tubulin (green) and DAPI. The graph shows the percentage of cells with G1 asters and oriented (□) or misoriented (▪) nuclear architecture. In addition, strains GA-3330 (TUB1-GFP pNOP1-CFP) and GA-3596 (lte1Δ TUB1-CFP pNOP1-CFP) were analysed by live microscopy (Tub1-GFP, green; Nop1-CFP, red). 100 cells were scored for misoriented or oriented nucleolus, as indicated. Bar=5 μm.
Discussion
Cell division can only proceed efficiently with a properly choreographed cycle of chromatin compaction and decompaction. Many studies have addressed the mechanisms that drive mitotic chromosome condensation, yet there have been very few attempts to understand the molecular events that coordinate chromatin decondensation with other events of the cell cycle. The yeast rDNA locus assumes a compact spiral structure at the onset of anaphase, reminiscent of the backbone of mitotic chromosomes in higher organisms (Maeshima and Laemmli, 2003; Lavoie et al, 2004). In yeast, this compact rDNA structure requires the binding of Condensin (Lavoie et al, 2002). Here, we show that the dispersion of Brn1 from this structured rDNA chromatin occurs in late telophase, starting in daughter cells just before spindle disassembly. It is correlated with a second wave of Cdc14 release. In lte1 and tem1 mutants, spindle disassembly was no longer correlated with Brn1 release, suggesting that divergent or unequal pathways control these two events. These bifurcating pathways may require different levels of Cdk inactivation. Using a special form of the Cdk, cdc28-as1, which is sensitive to an ATP analogue, we showed that in arrested cdc15-2 cells the inhibition of the mitotic Cdk can trigger both Brn1 release and spindle dissociation. This suggests that either the difference between the pathways is upstream of the final event of Cdk inhibition, or the two events are differentially sensitive to levels of Cdk inactivation. In other words, spindle disassembly may require a less complete inhibition of Cdk than rDNA decompaction. In either case, the coordination of these two events seems to be controlled by the MEN cascade, which regulates Cdc14 phosphatase release as well as Cdk inactivation. By perturbing the MEN pathway, and in particular Lte1, we could delay both Cdc14- and Brn1 release, generating cells that contain G1-phase nuclei with short microtubule asters and a compact rDNA structure.
Our ability to separate normally coincident cell-cycle events by mutation of a signalling cascade is reminiscent of early studies of the G1/S transition in budding yeast (Hartwell et al, 1974). The bud emergence cycle could be shown to be under control of a pathway distinct from that controlling the initiation and completion of DNA synthesis, although both were downstream of Cdc28 kinase activation (Hartwell et al, 1974). In this study, we conclude that the shift from mitotic to interphase spindle morphology can be temporally separated from rDNA decompaction by perturbation of the MEN pathway. Although we cannot rule out that other signals downstream of Tem1 contribute to the coordination of Brn1 release with spindle changes, our present study clearly implicates the Lte1-Tem1-Cdc15 pathway in controlling this event. To our knowledge this is the first analysis of the connections between the MEN pathway and the control of chromatin decompaction.
What triggers Brn1 release?
The question remains as to what might be the crucial targets of Cdk or Cdc14 phosphatase in the rDNA chromatin whose modification leads to decondensation. Although we can rule out massive degradation of Brn1 as the trigger for its release, Brn1 or another Condensin subunit could be the targets of dephosphorylation at the late telophase/early G1 transition. Moreover, such modification could arise either from Cdk inactivation, Cdc14 phosphatase activation, both or from the action of another kinase such as Cdc5, which also has a late anaphase arrest point (Hartwell et al, 1974). Brn1 itself does not have Cdk1 consenses, but could be modified by other kinases. It is beyond the scope of this study to identify the relevant targets of the enzymes that control late telophase, yet this could be approached by phosphoproteomic analyses in appropriate mutants.
The loss of Lte1 leads not only to an inefficient release of Brn1, but also to impaired release of Cdc14 phosphatase. We assume that impaired Cdc14 release correlates with incomplete activation, which could also explain the inefficient degradation of Clb2 observed in this mutant. Importantly, in vertebrates the phosphorylation of non-SMC subunits of Condensin seems to promote their initial association with chromatin (Kimura et al, 1998, 2001). Moreover, Aurora B kinase is required for compaction in S. cerevisiae (Lavoie et al, 2004) and for the loading of Condensin I onto mitotic chromosomes in man (Lipp et al, 2007). Finally, it was recently reported that in mammalian cells PP1 phosphatase is recruited to chromatin during anaphase (Trinkle-Mulcahy et al, 2006) to promote release of a ‘regulator of chromosome architecture' (RCA) (Vagnarelli et al, 2006). There is as yet no identified homologue of RCA in yeast, but the parallels between these phosphatase-dependent control mechanisms is striking. Other phosphatases may also be involved in the modulation of chromatin structure. We note that Cdc14 activation also promotes Condensin loading at the onset of anaphase (D'Amours et al, 2004; Sullivan et al, 2004), which suggests that Cdc14 plays a dual role, acting both as a positive and a negative regulator of chromatin compaction. This suggests that it targets different substrates at the very beginning and the very end of anaphase.
Lte1 contributes to the establishment of nuclear organization
In addition to the role of MEN in coordinating decompaction and spindle disassembly, we find that Lte1, or factors downstream of this GEF, also affect the early G1-phase rotation of the yeast nucleus. This rotation positions the nucleolus opposite the SPB and the site of bud emergence (Bystricky et al, 2005). Given that Lte1 relocates from the bud cortex during late mitosis (Bardin et al, 2000; Seshan et al, 2002), it is a likely regulator of both cytoskeletal elements and nucleolar position, possibly orienting the nucleus with respect to the SPB. Once released from the bud cortex, Lte1 would inevitably encounter the daughter nucleus before the maternal one, consistent with our finding that daughter nucleoli release Brn1 and decondense before the maternal nuclei are affected (Figure 1). It is possible that Lte1 forms a concentration gradient as it is released, contributing to this difference.
It is not excluded that Lte1 acts as a GEF for GTPases other than Tem1. Indeed, several lines of evidence implicate GTP gradients in the control of mitotic events. Recent studies implicate Ran GTPase in the assembly of the mitotic spindle, nuclear-envelope dynamics and the timing of cell-cycle transitions (reviewed in Clarke and Zhang, 2008). Moreover, another chromatin associated GEF in yeast, Prp20, influences non-rDNA chromatin structure and nuclear organization (Belhumeur et al, 1993). Thus, we hypothesize that a GTP-controlled crosstalk between yeast cytoskeleton and SPB promotes nuclear rotation in early G1 (Bystricky et al, 2005).
This study is the first to address the relationship of chromatin decompaction with signalling pathways that control exit from mitosis. We show that the MEN pathway controls Brn1/Condensin release in late telophase, and that this release coincides with long spindles and late mitotic Cdc14 activation. Indeed, Brn1 dispersion normally precedes the disassembly of the mitotic spindle, and perturbation of the MEN-controlled Cdc14 release selectively delays Brn1 release. Finally, we note that another Brn1 associated factor, Chd1, is a nucleosome-remodelling component that regulates transcription elongation. Future studies will address whether this factor also plays a role in post-mitotic chromosome decondensation, possibly being targeted to compact rDNA by interaction with Brn1.
Materials and methods
Strains and yeast methods
The strains used in this study are listed in Supplementary Table 1. All strains are derivatives of W303, with the exception of GA-4383 and GA-3717 (wt and tem1-1 mutant), which are isogenic, derived from the S228c background, and backcrossed twice to W303. Tagging and deletion were achieved using a PCR-based technique (Longtine et al, 1998). Standard yeast media were supplemented with 25 mg/l adenine. Nocodazole was added (10 μg/ml) to cultures adjusted to 1% DMSO.
Chromatin fractionation
Chromatin fractionation was performed as described in Pasero et al (1999) with slight modifications. After spheroplasting, cells were washed twice in 50 mM Hepes-KOH pH 7.5, 20 mM KCl, 2 mM EDTA-KOH, 0.05 mM spermine, 0.125 mM spermidine, 1 M sorbitol,1% Trasylol, and 1 mM PMSF. The pellet of spheroplasts (∼4 × 108cells) was then resuspended in 1 ml of 50 mM Hepes-KOH pH 7.5, 2.5 mM MgCl2, 10 mM glycerol 2-phosphate, 0.1 mM Na3VO4, 0.25% Triton X-100, 300 μg/ml benzamidine, 1 μg/ml pepstatin A, 2 μg/ml antipain, 0.5 μg/ml leupeptin, 100 μg/ml TPCK, 50 μg/ml TLCK. Micrococcal nuclease (1 U/ml) supplemented with 1 mM CaCl2 was used to digest genomic DNA at 37°C for 2 min, after which the reaction was stopped by 2 mM EGTA. Western blots were performed using HRP-conjugated secondary antibodies, and the signal was acquired with Quantity One software (BioRad). Antibody dilutions were: anti-Myc (9E10) 1/3000; anti-Top2 1/7500; anti-Mcm2 1/3000 (Santa Cruz, yN19); anti-HA (12CA5) 1/3000; and anti-Clb2 1/2000 (Santa Cruz, SC 9071).
Immunoprecipitation
Exponentially growing cultures of strains GA-180 (wt) and GA-1656 (Brn1-13myc) were blocked in nocodazole, harvested and washed with IP buffer (50 mM Hepes pH 7.5, 1% NP-40, 150 mM NaCl, 10 mM NaF, 60 mM β-glycerophosphate, 0.1 mM VaV03, 3 mM Na-pyrophosphate and complete protease inhibitors (Roche). Zirconia beads were added to the pellet (0.5 g) in 600 μl IP buffer, and cells were broken by bead beating. The whole cell extract was then treated with DNase I, centrifuged, and the supernatant incubated with antibodies. 9E10 antibodies 1:150 coupled with anti-mouse IgG-Dynal beads equilibrated in IP buffer were added to the supernatant and incubated for 2 h at 4°C. Beads were washed three times with IP buffer and boiled.
Mass spectrometry
Proteins were resolved in a 8% acrylamide gel and silver stained. The bands of interest were cut from the gel, destained according to Shevchenko et al (1996), and the proteins digested by trypsin. Extracted peptides were concentrated, mixed with the α-cyano-4hydroxycinnamic acid matrix, and spotted on a MALDI plate before being analysed by MALDI-TOF mass spectrometry (PE Biosystems Voyager System 2016 mass spectrometer). Settings were: mode of operation: reflector; extraction mode: delayed; polarity: positive; acquisition control: manual; accelerating voltage: 20 000 V; grid voltage: 56.5%; mirror voltage ratio: 1.12; guide wire 0: 0.05%; extraction delay time: 120 nsec; acquisition mass range: 700–5000 Da; number of laser shots: 128/spectrum; laser intensity: 1536; calibration matrix: α-cyano-4-hydroxycinnamic acid; low mass gate: 700 Da; timed ion selector: off; source pressure: 9.349E-008; mirror pressure: 7.691E-008 The analysis of mass spectra led to lists of M/Z values specific to Brn1 (masses common to both non-tagged and tagged Brn1 samples were discarded) matching with masses of putative peptides whose sequences were found in the ProFound database. Each protein was identified by at least five different peptides of at least six residues, with a peptide mass accuracy of ±0.3 Da. The procedure was performed four times to validate protein identity.
IF and Microscopy
Cells were fixed with 4% paraformaldehyde (PAF) for 20 min at 20°C, followed by spheroplasting with lyticase and Zymolyase. Cells were then spotted on a microscope slide, permeabilized, and processed IF as described (Gotta et al, 1996).
For live imaging stacks and time lapse, cultures were grown to 5–8 × 106 cells/ml. Cells were trapped on a concanavanin A-coated coverslip, in a Ludin chamber (Life Imaging Services) and imaged in SC media at 30°C using a Zeiss LSM510 confocal microscope as described (Bystricky et al, 2005). Stacks were taken with a step size of 200 nm. Images were acquired on multi-tracking mode using 458 nm (CFP) and 488 nm (GFP). Settings for the Zeiss were: argon laser 5.3 Amps; output 25%; detector gain 930–990; amplifier gain: 1; amplifier offset: 0.2–0.1 V; laser transmission 2–5%; scan speed 10 (1.28 μs/pixel); 2–4 averages/line using a 1.8 zoom (pixel size 100 × 100 nm) and a 100 × Plan-Apochromat objective (NA 1.4). For time course experiments, cells were fixed with 1% PAF and imaged with the LSM510 (Zeiss). IF imaging was performed on an LSM510Meta (Zeiss), with settings as described above except for detector gain (700–800); scan speed of 8 (2.45 μs/pixel); 8 averages/line using a 1.8–3.2 zoom, using lines 405 nm (BP 420–480), 488 nm (505–550), 543 nm (LP 560) and 633 nm (636–753).
Supplementary Material
Supplementary Movie S1
Supplementary Movie S2
Supplementary Information
Acknowledgments
We thank M Peter (ETH, Zurich) for the ATP analogue, and colleagues at ISREC, the University of Geneva, and the FMI imaging facility for technical support. We thank F Uhlmann for plasmids, strains and discussions. This study was supported by an EMBO long-term fellowship to DL, by grants from the Swiss Cancer League to DL, SMG and EV and from the Spanish Government (Ministerio de Educacion y Ciencia) to EV. We acknowledge the Novartis Research Foundation and the Frontiers in Genetics NCCR for generous support.
References
- Adames NR, Oberle JR, Cooper JA (2001) The surveillance mechanism of the spindle position checkpoint in yeast. J Cell Biol 153: 159–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardin AJ, Visintin R, Amon A (2000) A mechanism for coupling exit from mitosis to partitioning of the nucleus. Cell 102: 21–31 [DOI] [PubMed] [Google Scholar]
- Belhumeur P, Lee A, Tam R, DiPaolo T, Fortin N, Clark MW (1993) GSP1 and GSP2, genetic suppressors of the prp20-1 mutant in S. cerevisiae: GTP-binding proteins involved in the maintenance of nuclear organization. Mol Cell Biol 13: 2152–2161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhalla N, Biggins S, Murray AW (2002) Mutation of YCS4, a budding yeast Condensin subunit, affects mitotic and nonmitotic chromosome behavior. Mol Biol Cell 13: 632–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop AC, Ubersax JA, Petsch DT, Matheos DP, Gray NS, Blethrow J, Shimizu E, Tsien JZ, Schultz PG, Rose MD, Wood JL, Morgan DO, Shokat KM (2000) A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature 407: 395–401 [DOI] [PubMed] [Google Scholar]
- Bystricky K, Heun P, Gehlen L, Langowski J, Gasser SM (2004) Long-range compaction and flexibility of interphase chromatin in budding yeast analyzed by high-resolution imaging techniques. Proc Natl Acad Sci USA 101: 16495–16500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bystricky K, Laroche T, van Houwe G, Blaszczyk M, Gasser SM (2005) Chromosome looping in yeast: telomere pairing and coordinated movement reflect anchoring efficiency and territorial organization. J Cell Biol 168: 375–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciosk R, Zachariae W, Michaelis C, Shevchenko A, Mann M, Nasmyth K (1998) An ESP1/PDS1 complex regulates loss of sister chromatid cohesion at the metaphase to anaphase transition in yeast. Cell 93: 1067–1076 [DOI] [PubMed] [Google Scholar]
- Clarke PR, Zhang C (2008) Spatial and temporal coordination of mitosis by Ran GTPase. Nat Rev Mol Cell Biol 9: 464–477 [DOI] [PubMed] [Google Scholar]
- D'Ambrosio C, Kelly G, Shirahige K, Uhlmann F (2008) Condensin-dependent rDNA decatenation introduces a temporal pattern to chromosome segregation. Curr Biol 18: 1084–1089 [DOI] [PubMed] [Google Scholar]
- D'Amours D, Stegmeier F, Amon A (2004) Cdc14 and Condensin control the dissolution of Cohesin-independent chromosome linkages at repeated DNA. Cell 117: 455–469 [DOI] [PubMed] [Google Scholar]
- Denison C, Rudner AD, Gerber SA, Bakalarski CE, Moazed D, Gygi SP (2005) A proteomic strategy for gaining insights into protein sumoylation in yeast. Mol Cell Proteomics 4: 246–254 [DOI] [PubMed] [Google Scholar]
- Freeman L, Aragon-Alcaide L, Strunnikov A (2000) The Condensin complex governs chromosome condensation and mitotic transmission of rDNA. J Cell Biol 149: 811–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghiara JB, Richardson HE, Sugimoto K, Henze M, Lew DJ, Wittenberg C, Reed SI (1991) A cyclin B homolog in S. cerevisiae: chronic activation of the Cdc28 protein kinase by cyclin prevents exit from mitosis. Cell 65: 163–174 [DOI] [PubMed] [Google Scholar]
- Gotta M, Laroche T, Formenton A, Maillet L, Scherthan H, Gasser SM (1996) The clustering of telomeres and colocalization with Rap1, Sir3, and Sir4 proteins in wild-type S. cerevisiae. J Cell Biol 134: 1349–1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guacci V, Koshland D, Strunnikov A (1997) A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of MCD1 in S. cerevisiae. Cell 91: 47–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geymonat M, Spanos A, Smith SJ, Wheatley E, Rittinger K, Johnston LH, Sedgwick SG (2002) Control of mitotic exit in budding yeast. In vitro regulation of Tem1 GTPase by Bub2 and Bfa1. J Biol Chem 277: 28439–28445 [DOI] [PubMed] [Google Scholar]
- Hartwell LH, Culotti J, Pringle JR, Reid BJ (1974) Genetic control of cell division cycle in yeast. Science 183: 46–51 [DOI] [PubMed] [Google Scholar]
- Higuchi T, Uhlmann F (2005) Stabilization of microtubule dynamics at anaphase onset promotes chromosome segregation. Nature 433: 171–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T (2005) Condensins: organizing and segregating the genome. Curr Biol 15: R265–R275 [DOI] [PubMed] [Google Scholar]
- Jensen S, Geymonat M, Johnson AL, Segal M, Johnston LH (2002) Spatial regulation of the guanine nucleotide exchange factor Lte1 in S. cerevisiae. J Cell Sci 115: 4977–4991 [DOI] [PubMed] [Google Scholar]
- Kimura K, Cuvier O, Hirano T (2001) Chromosome condensation by a human Condensin complex in Xenopus egg extracts. J Biol Chem 276: 5417–5420 [DOI] [PubMed] [Google Scholar]
- Kimura K, Hirano M, Kobayashi R, Hirano T (1998) Phosphorylation and activation of 13S Condensin by Cdc2 in vitro. Science 282: 487–490 [DOI] [PubMed] [Google Scholar]
- Kratky O, Porod G (1994) Röntgenuntersuchung gelöster Fadenmoleküle. Rec Trav Chim Pays Bas 68: 1106–1123 [Google Scholar]
- Lavoie BD, Hogan E, Koshland D (2002) In vivo dissection of the chromosome condensation machinery: reversibility of condensation distinguishes contributions of Condensin and Cohesin. J Cell Biol 156: 805–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavoie BD, Hogan E, Koshland D (2004) In vivo requirements for rDNA chromosome condensation reveal two cell-cycle-regulated pathways for mitotic chromosome folding. Genes Dev 18: 76–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavoie BD, Tuffo KM, Oh S, Koshland D, Holm C (2000) Mitotic chromosome condensation requires Brn1p, the yeast homologue of Barren. Mol Biol Cell 11: 1293–1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SE, Jensen S, Frenz LM, Johnson AL, Fesquet D, Johnston LH (2001) The Bub2-dependent mitotic pathway in yeast acts every cell cycle and regulates cytokinesis. J Cell Sci 114: 2345–2354 [DOI] [PubMed] [Google Scholar]
- Lipp JJ, Hirota T, Poser I, Peters JM (2007) Aurora B controls the association of Condensin I but not Condensin II with mitotic chromosomes. J Cell Sci 120: 1245–1255 [DOI] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A III, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in S. cerevisiae. Yeast 14: 953–961 [DOI] [PubMed] [Google Scholar]
- Maeshima K, Laemmli UK (2003) A two-step scaffolding model for mitotic chromosome assembly. Dev Cell 4: 467–480 [DOI] [PubMed] [Google Scholar]
- Nasmyth K, Adolf G, Lydall D, Seddon A (1990) The identification of a second cell cycle control on the HO promoter in yeast: cell cycle regulation of SW15 nuclear entry. Cell 62: 631–647 [DOI] [PubMed] [Google Scholar]
- Ouspenski II, Cabello OA, Brinkley BR (2000) Chromosome condensation factor Brn1p is required for chromatid separation in mitosis. Mol Biol Cell 11: 1305–1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasero P, Duncker BP, Schwob E, Gasser SM (1999) A role for the Cdc7 kinase regulatory subunit Dbf4p in the formation of initiation-competent origins of replication. Genes Dev 13: 2159–2176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira G, Hofken T, Grindlay J, Manson C, Schiebel E (2000) The Bub2p spindle checkpoint links nuclear migration with mitotic exit. Mol Cell 6: 1–10 [PubMed] [Google Scholar]
- Queralt E, Lehane C, Novak B, Uhlmann F (2006) Downregulation of PP2A(Cdc55) phosphatase by separase initiates mitotic exit in budding yeast. Cell 125: 719–732 [DOI] [PubMed] [Google Scholar]
- Schleiffer A, Kaitna S, Maurer-Stroh S, Glotzer M, Nasmyth K, Eisenhaber F (2003) Kleisins: a superfamily of bacterial and eukaryotic SMC protein partners. Mol Cell 11: 571–575 [DOI] [PubMed] [Google Scholar]
- Schober H, Kalck V, Vega-Palas MA, Van Houwe G, Sage D, Unser M, Gartenberg MR, Gasser SM (2008) Controlled exchange of chromosomal arms reveals principles driving telomere interactions in yeast. Genome Res 18: 261–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seshan A, Bardin AJ, Amon A (2002) Control of Lte1 localization by cell polarity determinants and Cdc14. Curr Biol 12: 2098–2110 [DOI] [PubMed] [Google Scholar]
- Shevchenko A, Wilm M, Vorm O, Jensen ON, Podtelejnikov AV, Neubauer G, Shevchenko A, Mortensen P, Mann M (1996) A strategy for identifying gel-separated proteins in sequence databases by MS alone. Biochem Soc Trans 24: 893–896 [DOI] [PubMed] [Google Scholar]
- Shirayama M, Matsui Y, Tanaka K, Toh-e A (1994) Isolation of a CDC25 family gene, MSI2/LTE1, as a multicopy suppressor of ira1. Yeast 10: 451–461 [DOI] [PubMed] [Google Scholar]
- Shou W, Seol JH, Shevchenko A, Baskerville C, Moazed D, Chen ZW, Jang J, Shevchenko A, Charbonneau H, Deshaies RJ (1999) Exit from mitosis is triggered by Tem1-dependent release of the protein phosphatase Cdc14 from nucleolar RENT complex. Cell 97: 233–244 [DOI] [PubMed] [Google Scholar]
- Stegmeier F, Visintin R, Amon A (2002) Separase, polo kinase, the kinetochore protein Slk19, and Spo12 function in a network that controls Cdc14 localization during early anaphase. Cell 108: 207–220 [DOI] [PubMed] [Google Scholar]
- Strunnikov AV, Hogan E, Koshland D (1995) SMC2, a S. cerevisiae gene essential for chromosome segregation and condensation, defines a subgroup within the SMC family. Genes Dev 9: 587–599 [DOI] [PubMed] [Google Scholar]
- Sullivan M, Higuchi T, Katis VL, Uhlmann F (2004) Cdc14 phosphatase induces rDNA condensation and resolves Cohesin-independent cohesion during budding yeast anaphase. Cell 117: 471–482 [DOI] [PubMed] [Google Scholar]
- Toth A, Queralt E, Uhlmann F, Novak B (2007) Mitotic exit in two dimensions. J Theor Biol 248: 560–573 [DOI] [PubMed] [Google Scholar]
- Trinkle-Mulcahy L, Andersen J, Lam YW, Moorhead G, Mann M, Lamond AI (2006) Repo-Man recruits PP1 gamma to chromatin and is essential for cell viability. J Cell Biol 172: 679–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlmann F, Lottspeich F, Nasmyth K (1999) Sister-chromatid separation at anaphase onset is promoted by cleavage of the Cohesin subunit Scc1. Nature 400: 37–42 [DOI] [PubMed] [Google Scholar]
- Vagnarelli P, Hudson DF, Ribeiro SA, Trinkle-Mulcahy L, Spence JM, Lai F, Farr CJ, Lamond AI, Earnshaw WC (2006) Condensin and Repo-Man-PP1 co-operate in the regulation of chromosome architecture during mitosis. Nat Cell Biol 8: 1133–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma R, Annan RS, Huddleston MJ, Carr SA, Reynard G, Deshaies RJ (1997) Phosphorylation of Sic1p by G1 Cdk required for its degradation and entry into S phase. Science 278: 455–460 [DOI] [PubMed] [Google Scholar]
- Visintin R, Craig K, Hwang ES, Prinz S, Tyers M, Amon A (1998) The phosphatase Cdc14 triggers mitotic exit by reversal of Cdk-dependent phosphorylation. Mol Cell 2: 709–718 [DOI] [PubMed] [Google Scholar]
- Visintin R, Hwang ES, Amon A (1999) Cfi1 prevents premature exit from mitosis by anchoring Cdc14 phosphatase in the nucleolus. Nature 398: 818–823 [DOI] [PubMed] [Google Scholar]
- Wang Y, Hu F, Elledge SJ (2000) The Bfa1/Bub2 GAP complex comprises a universal checkpoint required to prevent mitotic exit. Curr Biol 10: 1379–1382 [DOI] [PubMed] [Google Scholar]
- Yang CH, Lambie EJ, Hardin J, Craft J, Snyder M (1989) Higher order structure is present in the yeast nucleus: autoantibody probes demonstrate that the nucleolus lies opposite the spindle pole body. Chromosoma 98: 123–128 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Movie S1
Supplementary Movie S2
Supplementary Information