Abstract
Most polycistronic genes are expressed in a single transcript, in which each cistron produces a fixed amount of protein. In this report, we show the first example of differential degradation of a polycistronic gene induced by a small regulatory RNA (sRNA). Our data show that the iron-responsive sRNA, RyhB, binds to the second cistron of the polycistronic mRNA, iscRSUA, which encodes the necessary machinery for biosynthesis of Fe–S clusters, and promotes the cleavage of the downstream iscSUA transcript. This cleavage gives rise to the remaining 5′-section of the transcript encoding IscR, a transcriptional regulator responsible for activation and repression of several genes depending on the cellular Fe–S level. Our data indicate that the iscR transcript is stable and that translation is active. The stability of the iscR transcript depends on a 111-nucleotide long non-translated RNA section located between iscR and iscS, which forms a strong repetitive extragenic palindromic secondary structure and may protect against ribonucleases degradation. This novel regulation shows how sRNAs and mRNA structures can work together to modulate the transcriptional response to a specific stress.
Keywords: iscRSUA, mRNA polarity, REP sequence, RNA degradosome, small RNA
Introduction
Numerous proteins, such as enzymes, metabolite transporters, and gene regulators, depend on prosthetic groups to achieve their normal functions. One important and probably the most ancient prosthetic group are Fe–S clusters, which regulate many metabolic functions, allow electron transport, control gene expression, and act as sensors of intracellular conditions. Virtually, all cells depend on Fe–S cofactors, as they are biosynthesised by Eubacteria, Archaea, and mitochondria in eukaryotes. Biosynthesis of Fe–S clusters is closely linked to haeme synthesis and regulation of cellular iron homoeostasis by the iron regulatory protein 1 in eukaryotes (reviewed in Lill and Muhlenhoff, 2006). As it shows remarkable similarities to the bacterial system, it appears that the eukaryotic Fe–S biosynthesis has probably been inherited from eubacteria.
Formation of Fe–S clusters in bacteria depends on three distinct and highly conserved protein machineries. The first machinery to be discovered, the nitrogen fixation system, is exclusive to the Fe–S cluster assembly of nitrogenase, which converts N2 into NH3 (reviewed in Rees and Howard, 2000; Dos Santos et al, 2004; Ayala-Castro et al, 2008). The second machinery, termed ISC (iron–sulphur cluster), is responsible for most of the cellular Fe–S proteins and, as such, performs the housekeeping Fe–S biosynthesis (Zheng et al, 1998; Takahashi and Nakamura, 1999; Outten et al, 2004). In Escherichia coli, the ISC components are encoded on the iscRSUA polycistronic transcript. The IscS product is responsible for providing selenium and sulphur for several modified tRNAs (Lauhon, 2002). Inactivation of iscS produces severe growth defects, indicating its biological importance (Takahashi and Nakamura, 1999; Schwartz et al, 2000). IscS is also extensively involved in the synthesis and repair of the Fe–S cluster (Takahashi and Nakamura, 1999; Schwartz et al, 2000). The third Fe–S cluster synthesis machinery, designated SUF (sulphur mobilisation), performs similar functions to the ISC system, although specifically under iron depletion and oxidative stress (Takahashi and Tokumoto, 2002; Outten et al, 2004). Interestingly, although the single inactivation of the isc or of the suf operon does not impair viability, inactivation of both isc and suf operons is lethal in E. coli. This indicates that these systems share similar essential functions (Takahashi and Tokumoto, 2002; Outten et al, 2004; Tokumoto et al, 2004).
Although both isc and suf operons are expressed during iron depletion, each operon responds to a different activation mechanism. During normal growth, in the presence of sufficient iron, transcription of the suf operon is directly repressed by the Fur (ferric uptake regulator) protein attached to the suf promoter (Outten et al, 2004). Under low iron conditions, however, Fur releases the transcriptional repression, which allows expression of the suf operon. Contrary to suf, the isc operon is not regulated by Fur but by IscR, a transcriptional repressor encoded as part of the iscRSUA polycistron (Schwartz et al, 2001; Giel et al, 2006). This iscRSUA autorepression depends on IscR loaded with Fe–S clusters (Holo-IscR), which are synthesised by IscS, IscU, and IscA (Schwartz et al, 2001). Therefore, when Fe is insufficient, or when IscSUA activity is limited, the IscR regulator, now free of the Fe–S cluster (Apo-IscR), relieves its repression on the isc promoter, thus allowing transcription (Schwartz et al, 2000; Outten et al, 2004). Furthermore, it was shown recently that Apo-IscR could function as a transcriptional activator of the suf operon (Giel et al, 2006; Yeo et al, 2006; Lee et al, 2008).
A recently characterised regulator of the isc polycistronic transcript is the small regulatory RNA (sRNA), RyhB (Massé et al, 2005). RyhB is a Fur-regulated sRNA that binds on target mRNAs to induce their degradation by the concerted action of the RNA degradosome (composed of RNase E, an RNA helicase, and the polynucleotide phosphorylase PNPase) and of the RNA chaperone, Hfq (Massé and Gottesman, 2002; Massé et al, 2003; Morita et al, 2005). The RNA chaperone, Hfq, is critical for RyhB stability, function, and pairing with its mRNA targets (Massé and Gottesman, 2002; Massé et al, 2003; Geissmann and Touati, 2004). In addition to this, Hfq is part of a complex with RNase E and RyhB, which initiates the target mRNA degradation (Morita et al, 2005). An earlier microarray study indicated that RyhB induces the degradation of at least 18 transcripts, encoding a total of 56 proteins (Massé et al, 2005). In this microarray study, the RyhB-induced repression was observable on the entire full-length transcripts (monocistronic and polycistronic alike), except for the iscRSUA transcript, which showed an unusual repression pattern. Indeed, RyhB represses the downstream part of the isc polycistron, iscSUA, without affecting the upstream part, iscR. This surprising observation suggests that RyhB induces a discoordination in the iscRSUA polycistronic transcript, which results in specific iscR expression during iron depletion. However, the mechanism of this sRNA-induced discoordination was not understood.
At least two polycistronic transcripts whose expressions are discoordinated by sRNAs have been described recently, each with a different mechanism. In the first mechanism, the sRNA Spot42 blocks the initiation of translation of the third gene in the polycistronic galETKM mRNA (Moller et al, 2002). This regulation is purely translational, as the levels of galETKM polycistron are unaffected by the action of Spot42 sRNA (Moller et al, 2002). The second mechanism of polycistronic discoordination relies on GlmY sRNA, which activates both the stability and the translation of glmS, a transcript that originates from a sRNA-independent RNase E cleavage between the open reading frames (ORFs) of the polycistronic glmUS mRNA (Kalamorz et al, 2007; Reichenbach et al, 2008; Urban and Vogel, 2008). In this example, however, the sRNA does not act on the polycistronic mRNA, but on the previously cleaved monocistronic glmS mRNA product.
We describe here an earlier uncharacterised mechanism of sRNA-induced discoordination of genes within the polycistronic iscRSUA transcript. By direct RyhB action, the downstream section encoding the Fe–S cluster formation machinery, iscSUA, is degraded, giving rise to the remaining fragment encoding IscR, a transcriptional regulator. We show that RyhB pairs with the ribosome-binding site (RBS) of iscS to promote the degradation of the iscSUA section by the RNA degradosome action. Although the iscSUA section decreases, the upstream iscR section remains protected by a strong secondary structure located between the iscR and iscS genes. This intergenic secondary structure is reminiscent of repetitive extragenic palindromic (REP) sequences (see the Discussion section for details). The 3′-end of the iscR fragment, generated after the RyhB-induced cleavage, has been mapped several nucleotides upstream of the RyhB pairing site on iscS. As expected, although the expression of IscS protein is significantly reduced, the expression of IscR remains intact. This novel mechanism of post-transcriptional processing, which modulates the activity and promoter specificity of the IscR transcription factor, shows the synergy between an sRNA and a REP sequence for optimal usage of polycistronic mRNAs.
Results
Expression of the sRNA RyhB reduces the iscSUA transcript level and increases iscR transcript under iron depletion
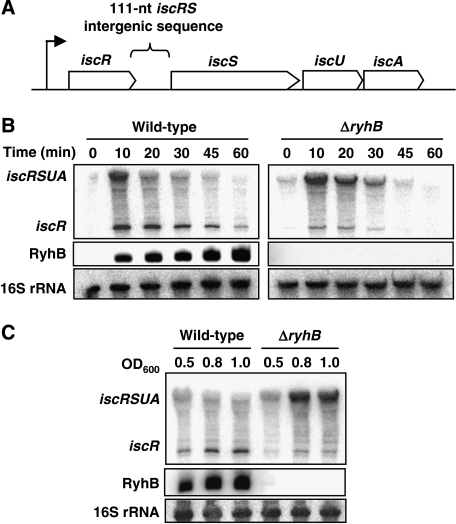
The sRNA RyhB promotes the degradation of a group of mRNAs encoding Fe-using proteins (Massé and Gottesman, 2002; Massé et al, 2003, 2005). In an earlier experiment designed to determine new mRNAs targeted by the sRNA, we observed that RyhB downregulated the 3′-section of the iscRSUA polycistronic mRNA (shown in Figure 1A), iscSUA, while leaving the upstream iscR section intact (Massé et al, 2005). We confirmed these results with quantitative real-time polymerase chain reaction (PCR), in which RyhB expression significantly decreased iscS transcript levels without affecting iscR transcript levels (data not shown). To investigate this mechanism of RyhB-induced iscRSUA polycistron discoordination, we extracted the total RNA from wild-type and ΔryhB cells grown in Luria-Bertani (LB) media in the presence of the iron chelator, 2,2′-dipyridyl (dip), which induces both RyhB and iscRSUA expressions (see the Introduction section; Massé and Gottesman, 2002). We then carried northern blots with probes specific to iscR and RyhB transcripts. As shown in Figure 1B middle panels, the sRNA RyhB is strongly expressed in wild-type cells after the addition of dip. In the northern blots carried out with the iscR-specific probe (Figure 1B, upper panels), two distinct bands are detected. The first is a low-molecular-weight band corresponding to iscR alone and a second band of higher molecular weight corresponding to the full-length iscRSUA. Co-migration with an RNA molecular weight marker shows the length difference between the two observed bands (Supplementary Figure S1C). Although the iscR band is quite significant in the wild-type cells, it is weak in the ΔryhB cells (compare left panel with right panel in Figure 1B). Whether in wild-type or ΔryhB cells, the level of iscRSUA transcript decreases after 10–20 min, presumably because of the recovery of intracellular iron homoeostasis and activated Holo-IscR repression (see Discussion for details). When we used the same RNA samples to carry out northern blot with an iscS-specific probe, only the full-length iscRSUA fragment was detected (as shown in Supplementary Figure S1B). This indicates that both iscR and iscRSUA mRNAs exist as individual molecules in the cell.
Figure 1.
RyhB sRNA induces a polarity in the iscRSUA polycistronic transcript. (A) Genomic organisation of the iscRSUA polycistron. (B) Northern blots on total RNA extracted from wild-type (EM1055) and ΔryhB (EM1238) cells, hybridised with an iscR-specific or RyhB-specific probe. Cells were grown in LB medium until an OD600 of 0.5 and dip was added (200 μM final) at time 0 to induce the expression of RyhB and the isc operon. Total RNA was extracted at the indicated times. (C) Northern blots on total RNA extracted from wild-type (EM1055) and ΔryhB (EM1238) cells, hybridised with an iscR-specific or RyhB-specific probe. Cells were grown in M63 medium (without iron) and total RNA was extracted at the indicated OD600. 16S rRNA was used as a loading control.
We then investigated the steady-state expression of the iscRSUA transcript in wild-type and ΔryhB cells grown in minimal M63 media without iron, which allows constitutive RyhB expression in wild-type cells (Figure 1C, middle panel). Figure 1C upper panel shows that, although the iscR transcript is more abundant in the wild-type strain (left panel), the full-length iscRSUA transcript becomes dominant in the ΔryhB mutant (right panel). Contrary to the results obtained in the LB medium, the iscRSUA expression is very stable in ΔryhB cells grown in M63 medium without iron (compare Figure 1B and C). This suggests that IscR remains under the Apo form and does not repress the isc operon. Taken together, these results indicate that RyhB promotes the specific expression of the iscR transcript and significantly reduces the full-length iscRSUA transcript level.
The stability of the iscRSUA transcript is decreased by the sRNA RyhB
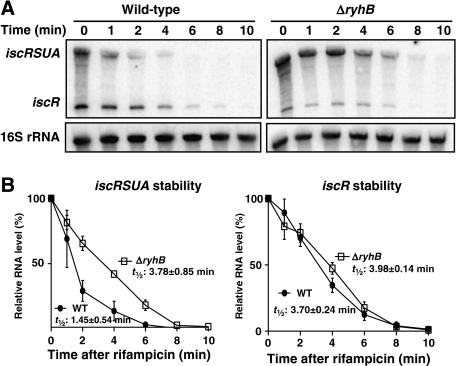
RyhB promotes the full degradation of many target mRNAs (Massé et al, 2003, 2005). However, our results with iscRSUA indicate that RyhB triggers the specific degradation of the downstream cistrons, iscS, iscU, and iscA, without affecting the iscR fragment. To investigate this, we determined the specific stability of iscRSUA and iscR mRNAs in the absence or presence of RyhB. In the experiment shown in Figure 2, we added dip in the culture for 10 min to induce ryhB and iscRSUA expression, followed by the addition of rifampicin to stop transcription. Total RNA was then extracted at different time points and the RNA was hybridised with an iscR-specific probe (Figure 2A). As shown in Figure 2B, the half-life of the iscR mRNA is almost the same whether RyhB is expressed (wild-type strain: 3.70 min) or not (ΔryhB mutant: 3.98 min). However, the half-life of the iscRSUA mRNA is significantly shorter in the wild-type strain (1.45 min) than that in the ΔryhB mutant (3.78 min). This result indicates that RyhB decreases the stability of the iscRSUA mRNA without affecting iscR mRNA.
Figure 2.
RyhB decreases the stability of the iscRSUA transcript, not iscR transcript. (A) Northern blots on total RNA extracted from wild-type (EM1055) and ΔryhB (EM1238) cells and hybridised with an iscR probe. Cells were grown in LB media to an OD600 of 0.5, at which point dip was added (200 μM final) for 10 min to induce the expression of RyhB and isc. Rifampicin was added (250 μg/ml final) at time 0 to block transcription. Total RNA was extracted at indicated times. 16S rRNA was used as a loading control. (B) Densitometry analysis of three northern blots carried out as in panel A.
The RNA degradosome and the RNA chaperone, Hfq, are essential for the RyhB-induced isc polarity
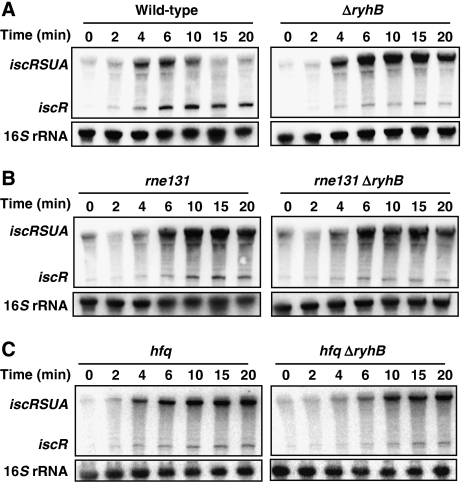
We showed earlier that RNase E and the RNA degradosome are involved in the RyhB-mediated degradation of target mRNAs (Massé et al, 2003). We tested for the potential role of the RNA degradosome in the RyhB-induced discoordination of the iscRSUA operon during iron depletion. As shown in Figure 3B, the inactivation of the RNA degradosome (rne131 mutant) results in an increased iscRSUA mRNA level (compare with wild-type in Figure 3A). Interestingly, the iscR fragment does not accumulate significantly in the rne131 mutant. This indicates that RyhB-dependent iscR accumulation depends on partial degradation of the iscRSUA fragment and not on blocked transcription. We do not observe a notable difference between the rne131 mutant and the rne131 ΔryhB double mutant, indicating that the sRNA has no effect without the RNA degradosome.
Figure 3.
The RNA degradosome and the RNA chaperone, Hfq, are essential for RyhB-induced differential degradation of the iscRSUA polycistron. (A) Northern blots on total RNA extracted from wild-type (EM1055) and ΔryhB (EM1238) cells and hybridised with an iscR-specific probe. Cells were grown in LB media to an OD600 of 0.5, at which point dip was added (200 μM final) at time 0 to induce the expression of RyhB and iscRSUA. Total RNA was extracted at indicated times. (B) Same as in panel A but total RNA extracted from RNA degradosome mutant (EM1377; rne131) and ΔryhB rne131 double mutant (GD010). (C) Same as in panel A but total RNA extracted from hfq mutant (EM1265) and hfq ΔryhB double mutant (KP111). 16S rRNA was used as a loading control.
The RNA chaperone Hfq is essential for both RyhB stability and function (Massé and Gottesman, 2002; Massé et al, 2003; Geissmann and Touati, 2004; Morita et al, 2005). To investigate the role of Hfq in the iscRSUA regulation, we compare the effect of RyhB induction between wild-type and hfq cells. As shown in Figure 3C, the absence of Hfq results in reduced iscR transcript level as compared with wild type (Figure 3A). It can be noted that the level of full-length iscRSUA (at time points 15–20 min) is significantly more in rne131 and hfq cells (independently from ryhB) than in wild-type cells, showing the critical effect of these factors on the differential degradation of the iscRSUA polycistron. In addition, these results indicate that when RyhB is absent (ΔryhB) or non-functional (rne131 and hfq), the isc operon does not self-repress efficiently as in wild type. This suggests that RyhB promotes the formation of Holo-IscR by iron-sparing. Eventually, this will result in transcriptional repression of the isc operon (see the Discussion section for details).
The intergenic region between iscR and iscS forms a strong secondary structure that is responsible for the RyhB-dependant accumulation of iscR
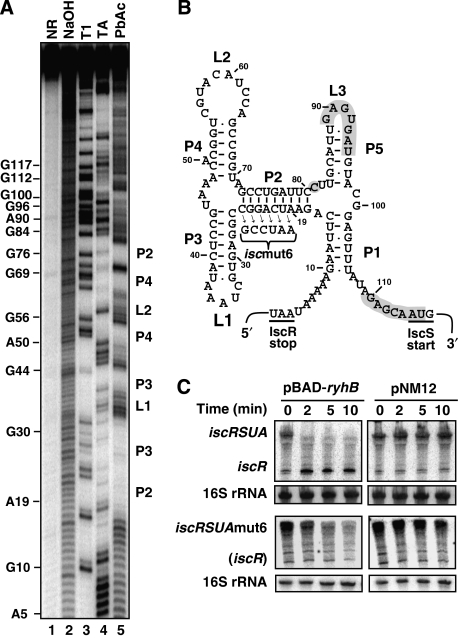
To explain the accumulation of iscR mRNA after the expression of RyhB, we sought for potential secondary structure in the 111-nucleotide long intergenic region between iscR and iscS. Using the mfold software (http://www.bioinfo.rpi.edu/applications/mfold), we found that this intergenic region can form a strong secondary structure (Figure 4B), which is very well conserved among Enterobacteriaceae as shown in Figure 5. This conservation of secondary structure suggests an important physiological role. The secondary structure was also determined in vitro by lead acetate (PbAc) probing (Figure 4A), which cleaves single-strand RNA molecule. We note that stems P2, P3, and P4 are protected against cleavage by Pb+ ions (Figure 4A and B), indicating that they are double stranded. Stems P1 and P5 are cleaved, suggesting they form weaker interactions. This structure is reminiscent of REP sequence (see the Discussion section for details).
Figure 4.
The iscRS intergenic region harbours a strong secondary structure (A). Secondary structure probing of the intergenic iscRS region (5′-end labelled). The iscS fragment used contains the intergenic region between iscR and iscS and the first 100 nucleotides of iscS coding sequence. Probing was carried out in denaturing conditions (lanes 3 and 4) or in native conditions (lane 5). The position of several G and A residues are given. NR, untreated RNA control; NaOH, alkaline ladder; T1, RNase T1; TA, RNase TA; PbAc, lead acetate. (B) Secondary structure of the intergenic region between iscR and iscS cistrons as determined by the mfold software (http://www.bioinfo.rpi.edu/applications/mfold/). The P2, P3, and P4 stems and the L1 and L2 loops are visible by secondary structure probing in panel A. The C81 (grey circle) is the last nucleotide in the RyhB-induced iscR transcript as determined by 3′-RACE (see text for details). (C) Northern blots on total RNA extracted from the wild-type strain (EM1455) and the iscRSUAmut6 mutant ((GD114) mutation shown in panel B and hybridised with an iscR probe. Both strains carry a plasmid allowing arabinose-dependent RyhB expression (pBAD-ryhB) or a control vector (pNM12). Cells were grown in M63 media to an OD600 of 0.5 and arabinose was added at time 0 (final concentration of 0.01%). Total RNA was extracted at the indicated time. 16S rRNA was used as a loading control.
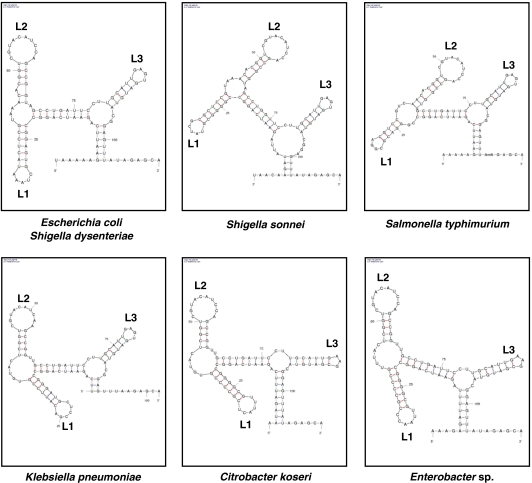
Figure 5.
Conservation of sequence and structure in the intergenic region between iscR and iscS among Enterobacteriacea. Sequence homologies were found using NCBI BLAST tool and the secondary structure was determined using the mfold software. All structures shown are localised in the intergenic region between iscR and iscS or their putative homologues in each bacterium. Localisation of loops L1, L2 and L3 are also indicated.
To assess the potential role of this secondary structure, we constructed a mutant (iscmut6; see Figure 4B for description), in which we disrupted the main stem (P2). We then analysed the effect of RyhB on the iscmut6 mutant using the pBAD-ryhB vector, which expresses RyhB from an arabinose-inducible promoter (Massé et al, 2003). Cells carrying either the pBAD-ryhB or control pNM12 plasmids (described in Materials and methods) were grown in minimal M63 medium, which allows the constitutive expression of the isc operon. Although the induction of RyhB (pBAD-ryhB) leads to a decrease of the full-length iscRSUA and iscmut6 transcripts, as shown in Figure 4C (left panels), accumulation of iscR fragments occurs only from the iscRSUA transcript. These results indicate that the secondary structure between iscR and iscS is necessary for the accumulation of an iscR fragment after the expression of RyhB.
We then characterised the 3′-end of the accumulating iscR RNA fragment after the expression of RyhB, by carrying out a 3′-RACE experiment (described in Materials and methods). To do this, we used the total RNA extracted from cells in which RyhB has been expressed for 30 min (see Supplementary Figure S6; pBAD-ryhB; 30 min). As shown in Figure 4B, the 3′-end of the iscR fragment is situated in the untranslated region between iscR and iscS, upstream of the pairing with RyhB (see below). Hence, the iscR RNA fragment resulting from the RyhB expression contains the entire ORF of iscR and thus is likely to be translated. It can be noted that the 3′-end of the iscR fragment is situated just downstream of the stronger stem (P2) of the structure described above.
Direct pairing of RyhB at the 5′-UTR of the iscS mRNA
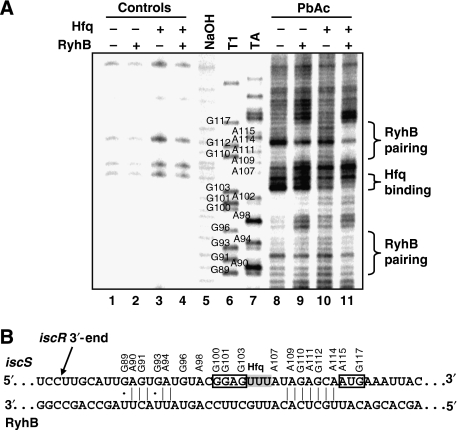
To investigate the effect of RyhB on the iscRSUA polycistron, we sought for a possible pairing site between both RNAs. Using bioinformatics tools, we found a putative pairing between RyhB and the 5′-UTR of iscS (Figures 4B and 6B), which covers from the region upstream of the RBS to the first codon of the target mRNA, a hallmark of negatively regulating sRNAs (Gottesman, 2004). To investigate the pairing between RyhB and iscS 5′-UTR, we carried out footprinting assays with a 5′-end radiolabelled iscS RNA in the absence and presence of RyhB RNA and the RNA chaperone, Hfq. We used PbAc probing to visualise the pairing between both RNAs, as Pb+ ions specifically cleave single-stranded RNA. As shown in Figure 6A, the addition of RyhB to iscS decreases the cleavage in the region of the start codon of iscS (compare lanes 8 and 9), indicating a pairing between both iscS and RyhB RNAs. The effect of the RNA chaperone, Hfq, was also addressed in this experiment. In the presence of Hfq, the protecting effect of RyhB on iscS is increased (compare lanes 9 and 11), indicating that the chaperone facilitates the pairing between both RNAs. The addition of Hfq alone protects the nucleotides between G103 and A107 (compare lanes 8 and 10), suggesting that it binds to the three Us located downstream of the iscS Shine–Dalgarno sequence. All these results are consistent with the predicted pairing shown in Figure 6B. Although it is likely that the region between G100 and U104 of iscS interacts with RyhB, there is no such evidence, as this region seems resistant to Pb+ cleavage (Figure 6A).
Figure 6.
Analysis of the RyhB pairing site on iscS. (A) Secondary structure probing of the 5′-end labelled iscS mRNA in the presence or absence of RyhB and Hfq. The position of several G and A residues are given. NaOH, alkaline ladder; T1, RNase T1; TA, RNase TA; PbAc, lead acetate. All experiments were carried out in native conditions except for NaOH, T1 and TA ladders. (B) Pairing between RyhB and the 5′-UTR of iscS. This pairing overlaps the putative ribosome-binding site and the translation start codon of iscS (boxed area). The 3′-end of the iscR mRNA that accumulates after the expression of RyhB was determined by 3′-RACE and is shown in panel B. It is situated in the 111-nucleotide long intergenic region between iscR and iscS cistrons.
We also tested the RyhB–iscS pairing in vivo using an iscRS′–lacZ transcriptional fusion inserted in single copy at the phage-λ integration site of the chromosome (illustrated in Supplementary Figure S7; see Materials and methods for details). As shown in Supplementary Figure S3, the β-galactosidase activity is decreased in the wild-type fusion after the expression of RyhB from an arabinose-inducible promoter. We constructed several mutants of iscS to disrupt the pairing between RyhB and iscS, and we measured the β-galactosidase activity in the absence or presence of RyhB. We had to mutate at least seven nucleotides in the iscS 5′-UTR region to disrupt the RyhB effect (iscRS′mut7-lacZ). However, the RyhB mutant that is compensatory to iscRS′mut7-lacZ could not restore the wild-type effect. This suggests that, as the mutated region is close to the Hfq-binding site determined for RyhB (Geissmann and Touati, 2004), the compensatory RyhB mutant becomes ineffective. Together, the in vitro and in vivo results indicate that RyhB pairs at the 5′-UTR of iscS, which is the first step to a decrease in full-length iscRSUA transcript and accumulation of an iscR transcript. The pairing is particularly strong on the iscS initiation codon region, correlating with our footprinting assays above (see Figure 6A). These results suggest that RyhB pairing could disturb translation initiation.
Physiological significance of the RyhB-induced discoordination of iscRSUA polycistron
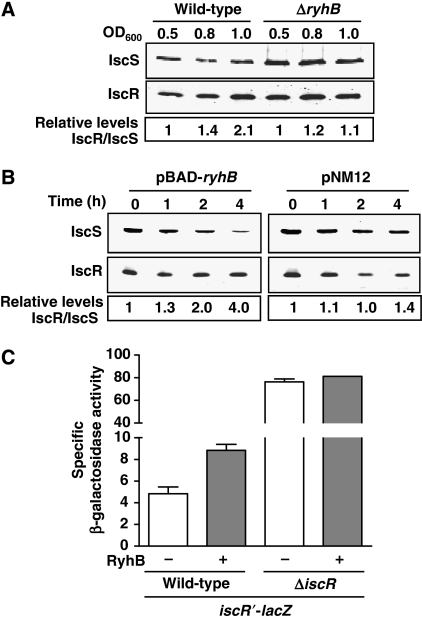
Our 3′-RACE experiment (Figure 4B) showed that the RyhB-dependent iscR fragment contains the entire ORF of iscR. However, it is unclear whether translation remains active or not. To address this question, we used quantitative western blots to measure the levels of IscR and IscS proteins after RyhB expression. We first measured the steady-state level from wild-type and ΔryhB cells grown in minimal M63 medium (similar to Figure 1C). The results in Figure 7A indicate a significant increase (over twofold) in the IscR/IscS ratio, as cells reached an OD600 of 1.0. We also carried out a similar experiment, but with RyhB expressed from the pBAD-ryhB vector. As shown in Figure 7B, the expression of RyhB (pBAD-ryhB) leads to a significant decrease in the protein levels of IscS. However, in same conditions, IscR levels remain stable even in the presence of RyhB. These results show a fourfold increase in the IscR/IscS ratio after 4 h of RyhB expression as compared with the control experiment with an empty vector (pNM12), in which the IscR and IscS levels are not significantly affected.
Figure 7.
Effect of RyhB on IscS and IscR proteins. (A) Quantitative western blots using IscR- and IscS-specific antibodies on total protein extracted from wild-type (EM1055) and ΔryhB (EM1238) cells. Cultures were grown in M63 media (without iron) and total proteins were extracted at the indicated OD600. (B) Quantitative western blots using IscR- and IscS-specific antibodies on total proteins extracted from EM1455 cells carrying pNM12 or pBAD-ryhB plasmids. Cells were grown in LB media to an OD600 of 0.5 and arabinose was added at time 0. Total proteins were extracted at indicated times. In both western blots experiments (panels A and B), IRDye 800CW-conjugated secondary antibodies were used, allowing quantification. (C) The activity of the isc promoter was measured by β-galactosidase assay using the transcriptional iscR′-lacZ fusion. Wild-type (GD020) and ΔiscR (GD021) cells carrying either pNM12 (−RyhB) or pBAD-ryhB (+RyhB) were grown in LB media, in which arabinose was added at an OD600 of 0.1.
IscS, IscU, and IscA are responsible for Fe–S cluster biogenesis and are thought to transfer these clusters to IscR (Schwartz et al, 2001). When IscR is bound to a Fe–S cluster (Holo-IscR), it represses the isc promoter as a feedback control (Schwartz et al, 2001 see Introduction for details). However, as the IscS level decreases after RyhB expression, we hypothesised that the IscR protein produced in these conditions is under the Apo form (without a Fe–S cluster). To test this, we measured the expression of the isc promoter using an iscR′-lacZ transcriptional fusion (illustrated in Supplementary Figure S7) inserted at the λ attachment site in a strain harbouring the endogenous iscRSUA operon. As shown in Figure 7C, the expression of RyhB from a pBAD-ryhB plasmid leads to a twofold increase in β-galactosidase activity as compared with the control plasmid (pNM12). However, if we delete the endogenous copy of the polycistron (ΔiscR), there is no significant difference in the β-galactosidase activities with or without RyhB. These results suggest that although RyhB downregulates IscS, the IscR transcriptional regulator remains expressed, however, under the Apo-IscR form.
Discussion
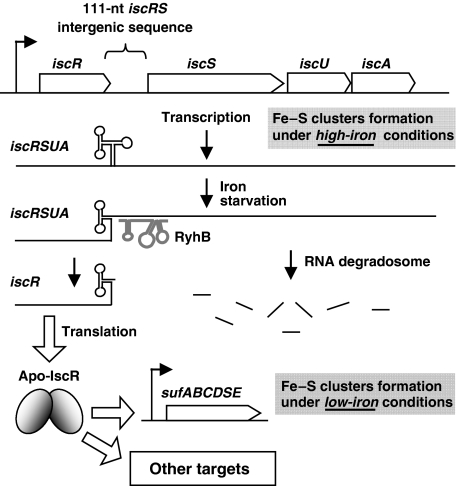
This work shows a novel sRNA-induced mechanism of transcript polarity through differential degradation of a polycistronic mRNA. In this mechanism, the sRNA RyhB initiates the degradation of a specific section of a target mRNA. Our working model (Figure 8) suggests that when RyhB is expressed during iron depletion, it pairs with the RBS and the translation start codon of iscS. This model is supported by data indicating that the pairing of RyhB with iscS results in a rapid (<2 min, Figure 4C) degradation of the 3′-region of the polycistron encoding IscS, IscU, and IscA. However, unlike any other sRNA target, the 5′-region of the iscRSUA polycistron is protected from the action of the RNA degradosome. We identified a REP sequence (Higgins et al, 1982; Newbury et al, 1987b) in the 111-nucleotide long intergenic region between iscR and iscS, which forms a strong secondary structure that likely protects the upstream iscR cistron from degradation. To our knowledge, this is the first time that a REP sequence is linked to the action of an sRNA.
Figure 8.
Working model of RyhB-induced partial degradation of the iscRSUA polycistron in Enterobacteriacea and its physiological significance. Under high iron conditions, RyhB is not expressed and the iscRSUA polycistron can be translated and assure the housekeeping Fe–S cluster biogenesis. When iron becomes insufficient, RyhB is expressed and pairs with the 5′-UTR of iscS. This leads to the recruitment of the RNA degradosome and the rapid degradation of iscS, iscU and iscA cistrons. However, a strong secondary structure in the intergenic region between iscR and iscS protects the iscR cistron from degradation. The IscR protein produced under these conditions is likely to be under the Apo form (lacking Fe-S) and may activate the expression of the suf operon, which would then overtake the Fe–S cluster biogenesis.
REP sequences occupy up to 1% of the E. coli genome mostly on non-translated RNA (Stern et al, 1984). When located downstream of an RNA, REP sequences have been shown to stabilise the transcript (Newbury et al, 1987b) and to be responsible for a differential rate of decay among parts of a polycistron (Belasco et al, 1985). Although it is not induced by an sRNA, an earlier demonstrated mechanism of discoordination based on REP sequence has been described for the polycistronic mRNA, rxcA, expressed in the photosynthetic bacterium Rhodopseudomonas capsulata. Although rxcA is expressed as a single transcript encoding five ORFs, the 3′-end of the mRNA is rapidly degraded, whereas the upstream RNA section encoding two ORFs remains stable (Belasco et al, 1985). This differential stability between both sections of the transcript results in a 10- to 30-fold excess in the polypeptides originating from the stable upstream section as compared with the rapidly decaying downstream section (Schumacher and Drews, 1978; Kaufmann et al, 1982). A REP sequence located in the intercistronic region is thought to be responsible for the protection of the upstream section of the mRNA (Belasco et al, 1985). Thus, the location of the REP structure and its effect on the discoordination of the rxcA mRNA is reminiscent of the iscRSUA mRNA described in this work.
Furthermore, the REP-dependent discoordination of the rxcA mRNA explains the generation of a small amount of iscR fragment in the absence of RyhB (Figures 1B and C, and 3A and B). As the normal turnover of the iscRSUA transcript (independently from RyhB) may start downstream of the REP sequence, in the iscSUA section, the progression of the 3′–5′ exonuclease of the RNA degradosome will stop at the REP structure, thereby generating an iscR fragment. Indeed, we have mapped an iscR fragment, extracted from ΔryhB cells, with the same 3′-end as the one described in Figure 4B. Thus, this suggests that although the 3′-end of iscR is not absolutely dependent on RyhB, it requires RyhB to accumulate to a significant amount.
An additional point to consider is the negative effect of RyhB on iscS translation initiation, which could be sufficient to initiate cleavage or transcriptional stop through the transcriptional terminator, Rho. Indeed, RyhB was shown to block translation in the absence of the RNA degradosome (Morita et al, 2006). However, our results in Figure 3B show that even though RyhB is expressed, there is no accumulation of the iscR fragment in the absence of the RNA degradosome. Therefore, although RyhB may block the translation initiation of iscS, it is not sufficient to induce a transcript cleavage (or a transcription termination) that would generate an iscR fragment.
Although the mechanism protecting iscR from degradation is fairly clear, we can speculate on two different pathways that explain the RyhB-initiated iscSUA degradation. First, RyhB pairs with iscRSUA and induces an endonucleolytic cleavage at the site identified by 3′-RACE (3′-end of iscR mRNA; Figure 4B). Such a cleavage close to the P2 stem would leave the P2, P3, and P4 stems intact, which would help stabilising the 3′-end of iscR. As the sequence CU situated between stems P2 and P3 is perfectly conserved among Enterobacteriaceae (Figure 5), this suggests that it may contribute to recruit or help in determining a specific cleavage site.
The second potential pathway implies that RyhB pairing induces an RNase E-dependent endonucleolytic cleavage within the iscS ORF, which is followed by 3′–5′ exonucleolytic degradation by the PNPase. This suggests that RyhB-induced degradation depends on a sequence within the iscS ORF recognised by RNAse E. To address this question, we developed an assay based on RyhB sensitivity of the iscS gene containing various deletions within the ORF. Our preliminary results suggest that the RyhB-induced initial cleavage occurs within the iscS ORF, downstream of RyhB pairing, and not in the iscRS intergenic structure, upstream of the RyhB pairing (G Desnoyers and E Massé, manuscript in preparation). Thus, in the light of these results, we favour the second mechanism, in which the iscRS intergenic REP structure blocks the upstream progression (3′–5′) of PNPase after an RNAse E-dependent cleavage in the iscS ORF. As we mapped the 3′-end of the iscR fragment close to (one nucleotide downstream) the P2 stem, we believe that the P2 stem is key to prevent the 3′–5′ exonucleolytic activity of PNPase to progress further upstream. This structure seems essential to protect the iscR cistron, as in the case of disruption of the P2 stem (Figure 4B and C) or complete deletion of the intergenic region (data not shown), the iscR RNA does not accumulate. As RyhB acts downstream of the REP sequence, this also explains the similar stability measured for the iscR fragment in the presence or absence of RyhB (Figure 2).
As suggested in our model, the intergenic REP sequence between iscRS is critical to stabilise iscR transcript. Nevertheless, the RNA degradosome contains a DEAD-box RNA helicase (RhlB) that unfolds structured RNA and potentially REP sequences. Indeed, RhlB has been shown to help in vitro degradation of REP-stabilised RNAs by an ATP-dependent mechanism (Py et al, 1996). However, it also has been reported that RhlB has no effect on accumulation of REP-stabilised mRNA in vivo (Khemici and Carpousis, 2004). This difference between in vitro and in vivo data suggests that a yet uncharacterised factor binds to the intergenic structure to stabilise it or to block the RNA helicase activity. We believe that the loop L2, which is perfectly conserved among Enterobacteriaceae (Figure 5, Supplementary S4, and S5), is potentially involved in binding such a factor. More generally, as REP sequences can be found within intergenic region of several polycistronic mRNAs (Bachellier et al, 1999), it seems likely that other sRNA-regulated polycistron, with similar regulation as iscRSUA, will be characterised.
As mentioned in the Introduction, a similar yet mechanistically different example of discoordination of the polycistronic glmUS mRNA has been reported lately (Kalamorz et al, 2007; Reichenbach et al, 2008; Urban and Vogel, 2008). In this case, the glmUS polycistron mRNA is first cleaved by RNAse E in the intergenic region, resulting in the appearance of glmS transcript. Then, in conditions of low glucosamine 6-phosphate (GlcN-6-P) concentration, the sRNA GlmZ is expressed and pairs with the newly formed glmS monocistronic RNA. This pairing of GlmZ activates glmS translation by freeing the RBS from an inhibitory secondary structure. The resulting increase in GlmS protein allows the expression of glucosamine 6-phosphate synthase GlmS, which replenishes the intracellular GlcN-6-P pool. Although this mechanism results in differential expression of a cistron, which originates from a polycistronic mRNA, it is fundamentally different from the RyhB-iscRSUA mechanism described in this work.
Another RyhB target, the sdhCDAB mRNA, has some analogy with the iscRSUA mRNA. The sdhCDAB transcript encodes four proteins and is downregulated by RyhB pairing at the translation start of the second cistron, sdhD (Massé and Gottesman, 2002). However, an attempt to detect the sdhC transcript, located upstream to the RyhB pairing, shown that sdhC was barely detectable in the presence of RyhB, and thus unlikely to be physiologically relevant (Massé and Gottesman, 2002). This observation reinforces the role of the iscRS intergenic structure for stabilising the upstream iscR transcript. An additional polycistronic gene with similarities to iscRSUA is malEFG, which encode maltose-binding receptor and transport proteins. The malEFG transcript harbours a REP sequence between malE and malF that was shown to stabilise malE against the RNA degradosome attack (Newbury et al, 1987a). Although no sRNA is known to regulate the malEFG transcript, we cannot rule out a similar mechanism as the iscRSUA regulated by RyhB sRNA.
Physiological significance
Physiologically, the mechanism described in this work may be important for the adaptation to iron depletion. Earlier, it has been suggested that iscRSUA expression increases during iron depletion (Outten et al, 2004). Our results show that it is mostly the level of iscR, and not that of iscS, iscU, and iscA, which is increased, because of the post-transcriptional action of RyhB (Figures 1B, C, and 4C). In conditions of dip treatment, there are two distinct mechanisms that contribute to iscR increase. The first is reduced Fe–S clusters, which contributes to generate Apo-IscR and the subsequent derepression of the isc operon. The second mechanism is accumulation of the iscR section consequently to the RyhB-induced cleavage of iscRSUA. To our knowledge, this is the first description of a system where the expression of both the sRNA and its target mRNA increase in the same conditions (iron depletion), although with distinct mechanisms.
We notice that when dip is used to induce the expression of both RyhB and isc operon, the expression of iscRSUA reaches a peak after 10 min, which is followed by a rapid decrease (Figures 1B and 3A). In contrast to this, in the absence of RyhB (ΔryhB) or its protein partners (hfq, rne131), the iscRSUA transcript accumulates significantly for a longer time period (Figures 1B and 3A–C). Besides, the direct effect of RyhB on the iscRSUA transcript, a second mechanism can partly explain the decrease in iscRSUA transcript in wild-type cells. When expressed, RyhB generates free intracellular iron by iron-sparing (Massé et al, 2005; Jacques et al, 2006), which would help the formation of Holo-IscR. In wild-type cells treated with dip, RyhB expression reduces the expression of non-essential iron-using proteins, leaving the available iron to essential proteins (such as IscR, repressing the iscRSUA transcription after 15-20 min). In the rne131 and hfq mutants, however, even though it is expressed, RyhB cannot function normally. We believe that in the absence of functional RyhB, the intracellular iron will be sequestered by non-essential proteins, which results in iron shortage for essential iron-dependent proteins, such as IscR. Contrary to wild-type cells, where IscR becomes active after 15–20 min of dip addition, the three other backgrounds (ΔryhB, rne131, and hfq) still express the isc operon as IscR lacks sufficient iron to act as a repressor (Apo-IscR).
The most important observation in our work is that by sRNA-induced differential degradation of the iscRSUA polycistron, the IscR protein is still produced even when iron is depleted. Recent work showed that IscR regulates about 40 genes involved in anaerobic respiration, oxidative stress response, Fe–S biogenesis, and other metabolic pathways (Giel et al, 2006). As iscR mRNA is protected from degradation during iron depletion, it suggests that the regulatory activity of IscR must be conserved during this stress. Interestingly, it has been reported that one of the promoters regulated by IscR is the suf promoter (Giel et al, 2006; Yeo et al, 2006; Lee et al, 2008). IscR contributes to the activation of the suf operon, but only under the Apo-IscR form. In addition to the fact that iron is limited when RyhB is expressed, our results suggest that the downregulation of iscS, iscU, and iscA by RyhB increases the ratio of Apo-IscR. This is confirmed in the experiments shown in Figure 7A and B. It is likely that under conditions of RyhB expression and iscSUA repression, the suf machinery becomes the main machinery for biogenesis of Fe–S clusters. Indeed, proteins encoded by the suf mRNA are similar in role to IscS, IscU, and IscA, but are thought to be more important during iron depletion (Outten et al, 2003). This hypothetical mechanism remains to be addressed in future work, as it is of interest to understand the global regulatory network of iron homoeostasis.
Materials and methods
Stains and plasmids
Strains used in this work are described in Table I. Derivatives of EM1055 were used in all experiments. Strains prepared by P1 transduction were selected for the appropriate antibiotic-resistant marker. Strain EM1451 was selected for growth on media without leucine and was screened for inability to grow with arabinose as the only carbon source. Strain GD114 was generated by three-step PCR mutagenesis. Briefly, a first PCR was carried out using oligos EM359 and EM619 as primers (see Table II for oligonucleotides description). A second PCR was carried out using EM361 and EM618 as primers. The two PCR products were mixed to serve as the template for the third PCR (primers EM359–EM361). The resulting PCR product was digested by BamH1 and ligated into BamH1-digested pFRΔ. The fusion was delivered in single copy into the bacterial chromosome at the λ att site as described earlier (Simons et al, 1987). Stable lysogens were screened for single insertion of recombinant λ by PCR (Powell et al, 1994).
Table 1.
Strains and plasmids used in this study
| Strain number | Relevant markers | Reference/source |
|---|---|---|
| EM1055 | MG1655 Δlac X174 | Massé and Gottesman (2002) |
| EM1238 | EM1055 ΔryhB∷cat | Massé and Gottesman (2002) |
| EM1377 | EM1055 rne-131 zce-726::Tn10 | Massé et al (2003) |
| GD010 | EM1055 rne-131 zce-726::Tn10 ΔryhB::cat | EM1377+P1 (EM1238) |
| EM1265 | EM1055 hfq-1∷Ω(kan;Bcl1) | Massé et al (2003) |
| KP111 | EM1055 hfq-1∷Ω(kan;Bcl1) ΔryhB∷cat | EM1265+P1 (EM1238) |
| EM1059 | EM1055 Δara714 leu∷Tn10 | Massé et al (2003) |
| EM1451 | EM1055 Δara714 leu+ | EM1059+P1 (EM1055) |
| EM1455 | EM1055 Δara714 leu+ ΔryhB∷cat | EM1451+P1 (EM1238) |
| PK5956 | BW25113 ΔiscR∷kan | Schwartz et al (2001) |
| PK6364 | RZ4500 λiscR′-lacZ | Schwartz et al (2001) |
| GD175 | EM1055 Δara714 leu+ ΔryhB∷cat ΔiscR∷kan | EM1455+P1 (PK5956) |
| GD020 | EM1055 Δara714 leu+ ΔryhB∷cat λiscR′-lacZ | EM1455+λPK6364 |
| GD021 | EM1055 Δara714 leu+ ΔryhB∷cat ΔiscR∷kan λiscR′-lacZ | GD175+λPK6364 |
| GD114 | EM1055 Δara714 leu+ ΔryhB∷cat ΔiscR∷kan λiscRSUAmut6 | GD175+λpFRΔ-iscRSUAmut6 |
| Plasmids | Description | Reference/source |
| pNM12 | pBAD24 derivative | Majdalani et al (1998) |
| pBAD-ryhB | pBAD24+RyhB (arabinose inducible promoter) | Massé et al (2003) |
| pFRΔ-iscRSUAmut6 | Mutated intergenic iscRS region | Present study |
Table 2.
Oligonucleotides used in this study
| Oligo number | Sequence 5′–3′ |
|---|---|
| EM138 | TAATACGACTCACTATAGGGAGACTGTTCAGATCCACGC CCTGC |
| EM139 | CTGAACGGTGACCTGGAACAC |
| EM144 | TAATACGACTCACTATAGGGAGAGCTGACGACCAGACAC ATCC |
| EM145 | ACAACTGTTTTCCCGTCTGCG |
| EM190 | TAATACGACTCACTATAGGGAGACAGCACCCGGCTGGC TAAG |
| EM191 | CGATCAGGAAGACCCTCGC |
| EM293 | TAATACGACTCACTATAGGGAGACGCTTTACGCCCAGTA ATTCC |
| EM294 | CTCCTACGGGAGGCAGCAGT |
| EM359 | TCAGCGGATCCCCGAATAACAGCCGTTGC |
| EM361 | AACTCGGATCCACGGTGGGGTTATC |
| EM385 | CAAAGGTTCCGTCCATCGTC |
| EM408 | TGTAATACGACTCACTATAGGGCACCCGCACACAAGACG |
| EM618 | TTAATAAAAAGATTCAGAAAATCCGCGGAGTGCTAAAT ACTCCGTA |
| EM619 | ACGGAGTATTTAGCACTCCGCGGATTTTCTGAATTCTTT TTATTAA |
RNA extraction and northern blot analysis
Cells were grown at 37°C on LB or M63 media and total RNA was extracted using hot phenol procedure (Aiba et al, 1981). When using M63 medium, the cells were washed prior extraction. Ampicillin was used at 50 μg/ml in the culture when cells carried plasmids. Arabinose (0.01%) and 2,2′-dipyridyl (200 μM) were added when indicated. Half-life determination of RNA was carried out by the addition of 250 μg/ml rifampicin. After total RNA extraction, 5 μg of total RNA were loaded on a polyacrylamide gel (4% acrylamide 29:1, 8 M urea). After migration, the RNA was transferred to a Hybond-XL membrane (Amersham Biosciences) and crosslinked with UV (1200 J). The membrane was prehybridised with 50% formamide, 5 × SSC, 5 × Denhardt reagent, 1% SDS, and 100 μg/ml sheared salmon sperm DNA for 4 h at 60°C. Then, the radiolabelled probe was added directly in the prehybridisation buffer with the membrane and incubated for 16 h at 60°C. Before exposure on a phosphor screen, the membrane was washed three times with 1 × SSC/0.1% SDS and once with 0.1 × SSC/0.1% SDS. The phosphor screen was shown on a Storm 860 (Molecular Dynamics) and quantification was carried out with ImageQuant software (Molecular Dynamics).
3′-RACE
3′-RACE assays were carried out essentially as described earlier (Argaman et al, 2001). Briefly, RNA was dephosphorylated with calf intestine alkaline phosphatase (New England Biolab) and ligated with a 3′-RNA adapter (5′-uucacuguucuuagcggccgcaugcuc-idT-3′; Dharmacon Research, Chicago, IL, USA). Reverse transcription was carried out with 100 pmol of a single primer complementary with the RNA adapter. The reverse transcription products were amplified by PCR and separated on agarose gels. Bands of interest were excised, purified on GFX Gel Band purification kit (GE Healthcare), cloned using the Zero Blunt TOPO kit (Invitrogen) and sequenced (DNA Landmarks, Canada). At least three independent assays were carried out.
Radiolabelled RNAs generated by in vitro RNA synthesis
The radiolabelled probes used for northern blot analysis were transcribed with T7 RNA polymerase (Roche, Germany) from a PCR product to generate the antisense transcript of the gene of interest. Transcription was carried out in the T7 transcription buffer (40 mM Tris-HCL at pH 8.0, 6 mM MgCl2, 10 mM dithiothreitol, 2 mM spermidine), 400 μM NTPs (A, C and G), 10 μM UTP, 3 μl of α-32P-UTP (3000 Ci/mmol), 20 U RNA guard, 20 U T7 RNA polymerase and 0.5 μg DNA template. After 4 h of incubation at 37°C, the mixture was treated with 2 U of Turbo DNAse (Ambion) and extracted once with phenol–chloroform. Non-incorporated nucleotides were removed with a G-50 Sephadex column. The primers used for generating DNA templates for in vitro RNA synthesis were EM144–EM145 (iscR), EM138–EM139 (iscS), EM190–EM191 (ryhB) and EM293–EM294 (16S rRNA). For the 5′-end labelling of iscRS RNA, we transcribed with T7 RNA polymerase from a PCR product (EM385–EM408) as described above. Trancripts were dephosphorylated with calf intestine phosphatase (New England Biolabs) and 5′-end labelled with γ-32P-ATP using T4 polynucleotide kinase (New England Biolabs) according to the manufacturer's protocol.
RNA secondary structure probing
Secondary structure probing was performed on 5′-end labelled iscRS RNA (see above). RyhB RNA was transcribed with T7 RNA polymerase from a PCR product (EM88–EM89). Hfq was purified as described earlier (Prévost et al, 2007). Final concentration of 0.25 μM iscRS, 1 μM RyhB, and 1 μM Hfq were used. Ribonucleases T1 (0.1 U) (Ambion) and TA (2.5 U) (Jena Biosciences) were used for 5 min at 37°C in the sequence buffer (Ambion). Alkaline ladder was carried out in the alkaline buffer (Ambion) for 5 min at 90°C. Lead acetate cleavages were carried out with 2.5 mM PbAc (Sigma-Aldrich) in the structure buffer (Ambion) with 0.1 mg/ml of yeast RNA (Ambion). Reactions were stopped by adding 10 μl of loading buffer II (Ambion). Samples were heated at 90°C for 1 min and separated on 5% polyacrylamide/7 M urea gel. Gels were dried and exposed on the phosphor screen (see above).
Western blot analysis
Cells were grown at 37°C in M63 media. When using pBAD-ryhB or pNM12 plasmids, ampicillin was used at a final concentration of 50 μg/ml and arabinose was added at mid-log phase at a final concentration of 0.01%. Total proteins were harvested by precipitation with trichloroacetic acid. Ice-cold trichloroacetic acid was added to 1 ml of culture at a final concentration of 10% and incubated on ice for 10 min. Proteins were spun down and pellets washed with ice-cold 80% acetone. Dried pellets were resuspended in sodium dodecyl sulfate-polyacrylamide gel electrophoresis buffer at volume normalised to the OD600 of cultures. The equivalent volume for 0.025 OD600 unit of culture was separated on a 12% bis-Tris gel and transferred to the nitrocellulose membrane. The anti-IscS (kind gift from Larry Vickery) and anti-IscR (kind gift from Patricia Kiley) were used at dilutions of 1:1000 and 1:10 000, respectively. The IRDye 800CW-conjugated goat anti-rabbit secondary antibody (Li-Cor Biosciences) was used at a dilution of 1:15 000. Western blots were showed on an Odyssey Infrared Imaging System (Li-Cor Biosciences) and quantification was carried out using the Odyssey Application Software.
β-Galactosidase assays
Kinetic assays for β-galactosidase activity were performed as described earlier (Prévost et al, 2007) using a SpectraMax 250 microtitre plate reader (Molecular Devices). Briefly, overnight bacterial culture was incubated in the LB media with ampicillin at a final concentration of 50 μg/ml at 37°C and diluted 1000-fold into 50 ml of fresh LB media with ampicillin at 37°C. Cultures were grown with agitation to an OD600 of 0.1 before inducing RyhB expression by adding arabinose to a final concentration of 0.01% (strains carrying pBAD-ryhB or the control vector pNM12). Specific β-galactosidase activities were calculated using the formula Vmax/OD600. The results reported represent data of at least three experimental trials.
Supplementary Material
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Figure S5
Supplementary Figure S6
Supplementary Figure S7
Review Process File
Acknowledgments
We thank Susan Gottesman, Peter D. Pawelek, Szabolcs Semsey and Cari Vanderpool for discussion and suggestions throughout this work. We acknowledge Patricia Kiley for helpful discussions and IscR antibodies, and Larry Vickery for IscS antibodies. We are grateful to all members of the laboratory for useful comments and discussions on the paper. This work was funded by an operating Grant MOP69005 from the Canadian Institute for Health Research (CIHR). EM is a Canadian Institutes for Health Research (CIHR) new investigator scholar.
References
- Aiba H, Adhya S, de Crombrugghe B (1981) Evidence for two functional gal promoters in intact Escherichia coli cells. J Biol Chem 256: 11905–11910 [PubMed] [Google Scholar]
- Argaman L, Hershberg R, Vogel J, Bejerano G, Wagner EG, Margalit H, Altuvia S (2001) Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr Biol 11: 941–950 [DOI] [PubMed] [Google Scholar]
- Ayala-Castro C, Saini A, Outten FW (2008) Fe–S cluster assembly pathways in bacteria. Microbiol Mol Biol Rev 72: 110–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachellier S, Clement JM, Hofnung M (1999) Short palindromic repetitive DNA elements in enterobacteria: a survey. Res Microbiol 150: 627–639 [DOI] [PubMed] [Google Scholar]
- Belasco JG, Beatty JT, Adams CW, von Gabain A, Cohen SN (1985) Differential expression of photosynthesis genes in R. capsulata results from segmental differences in stability within the polycistronic rxcA transcript. Cell 40: 171–181 [DOI] [PubMed] [Google Scholar]
- Dos Santos PC, Smith AD, Frazzon J, Cash VL, Johnson MK, Dean DR (2004) Iron–sulfur cluster assembly: NifU-directed activation of the nitrogenase Fe protein. J Biol Chem 279: 19705–19711 [DOI] [PubMed] [Google Scholar]
- Geissmann TA, Touati D (2004) Hfq, a new chaperoning role: binding to messenger RNA determines access for small RNA regulator. EMBO J 23: 396–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giel JL, Rodionov D, Liu M, Blattner FR, Kiley PJ (2006) IscR-dependent gene expression links iron–sulphur cluster assembly to the control of O-regulated genes in Escherichia coli. Mol Microbiol 60: 1058–1075 [DOI] [PubMed] [Google Scholar]
- Gottesman S (2004) The small RNA regulators of Escherichia coli: roles and mechanisms. Annu Rev Microbiol 58: 303–328 [DOI] [PubMed] [Google Scholar]
- Jacques JF, Jang S, Prévost K, Desnoyers G, Desmarais M, Imlay J, Massé E (2006) RyhB small RNA modulates the free intracellular iron pool and is essential for normal growth during iron limitation in Escherichia coli. Mol Microbiol 62: 1181–1190 [DOI] [PubMed] [Google Scholar]
- Higgins CF, Ames GF, Barnes WM, Clement JM, Hofnung M (1982) A novel intercistronic regulatory element of prokaryotic operons. Nature 298: 760–762 [DOI] [PubMed] [Google Scholar]
- Kalamorz F, Reichenbach B, Marz W, Rak B, Gorke B (2007) Feedback control of glucosamine-6-phosphate synthase GlmS expression depends on the small RNA GlmZ and involves the novel protein YhbJ in Escherichia coli. Mol Microbiol 65: 1518–1533 [DOI] [PubMed] [Google Scholar]
- Kaufmann N, Reidl H, Golecki JR, Garcia AF, Drews G (1982) Differentiation of the membrane system in cells of Rhodopseudomonas capsulata after transition from chemotrophic to phototrophic growth conditions. Arch Microbiol 131: 313–322 [Google Scholar]
- Khemici V, Carpousis AJ (2004) The RNA degradosome and poly(A) polymerase of Escherichia coli are required in vivo for the degradation of small mRNA decay intermediates containing REP-stabilizers. Mol Microbiol 51: 777–790 [DOI] [PubMed] [Google Scholar]
- Lauhon CT (2002) Requirement for IscS in biosynthesis of all thionucleosides in Escherichia coli. J Bacteriol 184: 6820–6829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KC, Yeo WS, Roe JH (2008) Oxidant-responsive induction of the suf operon, encoding a Fe-S assembly system, through Fur and IscR in Escherichia coli. J Bacteriol 190: 8244–8247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lill R, Muhlenhoff U (2006) Iron–sulfur protein biogenesis in eukaryotes: components and mechanisms. Annu Rev Cell Dev Biol 22: 457–486 [DOI] [PubMed] [Google Scholar]
- Majdalani N, Cunning C, Sledjeski D, Elliott T, Gottesman S (1998) DsrA RNA regulates translation of RpoS message by an anti-antisense mechanism, independent of its action as an antisilencer of transcription. Proc Natl Acad Sci USA 95: 12462–12467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massé E, Escorcia FE, Gottesman S (2003) Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev 17: 2374–2383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massé E, Gottesman S (2002) A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc Natl Acad Sci USA 99: 4620–4625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massé E, Vanderpool CK, Gottesman S (2005) Effect of RyhB small RNA on global iron use in Escherichia coli. J Bacteriol 187: 6962–6971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moller T, Franch T, Udesen C, Gerdes K, Valentin-Hansen P (2002) Spot 42 RNA mediates discoordinate expression of the E. coli galactose operon. Genes Dev 16: 1696–1706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita T, Maki K, Aiba H (2005) RNase E-based ribonucleoprotein complexes: mechanical basis of mRNA destabilization mediated by bacterial noncoding RNAs. Genes Dev 19: 2176–2186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita T, Mochizuki Y, Aiba H (2006) Translational repression is sufficient for gene silencing by bacterial small noncoding RNAs in the absence of mRNA destruction. Proc Natl Acad Sci USA 103: 4858–4863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newbury SF, Smith NH, Higgins CF (1987a) Differential mRNA stability controls relative gene expression within a polycistronic operon. Cell 51: 1131–1143 [DOI] [PubMed] [Google Scholar]
- Newbury SF, Smith NH, Robinson EC, Hiles ID, Higgins CF (1987b) Stabilization of translationally active mRNA by prokaryotic REP sequences. Cell 48: 297–310 [DOI] [PubMed] [Google Scholar]
- Outten FW, Djaman O, Storz G (2004) A suf operon requirement for Fe–S cluster assembly during iron starvation in Escherichia coli. Mol Microbiol 52: 861–872 [DOI] [PubMed] [Google Scholar]
- Outten FW, Wood MJ, Munoz FM, Storz G (2003) The SufE protein and the SufBCD complex enhance SufS cysteine desulfurase activity as part of a sulfur transfer pathway for Fe–S cluster assembly in Escherichia coli. J Biol Chem 278: 45713–45719 [DOI] [PubMed] [Google Scholar]
- Powell BS, Rivas MP, Court DL, Nakamura Y, Turnbough CL Jr (1994) Rapid confirmation of single copy lambda prophage integration by PCR. Nucleic Acids Res 22: 5765–5766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prévost K, Salvail H, Desnoyers G, Jacques JF, Phaneuf E, Massé E (2007) The small RNA RyhB activates the translation of shiA mRNA encoding a permease of shikimate, a compound involved in siderophore synthesis. Mol Microbiol 64: 1260–1273 [DOI] [PubMed] [Google Scholar]
- Py B, Higgins CF, Krisch HM, Carpousis AJ (1996) A DEAD-box RNA helicase in the Escherichia coli RNA degradosome. Nature 381: 169–172 [DOI] [PubMed] [Google Scholar]
- Rees DC, Howard JB (2000) Nitrogenase: standing at the crossroads. Curr Opin Chem Biol 4: 559–566 [DOI] [PubMed] [Google Scholar]
- Reichenbach B, Maes A, Kalamorz F, Hajnsdorf E, Gorke B (2008) The small RNA GlmY acts upstream of the sRNA GlmZ in the activation of glmS expression and is subject to regulation by polyadenylation in Escherichia coli. Nucleic Acids Res 36: 2570–2580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher A, Drews G (1978) The formation of bacteriochlorophyll.protein complexes of the photosynthetic apparatus of Rhodopseudomonas capsulata during early stages of development. Biochim Biophys Acta 501: 183–194 [DOI] [PubMed] [Google Scholar]
- Schwartz CJ, Djaman O, Imlay JA, Kiley PJ (2000) The cysteine desulfurase, IscS, has a major role in in vivo Fe–S cluster formation in Escherichia coli. Proc Natl Acad Sci USA 97: 9009–9014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz CJ, Giel JL, Patschkowski T, Luther C, Ruzicka FJ, Beinert H, Kiley PJ (2001) IscR, an Fe–S cluster-containing transcription factor, represses expression of Escherichia coli genes encoding Fe–S cluster assembly proteins. Proc Natl Acad Sci USA 98: 14895–14900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons RW, Houman F, Kleckner N (1987) Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene 53: 85–96 [DOI] [PubMed] [Google Scholar]
- Stern MJ, Ames GF, Smith NH, Robinson EC, Higgins CF (1984) Repetitive extragenic palindromic sequences: a major component of the bacterial genome. Cell 37: 1015–1026 [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Nakamura M (1999) Functional assignment of the ORF2-iscS-iscU-iscA-hscB-hscA-fdx-ORF3 gene cluster involved in the assembly of Fe–S clusters in Escherichia coli. J Biochem 126: 917–926 [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Tokumoto U (2002) A third bacterial system for the assembly of iron–sulfur clusters with homologs in archaea and plastids. J Biol Chem 277: 28380–28383 [DOI] [PubMed] [Google Scholar]
- Tokumoto U, Kitamura S, Fukuyama K, Takahashi Y (2004) Interchangeability and distinct properties of bacterial Fe–S cluster assembly systems: functional replacement of the isc and suf operons in Escherichia coli with the nifSU-like operon from Helicobacter pylori. J Biochem 136: 199–209 [DOI] [PubMed] [Google Scholar]
- Urban JH, Vogel J (2008) Two seemingly homologous noncoding RNAs act hierarchically to activate glmS mRNA translation. PLoS Biol 6: e64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo WS, Lee JH, Lee KC, Roe JH (2006) IscR acts as an activator in response to oxidative stress for the suf operon encoding Fe–S assembly proteins. Mol Microbiol 61: 206–218 [DOI] [PubMed] [Google Scholar]
- Zheng L, Cash VL, Flint DH, Dean DR (1998) Assembly of iron–sulfur clusters. Identification of an iscSUA-hscBA-fdx gene cluster from Azotobacter vinelandii. J Biol Chem 273: 13264–13272 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Figure S5
Supplementary Figure S6
Supplementary Figure S7
Review Process File