Abstract
RepB initiates plasmid rolling-circle replication by binding to a triple 11-bp direct repeat (bind locus) and cleaving the DNA at a specific distant site located in a hairpin loop within the nic locus of the origin. The structure of native full-length RepB reveals a hexameric ring molecule, where each protomer has two domains. The origin-binding and catalytic domains show a three-layer α–β–α sandwich fold. The active site is positioned at one of the faces of the β-sheet and coordinates a Mn2+ ion at short distance from the essential nucleophilic Y99. The oligomerization domains (ODs), each consisting of four α-helices, together define a compact ring with a central channel, a feature found in ring helicases. The toroidal arrangement of RepB suggests that, similar to ring helicases, it encircles one of the DNA strands during replication to confer processivity to the replisome complex. The catalytic domains appear to be highly mobile with respect to ODs. This mobility may account for the adaptation of the protein to two distinct DNA recognition sites.
Keywords: DNA-binding protein, nuclease, plasmid replication, replication initiator, X-ray crystal structure
Introduction
DNA replication is a central biological event that requires a mechanism that deals with the incapacity of DNA polymerases (DNAP) to start de novo DNA synthesis. Several mechanisms have evolved, of which rolling-circle replication (RCR) is used by a variety of genetic entities (transposons, bacterial plasmids, bacteriophages and viruses) that replicate autonomously in a wide range of organisms, from prokaryotes to humans (Campos-Olivas et al, 2002). RCR is initiated by a key triggering reaction that consists in the site-specific cleavage of one of the strands of the duplex within the origin of replication. This cleavage is catalysed by RCR initiator proteins, which thus provide a primer (the newly generated 3′-OH end) for DNAP to start synthesis. RCR initiators are also involved in termination of the replicative process, a step that requires the endonuclease and strand-transfer activities of these proteins (Novick, 1998).
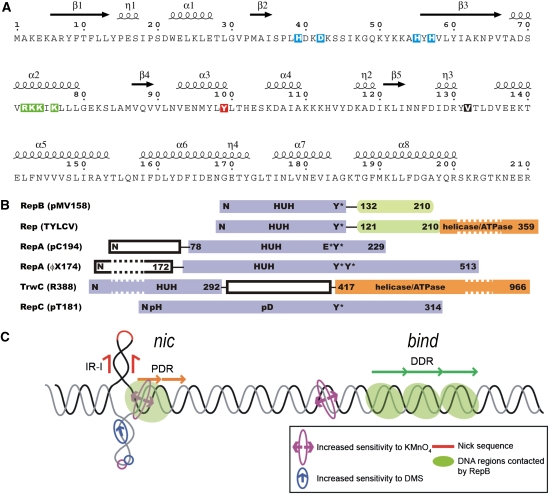
Initiators of RCR-like mechanisms constitute a vast superfamily, which includes proteins involved either in replication of bacteriophages, plasmids, and plant and animal viruses (the Rep class) or in the conjugal transfer of plasmid DNA (the Mob class) (Ilyina and Koonin, 1992). The entire superfamily appears to share a common endonucleolytic mechanism (Dyda and Hickman, 2003) based on a catalytic tyrosine and a divalent metal cation coordinated by, among other ligands, two histidine residues that surround a bulky hydrophobic amino-acid residue in the primary sequence (the His-hydrophobic-His (HUH) sequence motif, see Figure 1A and B). The three-dimensional (3D) structures of the nuclease domain of a few RCR initiators have been solved, all as monomers. Among these, there are representatives of the Mob class (Datta et al, 2003; Guasch et al, 2003; Boer et al, 2006; Monzingo et al, 2007) and the viral Rep proteins (Campos-Olivas et al, 2002; Hickman et al, 2002, 2004; Vega-Rocha et al, 2007a, 2007b). To date, there are no 3D structures of any of the RCR Rep proteins from plasmids or bacteriophages and no 3D structures encompassing both nuclease and additional domains of the Rep or Mob class proteins. Structurally, the RCR initiator domain superfamily is characterized by a five-stranded antiparallel β-sheet flanked by a variable number of α-helices. The active Tyr residue is located in one of these α-helices. The central three strands of the β-sheet have the same relative arrangement and topology in both classes of RCR initiator domains, with the middle strand bearing the HUH sequence motif. In contrast, the flanking strands of the β-sheet are provided by the C-terminal moiety of the domain in the Rep class of initiators, but by the N-terminal moiety in the Mob class (Dyda and Hickman, 2003). This difference in arrangement accounts for the reversed position of the HUH sequence motif relative to the catalytic Tyr in the primary sequences and indicates that a circular permutation event is at the origin of the evolutionary divergence of these proteins (Guasch et al, 2003; Russi et al, 2008). Interestingly, a 3D structure homologous to that of the RCR initiator domain superfamily is present in the DNA-binding domain of the non-RCR viral initiator proteins E1 (from bovine papillomavirus) (Enemark et al, 2002) and large T-antigen (from simian virus 40) (Hickman et al, 2002). The DNA-binding domain of these viral proteins also exhibits a three-layer α–β–α sandwich fold containing a central five-stranded antiparallel β-sheet, but lacks the active site and, therefore, the endonucleolytic activity that characterizes the RCR initiator domain superfamily (Gomis-Rüth and Coll, 2006).
Figure 1.
Secondary structure of RepB, domain organization of RepB and related proteins, and dso of pMV158. (A) The primary sequence of RepB, including the annotation of the secondary structure elements as found in the C2- and C3-structures, η denotes 310-helices. The residues involved in metal coordination, which include the histidines of the HUH sequence motif (residues H55, Y56 and H57), are boxed in blue. The catalytic tyrosine is boxed in red, the mutated residues are boxed in green and hinge V132 is shown as white on a black background. (B) Alignment of the tyrosine residues responsible for nicking activity in various RCR initiator proteins (adapted from Campos-Olivas et al, 2002). The catalytic tyrosine of TrwC, which is at the N-terminus due to a circular permutation in the sequence, cannot be aligned with the others. The reactive tyrosines are marked with Y*, and metal-binding sequence motifs are marked with ‘HUH'. Several poly His (pH) and poly Asp (pD) sequences were identified in the sequence of RepC from plasmid pT181, which could be involved in metal binding. Some of the N-terminal and C-terminal domains have been shortened (indicated by dashed boxes). (C) Schematic representation of the pMV158 dso, showing the approximate relative locations of the nick site, the proximal direct repeats (PDR) of the nic locus and the distal direct repeats (DDR) constituting the bind locus. The regions that are contacted by RepB and the regions with increased sensitivity to KMnO4 or dimethyl sulphate (DMS) on binding of RepB are also indicated.
In addition to the catalytic N-terminal domain, the Mob and viral Rep proteins have C-terminal domains with ATPase and DNA helicase activities. The helicase domains of the viral RCR Rep proteins have been classified among ATPases associated with diverse cellular activities (AAA+) that belong to superfamily 3 (SF3) of small DNA and RNA virus helicases, which often form hexameric rings (Hickman et al, 2004; Clerot and Bernardi, 2006). In contrast, RCR Rep proteins from plasmids and bacteriophages lack helicase activity and must recruit a separate bacterial helicase to carry out the progression of the replication fork (Takahashi et al, 1978; Petit et al, 1998; Bruand and Ehrlich, 2000; Chang et al, 2002).
Protein RepB is the initiator of DNA replication of streptococcal RCR plasmid pMV158 (de la Campa et al, 1990). RepB, which is purified as a homo-hexamer (Ruiz-Masó et al, 2004), binds with high affinity to a specific region (the bind locus consisting of three 11-bp tandem direct repeats) within the double strand origin of replication (dso, see Figure 1C). The protein shows endonucleolytic and strand-transfer activities and cleaves a specific single-stranded (ss) DNA sequence of the nic locus of the dso at a precise dinucleotide (the nick site) (Moscoso et al, 1995; Ruiz-Masó et al, 2007). The nick site is located on the hairpin loop of an extruded inverted repeat, 84 bp upstream from the bind locus. The nucleophilic attack on the scissile phosphodiester bond of the DNA is exerted by the catalytic Y99 of RepB (Moscoso et al, 1997). Like other RCR Rep initiators from plasmids and bacteriophages, RepB lacks ATPase and helicase activities (de la Campa et al, 1990; Moscoso et al, 1995). Thus, the unwinding of the double-stranded (ds) DNA as the replication fork advances and the cleaved strand is peeled off may rely on the recruitment of a host-encoded helicase, most probably PcrA.
Here, we present the 3D crystal structure of the full-length native RepB protein in two distinct forms. This constitutes the first example of a Rep protein structure from RCR plasmids and reveals the determinant role of the C-terminal domain in quaternary organization. The structure unveils that RepB is a toroidal homo-hexameric ring, each protomer comprising an N-terminal domain, which carries the catalytic active site, and a C-terminal hexamerization domain, which forms a tight cylinder with a six-fold symmetry in the hexamer. We further show the electron microscopy (EM) 3D reconstruction of RepB both unbound and bound to its high-affinity target, the DNA of the bind locus. Finally, we discuss the mechanistic implications of the hexameric form of the protein.
Results
X-ray crystal structures
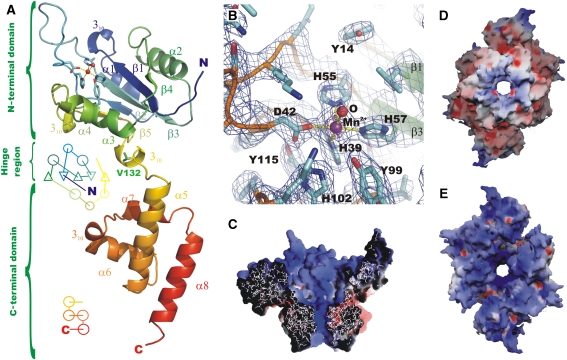
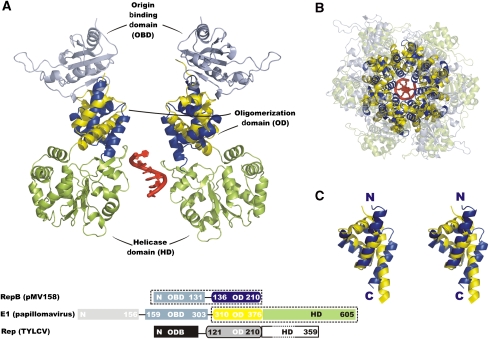
Two structures of the RepB hexamer (RepB6) were obtained from crystals belonging to a trigonal and a tetragonal form, respectively. In both, the protein shows a hexameric ring quaternary structure with a cup-like shape. Each monomer consists of two domains (Figure 2A) connected by a short hinge region, where the N-terminal origin-binding domain (OBD, see below) hosts the catalytic activity, and the C-terminal oligomerization domain (OD) is responsible for hexamerization.
Figure 2.
Structure of the RepB monomer, active site and electrostatic surface representations. (A) A cartoon drawing of a single protomer of RepB, indicating the topology and secondary structure elements (Kabsch and Sander, 1983) of the OD and OBD domains. V132, located in the hinge region between the two domains, is shown in green, the active site is indicated using a ball-and-stick representation of the metal and its first coordination sphere. (B) A close-up of the active site, including the σA-weighted 2Fo−Fc electron density map contoured at 1.2σ (blue) and 4σ (purple). The metal-binding residues and the coordinated water/hydroxide ligand (‘O') are indicated. (C) The electrostatic potential on the solvent-accessible surface of a cross-section of the C3-structure and of the C2-hexamer, viewed (D) from the C-termini and (E) from the N-termini. Blue, red and white represent positively charged, negatively charged and neutral surface, respectively.
The all-helical ODs (T133–R210) consist of α-helices 5–8. α5 and α8 run parallel, whereas the shorter α6 and α7, interconnected by a 310-helix, are perpendicular to the former two. The ODs form a toroidal ring in both structures with near six-fold symmetry through tight inter-OD interactions, burying ∼715 Å2 between residues D135–R203 for all pairs of consecutive domains in both structures. The r.m.s.d. in the Cα positions of the OD rings of the two structures is 0.49 Å, whereas the maximum pair-wise r.m.s.d. in the Cα positions of the ODs is 0.36 Å. The inner surface of the OD ring is formed by the C-terminal α8 helices and narrows down from a maximum diameter of ∼20 Å to a minimum diameter of ∼13 Å, delimited by the side chains of K191 pointing towards the lumen of the channel.
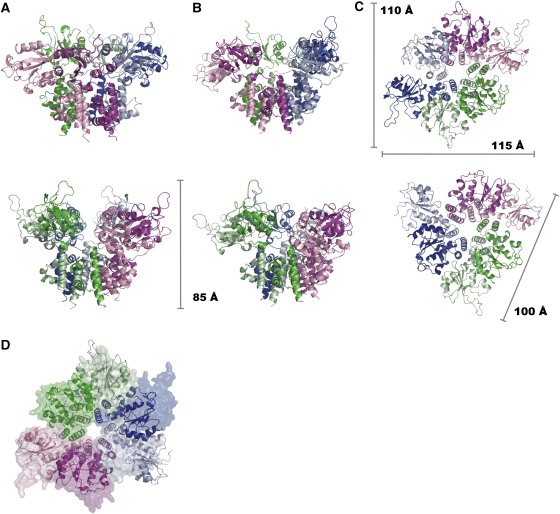
The OBDs (K3–Y131) consist of a central antiparallel five-stranded β-sheet flanked by helices on both sides. Helices α1 and α2, at one face of the sheet, are arranged diagonally with respect to it. The active site is on the opposite side of the sheet, where helix α3 provides the catalytic Y99. This helix is connected to strand β5 through the short helix α4 and a 310-helix. The OBDs do not follow the six-fold symmetry of the ODs: in the trigonal form, which will be referred to as the ‘C2-form', two sets of three OBDs each are related by a crystallographic two-fold symmetry (Figure 3A and C). The complex can be described as a dimer of trimers, where the OBD hexamer appears compressed in a direction perpendicular to the six-fold axis of the OD. The compression is a result of different orientations and locations of the OBDs with respect to the OD ring (Figure 4). The backbone of residues around V132 acts as a hinge between the two domains (Figure 2A), allowing differences of up to 55° in the orientation of the OBDs.
Figure 3.
Cartoon representations of the C2- and C3-structures of the RepB6 in different orientations, indicating the dimensions. Shown are distinct side views on the (A) C2- and (B) C3-structures. (C) View on the OBDs along the local axis of the C2 (top)- and C3 (bottom)-structures. (D) shows the superposition of the two structures, where C2 is represented by a molecular surface and C3 by ribbons.
Figure 4.
Movement of the OBDs. Cartoon drawing of the three non-equivalent monomers of the C2-structure in two orientations (left and right panels) after superpositioning of the ODs.
The positions of the OBDs with respect to the ODs also change significantly when comparing the two crystal forms. The tetragonal form will be referred to as the ‘C3-form', as the N-terminal domains exhibit a local near three-fold rather than two-fold symmetry. Here, the complex can be considered as a trimer of dimers, where the OBDs alternatively switch between two positions relative to the hexameric ring (Figure 3B and C). Optimal superposition of the C2- and C3-forms reveals a high positional similarity for three of the OBDs, but large differences in the other three (Figure 3D).
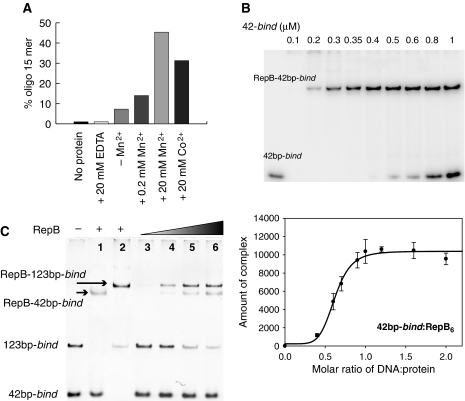
The active site of the catalytic domain is located on one of the faces of its central β-sheet, in a groove flanked by helices α3 and α4, by residues of strands β2 and β3 and by a long and flexible 21-residue long loop that connects β2 and β3 (Figure 2A). A metal ion is found at the active site in an octahedral-minus-one or square-based pyramidal coordination, which involves atoms H39 Nδ1, D42 Oδ1, H57 Nɛ2 and H55 Nɛ2 (in apical position) and a single-solvent molecule (Figure 2B). The resulting distorted trigonal bipyramidal coordination, involving five ligands, resembles that of manganese superoxide dismutases (e.g. PDB entry 1IXB). The Oη atom of the catalytically active Y99 is found at a distance of ∼5 Å to the metal ion. Further away, Y115 is involved in a hydrogen bond to the D42 carboxyl group. The RepB metal ion was determined to be a Mn2+ cation through X-ray absorption near-edge energy scans and anomalous density map calculations (see Supplementary Table I). These results are consistent with those of RepB activity assays in the presence of a series of divalent cations (Mn2+, Co2+, Ba2+, Ca2+, Zn2+ and Mg2+). Only Mn2+ and Co2+ were able to promote nicking-closing of supercoiled plasmid DNA (not shown) and cleavage of an ssDNA fragment containing the nick sequence (Figure 5A).
Figure 5.
DNA cleavage by RepB and stoichiometry of binding of RepB to the bind locus. (A) RepB-mediated cleavage of a 23-mer ssDNA containing the nick sequence in the presence of Mn2+ or Co2+. The cleavage activity is represented as the percentage of the reaction product (15-mer). A small basal cleavage level (∼5%) due to the presence of trace amounts of metal ions was abolished by the addition of 20 mM EDTA. (B) Electrophoresis gel (top) and a plot of the amount of complex (integrated peak area in arbitrary units) as a function of the DNA:protein ratio (bottom) obtained from the EMSA experiment. (C) EMSA analysis of complexes generated on addition of RepB to a mixture of the 42bp-bind and 123bp-bind DNAs. Positions of the free DNAs and of the RepB–DNA complexes are indicated. Lanes 1 and 2 show the complexes generated by binding of RepB to the separate fragments. RepB6 concentrations were: 0 (—), 0.35 (lanes 1, 2 and 5), 0.17 (lane 3), 0.25 (lane 4) and 0.44 (lane 6) μM.
EM and 3D reconstruction of RepB and DNA-bound RepB
RepB molecules were clearly observed in the electron micrographs with the aid of a staining agent; analysis of unstained specimens was unsuitable because the molecular weight of RepB6 is <150 kDa. A random conical tilt (RCT) approach (Radermacher, 1988) was used, as preliminary refinement experiments with DNA-free protein suggested that RepB bound to the support film within a limited range of orientations. An analysis of the rotational symmetry of the particles in the raw data set by XMIPP (Sorzano et al, 2004) revealed the presence of components with two-, three- and six-fold symmetries. To study the relative convergence, we compared reconstructions obtained by first imposing and then relaxing each of these symmetries. A consistent volume was obtained only when assuming a two-fold symmetry. Moreover, the three- and six-fold symmetries-relaxed reconstructions still displayed a strong bias towards two-fold. Furthermore, simultaneous refinement of the two-, three- and six-fold symmetrized reconstructions using a multi-reference refinement strategy implemented in EMAN showed that most of the particles correlated better with the two-fold symmetrized reconstruction. These results suggested that under our EM experimental conditions, most of the RepB6 particles in solution are characterized by a two-fold rotational symmetry.
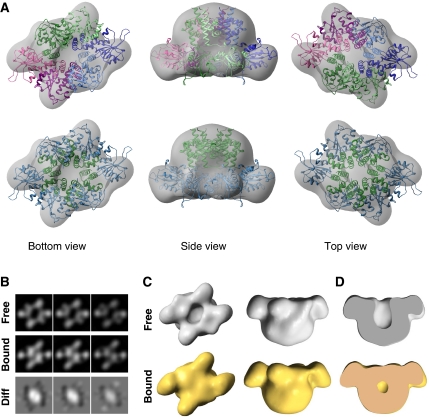
The final 3D reconstruction of RepB6 at 22 Å resolution (Figure 6A) shows a cup-shaped structure of two-fold symmetry with a narrow and a wide region enclosing a cavity. Furthermore, the two-fold symmetry of the wide region and the six-fold symmetry of the narrow region match the symmetries observed in the C2-structure. The characteristic crevice between the OBDs of the C2 X-ray structure shown in Figure 3A is also observed in the EM structure. The orientation of the C2-structure into the EM map was searched using unbiased computational fitting methods (Garzon et al, 2007). We found that the top score solution (cross-correlation=0.8) placed the atomic structure within the EM reconstruction, so that most of the EM density was occupied by the atomic model and only small segments of the atomic coordinates were placed out of the EM map. This confirmed the close agreement between the EM reconstruction and the C2-form (Figure 6A; Supplementary Figure S1). The remaining solutions revealed unsatisfactory superposition of both structures (see Supplementary Figure S1). The residual differences found in the N-terminal region can be attributed to the intrinsic flexibility and movement of these domains and to the limited resolution of the EM reconstruction.
Figure 6.
EM structure of RepB and DNA-bound RepB. (A) Several views of the 3D reconstruction of RepB6 (grey surface) superposed on the C2 X-ray structure. Top panels show the atomic model using a different colour for each protomer, whereas bottom panels display the same view coloured by domain (OD in green and OBD in blue). (B) Sectional views of the RepB and DNA-bound RepB reconstructions showing the crevice and channel. Bottom panel displays equivalent sections for the difference map of these two structures. (C) Comparative views of the 3D-structures of RepB (top, grey solid surfaces) and DNA-bound RepB (bottom, gold solid surfaces). (D) Cut view of the bound and unbound RepB structures. All 3D density maps were rendered with UCSF Chimera (Pettersen et al, 2004).
The EM reconstruction of a purified complex between RepB and the 42bp-bind DNA (see Materials and methods) at 24 Å resolution was obtained by equivalent strategies (Figure 6C, bottom panels). Overall, it is very similar to the reconstruction of DNA-free RepB (Figure 6C, top panels) and to the C2 X-ray structure (Supplementary Figure S1). However, in the presence of DNA, a significant additional density occludes the crevice (Figure 6D, compare top and bottom panels). The fact that stain exclusion occurs only when DNA is present in the complex and exclusively at the crevice is a strong support for the location of the DNA molecule at this site, although it cannot be excluded that a movement of the N-terminal domains of RepB on DNA binding also contributes. The additional density becomes evident by inspecting the sectional views around the crevice region (Figure 6B) and by difference mapping of bound and unbound reconstructions (Supplementary Figure S2). The presence of this density was confirmed by control experiments that used the RepB DNA-free reconstruction as initial reference for the refinement with DNA-bound data and vice versa.
Stoichiometry of the RepB-bind locus complex
On the basis of the high-resolution footprints of RepB on the bind locus, three identical DNA-binding motifs of RepB6 were postulated to bind to the three direct repeats constituting the target DNA (Ruiz-Masó et al, 2007). As RepB6 has at most six DNA-binding motifs, it could in principle house up to two DNA molecules harbouring the bind locus. To address the issue of the stoichiometry of the complex of RepB and the bind locus, increasing amounts of the 42bp-bind DNA were added to a fixed amount of protein at a concentration at which its hexameric configuration has been shown (Ruiz-Masó et al, 2004). This resulted in increasing amounts of the RepB-bind locus complex and no free DNA until saturation was reached at an approximately 1:1 molar ratio RepB6:DNA (Figure 5B). To confirm that a single DNA molecule containing the bind locus binds to one RepB6, we designed a different approach in which increasing concentrations of the protein were added to a mixture of the 42bp-bind and 123bp-bind DNAs and the resulting complexes were analysed by electrophoresis mobility shift assay (EMSA). Complexes with the same electrophoretic mobility as those generated by the binding of RepB to either the 42bp-bind or the 123bp-bind DNAs were observed exclusively (Figure 5C), excluding simultaneous binding of two DNA molecules to the same protein hexamer (which should have resulted in a complex migrating between the RepB-123bp-bind and RepB-42bp-bind complexes). This finding is consistent with the notion that RepB6 accommodates a single DNA molecule harbouring the bind locus.
The oligomeric state of RepB within Streptococcus pneumoniae cells
To study the physiological association state of RepB, the distribution of distinct RepB-containing products in exponential-phase pneumococcal cells harbouring pMV158 was analysed in vivo as a function of the concentration of the cross-linker bis(sulfosuccinimidyl) suberate (BS3) by immunoblotting of SDS–PAGE separated total protein (Supplementary Figure S3). In the absence of BS3, a product migrating like the RepB monomer (RepB1, 24.1 kDa) was mainly detected. Increasing concentrations of BS3 resulted in a decrease of the non-cross-linked material (RepB monomers) paralleled by the appearance and increase of oligomeric material, which, on the basis of its migration, corresponded to RepB homo-complexes ranging from RepB dimers (RepB2) to RepB6. At the highest BS3 concentrations, a decrease in RepB3−5 was observed, which was accompanied by an increase in RepB6, whereas the RepB1 and RepB2 fractions remained constant. At saturating BS3 concentrations, where the cross-linked material should reflect the intracellular association state of the protein, mainly monomers (∼12%), dimers (∼25%) and hexamers (∼52%) were observed. This observation suggests that RepB6 oligomers are present in vivo and coexist with monomeric and dimeric forms of RepB.
Discussion
The structures presented here constitute the first full-length RCR initiator protein ever solved. RepB arranges into a hexamer that has tight interactions between the C-terminal ODs, whereas the N-terminal OBD and catalytic domain interactions are variable and bury a smaller interaction area. The structures show that the ODs are responsible for the formation of the observed quaternary structure of the hexamer and provide the first evidence of the formation of a hexameric ring in plasmid-encoded RCR Rep initiators. To date, other proteins of this group have been purified as monomers or as dimers, as exemplified, respectively, by the Rep proteins of plasmids of the pC194 (Ozaki et al, 1994) and pT181 (Zhao et al, 1998) families. Preliminary experiments that involve purification of the separate OBD and OD domains confirm that the catalytic activity and the hexamerization potential of the protein can be uncoupled. The OBD purifies as a monomer that retains both the ability to bind to the bind locus and the endonucleolytic activity on the nick site, whereas the OD forms a hexamer and lacks the DNA binding and catalytic capacities (Supplementary Figure S4A and D). The present study also provides evidence that RepB6 occurs in vivo under physiological conditions, thereby suggesting that hexamerization of the protein is relevant for the replication of pMV158 (Supplementary Figure S3).
The organization and function of the domains of RCR initiators of phages, plasmids and viruses (Figure 1B) suggest that these distantly related proteins diverged through gene fusion and recruitment events. The evolutionary relation also extends to non-RCR viral initiators of SF3 as is evident from the 3D-similarity searches based on the two RepB domains. Thus, the search based on the OBD identified papillomavirus E1 helicase and SV40 T-antigen as highly similar structures (DALI Z-scores=7.8 and 6.6, respectively; MATRAS Z-scores=31.2 and 28.6, respectively). Using the OD only, high similarity was obtained for E1 helicase (DALI Z-score=3.8; MATRAS Z-score=12.2). Viral replication initiators contain an OBD and a helicase domain in their primary sequences, which are N-terminal and C-terminal, respectively, to an OD that is responsible for the hexameric organization of the proteins. RCR Rep proteins of plasmids and phages contain the OBD, but generally lack the OD and the ATPase/helicase domains. Although these proteins are functionally minimized compared with viral members, they essentially conserve the function, fold (see below) and DNA recognition and processing mechanism of the remaining domains. The observation that RepB from pMV158 contains both OBD and OD implies that it is an evolutionary intermediate linking the RCR Rep proteins of phages and plasmids with those of the viruses. The evolutionary link between RepB and hexameric SF3 helicases of viruses additionally supports a functional role for the hexameric form of RepB.
The N-terminal endonuclease domains of RepB are responsible for recognition of the plasmid replication origin
RepB specifically recognizes its cognate origin by binding to the bind locus of the pMV158 dso (del Solar et al, 1993). The bind locus consists of a triple 11-bp direct repeat and can be assumed to be a B-DNA duplex that severely bends on binding of RepB (Ruiz-Masó et al, 2007). Our studies show that RepB6 binds to a single dsDNA containing the triple direct repeat (Figure 5B and C) and that RepB1, RepB2 and RepB6 are the predominant species in vivo (Supplementary Figure S3). Assembly of RepB6 onto the plasmid origin may occur stepwise through the consecutive addition of monomers or dimers, similar to what has been proposed for AAV5 Rep (Hickman et al, 2004), the papillomavirus E1 helicase (Enemark et al, 2000; Schuck and Stenlund, 2005) and the SV40 T-antigen (Bochkareva et al, 2006). The C3-form reflects the tendency of RepB to form dimers (Supplementary Figure S3), as the structure can be considered a trimer of dimers. It may thus represent a conformation that is required during the assembly of RepB6, although it cannot be discarded that this particular OBD arrangement is the result of forces governing crystal packing. We propose that the bind site serves to recruit RepB and to thereby facilitate the formation of the proper structure and topology of the protein and/or the DNA for subsequent processing of the nick site. The ring-shaped arrangement may have additional functional significance as discussed below.
Consistent with binding to only one bind locus-containing DNA molecule, the electrostatic potential on the solvent-accessible surface of the RepB structures shows a single, large electropositive region that covers the outer surface (Figure 2E) and crevice at the N-terminal of the hexamer and extends into the central OD channel (Figure 2C). Co-crystallization experiments of RepB with DNA sequences representing the bind region showed additional weak electron density at interaction distance to the positively charged surface of the OBDs. Although this density could not be interpreted, the presence of DNA in the crystals was confirmed using fluorescence microscopy (Supplementary Figure S5). This is consistent with the observation that crystallization trials of RepB in the presence of dsDNA fragments shorter than 33 bp did not result in diffraction-quality crystals, whereas the best crystals were obtained using the 35-bp fragment described below. Furthermore, the EM reconstruction of the complex shows that indeed the crevice is occupied on exposure to the bind locus (Figure 6B–D). The dimension of the surface formed by the OBDs in the C2-structure is ∼120 Å in the elongated direction, which is consistent with the length of the bind locus, that is, a dsDNA region of ∼35 bp.
For the AAV5 Rep (Hickman et al, 2004) and the PCV2 Rep (Vega-Rocha et al, 2007a) RCR domain superfamily proteins, it was shown that OBD residues located far from the active site, at one edge of the central β-sheet, interact with the DNA. Interestingly, the origins of replication of pMV158 and AAV5 resemble each other in the sense that both comprise a direct repeat recognition site in the vicinity of a hairpin containing the nick site. The X-ray structure of the complex between the AAV5 Rep OBD and a duplex B-DNA that contains the protein-binding site revealed that loop β4–β5 and the N-terminal end of α-helix C, located at the aforementioned edge of the β-sheet, penetrate the DNA major and minor grooves, respectively (Hickman et al, 2004). A superposition of the OBD of RepB and that of DNA-bound AAV5 Rep shows that RepB helix α2 is equivalent to α-helix C of AAV5 Rep (Supplementary Figure S6A). The AAV5 loop β4–β5, which binds the major groove of the DNA, is not present in RepB. However, the N-terminal tail of RepB includes a number of positively charged residues such as K3, K5 and R7 and is well positioned in RepB6 to contact the DNA. Helix α2 includes a number of positively charged side chains, that is, R72, K73, K74 and K76, which are appropriately positioned to contact the DNA backbone phosphates or the bases deep in the DNA grooves. Additional superpositions of the RepB OBD with those of the SV40 T-antigen and the E1 helicase (Supplementary Figure S6B and C), both in complex with dsDNA, further support the hypothesis that the N-terminal end of RepB helix α2 interacts with the nucleic acid, although in this case by intruding the major groove of the dsDNA instead of the minor groove. Moreover, we present evidence that the capacity of RepB OBD to bind to the bind locus is severely impaired by mutations of R72 or K76 to alanines, and that simultaneous substitution of R72, K73, K74 and K76 by alanines abolishes the binding capacity of the OBD (Supplementary Figure S4A) without affecting its endonucleolytic activity (Supplementary Figure S4D) or its overall secondary structure (Supplementary Figure S4C). The distribution of the mutations in the six OBDs of the C2-form of RepB shows that these residues face the crevice formed by the OBD arrangement (Supplementary Figure S4B). The structural comparisons and the mutation experiments, therefore, support the association of the bind locus with the central crevice of the distorted RepB6 ring and corroborate the binding region we propose on the basis of the positively charged surface of the domain and the DNA density found in EM and X-ray analyses. RepB contains the long and flexible β2–β3 loop in the OBD, which is not present in AAV Rep or in the E1 helicase and is much shorter in the large T-antigen. This loop contains several positively charged residues that may interact with DNA, although to date there is no experimental evidence for its involvement. In fact, this loop is distant from the region of AAV5 Rep that binds the DNA in the superpositions shown in Supplementary Figure S6. There is a possibility that this loop is involved in interactions with the nic locus. Indeed, a superposition of RepB OBD with the relaxase domain of TrwC in complex with a DNA hairpin shows that this loop is located in a region that corresponds to a subdomain of TrwC that was designated ‘the fingers' (α8–α11), which grasps the ssDNA before its entrance into the active site and forms one of the walls of the active site (Guasch et al, 2003). The β2–β3 loop of RepB OBD may, therefore, have a function comparable to the α-helical ‘fingers' of TrwC, folding over the ssDNA substrate.
Extensive co-crystallization assays with DNA oligonucleotides representing the stem-loop of the nic locus did not result in diffraction-quality crystals. This locus, located upstream of the bind site, must have a completely different conformation from the ds helix of the bind region, but it is also recognized by the OBDs. The association of RepB6 with drastically changing and diverse DNA structures, such as dsDNA and stem-loop DNA, could require—besides the use of different interacting surfaces—an adaptive capacity that is likely to be facilitated by the large movements of the individual OBDs. Given the similarity in domain function and organization (see Figures 1 and 7 and above) and the evolutionary relation between pMV158 RepB and other RCR Rep proteins as well as the replication initiators of the SF3 helicase superfamily, our results suggest that OBD movement plays a pivotal role in the mechanisms of all of these proteins. In fact, a high degree of flexibility of the OBD relative to the helicase domain has to be assumed in the model proposed for AAV5, given the interaction between Rep OBDs and cognate DNA (Hickman et al, 2004).
Figure 7.
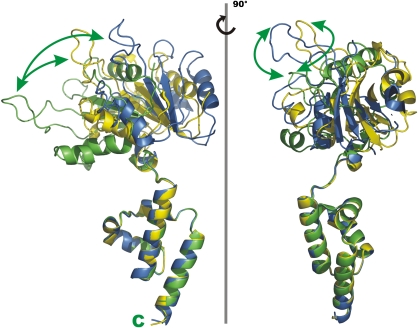
Comparison of the RepB and papillomavirus E1 helicase structures. The lower box diagram shows the alignment and domain organization of the full-length primary sequence of RepB, E1 and TYLCV Rep. Dashed boxes indicate the domains included in the figure, where the colouring of the domain diagram corresponds to the structure representations. The DNA fragment passing through the E1-structure is shown in red. (A) Superposition of two channel-flanking monomers of the hexamers of RepB and E1. The DNA fragment from the E1-structure is shown in red. (B) Top view of the superposition including all protomers of both structures, with the below-plane C-terminal domains of E1 and the above-plane N-terminal domains of RepB shown as transparent cartoon representations. (C) Stereo view of the superposition of the ODs of both structures showing the similarity in fold.
Catalytic mechanism of RepB
The active site includes a Mn2+ cation coordinated by three histidine residues (H39, H55 and H57) and an aspartate (D42, Figure 2B). As proposed earlier (Hickman et al, 2002; Boer et al, 2006), the metal ion probably coordinates one of the oxygen atoms of the scissile DNA phosphate, polarizing it and facilitating the nucleophilic attack of the hydroxyl group of Y99, which lies in close proximity and points in the appropriate direction (Figure 2B). There is experimental evidence showing that RepB, like filamentous phage gpII (Asano et al, 1999), forms a transient covalent complex with the 5′-P end of the cleaved DNA (Moscoso et al, 1995, 1997), and that generation of the circular ssDNA intermediate during termination of the plasmid leading strand synthesis requires that the 5′-P end be covalently attached to RepB (Moscoso et al, 1995). It is, therefore, likely that isoenergetic DNA strand-transfer reactions, in which the energy of the cleaved phosphodiester bond is stored as a phosphotyrosine linkage for the subsequent closing reaction, occur in vivo. A ‘flip-flop' mechanism that involves alternative nicking and nicking-closing reactions of two catalytic tyrosines of a single protomer has been proposed for the gene A protein of phage φX174 (Hanai and Wang, 1993; Grandoso et al, 2000; Datta et al, 2003). A mechanism analogous to the flip-flop scheme is also plausible for RepB. However, our biochemical and structural studies on RepB have not led to the identification of a suitable candidate for the second Tyr. Y115 and Y14 are in close proximity in the crystal structures, but were excluded because Y115 helps forming the metal-binding pocket and Y14 probably has limited freedom of movement as a result of its location on a central strand of the β-sheet. As an alternative, the second catalytic Tyr may be provided by the active sites of other monomers of RepB6, which would imply that substrates are transferred between OBDs during the termination reactions. The flexibility of the catalytic domain in the hexamer would facilitate the reorientation of the OBDs involved in catalysis, so that they come in close proximity for the second cleavage and religation reactions.
Evolutionary relations between Rep proteins and helicases support DNA enclosure by hexameric RepB
As discussed above, the resemblance in the domain organization of RepB and viral replication initiators points to an evolutionary link between the two protein families. The superposition of the α-helical hexamerization domain of RepB with the equivalent domain of papillomavirus E1 helicase, from a crystal structure of a construct comprising its OD and helicase domains (Enemark and Joshua-Tor, 2006), shows that they have the same fold (Figure 7C), although there is no obvious sequence similarity. A domain with a similar fold is expected to exist in TYLCV Rep as well. Its C-terminal ATPase/helicase domain is preceded by a domain that has been marked as an OD (Clerot and Bernardi, 2006) and that has a predicted secondary structure (Cuff and Barton, 2000) comprising four α-helices similar to the OD of RepB and E1.
The structural similarities between E1 helicase and RepB justify a comparison of the details of the functional mechanisms of the two proteins. Papillomavirus replication does not follow a rolling-circle mechanism and the E1 OBD does not cleave the DNA at the origin. Instead, a bubble is formed at the origin of replication, where two E1 initiator hexamers bind and melt the DNA. Each of the DNA strands is encircled by one hexamer, opening up the bubble in opposite directions (Fouts et al, 1999; Enemark et al, 2002). The structure of E1 complexed with a 13-base oligonucleotide shows the ssDNA in the central channel (Figure 7), with a helical conformation (Enemark and Joshua-Tor, 2006), thereby confirming earlier EM studies (Fouts et al, 1999). Similarly, DNA passage in the SV40 T-antigen has been shown to involve the central pore (VanLoock et al, 2002). The resemblance of the E1 and RepB hexameric rings (Figure 7) favours a mechanistic model with an ssDNA passage through the central channel of RepB.
Although a detailed model on the overall mechanism of RepB action awaits additional experiments, given the similarities between the plasmidic and viral systems, we speculate on the existence of various stages leading to the initiation of pMV158 replication. Initial binding of monomers or dimers of RepB to the bind locus may promote sequential assembly of a hexameric RepB to the plasmid origin. This process might involve not only the binding of three OBDs to the triple repeat bind locus, but also interaction of the remaining OBDs with regions of the nic locus that putatively include a pair of 7-bp directly repeated sequences, which share homology with the direct repeats of the bind locus (Ruiz-Masó et al, 2007), and the adjacent hairpin bearing the nick site. Next, the RepB ring might close around a DNA region that could have melted on assembly of the protein and/or cleavage at the nick site, thus encircling one of the plasmid strands within the central channel. Subsequent recruitment of a host helicase (perhaps PcrA) would allow further unwinding of the DNA and the concomitant progression of the hexamer along the plasmid. The strand enclosure may confer high processivity to the RepB/helicase/DNAP III replisome complex, thereby allowing replication of pMV158 in a broad range of bacterial hosts.
Materials and methods
Protein production
RepB was purified as described (Ruiz-Masó et al, 2004). Selenomethionine (SeMet)-labelled RepB was prepared in a similar way using strain B834(DE3).
Electrophoresis mobility shift assays
Reactions to measure binding stoichiometry were performed in buffer B (20 mM Tris–HCl pH 8.0, 1 mM EDTA, 5 mM DTT, 300 mM KCl) containing 0.5 μM of RepB6 and the indicated concentrations of a 32P-labelled 42-bp oligonucleotide (42bp-bind) carrying the bind locus (coordinates 529–570 of the pMV158 DNA sequence). After 20 min at 25°C, free and bound DNAs were separated by electrophoresis on native 5% PAA gels. Labelled DNA bands were detected by autoradiography and quantified with the storage phosphor technology, using an FLA-3000 (FUJIFILM) imaging system and the QuantityOne software (Bio-Rad).
In separate experiments, we studied binding reactions in buffer B of increasing amounts of RepB6 and a mixture of 0.35 μM of the 42bp-bind DNA and 0.07 μM of a 123-bp DNA fragment (123bp-bind), which also includes the bind locus (coordinates 498–620 of pMV158). Samples were analysed by electrophoresis as indicated above. Gels were stained with ethidium bromide and the DNA bands were visualized with the aid of a Gel-Doc documentation system (Bio-Rad).
Nicking activity of RepB on ss oligonucleotides
For cleavage assays, 0.9 pmol of the 5′-end-labelled 23-mer oligonucleotide 5′-TGCTTCCGTACTACG/ACCCCCCA-3′ (‘/' indicates the RepB nick-site) was incubated with RepB (1.3 pmol) for 10 min at 37°C in buffer B containing the indicated concentrations of divalent metal salts. Samples (20 μl) were treated with proteinase K and precipitated with ethanol. The products were separated on a 20% PAA sequencing gel, and detected and quantified as above.
In vivo chemical cross-linking
S. pneumoniae cultures carrying pMV158 were exponentially grown at 37°C in AGCH medium (Lacks, 1968) supplemented with 0.3% sucrose, 0.2% yeast extract and tetracycline (1 μg/ml) to OD650=0.3. Cells were washed with 50 mM Hepes pH 8.0 and concentrated 40-fold in buffer P (50 mM Hepes pH 8.0, 10 mM EDTA and 20% sucrose). The cross-linker BS3 (Pierce) was dissolved in 50 mM Hepes pH 8.0 just before use and added to the concentrated culture at a range of final concentrations. After incubation at 25°C for 20 min, Tris–HCl pH 7.5 was added at a final concentration of 150 mM to quench the reaction and cells were harvested by centrifugation. Total protein extracts were prepared as described by Espinosa et al (1984) and separated by SDS–tricine–PAGE. RepB complexes were detected by western blot with anti-RepB serum.
Cloning and purification of the OBD and OD domains
Experimental details can be found in the Supplementary data.
Construction and purification of various OBD mutants
Experimental details can be found in the Supplementary data.
Circular dichroism assays
Experimental details can be found in the Supplementary data.
Crystallography—trigonal (C2) form
Crystals of RepB were obtained in a vapour-diffusion setup in the presence of one equivalent of dsDNA bind locus fragments (coordinates 531–565 of the pMV158 DNA sequence, which is a subset of the 42bp-bind oligonucleotide also used for EMSA and EM) per RepB6, using 100 mM CaCl2, 50 mM Tris–HCl pH 8.5 and 10% PEG 8 K as buffer. These crystals did not grow in the absence of DNA. Native data to 2.7 Å were collected on parallelepiped crystals (0.4 × 0.4 × 0.2 mm) belonging to space group P3221 (C2 in the Supplementary Table). MAD experiments were conducted on similarly obtained crystals of RepB-SeMet derivatives (containing DNA fragments corresponding to pMV158 coordinates 532–565) and K2PtCl4-soaked crystals (containing DNA fragments corresponding to pMV158 coordinates 532–565), datasets Se34 and Pt35s1 in the Supplementary Table, respectively. Initial experimental phases, obtained from 8 Se positions using SHELXD (Sheldrick et al, 2001) and SHELXE (Sheldrick, 2002), were used to build an initial model comprising residues 135–202 of the ODs, aided by density modification using the NCS relationships of these domains with the program DM (Cowtan, 1994) of the CCP4 version 6.0 program suite (Collaborative Computational Project, 1994). The NCS relationships for the OBDs were obtained after identification of their β-sheets and were included in a subsequent density modification step using DMMULTI (Cowtan, 1994). The model was extended using improved density maps (Supplementary Figure S7), calculated using XtalView (McRee, 1999) from a combination of phases derived from the Pt MAD dataset, using SHELXD/E and the Se-derived phases. The resulting phases were improved against the C2-data using DMMULTI and the two sets of improved NCS relationships for both domains, respectively. A model comprising residues K3-R203 for all chains was then set up (Supplementary Table) using RESOLVE (Terwilliger, 2000) and REFMAC5 (Murshudov et al, 1997) runs, interspersed with careful manual building with Coot v1.4 (Emsley and Cowtan, 2004), applying TLS refinement to all respective domains and two sets of NCS restraints on the equivalent parts of the three copies of the OBD and OD domains. The final Rcryst and Rfree are 22.9 and 27.7, respectively.
Crystallography—tetragonal (C3) form
Crystals of tetragonal RepB were obtained from a vapour-diffusion crystallization setup in the presence of one equivalent of dsDNA bind locus fragments (pMV158 coordinates 531–565) per RepB6, using 200 mM MgCl2, 50 mM CHES pH 9, 10% PEG 4 K as crystallization buffer and belonged to space group P43. Native data were measured and processed to 3.6 Å (C3 in the Supplementary Table). Using molecular replacement methods, the C-terminal hexamer was placed using Phaser (McCoy et al, 2005), after which six of the N-terminal domains were found sequentially using Phaser, Amore (Navaza, 2001) and MOLREP (Vagin and Teplyakov, 1997) (Supplementary Figure S8). A model comprising residues 4–203 was refined with PHENIX v1.3 (Adams et al, 2004) (Supplementary Table), using NCS restraints for the six copies of each of the domains and refining a single isotropic B factor combined with TLS refinement for each of the OD and OBD domains, respectively. For three of the OBDs, loop β2–β3 (K43–K54) was disordered and not included in the refinement of the final model. In addition, residues E4–A6 and E81–M86 were disordered in one of the OBDs and were not included. The final Rcryst and Rfree are 22.3 and 25.7%, respectively.
Electron microscopy
A few microlitres of purified RepB and RepB–DNA complex, respectively, in buffer H (Tris–HCl 20 mM pH 8.0, 1 mM EDTA, 0.1 mM DTT, 450 mM KCl) were diluted to 60 nM and rapidly adsorbed to glow discharged carbon coated Cu/Pd grids and then negatively stained using 2% (m/w) uranyl acetate. DNA binding was carried out earlier by incubating RepB with a 10-fold molar excess of the 42bp-bind fragment (see above) for 20 min at 25°C in buffer B. The grids were observed in a JEOL 1230 transmission electron microscope operated at 100 kV and micrographs were taken at × 50 000 magnifications under low-dose conditions with the specimen holder tilted at 0°, 20° and 40°. In an independent experiment, untilted and tilted micrographs were sequentially taken from a particular area in the grid after an RCT data collection scheme (Radermacher, 1988). All micrographs were digitized using a Minolta Dimage Scan Multi PRO scanner at 10.6 μm and averaged to a final step size of 4.0 Å/pixel.
Image processing and 3D reconstructions
The RCT 3D reconstruction was performed with the XMIPP software (Sorzano et al, 2004), using 4519 pairs of manually extracted DNA-free protein particles followed by 2D reference-free alignment and classification of the untilted images using maximum-likelihood multi-reference methods (Scheres et al, 2005) as implemented in XMIPP. The resulting RCT reconstruction was low-pass filtered to 45 Å and used as unbiased initial reference model for angular refinement.
EMAN v1.7 (Ludtke et al, 1999) was used for image processing and 3D reconstruction using the low-dose micrographs. The 11 628 and 8659 particles were selected and extracted automatically from micrographs obtained from DNA-free and DNA-bound RepB, respectively, at 0°, 20° and 40° tilts. These were masked, band-pass filtered, centred, normalized and subjected to angular refinement. Volumes were iteratively refined reaching a final resolution of 22 and 24 Å for DNA-free and -bound reconstructions, respectively, estimated by the 0.5 Fourier shell correlation criteria. The handedness of the structures was determined by direct comparison with the crystal structure of the C2-form. Representation thresholds were chosen to account for the protein mass.
Fitting and difference mapping
Multi-resolution docking between the X-ray models and the EM maps was performed using ADP_EM (Garzon et al, 2007). An adaptation of this program was also used to align the DNA-free and DNA-bound maps. After alignment, the two structures were subtracted using SITUS (Wriggers et al, 1999).
Miscellaneous
Figure 1A was prepared using the EPSprit server (Gouet et al, 1999). The 3D-similarity searches were performed using DALI (Holm et al, 2008) and MATRAS (Kawabata, 2003). The molecular representations in Figures 2, 3, 7 and Supplementary Figure S5 were prepared with PyMol (DeLano, 2002). Graphs of the electrostatic potential surfaces were prepared with GRASP (Nicholls et al, 1991). The C2 and C3 X-ray structures have been deposited in the RCSB PDB as 3DKX and 3DKY, respectively.
Supplementary Material
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Figure S5
Supplementary Figure S6
Supplementary Figure S7
Supplementary Figure S8
Supplementary Table I
Supplementary Materials and Methods
Acknowledgments
This study was supported by the Spanish Ministerio de Ciencia e Innovación (Grants BFU2005-06758/BMC and BFU2008-02372 to MC, BIO2006-02668 to FXGR, BFU2007-63575 to GdS, SAF2005-00775/SAF2008-00451 to OL, BFU2007-65977 to PC and CSD00013 to ME), the Generalitat de Catalunya (Grant 2005SGR-00280 to MC), the Fundació La Marató de TV3 (Grant 052810 to MC), the Comunidad de Madrid (Grants S-BIO-0214-2006 to OL and PC and CM-BIO0260-2006 to ME), the Red Temática de Investigación Cooperativa en Cáncer (Grant RD06/0020/1001 to OL), the Human Frontiers Science Program (Grant RGP39/2008 to OL and PC) and the EU (Spine2-Complexes LSHG-2006-031220 and 3D-Repertoire LSHG-CT-2005-512028 projects). Synchrotron data collection was supported by the ESRF and the EU.
References
- Adams PD, Gopal K, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Pai RK, Read RJ, Romo TD, Sacchettini JC, Sauter NK, Storoni LC, Terwilliger TC (2004) Recent developments in the PHENIX software for automated crystallographic structure determination. J Synchrotron Radiat 11: 53–55 [DOI] [PubMed] [Google Scholar]
- Asano S, Higashitani A, Horiuchi K (1999) Filamentous phage replication initiator protein gpII forms a covalent complex with the 5′ end of the nick it introduced. Nucleic Acids Res 27: 1882–1889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochkareva E, Martynowski D, Seitova A, Bochkarev A (2006) Structure of the origin-binding domain of simian virus 40 large T antigen bound to DNA. EMBO J 25: 5961–5969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boer R, Russi S, Guasch A, Lucas M, Blanco AG, Perez-Luque R, Coll M, de la Cruz F (2006) Unveiling the molecular mechanism of a conjugative relaxase: the structure of TrwC complexed with a 27-mer DNA comprising the recognition hairpin and the cleavage site. J Mol Biol 358: 857–869 [DOI] [PubMed] [Google Scholar]
- Bruand C, Ehrlich SD (2000) UvrD-dependent replication of rolling-circle plasmids in Escherichia coli. Mol Microbiol 35: 204–210 [DOI] [PubMed] [Google Scholar]
- Campos-Olivas R, Louis JM, Clérot D, Gronenborn B, Gronenborn AM (2002) The structure of a replication initiator unites diverse aspects of nucleic acid metabolism. Proc Natl Acad Sci USA 99: 10310–10315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang TL, Naqvi A, Anand SP, Kramer MG, Munshi R, Khan SA (2002) Biochemical characterization of the Staphylococcus aureus PcrA helicase and its role in plasmid rolling circle replication. J Biol Chem 277: 45880–45886 [DOI] [PubMed] [Google Scholar]
- Clerot D, Bernardi F (2006) DNA helicase activity is associated with the replication initiator protein rep of tomato yellow leaf curl geminivirus. J Virol 80: 11322–11330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaborative Computational Project, N (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr 50: 760–763 [DOI] [PubMed] [Google Scholar]
- Cowtan K (1994) Joint CCP4 and ESF-EACBM Newsletter on Protein Crystallography 31: 34–38 [Google Scholar]
- Cuff JA, Barton GJ (2000) Application of multiple sequence alignment profiles to improve protein secondary structure prediction. Proteins 40: 502–511 [DOI] [PubMed] [Google Scholar]
- Datta S, Larkin C, Schildbach JF (2003) Structural insights into single-stranded DNA binding and cleavage by F factor TraI. Structure 11: 1369–1379 [DOI] [PubMed] [Google Scholar]
- de la Campa AG, del Solar GH, Espinosa M (1990) Initiation of replication of plasmid pLS1. The initiator protein RepB acts on two distant DNA regions. J Mol Biol 213: 247–262 [DOI] [PubMed] [Google Scholar]
- del Solar G, Moscoso M, Espinosa M (1993) Rolling circle-replicating plasmids from gram-positive and gram-negative bacteria: a wall falls. Mol Microbiol 8: 789–796 [DOI] [PubMed] [Google Scholar]
- DeLano WL (2002) The Pymol Molecular Graphics System on World Wide Web http://www.pymol.org
- Dyda F, Hickman AB (2003) A mob of reps. Structure 11: 1310–1311 [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Enemark EJ, Chen G, Vaughn DE, Stenlund A, Joshua-Tor L (2000) Crystal structure of the DNA binding domain of the replication initiation protein E1 from papillomavirus. Mol Cell 6: 149–158 [PubMed] [Google Scholar]
- Enemark EJ, Joshua-Tor L (2006) Mechanism of DNA translocation in a replicative hexameric helicase. Nature 442: 270–275 [DOI] [PubMed] [Google Scholar]
- Enemark EJ, Stenlund A, Joshua-Tor L (2002) Crystal structures of two intermediates in the assembly of the papillomavirus replication initiation complex. EMBO J 21: 1487–1496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinosa M, Lopez P, Lacks SA (1984) Transfer and expression of recombinant plasmids carrying pneumococcal mal genes in Bacillus subtilis. Gene 28: 301–310 [DOI] [PubMed] [Google Scholar]
- Fouts ET, Yu X, Egelman EH, Botchan MR (1999) Biochemical and electron microscopic image analysis of the hexameric E1 helicase. J Biol Chem 274: 4447–4458 [DOI] [PubMed] [Google Scholar]
- Garzon JI, Kovacs J, Abagyan R, Chacon P (2007) ADP_EM: fast exhaustive multi-resolution docking for high-throughput coverage. Bioinformatics 23: 427–433 [DOI] [PubMed] [Google Scholar]
- Gomis-Rüth FX, Coll M (2006) Cut and move: protein machinery for DNA processing in bacterial conjugation. Curr Opin Struct Biol 16: 744–752 [DOI] [PubMed] [Google Scholar]
- Gouet P, Courcelle E, Stuart DI, Metoz F (1999) ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics 15: 305–308 [DOI] [PubMed] [Google Scholar]
- Grandoso G, Avila P, Cayon A, Hernando MA, Llosa M, de la Cruz F (2000) Two active-site tyrosyl residues of protein TrwC act sequentially at the origin of transfer during plasmid R388 conjugation. J Mol Biol 295: 1163–1172 [DOI] [PubMed] [Google Scholar]
- Guasch A, Lucas M, Moncalian G, Cabezas M, Perez-Luque R, Gomis-Ruth FX, de la Cruz F, Coll M (2003) Recognition and processing of the origin of transfer DNA by conjugative relaxase TrwC. Nat Struct Biol 10: 1002–1010 [DOI] [PubMed] [Google Scholar]
- Hanai R, Wang JC (1993) The mechanism of sequence-specific DNA cleavage and strand transfer by phi X174 gene A* protein. J Biol Chem 268: 23830–23836 [PubMed] [Google Scholar]
- Hickman AB, Ronning DR, Kotin RM, Dyda F (2002) Structural unity among viral origin binding proteins: crystal structure of the nuclease domain of adeno-associated virus Rep. Mol Cell 10: 327–337 [DOI] [PubMed] [Google Scholar]
- Hickman AB, Ronning DR, Perez ZN, Kotin RM, Dyda F (2004) The nuclease domain of adeno-associated virus rep coordinates replication initiation using two distinct DNA recognition interfaces. Mol Cell 13: 403–414 [DOI] [PubMed] [Google Scholar]
- Holm L, Kaariainen S, Rosenstrom P, Schenkel A (2008) Searching protein structure databases with DaliLite v.3. Bioinformatics 24: 2780–2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilyina T, Koonin E (1992) Conserved sequence motifs in the initiator proteins for rolling circle DNA replication encoded by diverse replicons from eubacteria, eukaryotes and archaebacteria. Nucleic Acids Res 20: 3279–3285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W, Sander C (1983) Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22: 2577–2637 [DOI] [PubMed] [Google Scholar]
- Kawabata T (2003) MATRAS: a program for protein 3D structure comparison. Nucleic Acids Res 31: 3367–3369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacks SA (1968) Genetic regulation of maltosaccharide utilization in Pneumococcus. Genetics 60: 685–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludtke SJ, Baldwin PR, Chiu W (1999) EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol 128: 82–97 [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ (2005) Likelihood-enhanced fast translation functions. Acta Crystallogr D Biol Crystallogr 61: 458–464 [DOI] [PubMed] [Google Scholar]
- McRee DE (1999) XtalView/Xfit—a versatile program for manipulating atomic coordinates and electron density. J Struct Biol 125: 156–165 [DOI] [PubMed] [Google Scholar]
- Monzingo AF, Ozburn A, Xia S, Meyer RJ, Robertus JD (2007) The structure of the minimal relaxase domain of MobA at 2.1 A resolution. J Mol Biol 366: 165–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscoso M, del Solar G, Espinosa M (1995) Specific nicking-closing activity of the initiator of replication protein RepB of plasmid pMV158 on supercoiled or single-stranded DNA. J Biol Chem 270: 3772–3779 [DOI] [PubMed] [Google Scholar]
- Moscoso M, Eritja R, Espinosa M (1997) Initiation of replication of plasmid pMV158: mechanisms of DNA strand-transfer reactions mediated by the initiator RepB protein. J Mol Biol 268: 840–856 [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr 53: 240–255 [DOI] [PubMed] [Google Scholar]
- Navaza J (2001) Implementation of molecular replacement in AMoRe. Acta Crystallogr D Biol Crystallogr 57: 1367–1372 [DOI] [PubMed] [Google Scholar]
- Nicholls A, Sharp KA, Honig B (1991) Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins 11: 281–296 [DOI] [PubMed] [Google Scholar]
- Novick RP (1998) Contrasting lifestyles of rolling-circle phages and plasmids. Trends Biochem Sci 23: 434–438 [DOI] [PubMed] [Google Scholar]
- Ozaki E, Yasukawa H, Masamune Y (1994) Purification of pKYM-encoded RepK, a protein required for the initiation of plasmid replication. J Gen Appl Microbiol 40: 365–375 [Google Scholar]
- Petit MA, Dervyn E, Rose M, Entian KD, McGovern S, Ehrlich SD, Bruand C (1998) PcrA is an essential DNA helicase of Bacillus subtilis fulfilling functions both in repair and rolling-circle replication. Mol Microbiol 29: 261–273 [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004) UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25: 1605–1612 [DOI] [PubMed] [Google Scholar]
- Radermacher M (1988) Three-dimensional reconstruction of single particles from random and nonrandom tilt series. J Electron Microsc Tech 9: 359–394 [DOI] [PubMed] [Google Scholar]
- Ruiz-Masó JA, López-Zumel C, Menéndez M, Espinosa M, del Solar G (2004) Structural features of the initiator of replication protein RepB encoded by the promiscuous plasmid pMV158. Biochim Biophys Acta 1696: 113–119 [DOI] [PubMed] [Google Scholar]
- Ruiz-Masó JA, Lurz R, Espinosa M, del Solar G (2007) Interactions between the RepB initiator protein of plasmid pMV158 and two distant DNA regions within the origin of replication. Nucleic Acids Res 35: 1230–1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russi S, Boer R, Coll M (2008) Molecular machinery for DNA translocation. In Plasmids: Current Research and Future Trends, Lipps G (ed), pp 183–213. Norfolk, UK: Caister Academic Press [Google Scholar]
- Scheres SH, Valle M, Nunez R, Sorzano CO, Marabini R, Herman GT, Carazo JM (2005) Maximum-likelihood multi-reference refinement for electron microscopy images. J Mol Biol 348: 139–149 [DOI] [PubMed] [Google Scholar]
- Schuck S, Stenlund A (2005) Assembly of a double hexameric helicase. Mol Cell 20: 377–389 [DOI] [PubMed] [Google Scholar]
- Sheldrick G (2002) Macromolecular phasing with SHELXE. Z Kristallogr 217: 644–650 [Google Scholar]
- Sheldrick G, Hauptmann H, Weeks C, Miller R, Usón I (2001) Direct methods. In International Tables for Crystallography F, Arnold E, Rosmann M (eds), pp 333–351. Dordrecht/Boston/London: Kluwer Academic Publishers [Google Scholar]
- Sorzano CO, Marabini R, Velazquez-Muriel J, Bilbao-Castro JR, Scheres SH, Carazo JM, Pascual-Montano A (2004) XMIPP: a new generation of an open-source image processing package for electron microscopy. J Struct Biol 148: 194–204 [DOI] [PubMed] [Google Scholar]
- Takahashi S, Hours C, Iwaya M, Lane HED, Denhardt DT (1978) The Escherichia coli rep Gene in the Single-Stranded DNA Phages. New York: Cold Spring Harbor Laboratory Press [Google Scholar]
- Terwilliger TC (2000) Maximum-likelihood density modification. Acta Crystallogr D Biol Crystallogr 56: 965–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin A, Teplyakov A (1997) MOLREP: an automated program for molecular replacement. J Appl Crystallogr 30: 1022–1025 [Google Scholar]
- VanLoock MS, Alexandrov A, Yu X, Cozzarelli NR, Egelman EH (2002) SV40 large T antigen hexamer structure: domain organization and DNA-induced conformational changes. Curr Biol 12: 472–476 [DOI] [PubMed] [Google Scholar]
- Vega-Rocha S, Byeon IJ, Gronenborn B, Gronenborn AM, Campos-Olivas R (2007a) Solution structure, divalent metal and DNA binding of the endonuclease domain from the replication initiation protein from porcine circovirus 2. J Mol Biol 367: 473–487 [DOI] [PubMed] [Google Scholar]
- Vega-Rocha S, Gronenborn B, Gronenborn AM, Campos-Olivas R (2007b) Solution structure of the endonuclease domain from the master replication initiator protein of the nanovirus faba bean necrotic yellows virus and comparison with the corresponding geminivirus and circovirus structures. Biochemistry 46: 6201–6212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wriggers W, Milligan RA, McCammon JA (1999) Situs: a package for docking crystal structures into low-resolution maps from electron microscopy. J Struct Biol 125: 185–195 [DOI] [PubMed] [Google Scholar]
- Zhao AC, Ansari RA, Schmidt MC, Khan SA (1998) An oligonucleotide inhibits oligomerization of a rolling circle initiator protein at the pT181 origin of replication. J Biol Chem 273: 16082–16089 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Figure S5
Supplementary Figure S6
Supplementary Figure S7
Supplementary Figure S8
Supplementary Table I
Supplementary Materials and Methods