Abstract
Rising demand for food and bioenergy makes it imperative to breed for increased crop yield. Vegetative plant growth could be driven by resource acquisition or developmental programs. Metabolite profiling in 94 Arabidopsis accessions revealed that biomass correlates negatively with many metabolites, especially starch. Starch accumulates in the light and is degraded at night to provide a sustained supply of carbon for growth. Multivariate analysis revealed that starch is an integrator of the overall metabolic response. We hypothesized that this reflects variation in a regulatory network that balances growth with the carbon supply. Transcript profiling in 21 accessions revealed coordinated changes of transcripts of more than 70 carbon-regulated genes and identified 2 genes (myo-inositol-1-phosphate synthase, a Kelch-domain protein) whose transcripts correlate with biomass. The impact of allelic variation at these 2 loci was shown by association mapping, identifying them as candidate lead genes with the potential to increase biomass production.
Keywords: Arabidopsis, association mapping, biomass, metabolites, predictive
Plants use light energy to convert CO2 into carbohydrates. Although we might expect plant growth to be driven by the availability of carbohydrates and other central metabolites, recent studies point to a more complex interaction. Numerous free air CO2 elevation studies show that higher rates of photosynthesis do not lead to a commensurate increase in biomass and yield (1). Studies of natural genetic diversity reveal a negative correlation between the levels of metabolites and biomass or yield (2–4). Although biomass was only very weakly correlated with individual metabolites in an Arabidopsis recombinant inbred line (RIL) population, a highly significant prediction was obtained when multivariate analysis was used on the entire metabolite profile (3). These results indicate that much of the genetic variation for biomass production affects the balance between resource availability and developmental programs, which determine how rapidly these resources are used for growth.
Plants are exposed to a changeable environment and need to cope with continual changes in carbon (C) availability. One striking example is the daily alternation between a positive C balance in the light and a negative C balance in the dark. Growth nevertheless continues at night (5). This continued growth is possible because some newly fixed C accumulates as starch in the light and is remobilized at night to support respiration and growth. Starch is almost completely exhausted by the end of the night. If a change in the conditions (e.g., longer nights) leads to a temporary period of C starvation, the C budget is rebalanced (6–11) by increasing the rate of starch synthesis, decreasing the rate of starch breakdown, and decreasing the rate of growth (10, 11). Starchless mutants illustrate the importance of this buffer; they cannot grow in a light/dark cycle because they become C-starved every night, leading to an inhibition of growth that is not reversed for several hours into the next day (8, 12).
The following experiments test the hypothesis that starch turnover and C allocation occupy a central role in the network that coordinates metabolism with growth. We first investigate biomass and metabolite levels in 94 Arabidopsis accessions. This species-wide analysis reveals that starch content at the end of the day integrates many other metabolic traits and is negatively correlated with biomass. We then compare the expression of C-responsive transcripts in 21 accessions, identify candidate genes that may contribute to genetic variation in the regulation of metabolism and growth, and test their role by association mapping of sequence polymorphisms.
Results and Discussion
Many Metabolites Are Negatively Correlated to Biomass.
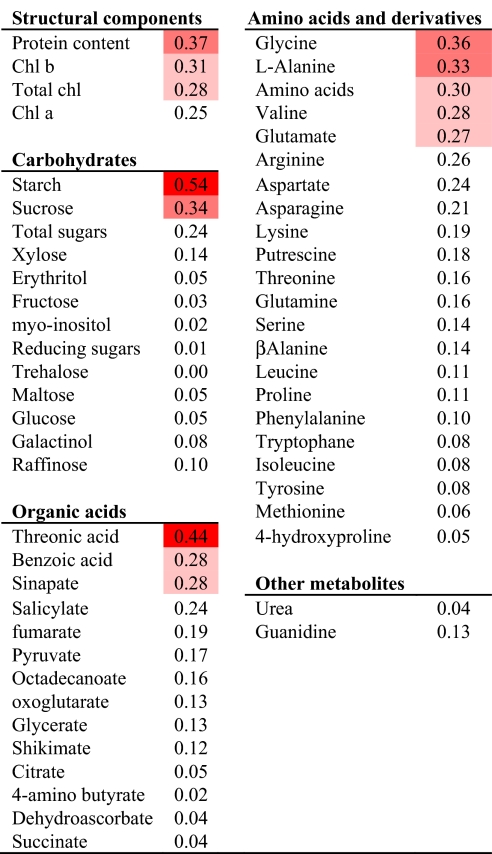
Over 400 Arabidopsis thaliana accessions were genotyped with 419 markers (13) to identify a genotypically diverse set of 94 accessions with maximized allelic richness (Table S1). The accessions were grown in short-day conditions (8 h light/16 h dark) in moderate light and well-fertilized soil to apply a moderate C deprivation. They were harvested at the end of the day, 5 weeks after germination when they were still in the vegetative growth phase. Rosette fresh weight (FW) was measured as an indicator of biomass. We have documented a very close relation between rosette FW and rosette dry weight (2). We analyzed starch, total protein, chlorophyll, and 48 low-molecular-weight metabolites, including individual amino acids, organic acids, sugars, lipids, and secondary metabolites (Table S1). Pair-wise Spearman's correlations were calculated for biomass against every metabolic trait (Table 1). Rosette biomass showed a high negative correlation to starch (R = −0.54); lower but significant negative correlations with protein (R = −0.37), chlorophyll (R = −0.31), and several low-molecular-weight metabolites (sucrose, total amino acids, glycine, alanine, glutamate, threonic acid, benzoic acid, sinapic acid); and nonsignificant negative correlations with other metabolites.
Table 1.
Spearman coefficients of metabolic traits against biomass
Correlations were calculated from mean data. Significant correlations at P < 0.00001, P < 0.001, and P < 0.01 are indicated by dark, medium, and light shading. Blue and red distinguish positive and negative correlations, respectively. The original data are in Table S1.
Partial Correlation Analysis to Remove Spurious Correlations.
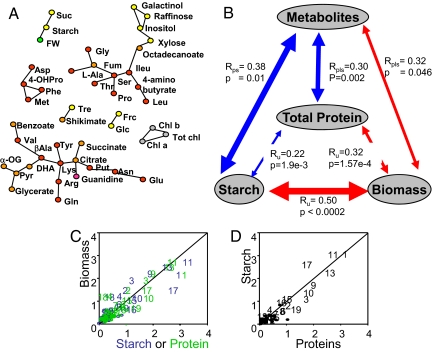
Because many metabolic traits correlate with each other (2), some of the correlations with biomass may be secondary. Partial Correlation Analysis was performed to correct for spurious secondary correlations (Fig. 1A). The analysis confirmed the link between biomass and starch but did not provide evidence for direct links of biomass to any other individual metabolic traits. Some links were found between metabolites; starch was linked to sucrose, glucose was linked to fructose but not to sucrose or starch, several amino acids were linked, and raffinose was linked to galactinol and myo-inositol, which are involved in its synthesis.
Fig. 1.
Multivariate analysis of the relations between biomass and metabolic traits. (A) Graphical Gaussian Model. Partial correlation was used to identify direct association between 2 metabolites and/or traits with the influence of all other ones removed. For clarity, the different classes of traits have been colored: green, biomass; gray, chlorophylls; yellow, sugars and sugar alcohols; orange, organic acids; red, amino acids; pink, other metabolites. (B) PLS regression analysis of the relation between 5 inputs. These include 3 univariate inputs (biomass, starch, total protein) and 1 multivariate input (all other metabolites). Linear regression was used to compare the univariate inputs, and PLS regression was used to predict each univariate class from the multivariate class. Cross-validation was used to determine regression coefficients (Rpls = regression coefficient obtained by PLS, Ru = regression coefficient obtained by univariate correlation with cross-validation) and their P values (values in italics are nonsignificant), with red and blue arrows indicating negative and positive relationships between inputs. (C–D) VIP values of metabolites in the PLS regression. Metabolites with high VIP values are indicated by numbers: 1, amino acids; 2, Arg; 3, l-alanine; 4, DHA; 5, Asn; 6, Glc; 7, Gln; 8, Glu; 9, Gly; 10, guanidine; 11, fumarate; 12, OHPro; 13, Pro; 14, raffinose; 15, red sugars; 16, sucrose; 17, total sugars; 18, threonate; 19, serine. (C) Comparison of loadings for the PLS prediction of starch and biomass (blue) or protein and biomass (green). (D) Comparison of loadings for the PLS prediction of starch and protein.
The negative correlation between biomass and starch was not due to population structure. Using Structure 2.1 (14) on 419 markers distributed across the whole genome, the smallest K value for highest posterior probability split the population into 7 subpopulations (Table S1). These 7 subpopulations had similar average values for biomass and starch. R values between starch and biomass were less than −0.63 in 3 subpopulations (containing 61 accessions), less than −0.42 in 2 subpopulations (containing 25 accessions), and less than −0.24 in the other 2 subpopulations (containing 11 accessions).
Partial Least Squares (PLS) Regression Reveals that Starch Integrates the Metabolic Status.
It has been shown that predictive power can be increased by using multivariate analysis to predict biomass from a linear combination of a set of low-molecular-weight metabolites (3). We investigated whether this was the case in our study. In datasets like ours, where the number of predictors (54) is close to the number of accessions (94), the predictive power of linear models is often improved by dimensionality-reduction methods like PLS regression. PLS identifies combinations of the original predictors that have the maximum covariance with the trait of interest. These orthogonal combinations are then used to predict the trait. The PLS prediction of biomass was significantly (F-test, P = 0.002) but only slightly improved when all of the metabolic traits were used as predictors, compared with the prediction provided by starch alone (R = 0.57 and 0.50, respectively; the value for starch was checked by cross-validation, hence the sign is absent and the value slightly lower than in Table 1).
To investigate the reasons for this lack of additivity, we divided the dataset into 3 univariate traits (biomass, starch, total protein) and 1 multivariate trait class (all other metabolites). The univariate traits were compared by using linear regression, and the multivariate trait class was used to predict each univariate trait using PLS regression (Fig. 1B). The predictive accuracy of each pair-wise comparison was assessed by cross-validation (see Materials and Methods). Starch and total protein showed a significant negative correlation with biomass (R = 0.50 and 0.32, respectively) and correlated weakly with each other (R = 0.22). PLS regression on the multivariate metabolite class allowed prediction of starch, protein, and biomass (R = 0.38, 0.30, and 0.32, respectively). Variance importance in the projection (VIP) gives an estimate of the contribution of a given predictor for a PLS regression. Starch, protein, and rosette biomass (Fig. 1 C and D) were predicted by the same metabolites, with remarkably similar VIP values (for a full list, see Table S2).
This analysis shows that starch and, to a lesser extent, total protein integrate metabolic status. It also indicates that the regulatory network that determines starch and protein levels contributes to the regulation of biomass. To provide functional information about this network, we subjected the reference genotype Col0 and 10 large and 10 small accessions (listed in Table S3) to a more detailed physiological and molecular analysis.
Ranking of Accessions for Biomass and Negative Correlation with Starch Retained in Different Growth Conditions.
The simplest explanation for the negative relation between biomass and starch would be that large accessions maximize growth at the expense of their C reserves. Such a strategy would be advantageous when excess C is available, but not when C is in short supply. We compared the biomass of the 21 accessions at 20 °C in an 8 h light/16 h dark regime with biomass at 20 °C in a much shorter (3 h light/12 h dark) or longer (12 h light/12 h dark) photoperiod (when C availability would be severely decreased and increased, respectively), and with biomass in an 8 h light/16 h dark photoperiod at 16 °C or 24 °C (when growth would be decreased and accelerated, respectively). A similar ranking was retained in all conditions (Table S4). Biomass was always negatively correlated to starch, and this relation was significant except in very short days (Table S4). These results indicate that large accessions gauge their growth to the C supply across a wide range of environmental conditions.
Sugar-Responsive Gene Network.
Changes in the C supply modify the transcript levels for hundreds of genes during the diurnal cycle (10, 11, 15–17). We asked whether C-responsive genes show coordinated changes of expression between the 21 accessions and whether any of their transcripts correlate with the levels of major metabolites or biomass. Two nonoverlapping sets of C-responsive genes were selected (Fig. S2) from published data, one including 52 genes whose transcripts change >log21.4 within 30 min of adding sucrose to C-starved seedlings (18) and one containing another 42 genes that show changes of expression during a diurnal cycle and an extended night (15, 17). Transcript levels were measured at the end of the night (Table S3), when changes in the C status have the largest impact (15, 17). Principal component analysis (PCA) generated a similar separation of the accessions (R = 0.93) in the first component, irrespective of which gene set was used (see Fig. S3). There was a significant correlation between the separation of accessions along the first component and biomass (R = 0.53, P = 0.013). The finding that transcript levels of C-regulated genes possess predictive power for biomass and that an almost identical result is given by 2 nonoverlapping sets of genes encouraged us to pursue the analysis.
Combined Network for Transcripts, Metabolites, and Biomass.
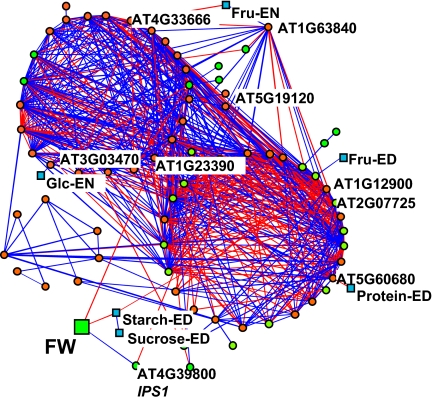
We used an algorithm that identifies functional modules within complex networks (19) to generate a correlation network that combines transcript levels, metabolite levels, and biomass (Fig. 2). This algorithm defines a module as a subset of nodes that are more connected to each other than to nodes in other modules. Starting from the initial state, in which each node represents a module, it performs iterations of merging, splitting, and transferring nodes between modules to maximize the interconnectivity of edges within modules and, thus, the modularity of the network. The resulting network contained 71 of the 94 genes investigated in this study. They were organized in 2 large, well-connected modules, a smaller module and several nodes that are only connected by 1 to 2 edges. One large module contains mainly C-repressed genes (23 of 28), and the second contains 16 C-repressed and 12 C-induced genes. Thus, most C-responsive genes show coordinated changes of expression across this set of 21 accessions.
Fig. 2.
Cartographic representation of the sugar-responsive gene network. Correlations were considered as significant for Rs >0.7 and P < 0.01 for gene–gene interactions and Rs >0.6 and P < 0.01 for gene–metabolites and metabolite–metabolite interactions, respectively. Genes are depicted as circles, with green and orange distinguishing between sugar-induced and sugar-repressed genes, respectively. Blue and red lines are for positive and negative correlations, respectively. Metabolic traits are depicted as squares. ED, end of the day; EN, end of the night.
Comparison with a Transcript Network Obtained by Perturbing C Status in Col0.
We compared the correlation network in Fig. 2 with a correlation network for the same 94 transcripts, which we generated from data obtained in earlier studies where we subjected 5-week-old rosettes of the reference accession Col0 to 23 treatments that alter endogenous C levels (15, 17, 18). The network that was obtained will be termed the ‘C-perturbation’ network. When we compared the R value for each gene–gene pair in the 2 correlation networks, data shuffling revealed a significant enrichment of shared positive (P = 6 × 10−5) and shared negative (P = 2.4 × 10−3) correlations (Fig. S4A). There was a lower, but still significant, enrichment of shared correlations with networks generated from a Col0 leaf developmental series (P = 3.5 × 10−3) (Fig. S4B) and a rosette abiotic stress series (P = 2 × 10−4) (Fig. S4C), but no significant enrichment in a root abiotic stress series (P = 0.21) (Fig. S4E).
We identified 26 genes where >30% of the significant gene–gene correlations are shared in the accession and C-perturbation networks (Table S5). They include many potential regulatory components, including bZIP transcription factors, F-box proteins, ATG8e, and two BTB/POZ proteins. Recent evidence implicates trehalose-6-phosphate (Tre6P) (17, 20, 21) and AKIN10/11 (17, 20) in C signaling. The 26 genes included 3 members of the trehalose-phosphate synthase gene family (TPS8, TPS10, TPS11) and 19 genes whose transcripts respond to AKIN10 overexpression, including bZIP1/AT5G49450, a direct target of AKIN10 (20). This overlap with known upstream components in sugar signaling supports the validity of our network approach. This set of genes represents a robust core of a C-signaling response. They show coordinated changes during perturbations of the C status in Col0 and between a diverse set of accessions.
Two Candidate Genes Whose Transcripts Correlate with Rosette Biomass.
Rosette biomass correlated with 2 genes in the network of Fig. 2, a Kelch repeat F-box protein (At1g23390) and myo-inositol-1-phosphate synthase 1 (IPS1/At4g39800) (R = 0.65 and −0.62 and P = 0.0016 and 0.0026, respectively). The Kelch repeat F-box protein At1g23390 is located in one of the large modules in the accession network (Fig. 2), and many of the connections are retained in the C-perturbation network (Table S5). IPS1 (At4g39800) is connected to many genes in the C-perturbation network (Table S5), but these connections are absent in the accession network (Fig. 2), except for a negative correlation with ATG8e (22).
Candidate Gene Allelic Variation.
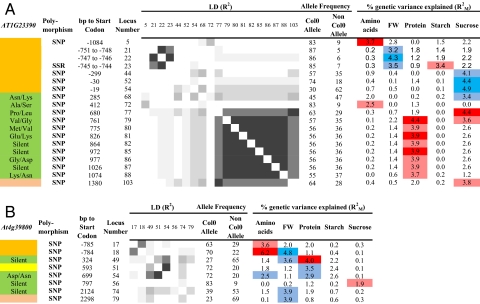
This correlation indicated that sequence diversity in IPS1/At4g39800 and At1g23390 might influence biomass. Genomic DNA, containing 1065 base pairs of the promotor and the entire transcribed region of At4g39800 and 1279 base pairs of the promotor and the entire transcribed region of At1g23390, was amplified and sequenced from 92 of the 94 accessions. We detected 95 SNPs, 9 insertion-deletion polymorphisms (InDels), and 1 simple sequence repeat (SSR) in Atlg23390 and 79 SNPs, 7 InDels, and 1 in SSR in At4g39800 (Table S6). The frequency of single, rare (<5), minor (<15), and major (≥15) polymorphisms was 34%, 19%, 15%, and 32% for At1g23390 and 38%, 28%, 17%, and 17% for At4g39800, respectively. Linkage disequilibrium (LD) was calculated by using Graphical Genotype 2.0 (23). Rare polymorphisms were excluded for the calculation. Both genes contained a set of polymorphisms in strong LD in the transcribed region and further polymorphisms in weaker LD in the promotor region (Table S6; a summary is provided in Fig. 3). The decay of LD to below 0.2 in 1–2 kB is in the range reported in ref. 24. Repeat number for the SSR in the At1g23390 promotor varied independently of loci in the remainder of the gene, probably because changes in repeat number can occur independently of, and more rapidly than, recombination.
Fig. 3.
Sequence polymorphisms in Kelch/At1g23390 (A) and IPS1/At4g39800 (B) that are significantly associated with the traits FW, starch, protein, sucrose or total amino acids. Full information about sequence polymorphisms and associations are given in Table S6. This display summarizes polymorphisms that show significant trait associations and is based on TAIR gene models At4g23390.1 and At4g39800. Gene regions are distinguished by coloring (orange, upstream sequence; green, exons; yellow, introns; salmon, downstream region). Significant (corrected P values <5%) polymorphisms are identified by their distance from the start codon, the locus number, and, for the ORF, the effect on the protein sequence. LD (expressed as R2 values) is classified in 6 classes (R2 < 0.1; 0.1 < R2 < 0.2; 0.2 < R2 < 0.4; 0.4 < R2 < 0.6; 0.6 < R2 < 0.8; 0.8 < R2) that are shaded from light to dark gray. For each polymorphism, the allele frequency is summed. The proportion of the genetic variance of the 5 traits explained by the marker main effect (RM2) are shaded to indicate the significance of the association (dark, P < 0.01; light, 0.01 < P < 0.05), with blue or red signifying a negative or positive effect of the Col0 allele, respectively.
Sequence diversity in these genes is not closely tied to population structure. When we K-clustered the accessions into 7 classes based on the polymorphisms in each of the genes and compared these with the 7 classes obtained after K-clustering (see Materials and Methods and Fig. S1), the overlap was only slightly larger than that expected by chance (2.7%).
Candidate Gene Association Mapping.
Using these polymorphisms and the trait values for rosette biomass, amino acid, starch, sucrose, and protein, marker trait association (Fig. 3) was determined in TASSEL (25) with a general linear model (GLM) using the population structure from Fig. S1 to control for population-structure effects (see Materials and Methods). Adjusted P values were calculated by using 10,000 permutations to correct for multiple testing.
For At1g23390, the SSR in the promoter (2 classes were formed with <12 and ≥12 repeats) was significantly associated with FW and starch. Ten SNPs in the transcribed region showed significant associations with protein and sucrose. They were in strong LD, and 7 caused nonsynonymous changes in the coding region (285, Asn/Lys; 680, Pro/Leu; 761, Val/Gly, 775, Met/Val; 826, Glu/Lys; 977, Gly/Asp; 1074, Lys/Asn). They were also in LD with 3 SNPs in the promotor (−1084, −845, −299) that showed associations with sucrose or amino acids and an SNP in the 3′UTR (position 1380) that associated with sucrose. For At4g39800, 3 adjacent SNPs in the promoter (−786, −785, and −784) associated with rosette biomass, amino acids, and starch. Another 4 SNPs in exon/intron regions of the gene and a SNP in the downstream region (2298) associated with rosette biomass, protein, and starch. These 5 SNPs were in strong LD, and one led to a nonsynonymous change in the coding sequence (699, Asp/Asn). As a control, we performed association mapping for the 419 markers used to genotype the population (see Materials and Methods), using the same trait and structure population datasets. No significant associations were found to any trait.
The large number of polymorphisms prevented association mapping against all individual haplotypes. Instead, we built haplotypes from the polymorphisms that showed associations and led to nonsynonymous changes. For At4g39800, we used position −784 (promotor) and position 699 (Asp/Asn). There were significant associations for FW (P = 0.0065; RM2 = 4.8%), starch (P = 0.043; RM2 = 3.1%), protein (P = 0.029; RM2 = 3.1%), and amino acids (P = 0.0003; RM2 = 7%). For At1g3200, we used the SSR in the promotor and a set of 9 SNPs in strong LD in the ORF (680, 761, 775, 826, 864, 972, 977, 1026, 1074). They generated 6 haplotypes, with significant associations for FW (P = 0.008; RM2 = 7.4%), protein (P = 0.048; RM2 = 4.9%), sucrose (P = 0.0004; RM2 = 9.7%), and, at a lower level of significance, starch (P = 0.085; RM2 = 4.8%). These results indicate interactive functions for the promotor and ORF of these 2 genes. In all cases where associations were found, a given allele or haplotype displayed an opposite effect on biomass and metabolic traits.
Finally, we checked whether any of the polymorphisms in the promotor correlated with transcript levels. Genotype information was available for 20 of the accessions in which transcript levels were determined. For At4g39800/IPS1, the minor (non-Col0) alleles at −785, −784, and −410 in the promoter region were found in 6, 8, and 2 of the accessions. They correlated with higher IPS1 transcript levels (P = 0.02, 0.05, and 0.02, respectively) (Table S6). For At1g23390, no significant correlations were found at P > 0.05, but several polymorphisms in the promoter region correlated at P < 0.1.
Concluding Remarks.
There is increasing interest in the possibility of using biomarkers to predict plant biomass. Meyer and colleagues showed that biomass can be predicted by a set of low-molecular-weight metabolites (3), but their study was restricted to a single biparental RIL population and did not reveal why this set of metabolites have predictive power. We show that metabolite levels change reciprocally to biomass across a large set of genotypically diverse Arabidopsis accessions. Further, and importantly, the changes of metabolites are integrated as changes in the level of starch and, to a lesser extent, protein.
This finding has the practical advantage that starch can be easily extracted and assayed. Using robotized systems (8), we can precisely measure starch levels in 400 samples per day. This will make it possible to identify genotypes where changes in biomass production are, and are not, connected to changes in central metabolism.
It also points to a biological explanation for the negative relation between biomass and metabolites, namely, that large accessions have a modified balance between the C supply and growth, which is integrated as a change in starch levels. In agreement, profiling of C-regulated transcripts revealed coordinated changes of many C-responsive transcripts between Arabidopsis accessions, including genes involved in Tre6P and AKIN10 signaling (16, 20, 21, 26). This hypothesis-driven approach also identified 2 candidate genes, encoding a myo-inositol-1-phosphate synthase and an unusual Kelch repeat-containing F-box family protein, whose transcript levels correlate with rosette biomass. Association mapping revealed polymorphisms in these genes that are related to rosette biomass and show opposite allelic effects on metabolites, including starch and protein. It has already been shown that antisense inhibition of a homolog to IPS1/At4g39800 in potato leads to increased levels of sucrose and starch, altered leaf morphology, precocious senescence, and decreased tuber yield (27), as expected if this enzyme or its products contribute to the regulation of C partitioning and growth.
Starch is a C-storage polymer without demonstrated regulatory activities. It is more likely that regulators of starch metabolism or signals derived from starch act as integrators of plant metabolism and growth. It is intriguing that starch and protein are correlated and are predicted by the same set of metabolites. A strong correlation between starch turnover, protein content, and biomass is also found when Col0 is grown in different photoperiods (9). The conserved correlation might reflect the large energy costs associated with protein synthesis and maintenance (see ref. 9 and references therein). The TOR/RSK pathway is known to regulate ribosome numbers, protein level, and growth in response to the nutrient status in yeast and animals (28, 29), and evidence is emerging for an analogous role in plants (30). It will be interesting to investigate whether this signaling pathway contributes to the close link between starch, protein, and biomass.
Thus, multilevel metabolic and molecular phenotyping can be used to systematically identify metabolic traits and genes that correlate species-wide with growth and, combined with deep genotyping, to identify allelic variation that underlies these relationships. This work identifies candidate genes and polymorphisms that may be used directly or through the isolation of homologs to modulate biomass production in crops and provides precedence for an efficient strategy for future use to identify (crop-) species-specific lead genes.
Materials and Methods
Plant Material and Growth.
A. thaliana accessions were obtained as in ref. 13 and grown in soil as in ref. 2. They were grown in at least 2 independent experiments. Each experiment contained 3 replicates of 5 pooled plants, with full randomization in growth cabinets to avoid microenvironmental effects. Material was harvested at the end of the light period. Samples typically contained 5 rosettes (≈800 mg of FW). They were powdered in liquid N2, subaliquoted, and stored at −80 °C.
Metabolite Assays.
Analysis of total amino acids, glucose, fructose, sucrose, starch, total protein, and chlorophyll was performed as in ref. 2 and GC-MS as in ref. 31, identifying metabolites by comparison with database entries of authentic standards (32).
Design and Validation of qRT-PCR Primers, RNA Preparation, and RT-PCR Assays.
Primers were designed and synthesized at MWG Biotech AG using the PRIME program of GCG Wisconsin Package, version 10.2. Global alignments of suggested primer sequences with genomic and transcript sequences were performed using NCBI-BLASTn (33) to ensure unique oligonucleotide sequences. All primers were checked for nonspecific signals arising from primer dimers or template contamination by measuring a water control. Sequences of primers are in Table S3. RNA preparation, real time PCR, data analysis, and procedures for cDNA synthesis were as in ref. 34. Cut-off CT values for all primers were set to 35 cycles. CT values were normalized to 4 reference genes (34), At2g28390 (SAND family protein), At3g53090 (HECT-domain-containing protein), At5g08290 (yellow-leaf specific protein 8), and At5g25760 (ubiquitin-conjugating enzyme), by subtracting the average CT value of the 4 reference genes from the CT value of the gene of interest for each accession. Data were normalized based on the gene-wise average of all accessions including Col0, so that ΔΔCT represents ΔCTA minus ΔCTAv. ΔΔCT values of technical and biological replicates within one accession were averaged, if they did not differ by more than 0.8 and 1.5, respectively. Genes were excluded if <11 accessions gave valid ΔΔCT values. Of the 92 genes in set 1 and set 2, 42 and 52 were retained, respectively. Average transcript levels for all genes in the accessions are provided in Table S3.
Genotyping and Analysis of Population Structure.
Selected accessions were genotyped with 460 SNP markers: 149 framework SNPs assembled in the frame of the A. thaliana “HapMap” project (J. Borevitz, personal communication; see http://naturalvariation.org/hapmap) and 311 SNPs with intermediate allele frequency selected by Warthmann et al. (35) (Table S1). Genotyping was carried out at Sequenom Inc. Population structure was analyzed by using Structure 2.1 (14), a model-based clustering method for inferring population structure that uses genotypic data from unlinked markers and accounts for the presence of LD by introducing population structure and attempting to find population groupings that are not in disequilibrium (14). An ancestry model allows population admixture. From the 460 SNP markers, 419 were used that had <25% missing data. Allele frequencies were assumed to be correlated (i.e., allele frequencies were likely to be similar due to shared ancestry or migration). The optimal number of subpopulations was simulated by setting K (number of subpopulation) from 1 to 15. The length of burn-in period as well as Markov Chain Monte Carlo iterations (MCMC) after burn-in were set to 100,000 for each run, and each run was iterated 10 times. When K was varied from 1 to 15, the posterior probability [Ln P(D)] improved steadily until K reached 7. The smallest K value for highest posterior probability was taken as optimal, splitting the entire population into 7 panels for associations mapping (Fig. S1). Accessions were assigned to the subpopulation or group to which they showed the highest probability of membership.
Spearman Regression Analysis and Partial Correlation Analysis.
Correlations were calculated from mean data for an accession across all replicates and experiments. The original data are given in Table S2. For Spearman correlation coefficients, Microsoft Office Excel was used. For partial correlation analysis, the data for trait levels were loaded into R (36), a graphical Gaussian model was fitted (37), the R package (38) was used to obtain a robust estimate of the partial correlation, a P value was inferred, and significant correlations between traits at a local FDR of 20% were extracted and depicted as edges between traits.
PLS Regression and Cross-Validation.
PLS identifies the combinations of the original predictor variables with maximum covariance with the response (39, 40). These orthogonal combinations replace the original data matrix and are used in a multivariate ordinary least squares regression to predict the response. The optimal number of components is determined by the maximum proportion of explained variance obtained in 5-fold cross-validation. The observations are divided into 5 subsets, a training set (4 subsets) is used to build a model that is applied to predict the response of the remaining subset, and this procedure is repeated 5 times to estimate the response for all observations. Cross-validation was also applied each time to the training set. The estimated vector is correlated with the measured response to obtain a measure of the predictive power of the predictor variables.
The weight of a predictor j in the linear combination resulting in PLS component i is denoted as wij. The VIP of each predictor j gives an estimate of the importance of that predictor for the PLS prediction using the h most important orthogonal components and is calculated as the sum of the wij (i = 1,… h) multiplied by the correlation of PLS component i with the response (41).
Cartographic Representation of the Sugar-Responsive Network.
The combined network (Fig. 2) was visualized using the algorithm developed by Guimera and Amaral (19), using a threshold for significant interactions of Rs >0.7 and P < 0.01 for gene–gene interactions and Rs >0.6 and P < 0.01 for gene–metabolites and metabolite–metabolite interactions, respectively. The network was generated from log2 transformation of the average data, given for metabolites and FW in Table S1 and transcript data in Table S3.
Sequencing.
Sequencing was performed using genomic DNA amplified from 92 of the 94 accessions, corresponding to 3745 base pairs of At4g39800 and 2668 base pairs of At1g23390. DNA was isolated by using a standard protocol (37). Genomic sequencing was performed on both strands using LargeDye terminator chemistry on ABI 3730 sequencers (Applied Biosystems) by the Automatic DNA Isolation and Sequencing unit at the Max Planck Institute for Plant Breeding. The sequences were assembled by using a Sequencher 4.8 (GeneCode) (see Table S6).
Candidate Gene Association Testing.
Association mapping was performed using the sequence-verified SNPs that occurred with a minimum allele frequency of 0.05 in the entire accession panel. The GLM function of the TASSEL (25) program was applied with the STRUCTURE results used to control for population structure, with 10,000 permutations to determine significance.
Supplementary Material
Acknowledgments.
We thank Dr. Nengyi Zhang for his valuable advice about the TASSEL software. This work was supported by a Netherlands Genomics Initiative Genomics Fellowship 050-72-412, the Max Planck Society, the European Commission under the 6th Framework Programme (contract LSHG-CT-2006-037704) and the Federal Ministry of Education and Research within the German Plant Genome Initiative (Genome Analysis of the Plant Biological System, grants 0313122B and 0315060E) and the Golm Forschungseinheiten zur Systembiologie program.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the EMBL Nucleotide Sequence database (accession nos. FM998553–FM998644 for At4g39800 and FM995274–FM995365 for At1g23390).
This article contains supporting information online at www.pnas.org/cgi/content/full/0903478106/DCSupplemental.
References
- 1.Rogers A, Ainsworth EA. In: Managed Ecosystems and CO2, Case studies, Processes and Perspectives. Nösberger J, editor. Berlin: Springer Verlag; 2006. [Google Scholar]
- 2.Cross JM, et al. Variation of enzyme activities and metabolite levels in 24 arabidopsis accessions growing in carbon-limited conditions. Plant Physiol. 2006;142:1574–1588. doi: 10.1104/pp.106.086629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meyer RC, et al. The metabolic signature related to high plant growth rate in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2007;104:4759–4764. doi: 10.1073/pnas.0609709104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schauer N, et al. Comprehensive metabolic profiling and phenotyping of interspecific introgression lines for tomato improvement. Nat Biotechnol. 2006;24:447–454. doi: 10.1038/nbt1192. [DOI] [PubMed] [Google Scholar]
- 5.Schurr U, Walter A, Rascher U. Functional dynamics of plant growth and photosynthesis — from steady-state to dynamics — from homogeneity to heterogeneity. Plant Cell Environ. 2006;29:340–352. doi: 10.1111/j.1365-3040.2005.01490.x. [DOI] [PubMed] [Google Scholar]
- 6.Chatterton NJ, Silvius JE. Photosynthate partitioning into leaf starch as affected by daily photosynthetic period duration in 6 species. Physiol Plant. 1980;49:141–144. [Google Scholar]
- 7.Geiger DR, Servaites JC, Fuchs MA. Role of starch in carbon translocation and partitioning at the plant level. Aust J Plant Physiol. 2000;27:571–582. [Google Scholar]
- 8.Gibon Y, et al. Adjustment of diurnal starch turnover to short days: Depletion of sugar during the night leads to a temporary inhibition of carbohydrate utilization, accumulation of sugars and post-translational activation of ADP-glucose pyrophosphorylase in the following light period. Plant J. 2004;39:847–862. doi: 10.1111/j.1365-313X.2004.02173.x. [DOI] [PubMed] [Google Scholar]
- 9.Gibon Y, Pyl E-T, Sulpice R, Höhne M, Stitt M. Adjustment of growth, starch turnover, protein content and central metabolism to a decrease of the carbon supply when Arabidopsis is grown in very short photoperiods. Plant Cell Environ. 2009 doi: 10.1111/j.1365-3040.2009.01965.x. doi: 10.1111/j.1365-3040.2009.01965.x. [DOI] [PubMed] [Google Scholar]
- 10.Smith AM, Stitt M. Coordination of carbon supply and plant growth. Plant Cell Environ. 2007;30:1126–1149. doi: 10.1111/j.1365-3040.2007.01708.x. [DOI] [PubMed] [Google Scholar]
- 11.Stitt M, Gibon Y, Lunn JE, Piques M. Multilevel genomics analysis of carbon signalling during low carbon availability: Coordinating the supply and utilisation of carbon in a fluctuating environment. Funct Plant Biol. 2007;34:526–549. doi: 10.1071/FP06249. [DOI] [PubMed] [Google Scholar]
- 12.Caspar T, Huber SC, Somerville C. Alterations in growth, photosynthesis, and respiration in a starchless mutant of Arabidopsis-thaliana (L) deficient in chloroplast phosphoglucomutase activity. Plant Physiol. 1985;79:11–17. doi: 10.1104/pp.79.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sulpice R, et al. Description and applications of a rapid and sensitive non-radioactive microplate-based assay for maximum and initial activity of d-ribulose-1,5-bisphosphate carboxylase/oxygenase. Plant Cell Environ. 2007;30:1163–1175. doi: 10.1111/j.1365-3040.2007.01679.x. [DOI] [PubMed] [Google Scholar]
- 14.Pritchard JK, Stephens M, Rosenberg NA, Donnelly P. Association mapping in structured populations. Am J Hum Genet. 2000;67:170–181. doi: 10.1086/302959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blasing OE, et al. Sugars and circadian regulation make major contributions to the global regulation of diurnal gene expression in Arabidopsis. Plant Cell. 2005;17:3257–3281. doi: 10.1105/tpc.105.035261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kolbe A, Tiessen A, Schluepmann H, Paul M, Ulrich S, Geigenberger P. Trehalose 6-phosphate regulates starch synthesis via posttranslational redox activation of ADP-glucose pyrophosphorylase. Proc Natl Acad Sci USA. 2005;102:11118–11123. doi: 10.1073/pnas.0503410102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Usadel B, et al. Global transcript levels respond to small changes of the carbon status during a progressive exhaustion of carbohydrates in Arabidopsis rosettes. Plant Physiol. 2008;146:1834–1861. doi: 10.1104/pp.107.115592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Osuna D, et al. Temporal responses of transcripts, enzyme activities and metabolites after adding sucrose to carbon-deprived Arabidopsis seedlings. Plant J. 2007;49:463–491. doi: 10.1111/j.1365-313X.2006.02979.x. [DOI] [PubMed] [Google Scholar]
- 19.Guimera R, Amaral LAN. Functional cartography of complex metabolic networks. Nature. 2005;433:895–900. doi: 10.1038/nature03288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baena-Gonzalez E, Rolland F, Thevelein JM, Sheen J. A central integrator of transcription networks in plant stress and energy signalling. Nature. 2007;448:938–U910. doi: 10.1038/nature06069. [DOI] [PubMed] [Google Scholar]
- 21.Schluepmann H, Pellny T, van Dijken A, Smeekens S, Paul M. Trehalose 6-phosphate is indispensable for carbohydrate utilization and growth in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2003;100:6849–6854. doi: 10.1073/pnas.1132018100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thompson AR, Vierstra RD. Autophagic recycling: Lessons from yeast help define the process in plants. Curr Opin Plant Biol. 2005;8:165–173. doi: 10.1016/j.pbi.2005.01.013. [DOI] [PubMed] [Google Scholar]
- 23.van Berloo R. GGT 2.0: Versatile software for visualization and analysis of genetic data. J Hered. 2008;99:232–236. doi: 10.1093/jhered/esm109. [DOI] [PubMed] [Google Scholar]
- 24.Clark RM, et al. Common sequence polymorphisms shaping genetic diversity in Arabidopsis thaliana. Science. 2007;317:338–342. doi: 10.1126/science.1138632. [DOI] [PubMed] [Google Scholar]
- 25.Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics. 2007;23:2633–2635. doi: 10.1093/bioinformatics/btm308. [DOI] [PubMed] [Google Scholar]
- 26.Lunn JE, et al. Sugar-induced increases in trehalose 6-phosphate are correlated with redox activation of ADPglucose pyrophosphorylase and higher rates of starch synthesis in Arabidopsis thaliana. Biochem J. 2006;397:139–148. doi: 10.1042/BJ20060083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keller R, Brearley CA, Trethewey RN, Muller-Rober B. Reduced inositol content and altered morphology in transgenic potato plants inhibited for 1D-myo-inositol 3-phosphate synthase. Plant J. 1998;16:403–410. [Google Scholar]
- 28.Edgar BA. How flies get their size: Genetics meets physiology. Nat Rev Genet. 2006;7:907–916. doi: 10.1038/nrg1989. [DOI] [PubMed] [Google Scholar]
- 29.Zaman S, Lippman SI, Zhao X, Broach JR. How Saccharomyces responds to nutrients. Annu Rev Genet. 2008;42:27–81. doi: 10.1146/annurev.genet.41.110306.130206. [DOI] [PubMed] [Google Scholar]
- 30.Deprost D, et al. The Arabidopsis TOR kinase links plant growth, yield, stress resistance and mRNA translation. Embo Reports. 2007;8:864–870. doi: 10.1038/sj.embor.7401043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lisec J, Schauer N, Kopka J, Willmitzer L, Fernie AR. Gas chromatography mass spectrometry-based metabolite profiling in plants. Nat Protoc. 2006;1:387–396. doi: 10.1038/nprot.2006.59. [DOI] [PubMed] [Google Scholar]
- 32.Schauer N, et al. GC-MS libraries for the rapid identification of metabolites in complex biological samples. FEBS Lett. 2005;579:1332–1337. doi: 10.1016/j.febslet.2005.01.029. [DOI] [PubMed] [Google Scholar]
- 33.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 34.Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 2005;139:5–17. doi: 10.1104/pp.105.063743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Warthmann N, Fitz J, Weigel D. MSQT for choosing SNP assays from multiple DNA alignments. Bioinformatics. 2007;23:2784–2787. doi: 10.1093/bioinformatics/btm428. [DOI] [PubMed] [Google Scholar]
- 36.R Development Core Team. Vienna: R Foundation for Statistical Computing; 2008. R: A language and environment for statistical computing. [Google Scholar]
- 37.Opgen-Rhein R, Strimmer K. From correlation to causation networks: A simple approximate learning algorithm and its application to high-dimensional plant gene expression data. Bmc Systems Biology. 2007;1:37. doi: 10.1186/1752-0509-1-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schafer J, Strimmer K. An empirical Bayes approach to inferring large-scale gene association networks. Bioinformatics. 2005;21:754–764. doi: 10.1093/bioinformatics/bti062. [DOI] [PubMed] [Google Scholar]
- 39.Wold H. Soft Modelling by Latent Variables. London: Academic; 1975. [Google Scholar]
- 40.Barker M, Rayens W. Partial least squares for discrimination. J Chemom. 2003;17:166–173. [Google Scholar]
- 41.Chong I-G, Jun C-H. Performance of some variable selection methods when multicollinearity is present. Chemometrics Intellig Lab Syst. 2005;78:103–112. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.