Abstract
Buforin II is a 21-aa potent antimicrobial peptide that forms, in a hydrophobic medium, an amphipathic structure consisting of an N-terminal random coil region (residues 1–4), an extended helical region (residues 5–10), a hinge (residue 11), and a C-terminal regular α-helical region (residues 12–21). To elucidate the structural features of buforin II that are required for its potent antimicrobial activity, we synthesized a series of N- and C-terminally truncated or amino acid-substituted synthetic buforin II analogs and examined their antimicrobial activity and mechanism of action. Deletion of the N-terminal random coil region increased the antibacterial activity ≈2-fold, but further N-terminal truncation yielded peptide analogs with progressively decreasing activity. Removal of four amino acids from the C-terminal end of buforin II resulted in a complete loss of antimicrobial activity. The substitution of leucine for the proline hinge decreased significantly the antimicrobial activity. Confocal fluorescence microscopic studies showed that buforin II analogs with a proline hinge penetrated the cell membrane without permeabilization and accumulated in the cytoplasm. However, removal of the proline hinge abrogated the ability of the peptide to enter cells, and buforin II analogs without a proline hinge localized on the cell surface, permeabilizing the cell membrane. In addition, the cell-penetrating efficiency of buforin II and its truncated analogs, which depended on the α-helical content of the peptides, correlated linearly with their antimicrobial potency. Our results demonstrate clearly that the proline hinge is responsible for the cell-penetrating ability of buforin II, and the cell-penetrating efficiency determines the antimicrobial potency of the peptide.
In addition to the highly specific cell-mediated immune response, vertebrates and other organisms have a defense system made up of distinct groups of broad-spectrum antimicrobial peptides (1). One major group of such peptides includes short linear polypeptides (≈40 aa or less) that have been isolated from diverse species such as insects and mammals (1, 2). The largest family includes those polypeptides that are positively charged and that adopt an amphipathic α-helical structure. Well-known examples of α-helical peptides are the cecropins of insects and mammals (1) and histatins from human saliva (3). In amphibians, which are rich in antimicrobial peptides, many amphipathic α-helical antimicrobial peptides [such as magainins (4), bombinins (5), buforins (6), and dermaseptin (7)] have been isolated from glands in the skin and gastrointestinal tract. These cationic α-helical peptides possess a broad range of antimicrobial activity against Gram-positive and Gram-negative bacteria and fungi, as well as protozoa (8–10). The precise mechanism of the broad-spectrum antimicrobial activity of these peptides is not yet fully understood. However, data revealed that these peptides attack the outer and inner membranes of bacteria, ultimately resulting in either disruption of the cell membrane (11) or cooperative permeabilization (12).
A 39-aa peptide, buforin I, was isolated from the stomach tissue of the Asian toad Bufo bufo garagrizans, and a more potent antimicrobial peptide of 21 amino acids, called buforin II, was derived from buforin I (6). Buforin II shows much stronger antimicrobial activity against a broad spectrum of microorganisms compared with other α-helical antimicrobial peptides (6). Interestingly, the action mechanism of buforin II has been suggested to be different from those of membrane-acting peptides, although the physicochemical properties of the peptide are similar to those of other α-helical peptides (13). Buforin II kills bacteria without cell lysis and has a strong affinity for DNA and RNA (13), suggesting the possibility that the target of buforin II is the intracellular nucleic acids, not the cell membranes. Buforin II has a helix-hinge-helix structure; the N-terminal extended helix includes residues 5 to 10, and the C-terminal helix includes residues 12 to 21. The helices are separated by a proline residue situated at amino acid position 11 (14). This two-helix organization and unique mode of action make buforin II a highly attractive candidate for deciphering the role of each structural element in conferring on buforin II its highly potent antimicrobial activity. In addition, structure-activity studies of buforin II may provide new insights into its unique molecular mechanism of antimicrobial action. In this study, we designed a series of structurally altered synthetic buforin II and determined their antimicrobial potencies and secondary structures. The mechanisms of bacterial killing action for buforin II analogs are also studied.
Materials and Methods
Design of Buforin II Analogs.
A series of truncated, amino acid-substituted, and laboratory-designed synthetic analogs of buforin II was synthesized to investigate the role of each structural element of buforin II in conferring highly potent antimicrobial activity. The ribbon model of buforin II is presented in Fig. 1, and the sequences of the peptides and their designations are given in Table 1. Buforin II was truncated progressively from its N-terminal random coil region to residues 5 to 21, 6 to 21, 7 to 21, 8 to 21, 9 to 21, 10 to 21, and 11 to 21 to evaluate the contribution of the N-terminal random coil and extended helical region to its antimicrobial activity. To examine the role of the C-terminal regular helix, C-terminally truncated analogs of buforin II corresponding to residues 1 to 17, 5 to 20, 5 to 19, and 5 to 18 of buforin II were synthesized. The importance of the proline hinge was investigated by using the peptide [L7]BUF (5–21), in which L was substituted for P-7 of BUF (5–21). However, removal of the proline hinge, which forms a kink in the structure, reduced the overall amphipathic structure of buforin II. To eliminate the unfavorable effect on the antimicrobial activity caused by the reduced amphipathicity from substituting leucine for proline, we synthesized the peptide [K2][K6][L7]BUF (5–21), in which L was substituted for P-7, and A-2 and F-6 were replaced with K to maintain its amphipathic structure. Designed synthetic buforin II analogs consisting of five repeats of the C-terminal regular α-helical motif RLLR or three repeats of RVHRLLR were also synthesized to evaluate the importance of the C-terminal amphipathic helix. In addition, to study the role of the extended α-helical region BUF (5–13) containing a proline hinge, we synthesized two hybrid peptides, BUF (5–13)[RLLR]3 and BUF (5–13)MG (1–14). BUF (5–13)[RLLR]3 contained BUF (5–13) and three repeats of the regular α-helical motif RLLR. BUF (5–13)MG (1–14) consisted of BUF (5–13) and the N-terminal α-helix [MG (1–14)] of magainin II.
Figure 1.
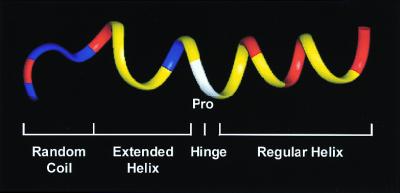
Ribbon-model representation of the backbone structure of buforin II in 50% trifluoroethanol. The N-terminal random coil, the extended helix, the hinge, and the C-terminal regular helix form an overall amphipathic structure. The amino acid residues are colored as follows: positively charged residues, red; other hydrophilic residues, blue; proline, white; other hydrophobic residues, yellow.
Table 1.
Amino acid sequences of buforin (BUF) II analogs
| Peptides | Amino acid sequences |
|---|---|
| Truncations | |
| BUF II | TRSSRAGLQFPVGRVHRLLRK |
| BUF(5–21) | RAGLQFPVGRVHRLLRK |
| BUF(6–21) | AGLQFPVGRVHRLLRK |
| BUF(7–21) | GLQFPVGRVHRLLRK |
| BUF(8–21) | LQFPVGRVHRLLRK |
| BUF(9–21) | QFPVGRVHRLLRK |
| BUF(10–21) | FPVGRVHRLLRK |
| BUF(11–21) | PVGRVHRLLRK |
| BUF(1–17) | TRSSRAGLQFPVGRVHR |
| BUF(5–20) | RAGLQFPVGRVHRLLR |
| BUF(5–19) | RAGLQFPVGRVHRLL |
| BUF(5–18) | RAGLQFPVGRVHRL |
| Amino acid substitutions* | |
| [L7]BUF(5–21) | RAGLQFLVGRVHRLLRK |
| [K2][K6][L7]BUF(5–21) | RKGLQKLVGRVHRLLRK |
| [RVHRLLR]3 | RVHRLLRRVHRLLRRVHRLLR |
| [RLLR]5 | RLLRRLLRRLLRRLLRRLLR |
| Hybridizations | |
| BUF(5–13)[RLLR]3 | RAGLQFPVGRLLRRLLRRLLR |
| BUF(5–13)MG(1–14) | RAGLQFPVGGIGKFLHSAKKFGK |
Amino acid substitutions are indicated by the single-letter amino acid designation and number of the substituted residue in brackets.
Peptide Preparation.
All peptides, with either free or biotinylated amino termini, were synthesized by Fmoc [N-(9-fluorenyl) methoxycarbonyl] chemistry by the Analytical Research Group at Korea Basic Science Institute by using a MilliGen 9050 (Millipore) peptide synthesizer. The synthetic peptides were purified (>95% homogeneity) by reverse-phase HPLC on a C18 column (3.9 × 300 mm, Delta Pak, Millipore) by using a linear gradient of 25 to 80% acetonitrile in 0.1% trifluoroacetic acid for 40 min. All peptides were characterized by matrix-assisted laser desorption ionization mass spectroscopy (MALDI II; Kratos Analytical Instruments), with the peptide content of lyophilized samples determined by quantitative amino acid analysis with a Pico-tag analysis system on a Beckman 121 MB amino acid analyzer (Beckman Coulter).
Antimicrobial Assay.
Antimicrobial activity of the peptides against 11 selected organisms, including Gram-positive and Gram-negative bacteria and fungi, was determined by the broth microdilution assay (15). Briefly, single colonies of bacteria and fungi were inoculated into culture medium (3% trypticase soy broth and Sabouraud's medium, respectively) and cultured overnight at 37°C (or 30°C for fungi). An aliquot of this culture was transferred to 50 ml of fresh medium and incubated for an additional 3 to 6 h at 37°C (bacteria) or 30°C (fungi) to obtain the cells in midlogarithmic phase. The cells in midlogarithmic phase were harvested by centrifugation, washed with 10 mM sodium phosphate buffer (NAPB), pH 7.4, and resuspended in 10 ml of the same buffer. The colony-forming units (cfus) per milliliter were quantitated by spreading serial dilutions of the cell suspension onto three separate trypticase agar plates (Sabouraud's agar plates for fungi). A 2-fold dilution series of peptides in 10 mM NAPB was prepared, and serial dilutions (50 μl) were added to 50 μl of 5 × 104 cfu in static 96-well microtiter plates (Corning). After incubation for 3 h at 37°C (bacteria) or 30°C (fungi), fresh medium was added to the mixture and incubated at 37°C or 30°C for 16 h. The inhibition of growth was determined by measuring absorbance at 620 nm with a Model 550 Microplate Reader (Bio-Rad). The lowest concentration of peptide that completely inhibited growth of the organisms was defined as the minimal inhibitory concentration (MIC). The MICs were the average values obtained in triplicates on three independent measurements.
CD.
CD experiments were performed by using a Jasco 720 spectropolarimeter (Jasco, Tokyo) to determine the secondary structure of buforin II and its analogs. The spectra were measured between 200 and 250 nm either in the presence or absence of 50% (vol/vol) trifluoroethanol in 50 mM NAPB. Five consecutive scans per sample were performed in a 1-mm cell at 25°C (16). The helicity of the peptides was determined from the mean residue helicity at 222 nm (17). All data are the mean of three independent measurements, which did not deviate more than 5%.
Confocal Laser-Scanning Microscopy.
Escherichia coli cells in midlogarithmic phase were prepared as described in the antimicrobial assay section of Materials and Methods. E. coli cells (105 cfu) in 10 mM NAPB were incubated with biotin-labeled peptides at 37°C for 30 min. After incubation, cells were washed with 10 mM NAPB and immobilized on a glass slide, as described by Park et al. (13). The cells were then treated briefly with 0.2% Triton X-100/NAPB. The biotin-labeled peptides were visualized with 20 μg/ml streptavidin-FITC (Boehringer Mannheim) and observed with a Carl Zeiss LSM 410 laser-scanning confocal microscope. Fluorescent images were obtained with a 488-nm bandpass filter for excitation of FITC. Software merging of images was carried out by using a comos software (Zeiss).
FITC Labeling of Peptides.
Peptides were labeled with FITC essentially as described by Lane and colleague (18). In brief, FITC was freshly dissolved in methyloxysulfate to 1 mg/ml, and 700 μl of FITC solution was added to 100 μl of a solution of peptides (2 mg/ml) in 50 mM potassium phosphate buffer (final pH 7.4) to give a final concentration of 25 μg/ml. The calculated molar ratio of FITC to peptide was 0.1. After incubation for 16 h in the dark at 4°C, 500 μl of 50 mM NH4Cl was added to inactivate the residual FITC. The solution was incubated in the dark for an additional 2 h at 4°C and stored in aliquots at −20°C. The FITC-labeled peptides were purified by reverse-phase HPLC on a C18 column (3.9 × 300 mm, Delta Pak, Millipore) to give final products that were ≥95% pure.
FACS Analysis.
The influx of propidium iodide (PI), a DNA-staining fluorescent probe, and FITC-labeled peptides into bacterial cells was investigated by using a dual laser fluorescence-activated cell sorter (FACS, Beckton Dickinson). The results were analyzed on a MacIntosh computer by using the software package cellquest, provided by Beckton Dickinson.
Results
Antimicrobial Activity of Buforin II Analogs.
Deletion of the N-terminal random coil region (residues 1 to 4) of buforin II increased the antibacterial activity ≈2-fold but did not affect the antifungal activity. Further N-terminal truncation of the peptide chain to residues 6 to 21, 7 to 21, 8 to 21, 9 to 21, 10 to 21, and 11 to 21 decreased progressively both the antibacterial and antifungal activities (Table 2). On the other hand, removal of four amino acids from the C terminus, which produced a peptide of 17 amino acids (residues 1 to 17), resulted in a complete loss of antimicrobial activity (Table 2). In addition, sequential removal of individual amino acids from the C terminus of BUF (5–21) decreased the antimicrobial activity more dramatically than sequential amino acid truncation from the N terminus. These results indicate that the C-terminal helical region (residues 18 to 21) is important for antimicrobial activity, whereas the N-terminal random coil region of buforin II does not play a significant role in its antimicrobial activity. In fact, the N-terminal random coil region decreased the antimicrobial activity of buforin II. The notion that the C-terminal regular α-helix region is important for antimicrobial activity was further strengthened by the finding that [RLLR]5, a synthetic buforin II analog consisting of five repeats of the C-terminal sequence motif RLLR, had potent antimicrobial activity (Table 3).
Table 2.
Antimicrobial activities of buforin II and its truncated analogs
| Microorganism | Minimal inhibitory concentration, μg/ml
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BUF II | BUF (5–21) | BUF (6–21) | BUF (7–21) | BUF (8–21) | BUF (9–21) | BUF (10–21) | BUF (11–21) | BUF (1–17) | BUF (5–20) | BUF (5–19) | BUF (5–18) | |
| Gram-positive bacteria | ||||||||||||
| Bacillus subtilis | 2 | 1 | 4 | 4 | 8 | 16 | 32 | 32 | >256 | 16 | 64 | 128 |
| Staphylococcus aureus | 4 | 2 | 8 | 8 | 16 | 64 | 32 | 64 | >256 | 64 | 128 | 256 |
| Streptococcus mutans | 2 | 1 | 4 | 4 | 8 | 32 | 32 | 32 | >256 | 32 | 64 | 256 |
| Streptococcus pneumoniae | 4 | 2 | 4 | 4 | 16 | 16 | 32 | 64 | >256 | 32 | 128 | 128 |
| Gram-negative bacteria | ||||||||||||
| Escherichia coli | 4 | 2 | 4 | 4 | 8 | 32 | 32 | 32 | >256 | 16 | 64 | 256 |
| Salmonella typhimurium | 1 | 1 | 2 | 2 | 4 | 16 | 16 | 32 | >256 | 16 | 32 | 128 |
| Serratia marcescens | 4 | 2 | 2 | 2 | 8 | 32 | 32 | 64 | >256 | 32 | 64 | 256 |
| Pseudomonas putida | 2 | 1 | 4 | 4 | 16 | 16 | 64 | 64 | >256 | 32 | 64 | 256 |
| Fungi | ||||||||||||
| Candida albicans | 1 | 1 | 8 | 8 | 32 | 64 | 32 | 64 | >256 | 64 | >256 | >256 |
| Cryptococcus neoformans | 1 | 1 | 8 | 8 | 64 | 64 | 64 | 128 | >256 | 64 | 128 | 256 |
| Saccharomyces cerevisiae | 1 | 1 | 8 | 8 | 32 | 64 | 64 | 128 | >256 | 128 | >256 | >256 |
Minimal inhibitory concentrations were the average values obtained in triplicates on three independent measurements.
Table 3.
Antimicrobial activities of buforin II, substituted analogs, designed synthetic analogs, and magainin II
| Microorganism | Minimal inhibitory concentration, μg/ml
|
|||||||
|---|---|---|---|---|---|---|---|---|
| BUF II | [L7] BUF(5–21) | [K2][K6][L7] BUF(5–21) | [RLLR]5 | [RVHRLLR]3 | BUF(5–13) [RLLR]3 | BUF(5–13) MG(1–14) | Magainin II | |
| Gram-positive bacteria | ||||||||
| Bacillus subtilis | 2 | 32 | 16 | 2 | 8 | 1 | 16 | 50 |
| Staphylococcus aureus | 4 | 32 | 16 | 1 | 64 | 1 | 16 | 50 |
| Streptococcus mutans | 2 | 32 | 32 | 2 | 32 | 0.5 | 16 | 100 |
| Streptococcus pneumoniae | 4 | 32 | 16 | 2 | 128 | 1 | 16 | 50 |
| Gram-negative bacteria | ||||||||
| Escherichia coli | 4 | 32 | 16 | 2 | 128 | 1 | 16 | 100 |
| Salmonella typhimurium | 1 | 16 | 4 | 1 | 4 | 1 | 8 | 25 |
| Serratia marcescens | 4 | 64 | 32 | 2 | 64 | 1 | 16 | 50 |
| Pseudomonas putida | 2 | 16 | 16 | 2 | 64 | 2 | 32 | 50 |
| Fungi | ||||||||
| Candida albicans | 1 | 16 | 16 | 8 | >256 | 2 | 4 | 25 |
| Cryptococcus neoformans | 1 | 16 | 8 | 8 | 128 | 1 | 4 | 12 |
| Saccharomyces cerevisiae | 1 | 16 | 4 | 8 | >256 | 4 | 4 | 25 |
Minimal inhibitory concentrations were the average values obtained in triplicates on three independent measurements.
We next investigated the role of the proline hinge. The peptide [L7]BUF (5–21), which was made by substituting L for P-7 of BUF (5–21), showed much weaker antimicrobial activity than did BUF (5–21). The unfavorable effect on antimicrobial activity appeared not to be caused by the reduced amphipathicity of [L7]BUF (5–21), as the peptide [K2][K6][L7]BUF (5–21), in which L was substituted for P-7, and A-2 and F-6 were replaced with K to maintain an amphipathic structure, also showed reduced activity. This result suggests that the proline hinge is important for the antimicrobial activity of buforin II (Table 3). All these results led us to synthesize a hybrid peptide BUF (5–13)[RLLR]3, which contains both the extended α-helical region containing a proline hinge, BUF (5–13), and three repeats of the regular α-helical motif RLLR. The hybrid peptide BUF (5–13)[RLLR]3 showed stronger antimicrobial activity than did BUF (5–21) (Table 3).
Relationship Between the α-Helical Content of Truncated Buforin II Analogs and Their Antimicrobial Activity.
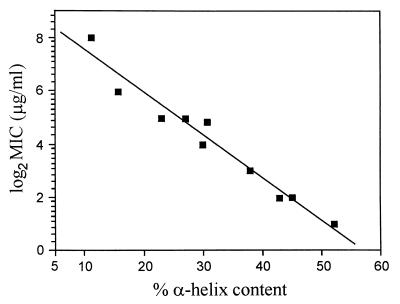
An increase in the antimicrobial activity of a linear α-helical antimicrobial peptide has been shown in several cases to correlate closely with an increase in α-helical secondary structure (19, 20). To examine the contribution of α-helical secondary structure to the antimicrobial activity of buforin II, CD spectra of buforin II and its analogs were obtained. Table 4 shows the α-helical content of buforin II analogs, as calculated by the method of Chen et al. (17). When the peptides were dissolved in aqueous buffer, no α-helicity was observed. In the presence of 50% trifluoroethanol, however, the peptides displayed 0 to 82% α-helical content. BUF (5–21) had a higher α-helical content (52%) than did the parent buforin II peptide (43%). The peptide analogs truncated sequentially from the N terminus of BUF (5–21) showed progressive decreases in α-helical content, and BUF (1–17) was completely devoid of the α-helical structure. The α-helical content of N- and C-terminally truncated buforin II analogs correlated linearly with their antimicrobial activities: the higher the α-helical content of the analogs, the stronger the antimicrobial activity (Fig. 2). However, the close linear relationship among proline hinge, α-helical content, and antimicrobial activity was observed with the truncated buforin II analogs containing a proline hinge but not with the synthetic and substituted peptides lacking the proline hinge (Table 4).
Table 4.
Structural data from CD of buforin II and its analogs
| Peptide | % α-helix
|
Peptide | % α-helix
|
||
|---|---|---|---|---|---|
| 50 mM NAPB* | 50% TFE† in 50 mM NAPB | 50 mM NAPB* | 50% TFE† in 50 mM NAPB | ||
| BUF II | 0 | 43 | BUF(5–20) | 0 | 30 |
| BUF(5–21) | 0 | 52 | BUF(5–19) | 0 | 15 |
| BUF(6–21) | 0 | 45 | BUF(5–18) | 0 | 12 |
| BUF(7–21) | 0 | 43 | [L7]BUF(5–21) | 0 | 57 |
| BUF(8–21) | 0 | 38 | [K2][K6][L7BUF(5–21) | 0 | 64 |
| BUF(9–21) | 0 | 32 | [RLLR]5 | 0 | 82 |
| BUF(10–21) | 0 | 27 | [RVHRLLR]3 | 0 | 80 |
| BUF(11–21) | 0 | 23 | BUF(5–13)[RLLR]3 | 0 | 55 |
| BUF(1–17) | 0 | 0 | BUF(5–13)MG(1–14) | 0 | 36 |
NAPB, sodium phosphate buffer, pH 7.4.
TFE, trifluoroethanol.
Figure 2.
Linear correlation between the MICs and the α-helical contents of the truncated buforin II analogs. The linear regression line is shown.
Confocal Laser-Scanning Microscopy.
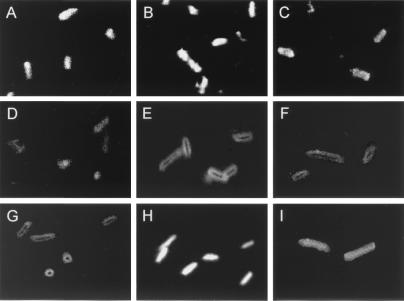
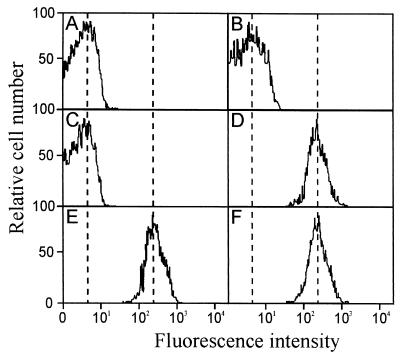
The importance of the proline hinge for the antimicrobial activity of buforin II prompted us to investigate further the antimicrobial mechanism of buforin II. To monitor the site of action of the peptides in E. coli, biotin-labeled buforin II analogs and magainin II were incubated with E. coli, and their localization within the bacterial cells was visualized by using streptavidin-FITC. At a concentration of 16 μg/ml, biotinylated BUF (5–21), BUF (8–21), and BUF (5–20) penetrated the bacterial cell membrane and accumulated in the cytoplasm of the cell, as is the case with biotinylated buforin II (Fig. 3). On the other hand, biotinylated [K2][K6][L7]BUF (5–21) and [RLLR]5 did not penetrate the cell membrane but remained associated with the cell membrane. The behavior of [K2][K6][L7]BUF (5–21) and [RLLR]5 is similar to that of magainin II, which is a membrane-permeabilizing α-helical antimicrobial peptide. Further support for these two different modes of action came from membrane integrity studies. Buforin II and its N- and C-terminally truncated analogs did not induce the influx of PI into E. coli cells, indicating nonmembrane permeabilizing killing action, whereas [K2][K6][L7]BUF (5–21), [RLLR]5, and magainin II did induce the influx of PI, which is indicative of cell membrane permeabilization (Fig. 4). Hybrid peptides BUF (5–13)[RLLR]3 and BUF (5–13)MG (1–14), in which the extended helix containing the proline hinge [BUF (5–13)] was fused to the noncell-penetrating peptide ([RLLR]3) and the N-terminal helix of magainin II [MG (1–14)], respectively, penetrated efficiently the bacterial cell membrane (Fig. 3). These results show clearly that the extended α-helical region, BUF (5–13), provided the noncell-penetrating peptides with the cell-penetrating ability.
Figure 3.
Confocal fluorescence microscopic images of E. coli cells. E. coli cells were treated with 16 μg/ml of biotinylated peptides at 37°C for 30 min and visualized with streptavidin-FITC. Microscopic pictures show the localization of the biotinylated peptides. Biotinylated buforin II (A), BUF (5–21) (B), BUF (8–21) (C), BUF (5–20) (D), BUF (5–13)[RLLR]3 (H), and BUF (5–13)MG (1–14) (I) penetrated the cell membrane and accumulated in the cytopalsm. Biotinylated [K2][K6][L7]BUF (5–21) (E), [RLLR]5 (F), and magainin II (G) remained on the cell surface.
Figure 4.
PI labeling of E. coli cells treated with magainin II or buforin II analogs. E. coli cells in midlogarithmic phase were washed with 10 mM NAPB and resuspended in the same buffer. E. coli cells (104 cfu) in 10 mM NAPB were incubated for 30 min at 25°C with 4 μg/ml of buforin II (A), 8 μg/ml of BUF (8–21) (B), 16 μg/ml of BUF (5–20) (C), 16 μg/ml of [K2][K6][L7]BUF (5–21) (D), 2 μg/ml of [RLLR]5 (E), or 100 μg/ml of the control peptide magainin II (F) in the presence of 9 μM of PI. The distribution of cells according to relative fluorescence intensities is shown.
We also examined the interactions of the buforin II analogs with the cell membrane of Bacillus subtilis, a Gram-positive bacterium, and obtained the same results as with E. coli (data not shown).
Relationship Between the α-Helical Content of N- and C-Terminally Truncated Buforin II Analogs and Their Cell-Penetrating Efficiency.
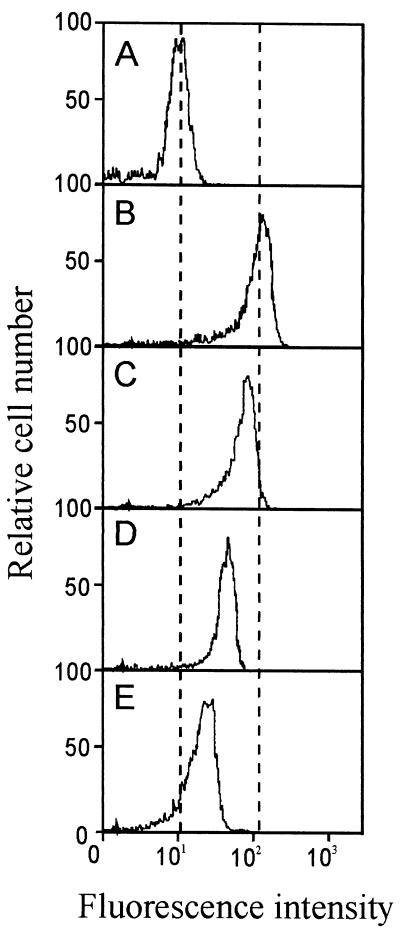
When an equal amount of the truncated buforin II peptide analogs was applied to E. coli, the cell-penetrating efficiency of the peptides varied depending on their α-helical content (Fig. 3). This phenomenon was further investigated by flow cytometric analysis. To evaluate the cell-penetrating efficiency of N- and C-terminally truncated buforin II analogs, we labeled the peptides with FITC and examined their influx into bacterial cells by using FACS analysis. Fig. 5 shows the results of the FACS analysis of cells incubated with FITC-labeled buforin II, BUF (5–21), BUF (8–21), and BUF (5–20) (16 μg/ml each). When E. coli cells (104 cfu) were treated with each of the FITC-labeled peptides (16 μg/ml each), the fluorescence intensity of the treated cells increased in accordance with the α-helical content of the treated FITC-labeled peptides, that is, the peptides with higher α-helical content penetrated the cells more efficiently, resulting in higher antimicrobial activity. This result suggests that the α-helical content of truncated buforin II analogs determines the cell-penetrating efficiency of the peptides and their antimicrobial activity.
Figure 5.
FACS analysis of E. coli cells treated with FITC-labeled buforin II analogs. E. coli cells in midlogarithmic phase were washed with 10 mM NAPB and resuspended in the same buffer. E. coli cells (104 cfu) in 10 mM NAPB were incubated for 30 min at 25°C with 10 mM NAPB (control) (A), 16 μg/ml of FITC-BUF (5–21) (B), FITC-buforin II (C), FITC-BUF (8–21) (D), or FITC-BUF (5–20) (E).
Discussion
The structure–activity relation of an antimicrobial peptide buforin II was studied to understand the structural requirements for its antimicrobial activity and the mechanism of bacterial killing action. The N-terminal random structure (residues 1 to 4) of buforin II does not contribute to its antimicrobial activity. In fact, the deletion of the random structure of buforin II increases the antibacterial activity ≈2-fold while increasing the α-helical content from 43 to 52%. However, the C-terminal α-helical region of buforin II is very critical for the antimicrobial activity. Even a single amino acid deletion from the C terminus of buforin II reduces dramatically the antimicrobial activity, and the deletion of four amino acids (residues 18 to 21) from the C terminus results in a complete loss of the antimicrobial activity and α-helical content. The C-terminal region seems to contribute to the antimicrobial activity of buforin II by providing an amphipathic stable α-helical structure.
Buforin II belongs to a class of linear α-helical antimicrobial peptides (2, 4, 5). However, the mechanism of killing action of buforin II is quite different from that of other linear α-helical peptides. Buforin II and its truncated analogs containing a proline hinge penetrate the cell membrane and kill microorganisms within a few minutes (13). Even though buforin II and its truncated analogs penetrate the cell membrane, they do not permeabilize it, as confirmed by FACS analysis of PI influx into cells. In contrast to buforin II and its truncated analogs containing a proline hinge in their structures, analogs [K2][K6][L7]BUF (5–21) and [RLLR]5, which do not have the proline hinge, function on the cell membrane, permeabilizing it, which is similar to the mechanism of action of magainin II, the well-known membrane permeabilizing antimicrobial peptide (13). The cell-penetrating efficiency of buforin II and its truncated analogs depends on their α-helical contents. It seems that α-helical content plays an important role in efficient translocation of buforin II and its truncated analogs. Therefore, it is clear that the proline hinge is definitely responsible for the cell-penetrating ability of buforin II and its truncated analogs, and the α-helical contents of the peptides determine the cell-penetrating efficiency, a key factor determining the antimicrobial activity. Matsuzaki (20) reported that proline-free magainin translocated across lipid bilayers as a model membrane system coupled with pore formation and lipid flip-flop. However, the proline-free magainin remains associated with the inner leaflet of the lipid bilayer after translocation of the artificial membrane (20), whereas buforin II penetrates the bacterial cell membrane and accumulates in the cytoplasm. Magainin tends to remain on bacterial cell membranes (Fig. 3) because its translocation efficiency (21), as well as its affinity for DNA and RNA (13), is very low. We found that F10W-buforin II, in which F at position 10 was replaced with W, crosses phosphatidylglycerol-based membranes much more effectively than F5W-magainin II (21). Interestingly, the buforin peptide induced neither membrane permeabilization nor lipid flip-flop on translocation of the artificial membrane (21), indicating that buforin has a new translocation mechanism. Furthermore, P11A substitution of buforin II, in which P at the position 11 was replaced with A, changed the mode of action to a magainin-type one (21). It is interesting to note that a substitution of a single amino acid for proline produces an antimicrobial peptide with a totally different mechanism of cell-killing action. The substitution of A for P of buforin II changed the properties of buforin II to a magainin-like one in terms of conformation, translocation efficiency, and leakage-coupled lipid flip-flop in lipid bilayer (21).
The actual role of the proline hinge in making buforin II penetrate the cell membrane remains to be studied. However, the distorted helical conformation of buforin II (residues 5–13) caused by proline may be related to efficient translocation. Buforin II conforms to a bent helix with the flexible N-terminal region in trifluoroethanol/buffer and probably in lipid bilayer (14). The unusual helical conformation of the proline-hinge region (residues 5–13) may provide a flexibility that is related to efficient translocation via an unknown mechanism caused by the distorted helical conformation with a larger diameter and a larger pitch. This proposed hypothesis was confirmed by the observation that when the proline-hinge region of BUF (5–13) was fused to the noncell-penetrating peptide [RLLR]3 and the N-terminal helix of magainin II [MG (1–14)], respectively, both hybrid peptides BUF (5–13)[RLLR]3 and BUF (5–13)MG (1–14) penetrated easily the bacterial cell membrane and accumulated in the cell cytoplasm with strong antimicrobial activity (Fig. 3). These hybrid peptides did not permeabilize the membrane, so that the influx of PI into cells was not detected by the FACS analysis (data not shown). We are currently investigating the detailed translocation mechanism of buforin II by using artificial membranes.
In summary, our results demonstrate clearly that buforins constitute a new class of antimicrobial peptides that target intracellular substances, most probably nucleic acids, without significantly permeabilizing the cell membrane. The systematic structure–activity relationship study of buforin II revealed that the cell-penetrating efficiency, which depends on α-helical content, is a critical factor for determining the antimicrobial potency of buforin II. Furthermore, the proline hinge is a key structural factor for the cell-penetrating property. Only a single amino acid substitution for proline changes buforin to a membrane-active magainin-like peptide, and conversely the insertion of a proline-hinge region can switch a membrane-permeabilizing peptide to a cell-penetrating one. Our findings provide important information in designing potent new peptidic antibiotics with different mechanisms.
Acknowledgments
This work was partially supported by grants from the Korea Science and Engineering Foundation (KOSEF-1999–1-20900–002-5), the Ministry of Education, the Ministry of Agriculture, Forestry and Fisheries of Korea/Special Grants Research Program (MAFF-SGRP), and the National Research Laboratory.
Abbreviations
- cfu
colony-forming unit
- PI
propidium iodide
- FACS
fluorescence-activated cell sorter
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.150518097.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.150518097
References
- 1.Boman H G. Annu Rev Immunol. 1995;13:61–92. doi: 10.1146/annurev.iy.13.040195.000425. [DOI] [PubMed] [Google Scholar]
- 2.Hoffmann J A, Hetru C. Immunol Today. 1992;13:411–415. doi: 10.1016/0167-5699(92)90092-L. [DOI] [PubMed] [Google Scholar]
- 3.Oppenheim F G, Xu T, McMillian F M, Levitz S M, Diamond R D, Offner G D, Troxler R F. J Biol Chem. 1988;263:7472–7477. [PubMed] [Google Scholar]
- 4.Zasloff M. Proc Natl Acad Sci USA. 1987;84:5449–5453. doi: 10.1073/pnas.84.15.5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gibson B W, Tang D, Mandrell R, Kelly M, Spindel E R. J Biol Chem. 1991;266:23103–23111. [PubMed] [Google Scholar]
- 6.Park C B, Kim M S, Kim S C. Biochem Biophys Res Comm. 1996;218:408–413. doi: 10.1006/bbrc.1996.0071. [DOI] [PubMed] [Google Scholar]
- 7.Mor A, Nicolas P. Eur J Biochem. 1994;219:145–154. doi: 10.1111/j.1432-1033.1994.tb19924.x. [DOI] [PubMed] [Google Scholar]
- 8.Berkowitz B A, Bevins C L, Zasloff M A. Biochem Pharmacol. 1990;39:625–629. doi: 10.1016/0006-2952(90)90138-b. [DOI] [PubMed] [Google Scholar]
- 9.Mor A, Nicolas P. J Biol Chem. 1994;269:1934–1939. [PubMed] [Google Scholar]
- 10.Schuster F L, Jacob L S. Antimicrob Agents Chemother. 1992;36:1263–1271. doi: 10.1128/aac.36.6.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boman H G. Cell. 1991;65:205–207. doi: 10.1016/0092-8674(91)90154-q. [DOI] [PubMed] [Google Scholar]
- 12.Shai Y. Trends Biochem Sci. 1995;20:460–464. doi: 10.1016/s0968-0004(00)89101-x. [DOI] [PubMed] [Google Scholar]
- 13.Park C B, Kim H S, Kim S C. Biochem Biophys Res Commun. 1998;244:253–257. doi: 10.1006/bbrc.1998.8159. [DOI] [PubMed] [Google Scholar]
- 14.Yi G S, Park C B, Kim S C, Cheong C. FEBS Lett. 1996;398:87–90. doi: 10.1016/s0014-5793(96)01193-3. [DOI] [PubMed] [Google Scholar]
- 15.Steinberg D, Lehrer R I. In: Methods in Molecular Biology 78: Antibacterial Peptide Protocols. Shafer W M, editor. Totowa, NJ: Humana Press; 1997. pp. 169–186. [Google Scholar]
- 16.Chen H, Brown J H, Morell J L, Huang C M. FEBS Lett. 1988;236:462–466. doi: 10.1016/0014-5793(88)80077-2. [DOI] [PubMed] [Google Scholar]
- 17.Chen Y H, Yang J T, Martinez H M. Biochemistry. 1972;11:4120–4131. doi: 10.1021/bi00772a015. [DOI] [PubMed] [Google Scholar]
- 18.Harlow E, Lane D. In: Antibodies: A Laboratory Manual. Harlow E, Lane D, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1988. p. 354. [Google Scholar]
- 19.Pathak N, Salas-Auvert R, Ruche G, Janna M, McCarthy D, Harrison R G. Proteins. 1995;22:182–186. doi: 10.1002/prot.340220210. [DOI] [PubMed] [Google Scholar]
- 20.Matsuzaki K. Biochim Biophys Acta. 1998;1376:391–400. doi: 10.1016/s0304-4157(98)00014-8. [DOI] [PubMed] [Google Scholar]
- 21.Matsuzaki K, Kobayashi S, Takeshima K, Kim S C. Biophys J. 2000;78:322A. [Google Scholar]