Abstract
TCDD is a reproductive toxicant and endocrine disruptor, yet the mechanisms by which it causes these reproductive alterations are not fully understood. In order to provide additional insight into the molecular mechanisms that underlie TCDD’s reproductive toxicity, we assessed TCDD-induced transcriptional changes in the ovary as they relate to previously described impacts on serum estradiol concentrations and altered follicular development in zebrafish. In-silico computational approaches were used to correlate candidate regulatory motifs with observed changes in gene expression. Our data suggest that TCDD inhibits follicle maturation via attenuated gonadotropin responsiveness and/or depressed estradiol biosynthesis, and that interference of estrogen-regulated signal transduction may also contribute to TCDD’s impacts on follicular development. TCDD may also alter ovarian function by disrupting various signaling pathways such as glucose and lipid metabolism, and regulation of transcription. Furthermore, events downstream from initial TCDD molecular-targets likely contribute to ovarian toxicity following chronic exposure to TCDD. Data presented here provide further insight into the mechanisms by which TCDD disrupts follicular development and reproduction in fish, and can be used to formulate new hypotheses regarding previously documented ovarian toxicity.
Keywords: TCDD, ovary, zebrafish, follicular development, microarray, endocrine disruptors
Introduction
2,3,7,8-Tetrachlorodibenzo-p-dioxin (TCDD) is a persistent environmental contaminant that is a known reproductive toxicant and endocrine disruptor in nearly all vertebrates. The effects of TCDD on reproduction and fertility have been studied extensively [1–3], and evidence suggests that TCDD compromises both ovarian function and follicular development. Female reproductive development is highly complex, and is synchronized by intricate and highly regulated signal transduction pathways that are integrated with the endocrine system. This complexity has made it particularly difficult to identify the molecular action of TCDD-induced ovarian toxicity. Although it is clear that TCDD impacts maturation and ovulation of ovarian follicles, as well as estradiol secretion [4–7], the mechanisms that underlie these reproductive toxicities are complicated and poorly understood.
It is generally accepted that TCDD toxicity is mediated by the aryl hydrocarbon receptor (AHR)-signaling cascade [8]. The ligand-bound AHR complex is translocated to the nucleus where it dimerizes with the aromatic hydrocarbon receptor nuclear translocator (ARNT) protein. This heteromeric complex binds to the aryl hydrocarbon-response element (AHRE) (TnGCGTG DNA motifs) located in the regulatory regions of several genes (e.g., , cyp1a1) and can initiate or suppress gene transcription [9–11]. While it is thought that such transcriptional regulation mediates the toxicity of TCDD, specific roles of these gene changes in dioxin-induced toxicity are not understood. Additionally, some actions of TCDD may be AHR-independent and/or result from downstream transcriptional changes.
Evidence suggests that disruptions in female reproduction by TCDD are likely the result of a direct effect at the ovary [12–17]. Since the AHR is expressed in the ovary, TCDD could disrupt critical cellular signals that regulate follicular development and/or estradiol biosynthesis via AHR-mediated alterations in gene transcription, thereby contributing to the observed decrease in ovarian development and reduced reproductive capacity. Alternatively, TCDD could interfere with the hypothalamic-pituitary-gonadal (HPG) axis or estradiol metabolism. For example, TCDD could negatively regulate estrogen signaling by inducing oxidative metabolism of estrogens via the AHR-pathway, by suppressing the expression and/or efficacy of the estrogen receptors, or by inhibiting estradiol-regulated gene expression [18–21].
Since the basic features of the HPG axis and AHR-signaling pathways in fish are fundamentally similar to other vertebrates [22–24], fish are excellent model systems with which to investigate the effects of endocrine-disrupting chemicals on vertebrate reproductive function. The zebrafish has proven to be an effective system for investigation into the teratogenic effects of TCDD [25–27]. Zebrafish are highly prolific with rapid follicular development, and many of the receptors, enzymes, and peptide growth factors involved in follicular development have been characterized (see Ge 2005 for review); therefore, it is particularly suited for investigating TCDD’s effects on the regulation of follicular development.
We have previously demonstrated that sublethal dietary exposure to TCDD alters follicular development, egg production, and serum 17β estradiol concentrations in zebrafish [4]. Here we investigate the transcriptional events in the ovary that precede these previously described histomorphologic and physiologic alterations. We used quantitative RT-PCR to assess the effects on the expression of several candidate genes important in the regulation of follicle development, oocyte maturation, and vitellogenesis, and used cDNA microarray technology to evaluate altered gene expression profiles to identify other cellular pathways potentially impacted by TCDD-exposure. Additionally, we used a functional genomics approach to examine candidate regulatory motifs in relation to different expression profiles in an effort to better clarify potential mechanisms of toxicity following chronic exposure to TCDD.
Materials and Methods
Experimental animals
Adult female (AB strain, Zebrafish International Resource Center) and male zebrafish (golden longfin, Ekwill Farms) were housed separately and acclimated for several weeks prior to the initiation of experiments. Fish were maintained at 26–28°C on a 14-hour light and 10-hour dark cycle in a flow-through buffered, de-chlorinated water system and were spawned once weekly during the experiment.
TCDD exposure and RNA extraction
Trout chow yielding a final concentration of approximately 0, 10, 40, or 100 ng TCDD/g food (ppb) was prepared as previously described [28]. Females were fed brine shrimp nauplii daily, and contaminated trout chow diet 5 of 7 days per week. Fish were fed en masse to satiation, and based upon the average food consumed per fish, received an estimated dose of 0, 0.08, 0.32, or 0.80 ng TCDD/female/day. Based upon our previous work, fish accumulated approximately 0, 0.6, 3, and 14 ng TCDD/g fish after 15 days, and ovaries accumulated approximately 0.4, 2, and 5 ng TCDD/g ovary (body burdens were not measured in this study). Following dietary exposure, five females from each treatment group were euthanized and ovaries were extracted and frozen in liquid nitrogen. Total RNA was isolated from individual ovaries using Trizol reagent (Invitrogen) and purified using an RNeasy MinElute cleanup kit (Qiagen) according to manufacturer’s instructions. Individual samples were analyzed by agarose gel electrophoresis to confirm the integrity of the 18S and 28S ribosomal RNAs, and quantified by UV spectrophotometry at 260 nm using a Nanodrop ND-1000 Spectrophotometer (Nanodrop Technologies).
Quantitative RT-PCR (QPCR)
QPCR was used to quantify selected gene transcripts important in both receptor- and non-receptor-mediated regulation of follicular development, as well as estradiol biosynthesis, included: luteinizing hormone receptor (lhr), follicle-stimulating hormone receptor (fshr), estrogen receptors (esr1, esr2a, esr2b), epidermal growth factor (egf), egf receptor (egfr), inhibins (inhbba, inhbb), steroidogenic acute regulatory protein (star), side chain cleavage enzyme (cyp11a1), and aromatase (cyp19a1a). cDNA was synthesized from 5 μg of total RNA from individual samples (n = 3 from each treatment group) using SuperscriptII reverse transcriptase (Invitrogen) and random hexamer primers following manufacturer’s instructions. QPCR was performed using the Stratagene Mx3000 system and the Full Velocity SYBR Green QPCR Master Mix with the manufacturer’s instructions (Stratagene) using gene-specific primers listed in Table 1. Agarose gel electrophoresis and melting curve analysis confirmed that specific products of expected size were amplified. All QPCR reactions were run in triplicate with negative (no template) controls. Relative gene expression data (fold-change) was calculated using the comparative quantification model [29] assuming 100% efficiency, using small nuclear ribonuclear protein D1 polypeptide (snrpd1) as the reference gene; snrpd1 is important for mRNA processing, and expression was unaltered by TCDD exposure. QPCR on individual samples (n = 3 from each treatment group) was also used to verify microarray expression profiles. The gene-specific primers for these transcripts (cluster 4: cyp1a1; cluster 2: sepp1a, lgals3l, and krt4; cluster 1: krml and vtg1; cluster 3: tfa) are also listed also in Table 1. The seven transcripts showed altered expression by at least 2-fold following exposure to at least one dose of TCDD, and were selected for microarray verification based upon their functional characterization, presence of candidate regulatory motifs, and to represent each of the clusters in gene expression (see details below and in Table 2).
Table 1.
Primer sequences used for QPCR. Gene names are listed in the text and Supplementary Tables.
| Gene symbol | Sense Primer | Antisense Primer |
|---|---|---|
| cyp1a1 | ACTGGTGCGACTGGTTAATATGAG | GTCTTCGCATGTGTTGATAAGAG |
| cyp11a1 | CAGGTTTTCACTGGAATCGG | CTGGTTAAAGATGCCATCCC |
| cyp19a1a | ACTTCCAGAAAAATGTTCCGAGTC | TCTGAGGATAAGCTGCACGC |
| egf | GCATCCAGAGAATGAACCTCGACGG | TGATTTGTCCTGTTTGTGTGTCCGC |
| egfr | CTTACAAAGCCTGGAACGAAGAGCC | CCTGACACACTTTCTTTTCCGGCG |
| esr1 | TCCACATCCACACAGTAGGC | CTTTCTTGATCAGGGTGGGG |
| esr2a | AAGCTGCTGTGTCTGCTGGACTCGG | TCATGCAGTGCAGGTGGTCCATGCC |
| esr2b | GGTGAAGTGTGTCATTTCTGTGCCG | GTGTGATTTTTGGGGCTGGGC |
| fshr | AGCCGATTTGGATGCTTTAAAAGGC | TCAGACAGATGTCAGTGCACTTGGG |
| inhbaa | GCACATTCAGAAGCCGACTGCC | TAAAGTCCGTCTGCCTGTGCGC |
| inhbb | CGAATTCGGTGGACAGACAAACGG | TCCGTTCATTATTAGGCTCGCGG |
| krml2.2 | TATAAACTCAAGTGCGAAAGGC | ACCAGCAAACAATCTTATGC |
| krt4 | CAAAACCAGTGTCACCACCG | CATCTCCTCATTAACAGGGG |
| lgals3l | GCTGTACAAGTGCATGTAAAGGG | CGTCTTTTATGCATGAAGCG |
| lhr | CAAAAAGGACGAGTCGCTGAAACGC | GCAGAAGAAAAACAAGAAGCAGGGC |
| seppa1 | AAATCTGACTTTAACTGGTCCAGTG | ATGTTACATGACCTTTGCCC |
| snrpd1 | CGTCACGATTGAGTGAAGAATGGC | TGAGATCTTCATCTCCTCGGCCC |
| star | ACCCACCTGTATTGTCATGCG | AATGGCTGCGTCTATACCCC |
| tfa | ATTAAGCACACTGTGGTCGG | AGCATGAACTGGCACTTGGG |
| vtg1 | GAGATTGAACTGACTGCAGCC | ATTCCACATGAACATAGGCC |
Table 2.
List of representative transcriptsa identified by microarray to be dysregulated greater than 2-fold in one or more treatment group. Presence of putative AHRE or ERE is noted. See Supplementary Tables 1–3 for complete gene list plus location of putative response elements.
| Gene symbol | 10 ppb | 40 ppb | 100 ppb | AHRE | ERE |
|---|---|---|---|---|---|
| Ovarian development | |||||
| fabp3 | −2.92 | −2.68 | −4.14 | ||
| tpte | −1.91 | −1.45 | −2.60 | * | |
| tph1l | −2.64 | −1.53 | 1.19 | * | |
| vtg1 | −7.38 | −1.62 | −1.83 | * | * |
| Detoxification/oxidative stress | |||||
| cyp1a | 6.93 | 14.87 | 24.68 | * | * |
| sepp1a | −1.71 | −1.21 | −2.48 | ||
| serpina1 | 29.44 | 1.51 | 3.07 | * | * |
| Lipid metabolism | |||||
| apoeb | −1.83 | −1.09 | −4.12 | * | |
| apom | 6.16 | 1.04 | 1.32 | ||
| lipf | −2.06 | −1.85 | −2.30 | * | * |
| Carbohydrate metabolism | |||||
| ldha | −1.78 | −1.45 | −2.59 | * | * |
| slc3a2 | −2.35 | −1.68 | −3.05 | ||
| Immune response/cellular repair | |||||
| anxa1a | −2.20 | −3.36 | −7.06 | * | * |
| cp | 24.49 | 1.06 | 2.16 | * | |
| fgb | 12.80 | −1.82 | −1.35 | * | |
| fgb | 5.66 | 1.43 | 1.61 | * | * |
| tfpib | −1.54 | −1.95 | −5.66 | ||
| timeless | −2.92 | 1.88 | 1.07 | * | |
| Structure | |||||
| krt4 | −1.96 | −2.93 | −5.65 | * | * |
| lgals3l | −1.89 | −2.68 | −2.56 | * | |
| Signal transduction/transcription/regulation of cell cycle | |||||
| rab5b | 2.45 | 1.34 | 2.13 | * | * |
| rad1 | −1.10 | 1.13 | −2.16 | ||
| rbp2a | −2.98 | −2.39 | −4.54 | * | * |
| Igfbp1 | 5.58 | −1.38 | −1.47 | * | |
| Junb | −2.21 | −1.88 | −4.39 | * | * |
| krml2.2 | −2.59 | −1.19 | −2.41 | * | * |
| lztfl1 | −1.12 | −1.12 | −2.33 | * | |
| sp4 | 1.51 | 2.32 | 1.96 | * | |
| ing3 | −2.29 | 1.17 | −1.32 | ||
| igfbp1 | 5.58 | −1.38 | −1.47 | * | |
| tfa | 11.64 | −2.09 | −1.32 | * | |
| ptena | −2.32 | 1.51 | 1.38 | * | |
Gene symbols are from Affymetrix probe identifiers and are organized into general functional groups. Transcripts in bold were verified by QPCR.
Microarray hybridization and analysis
Differential ovarian gene expression was determined using the Affymetrix Zebrafish Genome Array, which represents approximately 14,900 transcripts. Experiments were designed to comply with MIAME guidelines [30]. In an effort to reduce variability, target synthesis using pooled RNA (of equal quantity and comparable quality from 5 females per treatment group) [31–33], biotin-labeling, hybridizations, and staining were performed using standard Affymetrix reagents and methods (http://www.affymetrix.com/index.affx). Pooled samples were run in duplicate such that a total of eight arrays were used.
Images were extracted from TIFF files using Microarray Suite Version 5.0 (Affymetrix). Raw data can be viewed at the National Center for Biotechnology Information Gene Expression Omnibus website (http://www.ncbi.nlm.nih.gov/projects/geo/); series record GSE4859. Data were transformed and normalized using GeneSpring software (Agilent Technologies) and further analyzed using Bioconductor 1.6 [34]. Expressed data were obtained by selecting 8732 probes present in all eight chips. Differentially expressed probes were identified as those showing at least a 2-fold change in expression in both replicates of one or more of the treatment groups compared with control. The dysregulated transcripts were assigned to general functional groups based upon their Gene Ontology terms, or an annotated putative H. sapiens or M. musculus ortholog from Swiss-Prot (http://us.expasy.org/sprot/). The dysregulated transcripts were clustered using agglomerative hierarchical clustering [35] without performing mean centering of expression values, and using Ward’s method for merging clusters.
In-silico Transcription Factor Analysis
Possible cis-acting AHRE (GCGTG) and ERE (AGGTCAnnnTGACCT) sequences were identified within the regulatory region of differentially expressed genes as well as candidate genes determined by QPCR. The reference genes were 22877 RefSeq genes and Ensemble predictions mapped to the draft zebrafish genome, and the test sequences were RefSeqs overlapping with consensus sequences of differentially expressed Affymetrix probes. Of the dysregulated transcripts, 140 could be mapped to Ensemble genes and were used for subsequent analyses. The 5′ regulatory sequences of the reference and dysregulated genes were modeled by the 5′-UTR of each RefSeq (5000 bp upstream sequences from the transcriptional start site). Regulatory sequences were acquired from the June 2004 draft assembly of the zebrafish genome using the University of California – Santa Cruz table browser [36] and were scanned for possible transcription factor binding sites using TFBS [37], JASPAR [38] for AHRE sequences at a 100% scoring threshold, and TRANSFAC Public 7.0 [39] for ERE sequences at an 80% scoring threshold. To compare control and differentially expressed sequences, the hypergeometric distribution in R 2.1.0 was used to describe the probability of drawing a sample the same size as the differentially expressed sequences from the control sample with the same or fewer transcription factor binding sites identified. A chi-squared test was used to determine if there was a location bias of transcription factor binding sites with respect to identified clusters. The starting position distributions of AHREs and EREs within a cluster were compared with the corresponding distributions in the entire Ensemble known gene collection. Distributions for a cluster significantly different (p < 0.05) from control suggested putative regulatory patterns.
Data analysis
Statistical analysis of QPCR data was performed using Sigma-Stat 2.0 (SPSS, Inc) and presented as means ± standard error of the mean (SEM). Data were evaluated for homoscedasticity (Levene Median test) and one-way analysis of variance (ANOVA) was used to detect treatment-related effects on the expression of each candidate transcript. Where significant differences were indicated between treatment groups, and the data were homogeneous, pair-wise multiple comparisons were conducted using the Tukey test. When tests for homogeneous variance failed, the Kruskal-Wallis one way ANOVA on ranks was used, and significant differences were evaluated using the Dunn’s test. Pearson’s correlation was used to determine whether changes in gene expression identified by microarray were correlated with changes in expression of the seven transcripts determined by QPCR. For all analyses, significant differences were identified at p < 0.05.
Results
Effects on the regulation of follicular development and estradiol biosynthesis
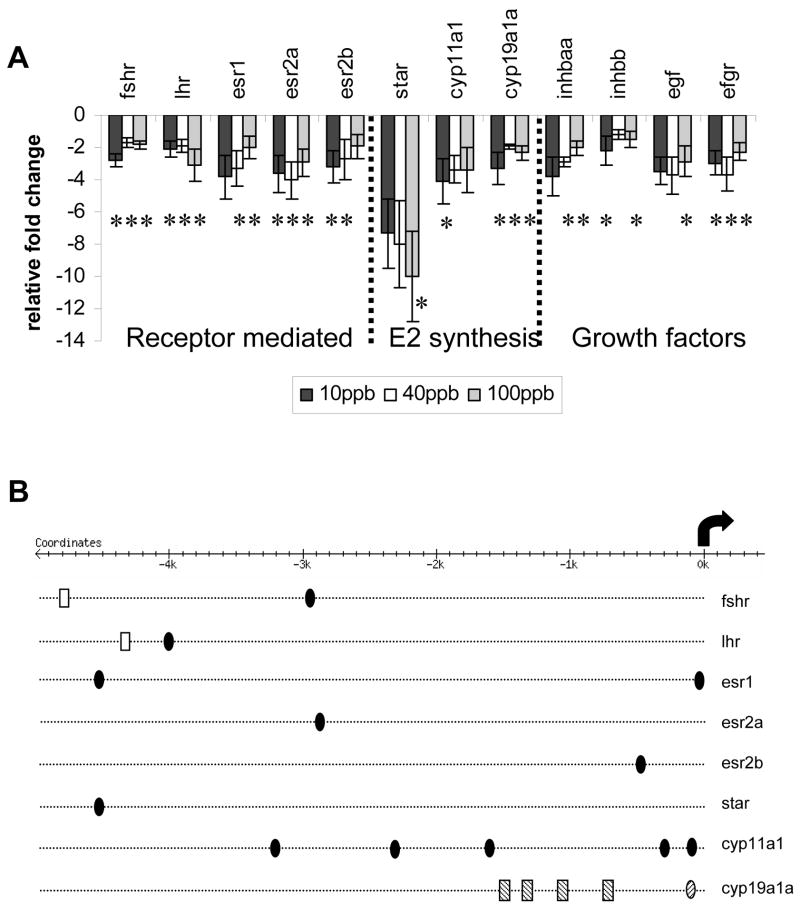
Dietary TCDD exposure suppressed the expression of all examined candidate transcripts that are important in both receptor- and non-receptor-mediated regulation of follicular development, as well as estradiol biosynthesis (Figure 1A). The candidate genes were down-regulated by at least 2-fold; overall changes in expression were not dose-dependent. Locations of putative AHREs and EREs within the regulatory region of the transcripts are illustrated in Figure 1B.
Figure 1.
A. Fold changes in expression of several genes important for follicular development determined by QPCR; normalized to snrpd1. *Denotes significant changes in expression compared with control (p < 0.05); dose-dependent changes were not observed. B. Location of putative AHREs (filled ovals, this study; lined ovals [86]; putative EREs (empty rectangles), and putative ½-EREs (lined rectangles [86] within upstream regulatory regions. Arrow represents the transcriptional start site.
Effects of TCDD on Global Ovarian Gene Expression
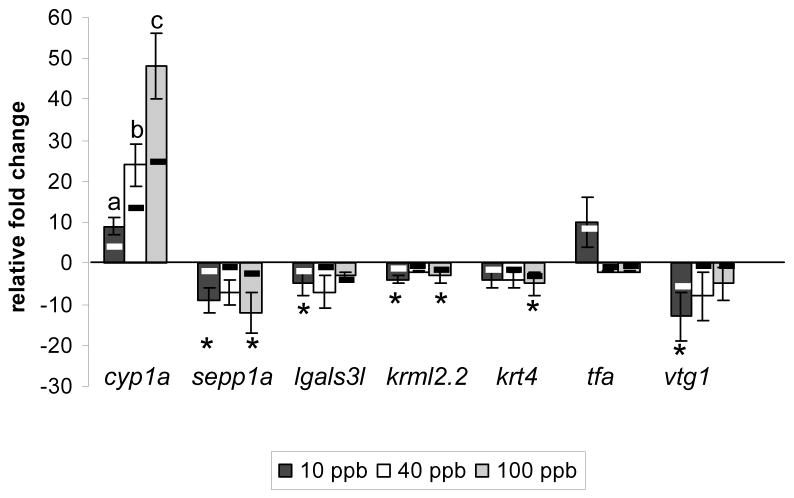
Two hundred thirty-five probe sets (representing 229 unique transcripts) were identified as being either induced or suppressed greater than 2-fold following exposure to one or more TCDD concentration. Overall, more transcripts were suppressed than were induced with 37% (85/229), 7% (16/229), and 40% (92/229)of the transcripts down-regulated and 11% (25/229), 4% (9/229), and 11% (25/229) of the transcripts up-regulated in the 10, 40, and 100 ppb treatment groups, respectively. QPCR using individual samples verified microarray results for seven transcripts (Figure 2). The direction and fold-change values for samples from individual animals were correlated with changes in expression determined by microarray analysis of pooled samples for all seven genes (p<0.01), demonstrating that our approach successfully identified changes in gene expression across treatment groups that could be validated with individual samples.
Figure 2.
Validation of microarray results. Bars represent mean relative fold changes in gene expression determined by QPCR; normalized to snrpd1. Letters or * denote significant differences for that gene compared with control. Black and white lines represent fold-changes in expression determined by array analysis. (Pearson’s correlation, p < 0.01).
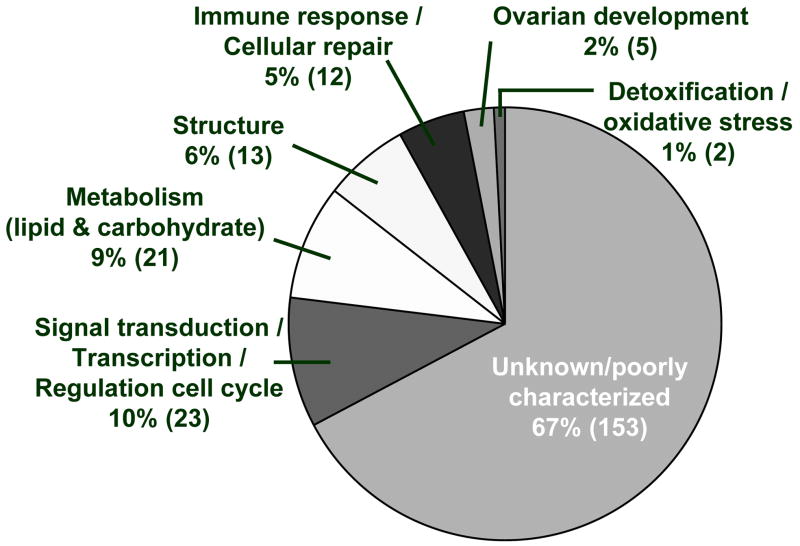
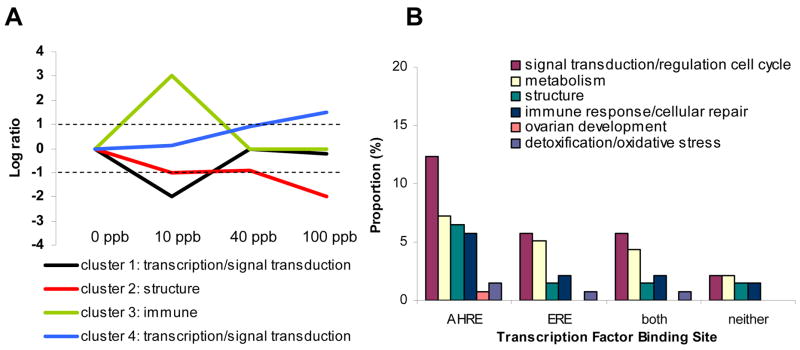
Differentially expressed transcripts with known functions (103/229) were assigned to general functional groups based upon the top three Gene Ontology (GO) functions and known biologic functions, and are important for signal transduction/transcription/regulation of the cell cycle, as well as for glucose and lipid metabolism, immune response/cellular repair, and structure (Table 2 and Figure 3). Hierarchical clustering grouped transcripts into one of five clusters (Figure 4A and Supplementary Table 1). On average, the 17 transcripts within the fifth cluster show minimal changes in gene expression across treatment groups and are therefore not shown in Figure 4 or discussed further (due to lack of statistical power). The majority of functionally annotated transcripts within clusters one and four regulate gene transcription or are important for signal transduction, while most functionally annotated transcripts within cluster two are important for maintaining structure, and those within cluster three play a role in immune function (see Figure 4A).
Figure 3.
Proportion of dysregulated ovarian transcripts within each general functional group. The number in parentheses represents the number of transcripts out of 229 within each functional group.
Figure 4.
A. Gene expression (expressed as the log ratio compared with control) patterns of four hierarchical gene clusters including the primary functional group represented within each cluster. B. Proportion of dysregulated ovarian transcripts within each general functional group that contain putative AHREs, ERES, both or neither.
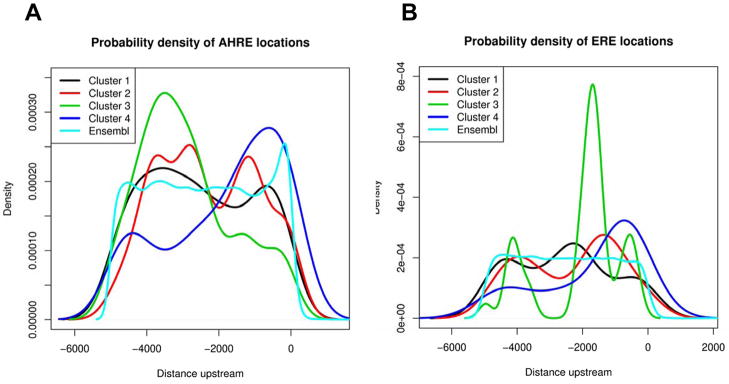
One hundred forty of the dysregulated transcripts were mapped to Ensemble genes, and 89% of these (125/140 corresponding to 113 unique transcripts) were found to have putative AHREs in the regulatory region of the gene (Figure 4B, Supplementary Tables 1 and 2), compared with 92% of the reference genes (21046/22877). Using hypergeometric distribution to calculate the probability of finding only 125 or fewer genes out of 140 that were drawn from 22877, our data suggest differentially expressed transcripts are not enriched for putative AHREs in (p = 0.14). Estimated probability density curves shown in Figure 5A illustrate the potential location bias of putative AHREs for each cluster along with the location of putative AHREs for all genes in the zebrafish genome for comparison. Upstream regions of genes within cluster 2 have putative AHRE distributions that are significantly different (p = 2.9 × 10−12) from those of the control gene set, with AHREs distributed 3000 – 4000 bp and 1500 bp upstream of the transcriptional start site (TSS). In cluster 3, AHREs tend to be further upstream (~3500 bp) from the TSS than control (p = 2.0 × 10−7), and in cluster 4, AHREs tend to be concentrated within 2000 bp upstream of the TSS (p = 0.01). Of the 113 unique transcripts that contain putative AHREs, 16% were upregulated and 51% were downregulated across treatment groups; many function as transcription factors and regulate the cell cycle (Figure 5).
Figure 5.
Probability density curves illustrate the potential location bias of putative AHREs (A) and EREs (B) with respect to the translational start site for the transcripts within each hierarchical cluster compared with reference. Note: Probability density curves represent the probability distribution in terms of integrals, illustrated as a line depicting the relative frequency that each response element is represented at that location.
Approximately 49% of the mapped dysregulated probes (68/140 corresponding to 58 unique transcripts) were found to have putative EREs in the regulatory region of the gene (Figure 4B, Supplementary Tables 1 and 3); 17% of these transcripts were upregulated and 55% were downregulated across treatment groups. Forty-two percent of the reference genes (9608/22877) contained putative EREs in the regulatory region; differentially expressed genes contained putative EREs at a much higher rate than would be expected (p = 0.05). Estimated probability density curves shown in Figure 5B illustrate the potential location bias of putative EREs for each cluster. In cluster 1, EREs tend to be centered around 2000 bp from the TSS (p = 3.0 × 10−7). In cluster 3, EREs are largely concentrated at about 1800–2000 bp upstream of the TSS (p = 8.2 × 10−7) and in cluster 4, EREs tend to be concentrated within 2000 bp upstream (p = 0.02). Many of these transcripts function in carbohydrate and lipid metabolism, or regulation of transcription or the cell cycle (Figure 4B).
Approximately 41% of the dysregulated probes contain both putative AHRE and ERE in the regulatory region (Figure 4B, Supplementary Table 1); approximately 20% of these transcripts were upregulated, while the majority of the transcripts (51%) were downregulated across treatment groups. Approximately 40% of the reference genes (9039/22877) contain putative AHREs and EREs in the regulatory region; the dysregulated transcripts were not significantly enriched for both AHREs and EREs (p = 0.35). As with those with putative EREs, many of these transcripts are important for carbohydrate and lipid metabolism or function as transcription factors and regulate the cell cycle (Figure 4B).
Discussion
TCDD has been shown to perturb the regulation of vertebrate follicular development and ovulation, as well as steroidogenesis in the ovary [3–6;40]. Regulation of these processes is complex, integrating receptor-mediated hormonal signals from the pituitary with locally produced factors to form an intimate regulatory network within and between follicles. This poses a challenge for identifying the molecular mechanisms that regulate TCDD’s reproductive toxicity. Furthermore, the sometimes subtle secondary and tertiary effects on gene expression following chronic dioxin exposure can be difficult to ascertain and to correlate with observed toxicities. This study represents one of the first attempts to characterize the effects on ovarian gene expression following exposure of fish to chronic, sublethal concentrations of TCDD. Using a candidate-gene approach in conjunction with a genomics approach, we have identified several novel dioxin-responsive genes that with further study may better clarify TCDD’s ovarian toxicity.
Effects on the regulation of follicular development and estradiol biosynthesis
All of the candidate genes selected for QPCR analysis (those important for receptor-and non-receptor mediated regulation of follicular development and estradiol biosynthesis) show peak expression in follicles that are in the mid-to-late stages of vitellogenesis, and should therefore be highly expressed in whole ovary of cycling females under normal conditions [41–44]. Gonadotropins are important regulators of follicular development, and in zebrafish, have been shown to induce the expression of several other genes important for oocyte development [41;45]. Suppression of both gonadotropin receptors suggests that impaired follicular development may be the result of this suppression. Similar effects on ovarian gonadotropin receptor expression have been shown in mammals [6;46–48] and others suggest that TCDD’s anovulatory effects are likely mediated directly at the ovary in an AHR-dependent manner [49;50]. Collectively, this suggests that TCDD primarily acts at the ovary to suppress expression of gonadotropin receptors.
Exposure to TCDD also downregulates the expression of several genes important in estradiol biosynthesis (star, cyp11a1, and cyp19a1a), and likely contributes to the previously observed reduced serum estradiol concentrations [4]. While it has been proposed that reduced estradiol concentrations following exposure to TCDD may result from suppression of aromatase activity, the mechanism by which TCDD disrupts ovarian steroidogenesis is not clear. Baba et al [51] show that exposure to DMBA, an AHR agonist, is correlated with increased estradiol concentrations and increased aromatase expression, while others suggest that the target of TCDD-induced suppression of estradiol is upstream of aromatase in the steroidogenic pathway [52]. However, other in vitro and in vivo studies demonstrate that exposure to TCDD decreases the expression and activity of aromatase, and is correlated with decreased estradiol concentrations [53–55]. While our data support findings in mammalian systems that suggest TCDD-induced reductions in serum estradiol may result from diminished aromatase expression/activity, impairing estradiol biosynthesis, we cannot rule out star and cyp11a1 as potential targets. Others have demonstrated that expression of star and cyp11a1 is suppressed in the interregnal gland following exposure to βNF, lending support for these transcripts as other potential targets for endocrine disruption by AHR-ligands [56].
While not as well characterized in fish, estradiol also plays a role in regulating follicle development and ovulation via ERs in the ovary. Three forms of the ER (esr1, esr2a, esr2b) are expressed in the zebrafish ovary, and all are capable of initiating transcription of genes [57;58]. Here we show that all three forms of the ER are downregulated following chronic exposure to TCDD, as has been demonstrated for mammals [59;60]. By suppressing the expression of ERs in the ovary, follicles may be unable to respond to estrogen signaling and/or induce the transcription of estrogen-responsive genes important for ovarian development [61–64]. Our array data further support this hypothesis in that more than half of transcripts with putative EREs were downregulated in the ovaries of TCDD-treated fish.
Finally, while follicular development is primarily controlled by gonadotropin hormones, various local factors released from granulosa and theca cells as well as from oocytes, also mediate gonadotropin signaling. Peptide growth factors such as inhibins and follistatins potentiate the action of gonadotropins and maturation-inducing steroid in the induction of final oocyte maturation [44]. Activin βA (Inhbaa) promotes ovary and follicle growth, whereas activin βB (Inhbb) exerts a tonic role throughout follicle development, and becomes critical at the late stage of oocytes maturation and/or ovulation [65]. Epidermal growth factor enhances the rate of oocyte maturation via gonadotropin signaling, inhibits apoptosis, stimulates follicle cell proliferation, and plays a role in controlling follicle survival and steroidogenesis [66;67]. TCDD exposure suppressed the expression of both forms of inhibin, as well as egf and its receptor, although not significantly at all doses, suggesting that impacts further upstream (e.g., expression of gonadotropin receptors) may have a greater impact on follicular development.
Effects of TCDD on Global Ovarian Gene Expression
While our microarray experimental design does not allow us to make direct conclusions about the mechanisms that underlie TCDD’s ovarian toxicity, we are still able to offer valuable insights into additional cellular pathways and signal cascades that are altered by chronic exposure to TCDD. TCDD disrupts several integrated cellular pathways (including structure, glucose and lipid metabolism, immune response, and regulation of transcription) reaffirming the complexity of TCDD toxicity and identifying several new avenues for further study. For example, chronic exposure to 10 ppb TCDD shows a trend for greater alterations in the expression of genes important for transcription/signal transduction (cluster 1) and immune response genes (cluster 3) compared to chronic exposure to 40 and 100 ppb TCDD. Exposure to 10 ppb TCDD for 15 days resulted in reduced estradiol concentrations in zebrafish, but follicular development, overall egg production and spawn success were not altered; follicular development, egg production and spawn success was reduced following exposure to 40 and 100 ppb TCDD for 15 days [4]. Therefore, the roles that such cellular pathways play in the modulation by TCDD of follicular development and reproductive success by TCDD warrants further study.
Following chronic exposure to TCDD, we show that transcripts such as keratins (krt 4, 8, and 18), collagens (col1a2, col5a2l), lectins (lgals3l and lgals1l2), and actins (acta2) necessary for maintaining structural integrity were suppressed. We verified the expression profiles for two of these transcripts (krt 4 and lgals3l); expression profiles for similar transcripts (cluster 2) suggest a trend for a dose-dependent suppression of structure-related transcripts. TCDD has also been shown to impact the expression of genes important for maintaining and metabolizing the extracellular matrix in the regenerating fin of zebrafish [68] and in the liver of medaka [69]. Since TCDD exposure is associated with wasting-syndrome, decreased ovarian somatic index, and inhibited regenerative growth of the fin and liver, the impacts of TCDD on the expression of structural proteins warrants further consideration as an integral component of the sublethal toxic response to TCDD.
Many of the ovarian transcripts disrupted by TCDD are important for regulation of the cell cycle and signal transduction. Genes such as cyp1a1 (cluster 4) show a trend for a dose-dependent increase in their suppression, while others such as the large Maf protein krml2.2 (cluster 1) are downregulated following chronic exposure to 10 ppb TCDD but not 40 or 100 ppb TCDD. Most of the genes within cluster 4 (15/17) have putative AHREs, and cyp1a1 is known to be induced by exposure to TCDD in an AHRE-mediated manner; therefore, investigation into whether the expression of such transcripts is induced by TCDD in an AHRE-mediated manner warrants further study. Large Maf proteins such as krml2.2 are important for regulation of cell differentiation. While the function of large Maf proteins in fish ovarian development is not known, krml2.2 plays a role in the differentiation of cell lineages in developing embryos [70], and large Maf proteins have been shown to be key regulators of gonad morphogenesis in Drosophila [71]. Expression of another gene, tfa (cluster 3) is also increased following chronic exposure to 10 ppb TCDD. In mammals, transferrins have an inhibitory effect on FSH-induced differentiation of granulosa cells [72;73]. Alterations in such transcripts may account for observed alterations in follicular development, and warrant further investigation. Similar impacts on the expression of transcripts important for signal transduction and regulation of cellular differentiation have been noted by others to occur in embryonic heart and regenerating tail of zebrafish [68;74;75] as well as liver, brain, and testis of medaka [76]. While the specific transcriptional profiles within these different tissue types following exposure to TCDD reflect tissue-specific targets of TCDD, overall impacts on the regulation of important signaling molecules and transcription factors likely constitute a common denominator in TCDD’s toxicity.
Our work also identifies several novel pathways by which chronic exposure to TCDD may alter ovarian development. For example, both glucose and lipid metabolism may be altered in the ovary as a result of TCDD exposure. Since glucose and lipids play active roles in oogenesis and egg quality in fish [77], perhaps TCDD-induced disruption of these pathways contributes to the observed impacts on ovarian development [4] as well as offspring survival [28]; this hypothesis warrants further consideration. Several genes important for immune response also appear altered by TCDD exposure. Alterations in the expression of transcripts involved in immune function could induce an inflammatory response, negatively impacting steroidogenesis and follicular development [78], or perhaps interfere with normal ovarian development by restricting the removal of apoptotic cells or cellular debris [79]. Such transcriptional changes could also reflect responses from immune cells as the result of tissue repair, as suggested by Volz et al. [69]. Additional studies are necessary to elucidate the impacts of altered immune response on ovarian development in fishes.
Insights into the mechanisms of TCDD-induced ovarian toxicity
In-silico analysis of the regulatory region of dysregulated transcripts have enabled us to further support hypotheses regarding the mechanisms by which chronic exposure to TCDD induces ovarian toxicity and to identify new avenues for continued research into TCDD’s sublethal toxic response in the ovary. The AHR-pathway remains active in zebrafish ovary following chronic exposure to TCDD, evidenced by increased cyp1a1 expression compared with control. While AHR-mediated changes in gene expression likely contribute to TCDD-induced ovarian toxicity, dysregulated transcripts identified by microarray do not appear to be enriched for AHREs compared with reference genes. While we cannot be sure which of these AHREs are functional, others have reported similar findings in that not all TCDD-induced changes in gene expression could be correlated with the presence of AHREs either by promoter analysis or determination of AHR-dependence [69;80;81]. Collectively, these studies suggest that TCDD-induced histopathologic alterations likely involve changes in the expression of genes downstream from initial AHRE-activated transcription in addition to direct AHRE-mediated changes in gene expression (e.g., by altering the expression of transcription factors and signaling proteins via the AHRE, leading to an alteration in the expression of their target genes in a tissue-specific manner).
Our data also suggest that impacts on the expression of estradiol-regulated genes may also be important for TCDD-induced ovarian toxicity. Several propose that interactions between AHR and ER pathways are inhibitory, and that AHR-ligands could suppress expression of genes via the AHRE [19;82;83]. While dysregulated transcripts identified by microarray are enriched for putative EREs, presence of both AHRE and ERE was not different than for reference genes; therefore no conclusions regarding potential interaction/interference between the two signaling pathways can be drawn from these data. Others suggest that when estrogen is present, AHR ligands can attenuate estrogen signaling at an ERE site, and that while the AHR is involved, the mechanism of action is independent from AHRE-mediated signaling [20;84;85]. Our findings here lend support to this hypothesis. Putative EREs were identified in the promoter regions of lhr, fshr, and cyp19a1a, of which only cyp19a1a has been demonstrated to be estrogen responsive in zebrafish [84]. Further, dysregulated transcripts identified by microarray appear to be enriched for EREs compared with reference genes. Collectively these studies suggest that the antiestrogenic actions of TCDD at the ovary entail multiple and perhaps gene/promoter-specific molecular mechanisms involving ER/AHR cross-talk, and warrant further study.
Conclusions
Environmental compounds that disrupt hormone signaling can exert a profound effect on reproduction. Our data suggest that TCDD inhibits expression of key genes that regulate follicular development and estradiol biosynthesis, and provide further evidence for a mechanistic link for impairment of reproduction in fish exposed to TCDD. Suppression of all of the candidate genes in this study following chronic exposure to TCDD is consistent with previously described reductions in serum 17β estradiol concentrations and attenuated follicular development [4], and suggests that TCDD inhibits the transition of pre-vitellogenic follicles to vitellogenic follicles via attenuated gonadotropin responsiveness and/or reduced estradiol biosynthesis. Overall lack of a dose response suggests that the ovary is highly sensitive to TCDD, and that accumulations of less than 1 ng TCDD/g female are sufficient to reduce reproductive capacity of female zebrafish. Furthermore, TCDD appears to impact several integrated cellular pathways, illustrating the complex and profound effects it has on the reproductive system. Our data also suggest that ovarian toxicities following chronic exposure to TCDD result from downstream effects of AHR-mediated signal transduction pathways, or from feedback reactions to cellular changes induced by TCDD. Further examination of the relationships among altered structural integrity, glucose and lipid metabolism, immune response, and regulation of transcription in response to ovarian toxicity will better clarify the mechanisms by which TCDD exerts its reproductive toxicity, particularly its low-dose, long-term effects.
Supplementary Material
Acknowledgments
We gratefully acknowledge Lisa Meyer for her technical assistance. Research was supported in part by: USEPA (TKH, GRO MA916290), UWM Institute of Environmental Health (TKH/RJH), Greater Milwaukee Foundation Shaw Scientist Award (MJC), National Institute of Biomedical Imaging and Bioengineering (MJH, R01EB001421), and the UWM Marine and Freshwater Biomedical Center (P30ES004184).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- 1.Peterson RE, Theobald HM, Kimmel GL. Developmental and reproductive toxicity of dioxins and related compounds: cross-species comparisons. Crit Rev Toxicol. 1993;23:283–335. doi: 10.3109/10408449309105013. [DOI] [PubMed] [Google Scholar]
- 2.Walker MK, Peterson RE. Toxicity of polychlorinated dibenzo-p-dioxins, dibenzofurans, and biphenyls during fish early development. In: Colborn T, Clement C, editors. Chemically Induced Alterations in Sexual and Functional Development: The Wildlife/Human Connection. Princeton, New Jersey: Princeton Scientific Publishing, Co., Inc.; 1992. pp. 195–202. [Google Scholar]
- 3.Pocar P, Fischer B, Klonisch T, Hombach-Klonisch S. Molecular interactions of the aryl hydrocarbon receptor and its biological and toxicological relevance for reproduction. Reproduction. 2005;129:379–89. doi: 10.1530/rep.1.00294. [DOI] [PubMed] [Google Scholar]
- 4.King Heiden T, Carvan MJ, III, Hutz RJ. Inhibition of Follicular Development, Vitellogenesis, and Serum 17{beta}-Estradiol Concentrations in Zebrafish Following Chronic, Sublethal Dietary Exposure to 2,3,7,8-Tetrachlorodibenzo-p-Dioxin. Toxicol Sci. 2006;90:490–9. doi: 10.1093/toxsci/kfj085. [DOI] [PubMed] [Google Scholar]
- 5.Pocar P, Brevini TA, Fischer B, Gandolfi F. The impact of endocrine disruptors on oocyte competence. Reproduction. 2003;125:313–25. doi: 10.1530/rep.0.1250313. [DOI] [PubMed] [Google Scholar]
- 6.Roby KF. Alterations in follicle development, steroidogenesis, and gonadotropin receptor binding in a model of ovulatory blockade. Endocrinology. 2001;142:2328–35. doi: 10.1210/endo.142.6.7993. [DOI] [PubMed] [Google Scholar]
- 7.Munkittrick KR, Portt CB, Van der Kraak GJ, Smith IR, Rokosh DA. Impact of bleached kraft mill effluent on population characteristics, liver MFO activity, and serum steroid levels of a Lake Superior white sucker (Catostomus commersoni) population. Can J Fish Aquat Sci. 1991;48:1371–80. [Google Scholar]
- 8.Schmidt JV, Bradfield CA. Ah receptor signaling pathways. Annu Rev Cell Dev Biol. 1996;12:55–89. doi: 10.1146/annurev.cellbio.12.1.55. [DOI] [PubMed] [Google Scholar]
- 9.Safe S, Krishnan V. Cellular and molecular biology of aryl hydrocarbon (Ah) receptor-mediated gene expression. Arch Toxicol Suppl. 1995;17:99–115. doi: 10.1007/978-3-642-79451-3_8. [DOI] [PubMed] [Google Scholar]
- 10.Denison DL, Gottlieb MB, Whitlock JP. The DNA recognition site for the dioxin-Ah receptor complex. Nucleotide sequence and functional analysis. J Biol Chem. 1988;263:17221–4. [PubMed] [Google Scholar]
- 11.Krishnan V, Porter W, Santostefano M, Wang X, Safe S. Molecular mechanism of inhibition of estrogen-induced cathepsin D gene expression by 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) in MCF-7 cells. Mol Cell Biol. 1995;15:6710–9. doi: 10.1128/mcb.15.12.6710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van der Kraak GJ, Munkittrick KR, McMaster ME, Portt CB, Chang JP. Exposure to bleached kraft pulp mill effluent disrupts the pituitary-gonadal axis of white sucker at multiple sites. Toxicol Appl Pharmacol. 1992;115:224–33. doi: 10.1016/0041-008x(92)90327-o. [DOI] [PubMed] [Google Scholar]
- 13.Gagnon MM, Dodson JJ, Hodson PV. Ability of BKME (bleached kraft mill effluent) exposed white suckers (Catostomus commersoni) to synthesize steroid hormones. Comp Biochem Physiol C. 1994;107:265–73. [Google Scholar]
- 14.Hutz RJ. Reproductive endocrine disruption by environmental xenobiotics that modulate the estrogen-signaling pathway, particularly tetrachlorodibenzo-p-dioxin (TCDD) Journal of Reproduction and Development. 1999;45:1–12. [Google Scholar]
- 15.Son DS, Ushinohama K, Gao X, et al. 2,3,7,8-Tetrachlorodibenzo-p-dioxin (TCDD) blocks ovulation by a direct action on the ovary without alteration of ovarian steroidogenesis: lack of a direct effect on ovarian granulosa and thecal-interstitial cell steroidogenesis in vitro. Reprod Toxicol. 1999;13:521–30. doi: 10.1016/s0890-6238(99)00048-9. [DOI] [PubMed] [Google Scholar]
- 16.Trewin AL, Woller MJ, Wimpee BA, Conley LK, Baldridge MG, Hutz RJ. Short-Term Hormone Release from Adult Female Rat Hypothalamic and Pituitary Explants is not Altered by 2,3,7,8-Tetrachlorodibenzo-p-dioxin. J Reprod Dev. 2007 doi: 10.1262/jrd.18101. [DOI] [PubMed] [Google Scholar]
- 17.Salisbury TB, Marcinkiewicz JL. In utero and lactational exposure to 2,3,7,8-tetrachlorodibenzo-p-dioxin and 2,3,4,7,8-pentachlorodibenzofuran reduces growth and disrupts reproductive parameters in female rats. Biol Reprod. 2002;66:1621–6. doi: 10.1095/biolreprod66.6.1621. [DOI] [PubMed] [Google Scholar]
- 18.Safe S, Wormke M, Samudio I. Mechanisms of inhibitory aryl hydrocarbon receptor-estrogen receptor crosstalk in human breast cancer cells. J Mammary Gland Biol Neoplasia. 2000;5:295–306. doi: 10.1023/a:1009550912337. [DOI] [PubMed] [Google Scholar]
- 19.Safe S, Wormke M. Inhibitory aryl hydrocarbon receptor-estrogen receptor alpha cross-talk and mechanisms of action. Chem Res Toxicol. 2003;16:807–16. doi: 10.1021/tx034036r. [DOI] [PubMed] [Google Scholar]
- 20.Ohtake F, Takeyama K, Matsumoto T, et al. Modulation of oestrogen receptor signalling by association with the activated dioxin receptor. Nature. 2003;423:545–50. doi: 10.1038/nature01606. [DOI] [PubMed] [Google Scholar]
- 21.Safe S, Krishnan V. Chlorinated hydrocarbons: estrogens and antiestrogens. Toxicol Lett. 1995;82–83:731–6. doi: 10.1016/0378-4274(95)03591-5. [DOI] [PubMed] [Google Scholar]
- 22.Lict P. Suitability of the mammalian model in comparative reproductive endocrinology. In: Ralph C, editor. Comparative Endocrinology: Developments and directions. New York: Alan R Liss, Inc; 1986. pp. 95–114. [PubMed] [Google Scholar]
- 23.Tanguay RL, Andreasen E, Walker MK, Peterson RE. Dioxin toxicity and aryl hydrocarbon receptor signaling in fish. 2003. [Google Scholar]
- 24.Hahn ME, Karchner SI, Shapiro MA, Perera SA. Molecular evolution of two vertebrate aryl hydrocarbon (dioxin) receptors (AHR1 and AHR2) and the PAS family. Proc Natl Acad Sci U S A. 1997;94:13743–8. doi: 10.1073/pnas.94.25.13743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carvan MJ, III, King Heiden T, Tomasiawicz H. The Utility of Zebrafish as a Model for Toxicological Research. In: Mommsen TP, Moon TW, editors. Biochemistry and Molecular Biology of Fishes. Vol. 6. Elsevier BV; 2005. pp. 3–41. [Google Scholar]
- 26.Teraoka H, Dong W, Hiraga T. Zebrafish as a novel experimental model for developmental toxicology. Congenit Anom (Kyoto) 2003;43:123–32. doi: 10.1111/j.1741-4520.2003.tb01036.x. [DOI] [PubMed] [Google Scholar]
- 27.Spitsbergen JM, Kent ML. The state of the art of the zebrafish model for toxicology and toxicologic pathology research--advantages and current limitations. Toxicol Pathol. 2003;31 (Suppl):62–87. doi: 10.1080/01926230390174959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.King Heiden T, Hutz RJ, Carvan MJ., III Accumulation, Tissue Distribution, and Maternal Transfer of Dietary 2,3,7,8,-Tetrachlorodibenzo-p-Dioxin: Impacts on Reproductive Success of Zebrafish. Toxicol Sci. 2005;87:497–507. doi: 10.1093/toxsci/kfi201. [DOI] [PubMed] [Google Scholar]
- 29.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brazma A, Hingamp P, Quackenbush J, et al. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat Genet. 2001;29:365–71. doi: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]
- 31.Kendziorski CM, Zhang Y, Lan H, Attie AD. The efficiency of pooling mRNA in microarray experiments. Biostatistics. 2003;4:465–77. doi: 10.1093/biostatistics/4.3.465. [DOI] [PubMed] [Google Scholar]
- 32.Shih JH, Michalowska AM, Dobbin K, Ye Y, Qiu TH, Green JE. Effects of pooling mRNA in microarray class comparisons. Bioinformatics. 2004;20:3318–25. doi: 10.1093/bioinformatics/bth391. [DOI] [PubMed] [Google Scholar]
- 33.Zhang W, Carriquiry A, Nettleton D, Dekkers JC. Pooling mRNA in Microarray Experiments and its Effect on Power. Bioinformatics. 2007 doi: 10.1093/bioinformatics/btm081. [DOI] [PubMed] [Google Scholar]
- 34.Gentleman RC, Carey VJ, Bates DM, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95:14863–8. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karolchik D, Hinrichs AS, Furey TS, et al. The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 2004;32:D493–D496. doi: 10.1093/nar/gkh103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lenhard B, Wasserman WW. TFBS: Computational framework for transcription factor binding site analysis. Bioinformatics. 2002;18:1135–6. doi: 10.1093/bioinformatics/18.8.1135. [DOI] [PubMed] [Google Scholar]
- 38.Sandelin A, Alkema W, Engstrom P, Wasserman WW, Lenhard B. JASPAR: an open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res. 2004;32:D91–D94. doi: 10.1093/nar/gkh012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matys V, Fricke E, Geffers R, et al. TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 2003;31:374–8. doi: 10.1093/nar/gkg108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Munkittrick KR, McMaster ME, McCarthy LH, Servos MR, Van der Kraak GJ. An overview of recent studies on the potential of pulp-mill effluents to alter reproductive parameters in fish. J Toxicol Environ Health B Crit Rev. 1998;1:347–71. doi: 10.1080/10937409809524558. [DOI] [PubMed] [Google Scholar]
- 41.Ge W. Intrafollicular paracrine communication in the zebrafish ovary: the state of the art of an emerging model for the study of vertebrate folliculogenesis. Mol Cell Endocrinol. 2005;237:1–10. doi: 10.1016/j.mce.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 42.Kwok HF, So WK, Wang Y, Ge W. Zebrafish gonadotropins and their receptors: I. Cloning and characterization of zebrafish follicle-stimulating hormone and luteinizing hormone receptors--evidence for their distinct functions in follicle development. Biol Reprod. 2005;72:1370–81. doi: 10.1095/biolreprod.104.038190. [DOI] [PubMed] [Google Scholar]
- 43.So WK, Kwok HF, Ge W. Zebrafish gonadotropins and their receptors: II. Cloning and characterization of zebrafish follicle-stimulating hormone and luteinizing hormone subunits--their spatial-temporal expression patterns and receptor specificity. Biol Reprod. 2005;72:1382–96. doi: 10.1095/biolreprod.104.038216. [DOI] [PubMed] [Google Scholar]
- 44.Wu T, Patel H, Mukai S, et al. Activin, inhibin, and follistatin in zebrafish ovary: expression and role in oocyte maturation. Biol Reprod. 2000;62:1585–92. doi: 10.1095/biolreprod62.6.1585. [DOI] [PubMed] [Google Scholar]
- 45.Pang Y, Ge W. Gonadotropin and activin enhance maturational competence of oocytes in the zebrafish (Danio rerio) Biol Reprod. 2002;66:259–65. doi: 10.1095/biolreprod66.2.259. [DOI] [PubMed] [Google Scholar]
- 46.Hirakawa T, Minegishi T, Abe K, et al. Effect of 2,3,7,8-tetrachlorodibenzo-p-dioxin on the expression of follicle-stimulating hormone receptors during cell differentiation in cultured granulosa cells. Endocrinology. 2000;141:1470–6. doi: 10.1210/endo.141.4.7424. [DOI] [PubMed] [Google Scholar]
- 47.Hirakawa T, Minegishi T, Abe K, Kishi H, Ibuki Y, Miyamoto K. Effect of 2,3,7,8-tetrachlorodibenzo-p-dioxin on the expression of luteinizing hormone receptors during cell differentiation in cultured granulosa cells. Arch Biochem Biophys. 2000;375:371–6. doi: 10.1006/abbi.1999.1678. [DOI] [PubMed] [Google Scholar]
- 48.Petroff BK, Roby KF, Gao X, et al. A review of mechanisms controlling ovulation with implications for the anovulatory effects of polychlorinated dibenzo-p-dioxins in rodents. Toxicology. 2001;158:91–107. doi: 10.1016/s0300-483x(00)00367-x. [DOI] [PubMed] [Google Scholar]
- 49.Fukuzawa NH, Ohsako S, Wu Q, et al. Testicular cytochrome P450scc and LHR as possible targets of 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) in the mouse. Mol Cell Endocrinol. 2004;221:87–96. doi: 10.1016/j.mce.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 50.Petroff BK, Gao X, Rozman KK, Terranova PF. Interaction of estradiol and 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) in an ovulation model: evidence for systemic potentiation and local ovarian effects. Reprod Toxicol. 2000;14:247–55. doi: 10.1016/s0890-6238(00)00075-7. [DOI] [PubMed] [Google Scholar]
- 51.Baba T, Mimura J, Nakamura N, et al. Intrinsic function of the aryl hydrocarbon (dioxin) receptor as a key factor in female reproduction. Mol Cell Biol. 2005;25:10040–51. doi: 10.1128/MCB.25.22.10040-10051.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moran FM, VandeVoort CA, Overstreet JW, Lasley BL, Conley AJ. Molecular target of endocrine disruption in human luteinizing granulosa cells by 2,3,7,8-tetrachlorodibenzo-p-dioxin: inhibition of estradiol secretion due to decreased 17alpha-hydroxylase/17,20-lyase cytochrome P450 expression. Endocrinology. 2003;144:467–73. doi: 10.1210/en.2002-220813. [DOI] [PubMed] [Google Scholar]
- 53.Drenth HJ, Bouwman CA, Seinen W, van den BM. Effects of some persistent halogenated environmental contaminants on aromatase (CYP19) activity in the human choriocarcinoma cell line JEG-3. Toxicol Appl Pharmacol. 1998;148:50–5. doi: 10.1006/taap.1997.8307. [DOI] [PubMed] [Google Scholar]
- 54.Myllymaki SA, Haavisto TE, Brokken LJ, Viluksela M, Toppari J, Paranko J. In utero and lactational exposure to TCDD; steroidogenic outcomes differ in male and female rat pups. Toxicol Sci. 2005;88:534–44. doi: 10.1093/toxsci/kfi308. [DOI] [PubMed] [Google Scholar]
- 55.Dasmahapatra AK, Wimpee BA, Trewin AL, Wimpee CF, Ghorai JK, Hutz RJ. Demonstration of 2,3,7,8-tetrachlorodibenzo-p-dioxin attenuation of P450 steroidogenic enzyme mRNAs in rat granulosa cell in vitro by competitive reverse transcriptase-polymerase chain reaction assay. Mol Cell Endocrinol. 2000;164:5–18. doi: 10.1016/s0303-7207(00)00245-8. [DOI] [PubMed] [Google Scholar]
- 56.Aluru N, Vijayan MM. Aryl hydrocarbon receptor activation impairs cortisol response to stress in rainbow trout by disrupting the rate-limiting steps in steroidogenesis. Endocrinology. 2006;147:1895–903. doi: 10.1210/en.2005-1143. [DOI] [PubMed] [Google Scholar]
- 57.Menuet A, Pellegrini E, Anglade I, et al. Molecular characterization of three estrogen receptor forms in zebrafish: binding characteristics, transactivation properties, and tissue distributions. Biol Reprod. 2002;66:1881–92. doi: 10.1095/biolreprod66.6.1881. [DOI] [PubMed] [Google Scholar]
- 58.Menuet A, Le Page Y, Torres O, Kern L, Kah O, Pakdel F. Analysis of the estrogen regulation of the zebrafish estrogen receptor (ER) reveals distinct effects of ERalpha, ERbeta1 and ERbeta2. J Mol Endocrinol. 2004;32:975–86. doi: 10.1677/jme.0.0320975. [DOI] [PubMed] [Google Scholar]
- 59.Tian Y, Ke S, Thomas T, Meeker RJ, Gallo MA. Transcriptional suppression of estrogen receptor gene expression by 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) J Steroid Biochem Mol Biol. 1998;67:17–24. doi: 10.1016/s0960-0760(98)00067-3. [DOI] [PubMed] [Google Scholar]
- 60.Tian Y, Ke S, Thomas T, Meeker RJ, Gallo MA. Regulation of estrogen receptor mRNA by 2,3,7,8-tetrachlorodibenzo-p-dioxin as measured by competitive RT-PCR. J Biochem Mol Toxicol. 1998;12:71–7. doi: 10.1002/(sici)1099-0461(1998)12:2<71::aid-jbt1>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 61.Wormke M, Stoner M, Saville B, Safe S. Crosstalk between estrogen receptor alpha and the aryl hydrocarbon receptor in breast cancer cells involves unidirectional activation of proteasomes. FEBS Lett. 2000;478:109–12. doi: 10.1016/s0014-5793(00)01830-5. [DOI] [PubMed] [Google Scholar]
- 62.DeVito MJ, Thomas T, Martin E, Umbreit TH, Gallo MA. Antiestrogenic action of 2,3,7,8-tetrachlorodibenzo-p-dioxin: tissue-specific regulation of estrogen receptor in CD1 mice. Toxicol Appl Pharmacol. 1992;113:284–92. doi: 10.1016/0041-008x(92)90126-d. [DOI] [PubMed] [Google Scholar]
- 63.Wang X, Porter W, Krishnan V, Narasimhan TR, Safe S. Mechanism of 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD)-mediated decrease of the nuclear estrogen receptor in MCF-7 human breast cancer cells. Mol Cell Endocrinol. 1993;96:159–66. doi: 10.1016/0303-7207(93)90106-t. [DOI] [PubMed] [Google Scholar]
- 64.Tijet N, Boutros PC, Moffat ID, Okey AB, Tuomisto J, Pohjanvirta R. Aryl hydrocarbon receptor regulates distinct dioxin-dependent and dioxin-independent gene batteries. Mol Pharmacol. 2006;69:140–53. doi: 10.1124/mol.105.018705. [DOI] [PubMed] [Google Scholar]
- 65.Wang Y, Ge W. Developmental profiles of activin betaA, betaB, and follistatin expression in the zebrafish ovary: evidence for their differential roles during sexual maturation and ovulatory cycle. Biol Reprod. 2004;71:2056–64. doi: 10.1095/biolreprod.104.032649. [DOI] [PubMed] [Google Scholar]
- 66.Janz DM, Van Der Kraak G. Suppression of apoptosis by gonadotropin, 17beta-estradiol, and epidermal growth factor in rainbow trout preovulatory ovarian follicles. Gen Comp Endocrinol. 1997;105:186–93. doi: 10.1006/gcen.1996.6820. [DOI] [PubMed] [Google Scholar]
- 67.Pati D, Balshaw K, Grinwich DL, Hollenberg MD, Habibi HR. Epidermal growth factor receptor binding and biological activity in the ovary of goldfish, Carassius auratus. Am J Physiol. 1996;270:R1065–R1072. doi: 10.1152/ajpregu.1996.270.5.R1065. [DOI] [PubMed] [Google Scholar]
- 68.Andreasen EA, Mathew LK, Tanguay RL. Regenerative Growth is Impacted by TCDD: Gene Expression Analysis Reveals Extracellular Matrix Modulation. Toxicol Sci. 2006 doi: 10.1093/toxsci/kfj118. [DOI] [PubMed] [Google Scholar]
- 69.Volz DC, Hinton DE, Law JM, Kullman SW. Dynamic Gene Expression Changes Precede Dioxin-Induced Liver Pathogenesis in Medaka Fish. Toxicol Sci. 2005 doi: 10.1093/toxsci/kfj033. [DOI] [PubMed] [Google Scholar]
- 70.Kajihara M, Kawauchi S, Kobayashi M, Ogino H, Takahashi S, Yasuda K. Isolation, characterization, and expression analysis of zebrafish large Mafs. J Biochem (Tokyo) 2001;129:139–46. doi: 10.1093/oxfordjournals.jbchem.a002825. [DOI] [PubMed] [Google Scholar]
- 71.Li MA, Alls JD, Avancini RM, Koo K, Godt D. The large Maf factor Traffic Jam controls gonad morphogenesis in Drosophila. Nat Cell Biol. 2003;5:994–1000. doi: 10.1038/ncb1058. [DOI] [PubMed] [Google Scholar]
- 72.Yu JH, Guo J, Guo J, Zeng FX, Tang GH. The inhibitory effect and its mechanism of transferrin on FSH-induced differentiation of granulosa cells. Sheng Li Xue Bao. 1994;46:209–16. [PubMed] [Google Scholar]
- 73.Yu JH, Findlay JK. An inhibitory effect of transferrin on differentiation of rat granulosa cells in vitro. Endocrinology. 1991;128:1841–8. doi: 10.1210/endo-128-4-1841. [DOI] [PubMed] [Google Scholar]
- 74.Handley-Goldstone HM, Grow MW, Stegeman JJ. Cardiovascular gene expression profiles of dioxin exposure in zebrafish embryos. Toxicol Sci. 2005;85:683–93. doi: 10.1093/toxsci/kfi116. [DOI] [PubMed] [Google Scholar]
- 75.Carney SA, Chen J, Burns CG, Xiong KM, Peterson RE, Heideman W. AHR Activation Produces Heart-Specific Transcriptional and Toxic Responses in Developing Zebrafish. Mol Pharmacol. 2006 doi: 10.1124/mol.106.025304. [DOI] [PubMed] [Google Scholar]
- 76.Volz DC, Bencic DC, Hinton DE, Law JM, Kullman SW. 2,3,7,8-Tetrachlorodibenzo-p-dioxin (TCDD) induces organ- specific differential gene expression in male Japanese medaka (Oryzias latipes) Toxicol Sci. 2005;85:572–84. doi: 10.1093/toxsci/kfi109. [DOI] [PubMed] [Google Scholar]
- 77.Mishra A, Joy KP. Ovarian monosaccharides (glucose and fructose): hormonal effects and their role in final oocyte maturation and egg quality in catfish Heteropneustes fossilis, Bloch. Indian J Exp Biol. 2004;42:1084–90. [PubMed] [Google Scholar]
- 78.Bornstein SR, Rutkowski H, Vrezas I. Cytokines and steroidogenesis. Mol Cell Endocrinol. 2004;215:135–41. doi: 10.1016/j.mce.2003.11.022. [DOI] [PubMed] [Google Scholar]
- 79.von Schalburg KR, Rise ML, Brown GD, Davidson WS, Koop BF. A comprehensive survey of the genes involved in maturation and development of the rainbow trout ovary. Biol Reprod. 2005;72:687–99. doi: 10.1095/biolreprod.104.034967. [DOI] [PubMed] [Google Scholar]
- 80.Puga A, Maier A, Medvedovic M. The transcriptional signature of dioxin in human hepatoma HepG2 cells. Biochem Pharmacol. 2000;60:1129–42. doi: 10.1016/s0006-2952(00)00403-2. [DOI] [PubMed] [Google Scholar]
- 81.Pande K, Moran SM, Bradfield CA. Aspects of dioxin toxicity are mediated by interleukin 1-like cytokines. Mol Pharmacol. 2005;67:1393–8. doi: 10.1124/mol.105.010983. [DOI] [PubMed] [Google Scholar]
- 82.Chen I, Hsieh T, Thomas T, Safe S. Identification of estrogen-induced genes downregulated by AhR agonists in MCF-7 breast cancer cells using suppression subtractive hybridization. Gene. 2001;262:207–14. doi: 10.1016/s0378-1119(00)00530-8. [DOI] [PubMed] [Google Scholar]
- 83.Kharat I, Saatcioglu F. Antiestrogenic effects of 2,3,7,8-tetrachlorodibenzo-p-dioxin are mediated by direct transcriptional interference with the liganded estrogen receptor. Cross-talk between aryl hydrocarbon- and estrogen-mediated signaling. J Biol Chem. 1996;271:10533–7. doi: 10.1074/jbc.271.18.10533. [DOI] [PubMed] [Google Scholar]
- 84.Cheshenko K, Brion F, Le Page Y, et al. Expression of Zebra Fish Aromatase cyp19a and cyp19b Genes in Response to the Ligands of Estrogen Receptor and Aryl Hydrocarbon Receptor. Toxicol Sci. 2007;96:255–67. doi: 10.1093/toxsci/kfm003. [DOI] [PubMed] [Google Scholar]
- 85.Bemanian V, Male R, Goksoyr A. The aryl hydrocarbon receptor-mediated disruption of vitellogenin synthesis in the fish liver: Cross-talk between AHR-and ERalpha-signalling pathways. Comp Hepatol. 2004;3:2. doi: 10.1186/1476-5926-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tong SK, Chung BC. Analysis of zebrafish cyp19 promoters. J Steroid Biochem Mol Biol. 2003;86:381–6. doi: 10.1016/s0960-0760(03)00347-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.