Abstract
Purpose
We comprehensively evaluated genetic variants in DNA repair genes with premenopausal breast cancer risk.
Methods
In this nested case-control study of 239 prospectively ascertained premenopausal breast cancer cases and 477 matched controls within the Nurses’ Health Study II, we evaluated 1,463 genetic variants in 60 candidate genes across 5 DNA repair pathways, along with DNA polymerases, Fanconi Anemia complementation groups, and other related genes.
Results
Four variants were associated with breast cancer risk with a significance level of <0.01; two in the XPF gene and two in the XRCC3 gene. An increased risk was found in those harboring a greater number of missense putative risk alleles (a priori defined in an independent study) in the non-homologous end-joining repair pathway of double-strand breaks (odds ratio per risk allele, 1.37 (95%confidence interval, 1.03–1.82), P trend, 0.03).
Conclusions
This study implicates variants of genes in the double-strand break repair pathway in the etiology of premenopausal breast cancer.
Keywords: polymorphism, DNA repair, breast cancer, premenopausal women
Introduction
Breast cancer is the most common cancer and the second leading cause of cancer death among women in the United States. Epidemiological studies have shown that familial breast cancer constitutes only about 5–10% of total breast cancer, and only 15–20% of the observed familial clustering of breast cancer is attributable to strongly predisposing BRCA1 and BRCA2 mutations [1]. Most of the genetic variants that contribute to the risk of developing sporadic breast cancer remain unknown [2].
Deficient DNA repair capacity has been suggested as a predisposing factor in familial and sporadic breast cancer [2–5]. Reduced DNA repair capacity among breast cancer cases has been observed in mutagen (X-rays, bleomycin, and BPDE [benzopyrene dihydrodiol epoxide]) sensitivity assays conducted in human peripheral blood lymphocytes [5–9] and in host cell reactivation assays with BPDE- or UV-induced damage [10, 11]. The wide range of carcinogens used in these assays suggests that defects in global DNA repair capacity, rather than a single substrate-specific DNA repair pathway, underlie cancer risk. The spectrum of p53 gene mutations in breast cancer suggests the involvement of multiple genotoxic compounds and DNA repair abnormalities in breast cell mutagenesis [12, 13]. The importance of DNA repair in breast cancer development is further supported by the involvement of BRCA1 and BRCA2 in many critical cellular processes including multiple DNA repair pathways and apoptosis through protein-protein interactions and transcriptional regulation. One mechanism that may lead to inter-individual variation in DNA repair capacity is germline variation in DNA repair genes [14–16]. Even though a variety of factors modulate the path from genotype to phenotype, there are substantial correlations between DNA repair gene variants and DNA repair capacity [17]. A deficient DNA repair capacity may be attributable to multiple polymorphisms in multiple DNA repair pathways.
Breast cancer in premenopausal women is more aggressive, with a poorer prognosis than postmenopausal breast cancer. The etiology for premenopausal breast cancer may differ from that for postmenopausal women, and involve a relatively stronger component of inherited predisposition. In this study of 239 cases and 477 matched controls among premenopausal predominantly Caucasian women in a nested case-control study within the Nurses’ Health Study II, we comprehensively and systematically evaluated genetic variation in 60 DNA repair genes in relation to breast cancer risk. These pathways/genes included direct reversion repair (MGMT), base excision repair (BER) (APE1, LIG3, NEIL1, NEIL2, OGG1, PARP1, XRCC1, FEN1), nucleotide excision repair (NER) (XPA, ERCC3, XPC, ERCC2, ERCC4, ERCC5, ERCC1, LIG1, ERCC6, ERCC8, RPA1, RPA2, RPA3), double-strand break (DSB) repair via a) homologous recombination (HR) (RAD50, RAD51, RAD52, XRCC2, XRCC3, NBN, MRE11A, ATM, ATR) or b) non-homologous end-joining (NHEJ) (XRCC4, XRCC5, XRCC6, ARTEMIS, PRKDC, LIG4), mismatch repair (MMR) (MSH2, MSH3, MSH6, MLH1, MLH3, PMS1, PMS2), DNA polymerases (POLB, POLD1, POLE, POLI, POLK), Fanconi Anemia complementation groups (FANCA, FANCC, FANCD2, FANCE, FANCF, FANCG), and other related genes (CHEK1, CHEK2, TP53, PCNA, BLM).
Materials and Methods
Study Population
The Nurses’ Health Study II was established in 1989 when 116,609 female registered nurses, ages 25 to 42 years, completed andreturned a mailed questionnaire. The cohort has been followed biennially to update exposures and ascertain newly diagnosed diseases. Between 1996 and 1999, 29,611 cohort members who werecancer-free and between the ages of 32 and 54 years providedblood samples [18]. Briefly, participants were sent a short questionnaire and a blood collection kit containing necessary supplies to have blood samples drawn by a local laboratoryor a colleague. Premenopausal women who had not taken oral contraceptives, been pregnant, or breast-fed within 6 months (n = 18,521) providedblood samples drawn on the 3rd to 5th day of their menstrualcycle (follicular draw) and 7 to 9 days before the anticipated start of their next cycle (luteal draw). All other women (n = 11,090) provided a single 30-mL, untimed blood sample. These samples were collected in a similar manner, shipped viaovernight courier with an ice pack to our laboratory, and separatedinto plasma, RBC, and WBC components. Samples have been storedin liquid nitrogen freezers since collection. Menopausal status determination for women providing untimed samples has been described previously [18]. Follow-up of the blood cohort was 98% in 2003. The study was approved by the Committee on the Use of Human Subjects in Research at Harvard School of Public Health andBrigham and Women’s Hospital.
Breast cancer cases were identified on biennial questionnaires;the National Death Index was searched for nonresponders. Caseshad no previously reported cancer diagnosis and were diagnosedwith breast cancer after blood collection but before June 1,2003. Each of the 239 premenopausal cases of breast cancerwas matched to two premenopausal controls (one pair with only 1:1 matching) (total n = 477) on age (±2years), month/year of blooddraw (±2 months), and race/ethnicity (Caucasian, African American, Asian, Hispanic, Other) (>93% of cases and controls are Caucasian), and for each blood collection, time of day (±2 hours), and fasting status (<2 h, 2–4, 5–7, 8–11,≥12). For each matching variable, >90% of matches were exact.
Single nucleotide polymorphism (SNP) selection
The characterization of common genetic variation in candidate DNA repair and related genes was conducted by genotyping a high density of common SNPs across the promoter, untranslated regions (UTRs), and coding and non-coding regions of 60 DNA repair genes [19]. Briefly, genotype data were collected from seven population samples, including 20 CEPH trios (60 individuals in total), which are a subset of the 30 trios used in the HapMap and 70 White subjects from the Multiethnic Cohort (MEC) study [20]. In total, about 3,000 SNPs have been genotyped across these 60 genes, including a high density of common SNPs (n > 2,700, minor allele frequency ≥ 5%) selected from the public dbSNP database and all known missense SNPs (>300, minor allele frequency ≥ 1%) identified through gene resequencing from the Environmental Genome Project (http://egp.gs.washington.edu/); the average spacing of common SNPs across each locus is 1.7 kb. Tag-SNPs were selected by the Tagger approach [21], which combines pairwise r2 methods [22] with the potential efficiency of multi-marker approaches [23]. In the selection of tag-SNPs for Caucasians (r2 >0.8), these SNPs genotyped in-house in the 20 CEPH trios and the HapMap phase I data of the same 60 Caucasians were combined to achieve a much higher density of SNP markers. The patterns of linkage disequilibrium (LD) in these individuals should provide an accurate estimate of the patterns in our study population [24]. The detailed description of the tag SNP selection for predicting untyped SNPs was presented elsewhere [19]. In brief, 91% of HapMap phase II SNPs are predicted by this panel with 80% or greater multi-allelic r2.
SNP Genotyping
High-throughput genotyping was performed using the Illumina high-multiplex BeadArray genotyping system at the MIT Broad Institute, Center for Genotyping and Analysis. The assay employs allele-specific extension methods and universal PCR amplification reactions conducted at 1,536 loci. DNA samples were processed through the highly multiplexed GoldenGate protocol using bar-coded microwell plates and robust automation systems. Among the 1,536 SNPs, there are 1,463 SNPs in 60 DNA repair genes, as described above.
The initial set of SNPs was chosen to include tag-SNPs for other ethnicities. Excluding 98 non-Caucasian SNPs, 1263 (88%) SNPs had a genotyping success rate >95%, and 1322 (92%) SNPs had a genotyping success rate ≥80%. SNPs with a genotyping success rate <80% were excluded from further analysis. Eight pairs of blinded duplicate samples were included. Analysis of 10072 pair tests revealed a 99.95% overall concordance rate. Five SNPs that failed the concordance test were excluded. Among these 1317 SNPs, there remained 1256 SNPs in the DNA repair genes for further analysis. There were 1088 out of the 1256 SNPs with minor allele frequency >0.01 in controls of our study. Among the controls, 38 loci had Hardy-Weinberg equilibrium χ2 p-values < 0.01 and were excluded. Hence, the final analysis included 1050 SNPs in the DNA repair genes.
Statistical Analysis
Analysis of main effect
Conditional logistic regression was employed to calculate odds ratios (ORs) and 95% confidence intervals (CIs). The test for main effects of SNPs was based on the additive model, treating genotype as an ordinal variable (wildtype coded as 0, heterozygote as 1, and homozygotes variant as 2). All P values were two-sided.
SNP spectral decomposition (SNPSpD) for correction of multiple testing
The Bonferroni correction, which is the most commonly used method to adjust type I error, α, treats every single-SNP test as an independent test and is overly conservative for SNPs that are in LD, because the Bonferroni correction ignores the correlation among SNPs. To address this limitation, we calculated the effective number of independent SNPs, Meff,i, for each candidate gene i, on the basis of the spectral decomposition (SpD) of matrices of pair-wise LD between SNPs [25, 26]. Meff provides a simple correction for multiple testing of non-independent SNPs in LD with each other. For each SNP for candidate gene i, the multiplicity-adjusted point-wise α (αp) was then calculated as α/Meff,i.
Interaction and subgroup analyses
Analysis of interactions between genetic variants and family history of breast cancer and subgroup analysis according to estrogen receptor (ER) and progesterone receptor (PR) status were restricted to those variants with P values <0.05 in the analysis of main effect. Unconditional logistic regression was used in these analyses. We modeled family history of breast cancer as a dichotomous variable (yes/no) and genotypes as carriers of variants vs non-carriers in the interaction analysis. We used a likelihood ratio test (LRT) to compare nested models that included terms for all combinations of the genotype and family history in the models with indicator variables for the main effects only. In subgroup analysis, each subtype of cases was compared with the common controls.
Selection of missense SNPs
In the final panel of 1,050 SNPs after exclusion criteria (refer to Results section), 65 SNPs were missense SNPs. Among them, 4 SNPs (NEIL2 rs8191664, CSB rs2228529, CSB rs2228526, and XPD rs1799793) were in high LD (defined as r2>0.90) with another missense SNP in the same gene and were excluded. Eight women had missing genotype data at > 10 loci and were removed. Hence, the analysis of missense SNPs was restricted to 61 SNPs in 31 genes among 708 women. We used the Partition-Ligation Expectation-Maximization (PLEM) algorithm [27] to impute the missing genotypes based on the estimated haplotype frequencies within each gene. In the event of only one single SNP in a candidate gene, missing genotypes were imputed by using the most common genotype for that SNP (User Manual of open source Java software Multifactor Dimensionality Reduction (MDR) 1.0.0 (http://sourceforge.net/projects/mdr/)) [28, 29].
Combined risk allele analysis of multiple missense SNPs
To test the hypothesis that multiple missense SNPs in the same pathway have an additive effect on breast cancer risk, we estimated the combined effect of the risk alleles for these SNPs in each pathway. First, we evaluated the main effect associated with each minor allele in an independent dataset, a set of 45 cases and 90 controls in premenopausal Caucasian women in the Multiethnic Cohort study [19]. If the minor allele was associated with an increased risk of breast cancer, we designated the minor allele as the risk allele. If the minor allele was found to be inversely associated with risk, we designated the common allele as the risk allele. We applied this a priori definition of risk allele for each locus from this independent dataset to risk allele designation in our study population. We summed the number of risk alleles of each pathway for each individual and evaluated the risk associated with the increasing number of risk alleles.
Results
Participants were 32 to 52 years old (mean, 44 years) at blood collection (Table 1). Differences between cases and controls for age at menarche, parity, and BMI at blood draw generally were small. A higher percentage of cases versus controls had a family history of breast cancer (19.3% versus 12.3%, respectively) and a history of benign breast disease (22.2% versus 16.1%, respectively).
Table 1.
Characteristics at blood collection of cases and their matched controls from the NHSII
| Cases (n =239) | Controls (n = 477) | |
|---|---|---|
| Age (y), mean (SD) | 44.1 (4.0) | 43.8 (3.9) |
| Parity,* mean (SD) | 2.1 (0.8) | 2.3 (1.0) |
| BMI at age 18 (kg/m2), mean (SD) | 20.9 (3.1) | 21.0 (2.6) |
| BMI at blood draw (kg/m2), mean (SD) | 24.9 (5.0) | 25.1 (5.5) |
| Family history of breast cancer, % | 19.3 | 12.3 |
| History of benign breast disease, % | 22.2 | 16.1 |
| Age at menarche >14 y, % | 15.8 | 17.5 |
| Ever used oral contraceptives, % | 82.9 | 85.6 |
Among parous women only.
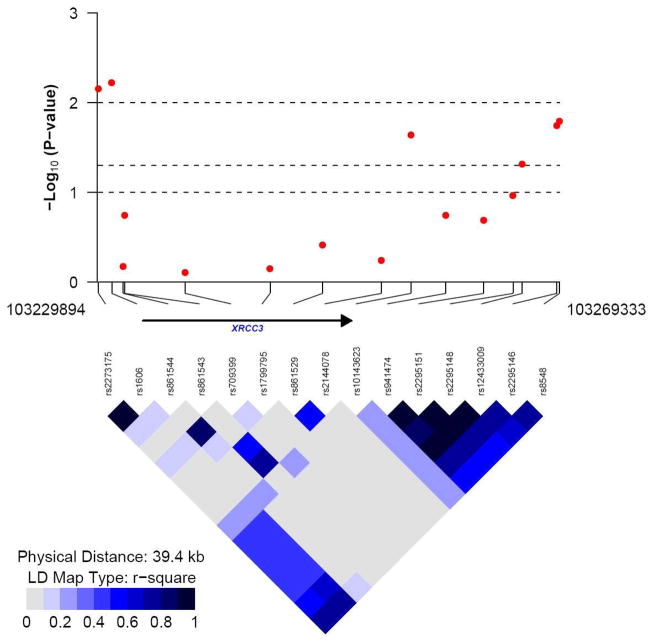
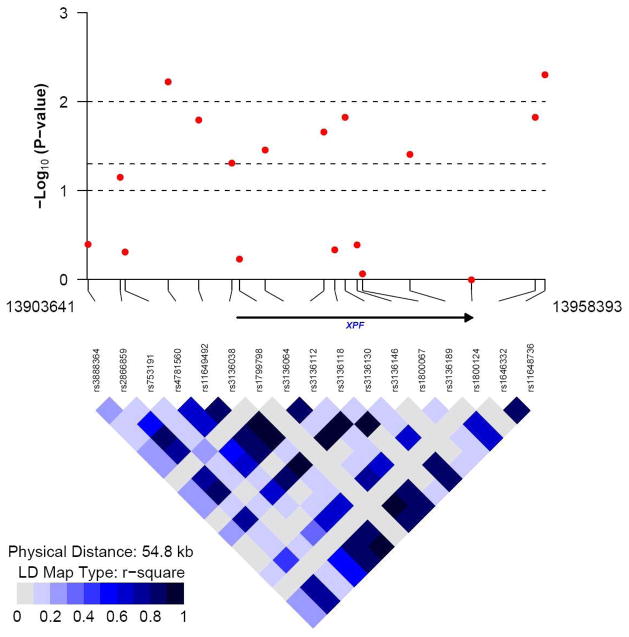
Forty-four SNPs were associated with altered pre-menopausal breast cancer risk in our study (Table 2), with P value <0.05 in the additive model. These 44 SNPs were located in 18 DNA repair genes with 1–3 SNPs per gene except for the XPF and XRCC3 genes. There were 9 SNPs in XPF and 6 in XRCC3. Among the 44 SNPs, four SNPs showed a significance level of <0.01; two SNPs in the XPF gene (R2=0.88) and two SNPs in the XRCC3 gene (R2=0.99). The LD plots for these two genes are displayed in Figure 1.
Table 2.
Main effects of 44 SNPs on pre-menopausal breast cancer risk in the additive model with P value <0.05
| Wildtype | Heterozygote | Homozygous variant | ||||
|---|---|---|---|---|---|---|
| Gene | SNP | case/control | case/control | case/control | Additive model OR (95%CI) | P, trend |
| XPF | RS11648736 | 127/193 | 89/222 | 22/58 | 0.71 (0.56–0.90) | 0.005 |
| XRCC3 | RS1606 | 127/215 | 95/195 | 15/60 | 0.70 (0.55–0.90) | 0.006 |
| XPF | RS4781560 | 144/236 | 80/200 | 14/39 | 0.69 (0.53–0.90) | 0.006 |
| XRCC3 | RS2273175 | 127/218 | 95/192 | 16/64 | 0.71 (0.56–0.91) | 0.007 |
| CHEK2 | RS10854805 | 161/282 | 70/164 | 7/28 | 0.69 (0.52–0.92) | 0.01 |
| RPA3 | RS2057931 | 69/165 | 98/193 | 43/54 | 1.38 (1.07–1.78) | 0.013 |
| XPF | RS3136130 | 121/189 | 89/223 | 25/59 | 0.74 (0.58–0.94) | 0.015 |
| XPF | RS1646332 | 117/187 | 95/220 | 24/65 | 0.74 (0.59–0.94) | 0.015 |
| RPA3 | RS6967126 | 73/182 | 116/221 | 45/64 | 1.33 (1.06–1.68) | 0.015 |
| XRCC3 | RS8548 | 120/205 | 98/203 | 20/66 | 0.74 (0.58–0.94) | 0.016 |
| XPF | RS11649492 | 114/185 | 99/226 | 24/64 | 0.74 (0.59–0.95) | 0.016 |
| XRCC3 | RS2295146 | 104/173 | 105/213 | 29/85 | 0.75 (0.60–0.95) | 0.018 |
| POLK | RS3213801 | 130/303 | 99/153 | 10/16 | 1.40 (1.06–1.85) | 0.018 |
| RPA1 | RS5030740 | 137/315 | 92/146 | 10/13 | 1.39 (1.06–1.84) | 0.019 |
| POLK | RS5744533 | 128/301 | 97/148 | 9/16 | 1.41 (1.06–1.88) | 0.019 |
| PARP1 | RS10915985 | 66/179 | 128/219 | 43/73 | 1.31 (1.04–1.64) | 0.02 |
| XPC | RS2733536 | 117/277 | 101/170 | 21/29 | 1.32 (1.04–1.68) | 0.021 |
| MSH3 | RS1650697 | 140/245 | 67/163 | 12/35 | 0.72 (0.54–0.95) | 0.022 |
| RPA3 | RS6966464 | 96/219 | 108/209 | 31/35 | 1.33 (1.04–1.70) | 0.022 |
| XPF | RS3136112 | 120/194 | 92/219 | 23/60 | 0.76 (0.59–0.96) | 0.022 |
| XRCC3 | RS10143623 | 90/212 | 103/197 | 44/60 | 1.29 (1.04–1.62) | 0.023 |
| RPA1 | RS12727 | 138/311 | 89/151 | 11/12 | 1.38 (1.04–1.82) | 0.025 |
| NEIL2 | RS8191649 | 132/304 | 93/149 | 14/21 | 1.35 (1.04–1.74) | 0.025 |
| CHEK2 | RS5752777 | 170/305 | 63/146 | 6/25 | 0.72 (0.54–0.96) | 0.026 |
| NEIL2 | RS8191642 | 130/301 | 95/152 | 14/22 | 1.34 (1.03–1.73) | 0.028 |
| FANCG | RS634801 | 52/144 | 121/222 | 62/105 | 1.29 (1.02–1.62) | 0.03 |
| CHEK1 | RS3731459 | 218/408 | 19/63 | 0/1 | 0.56 (0.33–0.94) | 0.03 |
| CHEK2 | RS6519761 | 174/314 | 59/139 | 6/23 | 0.72 (0.54–0.97) | 0.033 |
| MSH3 | RS380691 | 99/219 | 107/214 | 33/39 | 1.30 (1.02–1.65) | 0.033 |
| CHEK1 | RS7104660 | 220/413 | 18/60 | 0/1 | 0.56 (0.33–0.96) | 0.034 |
| XPF | RS3136064 | 118/195 | 96/213 | 24/65 | 0.77 (0.61–0.98) | 0.035 |
| APE1 | RS11160682 | 76/179 | 120/238 | 36/49 | 1.28 (1.01–1.62) | 0.037 |
| FANCC | RS356664 | 105/163 | 97/237 | 33/73 | 0.79 (0.63–0.99) | 0.038 |
| FANCC | RS554879 | 106/161 | 97/238 | 34/74 | 0.79 (0.63–0.99) | 0.039 |
| XPF | RS3136189 | 138/235 | 85/193 | 16/46 | 0.77 (0.60–0.99) | 0.039 |
| ATR | RS2227928 | 88/136 | 94/221 | 42/91 | 0.78 (0.62–0.99) | 0.039 |
| POLD | RS3218772 | 236/457 | 2/18 | 0/0 | 0.22 (0.05–0.96) | 0.044 |
| Ku70 | RS6002421 | 235/458 | 1/14 | 0/1 | 0.12 (0.01–0.95) | 0.044 |
| POLK | RS3756558 | 181/332 | 53/124 | 3/16 | 0.73 (0.53–0.99) | 0.046 |
| XRCC4 | RS10057194 | 217/404 | 18/63 | 2/4 | 0.61 (0.37–0.99) | 0.047 |
| XPC | RS2470352 | 128/295 | 93/157 | 13/19 | 1.30 (1.00–1.69) | 0.048 |
| XRCC3 | RS12433009 | 83/138 | 107/214 | 45/113 | 0.80 (0.64–1.00) | 0.048 |
| XPF | RS3136038 | 119/193 | 94/221 | 23/52 | 0.78 (0.61–1.00) | 0.049 |
| XPC | RS2733537 | 89/210 | 112/217 | 34/46 | 1.26 (1.00–1.59) | 0.049 |
The data on all 1050 SNPs in final analysis are listed in supplementary Table 1.
Figure 1.
The −log10 (P value for the association with breast cancer risk) and LD R2 plot generated for (a) XRCC3 gene (15 SNPs, 477 control subjects), and (b) XPF (ERCC4) gene (17 SNPs, 477 control subjects) respectively.
The data on the main effect of 1050 SNPs are provided in Supplementary Table 1. We performed analysis on interactions between genetic variants and family history of breast cancer and subgroup analysis according to estrogen receptor/progesterone receptor (ER/PR) status. These analyses were restricted to those variants with P value <0.05 in the analysis of main effect. The data are provided in Supplementary Tables 2–3.
We calculated the Meff value by SNPSpD for each of the 60 candidate genes (Table 3). On average, each candidate gene has 17.5±14.18 (Mean±SD) SNPs (range: 5 [NEIL1] - 69 [MGMT] SNPs). Because of the linkage disequilibrium (LD) among SNPs within each gene, on average, the value of Meff of each candidate gene is 14.18±10.01 (range: 3.44 [NEIL1] - [MGMT] 63.12). The percentage of reduction (i.e. how much the use of SNPSpD has “compressed” the total number of SNPs for a candidate gene i, defined as ) is 21.23±7.63% (range: 8.52% [MGMT, 69 SNPs, Meff = 63.12] - 45.97% [MLH3, 9 SNPs, Meff = 4.86]). We used the Meff value for correcting for multiple comparisons for each gene. As shown in Table 3, for all genes, the smallest P value for individual SNP was larger than the significance threshold adjusted by Meff value.
Table 3.
Gene-by-gene correction for multiple testing of SNPs that are in linkage disequilibrium (LD), based on the spectral decomposition (SpD) of pairwise LD matrices for SNP pairs.1
| Gene Symbol | Number of SNPs | M 2 | Meff3 | Adjusted threshold 4 | Smallest P value for individual SNP |
|---|---|---|---|---|---|
| XPF | 17 | 17 | 11.83 | 0.004 | 0.005 |
| XRCC3 | 15 | 15 | 11.48 | 0.004 | 0.006 |
| CHEK2 | 20 | 20 | 16.99 | 0.003 | 0.01 |
| RPA3 | 39 | 39 | 33.62 | 0.001 | 0.013 |
| POLK | 13 | 13 | 9.89 | 0.005 | 0.018 |
| RPA1 | 31 | 31 | 26.83 | 0.002 | 0.019 |
| PARP1 | 13 | 13 | 9.26 | 0.005 | 0.02 |
| XPC | 17 | 17 | 12.89 | 0.004 | 0.021 |
| MSH3 | 42 | 42 | 36.17 | 0.001 | 0.022 |
| NEIL2 | 37 | 37 | 30.71 | 0.002 | 0.025 |
| FANCG | 12 | 12 | 9.15 | 0.005 | 0.03 |
| CHEK1 | 12 | 12 | 10.16 | 0.005 | 0.03 |
| APE1 | 17 | 17 | 13.59 | 0.004 | 0.037 |
| FANCC | 14 | 14 | 10.58 | 0.005 | 0.038 |
| ATR | 14 | 14 | 12.11 | 0.004 | 0.039 |
| POLD | 13 | 13 | 11.61 | 0.004 | 0.044 |
| Ku70 | 7 | 7 | 5.40 | 0.009 | 0.044 |
| XRCC4 | 42 | 42 | 35.63 | 0.001 | 0.047 |
| PMS1 | 20 | 20 | 16.44 | 0.003 | 0.064 |
| Artemis | 19 | 19 | 15.08 | 0.003 | 0.068 |
| XPA | 14 | 14 | 11.87 | 0.004 | 0.069 |
| LIG1 | 25 | 25 | 19.39 | 0.003 | 0.073 |
| LIG4 | 17 | 17 | 13.84 | 0.004 | 0.076 |
| MGMT | 69 | 69 | 63.12 | 0.001 | 0.081 |
| XRCC2 | 19 | 19 | 16.42 | 0.003 | 0.091 |
| DNA-PK | 17 | 17 | 14.20 | 0.004 | 0.096 |
| XRCC1 | 14 | 14 | 11.95 | 0.004 | 0.10 |
| FANCE | 16 | 16 | 13.10 | 0.004 | 0.101 |
| RAD52 | 21 | 21 | 18.51 | 0.003 | 0.101 |
| BLM | 37 | 37 | 31.51 | 0.002 | 0.101 |
| MSH6 | 17 | 17 | 13.66 | 0.004 | 0.11 |
| OGG1 | 19 | 19 | 15.23 | 0.003 | 0.116 |
| PMS2 | 19 | 19 | 16.83 | 0.003 | 0.118 |
| TP53 | 10 | 10 | 8.82 | 0.006 | 0.123 |
| PCNA | 14 | 14 | 12.03 | 0.004 | 0.123 |
| FANCD2 | 7 | 7 | 4.42 | 0.011 | 0.127 |
| NBS1 | 14 | 14 | 10.14 | 0.005 | 0.138 |
| POLB | 10 | 10 | 7.76 | 0.006 | 0.145 |
| XPD | 12 | 12 | 9.51 | 0.005 | 0.15 |
| NEIL1 | 5 | 5 | 3.44 | 0.015 | 0.16 |
| MLH1 | 8 | 8 | 5.18 | 0.010 | 0.179 |
| LIG3 | 11 | 11 | 8.63 | 0.006 | 0.181 |
| XPG | 16 | 16 | 13.39 | 0.004 | 0.199 |
| CSB | 23 | 23 | 18.43 | 0.003 | 0.202 |
| FANCF | 8 | 8 | 6.95 | 0.007 | 0.225 |
| ERCC1 | 11 | 11 | 7.77 | 0.006 | 0.236 |
| RAD51 | 7 | 7 | 5.55 | 0.009 | 0.239 |
| POLE | 15 | 15 | 11.34 | 0.004 | 0.244 |
| MRE11 | 13 | 13 | 8.63 | 0.006 | 0.251 |
| Ku80 | 26 | 26 | 21.91 | 0.002 | 0.263 |
| FEN1 | 8 | 8 | 5.83 | 0.009 | 0.267 |
| MSH2 | 24 | 24 | 17.37 | 0.003 | 0.286 |
| CSA | 11 | 11 | 8.22 | 0.006 | 0.288 |
| RPA2 | 6 | 6 | 4.01 | 0.012 | 0.305 |
| XPB | 12 | 12 | 9.55 | 0.005 | 0.313 |
| RAD50 | 8 | 8 | 5.24 | 0.010 | 0.316 |
| FANCA | 19 | 19 | 13.10 | 0.004 | 0.35 |
| ATM | 15 | 15 | 11.60 | 0.004 | 0.357 |
| POLI | 10 | 10 | 8.20 | 0.006 | 0.413 |
| MLH3 | 9 | 9 | 4.86 | 0.010 | 0.518 |
Calculated using the SNPSpD method available at: http://genepi.qimr.edu.au/general/daleN/SNPSpD/
M: Original (total) number of marker loci after removing redundant (collinear) SNPs
Meff: Effective number of independent marker loci (caclulated using the formula: Meff = 1+(M−1) (1−Var(λobs)/M). The genome-wide significance threshold after Bonferoni correction would be αnominal/Meff = 0.05/850.92 = 5.88 × 10−5.
Adjusted threshold for significance for each gene, which is 0.05/Meff.
We evaluated the effect of multiple missense SNPs on premenopausal breast cancer risk. We first evaluated the main effect associated with each minor allele in a set of 45 cases and 90 controls in premenopausal Caucasian women in the Multiethnic Cohort study. We used the direction of the associations observed in this independent dataset as a priori definition of risk allele for each locus to assign risk allele in our study population. We summed the number of risk alleles of each pathway for each individual and evaluated the risk associated with the increasing number of risk alleles. The associations between the number of putative risk alleles carried in each pathway and breast cancer risk are presented in Table 4. A trend toward increased risk of breast cancer was found among women carrying a greater number of putative risk alleles in the DSB-NHEJ pathway. The OR associated with an additional risk allele in this pathway was 1.37 (95%CI, 1.03–1.82; P for trend, 0.03). Compared with women with 2–3 risk alleles, those with 4 risk alleles had OR of 1.69 (95%CI, 1.08–2.64) and those with 5–6 risk alleles had OR of 1.92 (95%CI, 1.02–3.60). No significant trend was observed for other pathways.
Table 4.
Combined effects of risk alleles of missense SNPs in each pathway
| Pathway | No. of risk alleles | Cases (%) | Controls (%) | OR (95% CI) | P, trend |
|---|---|---|---|---|---|
| BER | 3–5 | 58 (24.5) | 102 (21.7) | 1.00 | |
| 6 | 88 (37.1) | 152 (32.3) | 1.00 (0.66 – 1.52) | ||
| 7 | 64 (27.0) | 142 (30.1) | 0.80 (0.52 – 1.24) | ||
| 8–9 | 27 (11.4) | 75 (15.9) | 0.63 (0.36 – 1.09) | ||
| Per allele | 0.88 (0.76 – 1.02) | 0.09 | |||
|
| |||||
| NER | 5– 9 | 93 (39.2) | 190 (40.3) | 1.00 | |
| 10 | 57 (24.1) | 96 (20.4) | 1.21 (0.80 – 1.84) | ||
| 11 | 41 (17.3) | 102 (21.7) | 0.82 (0.53 – 1.28) | ||
| 12–16 | 46 (19.4) | 83 (17.6) | 1.16 (0.74 – 1.80) | ||
| Per allele | 1.01 (0.92 – 1.11) | 0.83 | |||
|
| |||||
| DSB-HR | 3–5 | 74 (31.2) | 138 (29.3) | 1.00 | |
| 6 | 67 (28.3) | 145 (30.8) | 0.86 (0.57 – 1.30) | ||
| 7 | 46 (19.4) | 107 (22.7) | 0.80 (0.51 – 1.25) | ||
| 8–12 | 50 (21.1) | 81 (17.2) | 1.15 (0.73 – 1.81) | ||
| Per allele | 0.99 (0.89 – 1.11) | 0.84 | |||
|
| |||||
| DSB-NHEJ | 2–3 | 31 (13.1) | 96 (20.4) | 1.00 | |
| 4 | 179 (75.5) | 329 (69.9) | 1.69 (1.08 – 2.64) | ||
| 5–6 | 27 (11.4) | 46 (9.8) | 1.92 (1.02 – 3.60) | ||
| Per allele | 1.37 (1.03 – 1.82) | 0.03 | |||
|
| |||||
| MMR | 2–4 | 82 (34.6) | 188 (39.9) | 1.00 | |
| 5 | 72 (30.4) | 114 (24.2) | 1.47 (0.99 – 2.18) | ||
| 6 | 46 (19.4) | 87 (18.5) | 1.23 (0.79 – 1.92) | ||
| 7–10 | 37 (15.6) | 82 (17.4) | 1.03 (0.65 – 1.65) | ||
| Per allele | 1.07 (0.96 – 1.18) | 0.22 | |||
|
| |||||
| DNA Polymerase | 3–4 | 38 (16.0) | 70 (14.9) | 1.00 | |
| 5 | 89 (37.6) | 179 (38.0) | 0.92 (0.57 – 1.47) | ||
| 6 | 95 (40.1) | 172 (36.5) | 1.01 (0.63 – 1.62) | ||
| 7–8 | 15 (6.3) | 50 (10.6) | 0.55 (0.27 – 1.11) | ||
| Per allele | 0.94 (0.78 – 1.13) | 0.50 | |||
|
| |||||
| 0–3 | 70 (29.5) | 130 (27.6) | 1.00 | ||
| Fanconi Anemia groups | 4 | 96 (40.5) | 197 (41.8) | 0.88 (0.60 – 1.29) | |
| 5 | 59 (24.9) | 119 (25.3) | 0.91 (0.59 – 1.39) | ||
| 6 | 12 (5.1) | 25 (5.3) | 0.86 (0.40 – 1.82) | ||
| Per allele | 0.97 (0.83 – 1.14) | 0.69 | |||
Discussion
Despite evidence of the role of high-penetrance mutations in BRCA1/2 in breast cancer, the importance of common inherited variants in DNA repair pathways and their interactions with environmental factors in causing breast cancer are relatively unknown. There are some published data on select genetic polymorphisms in DNA repair genes and breast cancer risk. However, previous studies have not given extensive consideration to multiple genes and polymorphisms in the pathways. We evaluated in considerably more detail the common variants in DNA repair and related genes using both missense-SNP and tag-SNP approaches among premenopausal women.
Specific DNA repair pathways are responsible for the repair of different types of DNA damage. (1) The BER is responsible for a wide variety of non-bulky exogenous and endogenous oxidative DNA damage and single strand breaks [30]. (2) The NER is a versatile repair system to remove a wide variety of bulky, helix-distorting lesions and adducts induced by environmental chemicals or endogenous metabolites [31, 32]. (3) The HR and NHEJ are two distinct mechanisms in the repair of DSB in mammalian cells. DSBs can be induced by other exogenous agents and endogenous reactive oxygen species. DSBs can also be generated as products of blocked replication forks and programmed rearrangements [33, 34]. (4) The MMR is responsible for the repair of base mispair and insertion/deletion mispair. Mutations in genes involved in mismatch repair (MSH2, MLH1, PMS1, and PMS2) result in microsatellite instability and replication errors. (5) The O6-methylguanine DNA methyltransferase (MGMT) is the gene involved in the direct reversal DNA repair that removes alkyl or methyl adducts from the O6 position of guanine. (6) Other candidates include Fanconi Anemia complementation groups and DNA polymerases [35]. Fanconi anaemia genes interact with DNA-damage-response proteins and other proteins related to cellular responses to carcinogenic stress and to caretaker and gatekeeper functions. Many different DNA polymerases found in human cells are specialized for operation in distinct DNA repair pathways, or for bypass of specific classes of adducts in DNA [36].
A complex disease such as breast cancer occurs through an intricate multifactorial interaction of genetic risk factors. In the analysis of main effect of 1,050 SNPs, two SNPs in the XRCC3 gene and two in the XPF gene were associated with altered breast cancer risk with P <0.01. There were 6 SNPs in the XRCC3 gene and 9 SNPs in the XPF gene with P <0.05. The XRCC3 gene is involved in DSB repair and the XPF gene is involved in NER pathway. Further work is needed to replicate these findings and identify variants across both loci to determine the optimal candidates for epidemiological and functional studies.
A dose-response relation between the increasing number of risk alleles in DNA repair genes and the decreased DNA repair capacity at the individual level has been shown [37]. We thus analyzed combined missense SNPs in each pathway. We defined risk alleles for missense SNPs on the basis of an independent external dataset of premenopausal Caucasian breast cancer cases and controls and evaluated the combined effect of these risk alleles in each pathway in our study. We found a significant trend of increased risk with increasing numbers of risk alleles in the DSB-NHEJ pathway. No such trend was observed for other pathways, which suggests differential contribution of each DNA repair pathway to breast cancer risk. The importance of DSB repair in breast cancer development is further supported by the involvement of BRCA1 and BRCA2 in the repair process of DSB. It has been shown that breast epithelium uniquely lacks redundant systems of DSB repair that are present in other tissues [38, 39], which suggests defects in the repair of DSB may be particularly important for breast cancer development. The NHEJ is the predominant mechanism in the repair of DSB in mammalian cells and is an error-prone repair process. Our data suggest the additive or synergistic effect of multiple DNA repair variants in the NHEJ pathway on premenopausal breast cancer risk and highlight the importance of a pathway-based approach to analyze multiple genes and polymorphisms for risk assessment. Further research is warranted to confirm these findings in premenopausal Caucasian women.
Supplementary Material
Acknowledgments
We thank Dr. Paul de Bakker at the Broad Institute of Harvard and MIT for selecting tagging SNPs. We thank Pati Soule for her laboratory assistance, Dr. Daniel B. Mirel at the Broad Institute Center for Genotyping and Analysis for his coordination, and Dr. Fredrick Schumacher for generating the LD plots. We also thank the participants in the Nurses’ Health Study for their dedication and commitment. The work is supported by NIH grants CA098233, CA118447, CA067262, and CA050385. The Broad Institute Center for Genotyping and Analysis is supported by grant U54 RR020278-01 from the National Center for Research Resources.
Abbreviations
- BER
base excision repair
- CI
confidence interval
- DSB
double strand break
- ER
estrogen receptor
- HR
homologous recombination
- LD
linkage disequilibrium
- MMR
mismatch repair
- NER
nucleotide excision repair
- NHEJ
non-homologous end-joining
- OR
odds ratio
- PR
progesterone receptor
- SNP
single nucleotide polymorphism
References
- 1.Ponder BA. Cancer genetics. Nature. 2001;411(6835):336–341. doi: 10.1038/35077207. [DOI] [PubMed] [Google Scholar]
- 2.Balmain A, Gray J, Ponder B. The genetics and genomics of cancer. Nat Genet. 2003;33(Suppl):238–244. doi: 10.1038/ng1107. [DOI] [PubMed] [Google Scholar]
- 3.Helzlsouer KJ, Harris EL, Parshad R, Fogel S, Bigbee WL, Sanford KK. Familial clustering of breast cancer: possible interaction between DNA repair proficiency and radiation exposure in the development of breast cancer. Int J Cancer. 1995;64(1):14–17. doi: 10.1002/ijc.2910640105. [DOI] [PubMed] [Google Scholar]
- 4.Helzlsouer KJ, Harris EL, Parshad R, Perry HR, Price FM, Sanford KK. DNA repair proficiency: potential susceptiblity factor for breast cancer. J Natl Cancer Inst. 1996;88(11):754–755. doi: 10.1093/jnci/88.11.754. [DOI] [PubMed] [Google Scholar]
- 5.Parshad R, Price FM, Bohr VA, Cowans KH, Zujewski JA, Sanford KK. Deficient DNA repair capacity, a predisposing factor in breast cancer. Br J Cancer. 1996;74(1):1–5. doi: 10.1038/bjc.1996.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jyothish B, Ankathil R, Chandini R, Vinodkumar B, Nayar GS, Roy DD, Madhavan J, Nair MK. DNA repair proficiency: a potential marker for identification of high risk members in breast cancer families. Cancer Lett. 1998;124(1):9–13. doi: 10.1016/s0304-3835(97)00419-9. [DOI] [PubMed] [Google Scholar]
- 7.Kovacs E, Stucki D, Weber W, Muller H. Impaired DNA-repair synthesis in lymphocytes of breast cancer patients. Eur J Cancer Clin Oncol. 1986;22(7):863–869. doi: 10.1016/0277-5379(86)90375-5. [DOI] [PubMed] [Google Scholar]
- 8.Motykiewicz G, Faraglia B, Wang LW, Terry MB, Senie RT, Santella RM. Removal of benzo(a)pyrene diol epoxide (BPDE)-DNA adducts as a measure of DNA repair capacity in lymphoblastoid cell lines from sisters discordant for breast cancer. Environ Mol Mutagen. 2002;40(2):93–100. doi: 10.1002/em.10095. [DOI] [PubMed] [Google Scholar]
- 9.Roy SK, Trivedi AH, Bakshi SR, Patel SJ, Shukla PH, Bhatavdekar JM, Patel DD, Shah PM. Bleomycin-induced chromosome damage in lymphocytes indicates inefficient DNA repair capacity in breast cancer families. J Exp Clin Cancer Res. 2000;19(2):169–173. [PubMed] [Google Scholar]
- 10.Ramos JM, Ruiz A, Colen R, Lopez ID, Grossman L, Matta JL. DNA repair and breast carcinoma susceptibility in women. Cancer. 2004;100(7):1352–1357. doi: 10.1002/cncr.20135. [DOI] [PubMed] [Google Scholar]
- 11.Shi Q, Wang LE, Bondy ML, Brewster A, Singletary SE, Wei Q. Reduced DNA repair of benzo(a)pyrene diol epoxide-induced adducts and common XPD polymorphisms in breast cancer patients. Carcinogenesis. 2004;16:16. doi: 10.1093/carcin/bgh167. [DOI] [PubMed] [Google Scholar]
- 12.Biggs PJ, Warren W, Venitt S, Stratton MR. Does a genotoxic carcinogen contribute to human breast cancer? The value of mutational spectra in unravelling the aetiology of cancer. Mutagenesis. 1993;8(4):275–283. doi: 10.1093/mutage/8.4.275. [DOI] [PubMed] [Google Scholar]
- 13.Greenblatt MS, Chappuis PO, Bond JP, Hamel N, Foulkes WD. TP53 mutations in breast cancer associated with BRCA1 or BRCA2 germ-line mutations: distinctive spectrum and structural distribution. Cancer Res. 2001;61(10):4092–4097. [PubMed] [Google Scholar]
- 14.Mohrenweiser HW, Jones IM. Variation in DNA repair is a factor in cancer susceptibility: a paradigm for the promises and perils of individual and population risk estimation? Mutat Res. 1998;400(1–2):15–24. doi: 10.1016/s0027-5107(98)00059-1. [DOI] [PubMed] [Google Scholar]
- 15.Mohrenweiser HW, Jones IM. Uncertainty of response to ionizing radiation due to genotype: potential role for variation in DNA repair genes. Radiat Res. 2000;154(6):722–723. discussion 723–724. [PubMed] [Google Scholar]
- 16.Mohrenweiser HW, Xi T, Vazquez-Matias J, Jones IM. Identification of 127 amino acid substitution variants in screening 37 DNA repair genes in humans. Cancer Epidemiol Biomarkers Prev. 2002;11(10 Pt 1):1054–1064. [PubMed] [Google Scholar]
- 17.Spitz MR, Wei Q, Dong Q, Amos CI, Wu X. Genetic susceptibility to lung cancer: the role of DNA damage and repair. Cancer Epidemiol Biomarkers Prev. 2003;12(8):689–698. [PubMed] [Google Scholar]
- 18.Tworoger SS, Sluss P, Hankinson SE. Association between plasma prolactin concentrations and risk of breast cancer among predominately premenopausal women. Cancer Res. 2006;66(4):2476–2482. doi: 10.1158/0008-5472.CAN-05-3369. [DOI] [PubMed] [Google Scholar]
- 19.Haiman C, Hsu C, de Bakker P, Frasco M, Sheng X, Van Den Berg D, Casagrande J, Kolonel L, Le Marchand L, Hankinson SE, et al. A comprehensive assessment of genetic variation in DNA repair pathway genes in relationship with breast cancer risk. Hum Mol Genet provisionally accepted. 2007 doi: 10.1093/hmg/ddm354. [DOI] [PubMed] [Google Scholar]
- 20.Kolonel LN, Henderson BE, Hankin JH, Nomura AM, Wilkens LR, Pike MC, Stram DO, Monroe KR, Earle ME, Nagamine FS. A multiethnic cohort in Hawaii and Los Angeles: baseline characteristics. Am J Epidemiol. 2000;151(4):346–357. doi: 10.1093/oxfordjournals.aje.a010213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Bakker PI, Yelensky R, Pe’er I, Gabriel SB, Daly MJ, Altshuler D. Efficiency and power in genetic association studies. Nat Genet. 2005;37(11):1217–1223. doi: 10.1038/ng1669. Epub 2005 Oct 1223. [DOI] [PubMed] [Google Scholar]
- 22.Carlson CS, Eberle MA, Rieder MJ, Yi Q, Kruglyak L, Nickerson DA. Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am J Hum Genet. 2004;74(1):106–120. doi: 10.1086/381000. Epub 2003 Dec 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stram DO, Leigh Pearce C, Bretsky P, Freedman M, Hirschhorn JN, Altshuler D, Kolonel LN, Henderson BE, Thomas DC. Modeling and E-M estimation of haplotype-specific relative risks from genotype data for a case-control study of unrelated individuals. Hum Hered. 2003;55(4):179–190. doi: 10.1159/000073202. [DOI] [PubMed] [Google Scholar]
- 24.Mueller JC, Lohmussaar E, Magi R, Remm M, Bettecken T, Lichtner P, Biskup S, Illig T, Pfeufer A, Luedemann J, et al. Linkage Disequilibrium Patterns and tagSNP Transferability among European Populations. Am J Hum Genet. 2005;76:3. doi: 10.1086/427925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li J, Ji L. Adjusting multiple testing in multilocus analyses using the eigenvalues of a correlation matrix. Heredity. 2005;95(3):221–227. doi: 10.1038/sj.hdy.6800717. [DOI] [PubMed] [Google Scholar]
- 26.Nyholt DR. A simple correction for multiple testing for single-nucleotide polymorphisms in linkage disequilibrium with each other. Am J Hum Genet. 2004;74(4):765–769. doi: 10.1086/383251. Epub 2004 Mar 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qin ZS, Niu T, Liu JS. Partition-ligation-expectation-maximization algorithm for haplotype inference with single-nucleotide polymorphisms. Am J Hum Genet. 2002;71(5):1242–1247. doi: 10.1086/344207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moore JH. Computational analysis of gene-gene interactions using multifactor dimensionality reduction. Expert Rev Mol Diagn. 2004;4(6):795–803. doi: 10.1586/14737159.4.6.795. [DOI] [PubMed] [Google Scholar]
- 29.Ritchie MD, Hahn LW, Roodi N, Bailey LR, Dupont WD, Parl FF, Moore JH. Multifactor-dimensionality reduction reveals high-order interactions among estrogen-metabolism genes in sporadic breast cancer. Am J Hum Genet. 2001;69(1):138–147. doi: 10.1086/321276. Epub 2001 Jun 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Memisoglu A, Samson L. Base excision repair in yeast and mammals. Mutat Res. 2000;451(1–2):39–51. doi: 10.1016/s0027-5107(00)00039-7. [DOI] [PubMed] [Google Scholar]
- 31.de Boer J, Hoeijmakers JH. Nucleotide excision repair and human syndromes. Carcinogenesis. 2000;21(3):453–460. doi: 10.1093/carcin/21.3.453. [DOI] [PubMed] [Google Scholar]
- 32.Wood RD. Nucleotide excision repair in mammalian cells. J Biol Chem. 1997;272(38):23465–23468. doi: 10.1074/jbc.272.38.23465. [DOI] [PubMed] [Google Scholar]
- 33.Karran P. DNA double strand break repair in mammalian cells. Curr Opin Genet Dev. 2000;10(2):144–150. doi: 10.1016/s0959-437x(00)00069-1. [DOI] [PubMed] [Google Scholar]
- 34.Khanna KK, Jackson SP. DNA double-strand breaks: signaling, repair and the cancer connection. Nat Genet. 2001;27(3):247–254. doi: 10.1038/85798. [DOI] [PubMed] [Google Scholar]
- 35.Wood RD, Mitchell M, Lindahl T. Human DNA repair genes, 2005. Mutat Res. 2005;577(1–2):275–283. doi: 10.1016/j.mrfmmm.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 36.Rothwell PJ, Waksman G. Structure and mechanism of DNA polymerases. Adv Protein Chem. 2005;71:401–440. doi: 10.1016/S0065-3233(04)71011-6. [DOI] [PubMed] [Google Scholar]
- 37.Matullo G, Peluso M, Polidoro S, Guarrera S, Munnia A, Krogh V, Masala G, Berrino F, Panico S, Tumino R, et al. Combination of DNA repair gene single nucleotide polymorphisms and increased levels of DNA adducts in a population-based study. Cancer Epidemiol Biomarkers Prev. 2003;12(7):674–677. [PubMed] [Google Scholar]
- 38.Scully R, Livingston DM. In search of the tumour-suppressor functions of BRCA1 and BRCA2. Nature. 2000;408(6811):429–432. doi: 10.1038/35044000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Welcsh PL, Schubert EL, King MC. Inherited breast cancer: an emerging picture. Clin Genet. 1998;54(6):447–458. doi: 10.1111/j.1399-0004.1998.tb03764.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.