Abstract
Recognition of the nucleic acid bases within the DNA scaffold comprises the basis for transmission of genetic information, dictating protein and cell assembly, organismal development, and evolution. Driven in part by the need to test our current understanding of this information transfer, chemists have begun to design and synthesize nonnatural bases and base pair structures to mimic the function of DNA without relying on Nature’s purine-pyrimidine base pair scaffold. Multiple examples have been recently described of new genetic architectures that self-assemble stably and sequence specifically in vitro, and experiments with polymerases in vitro show that at least isolated unnatural base pairs can be replicated as well. Moreover, recent experiments with unnatural bases in bacterial cells have demonstrated surprisingly efficient reading of the chemical information. This suggests the future possibility of redesigning and replacing the chemical information of an evolving cell while retaining biological function. Such unnatural base pair components, whether recognized by nature or not, are already proving useful in biotechnology. In addition to practical applications, this chemical approach also gives basic insight into the forces and mechanisms that govern DNA structure and replication.
What I cannot build, I cannot understand – Richard P. Feynman
The exquisitely organized complexity of biological systems has been a longstanding curiosity both of biologists and chemists. However, the two disciplines have commonly taken different approaches to studying the machinery that gives cells their function. Biologists have often taken a “top down” approach, for example by taking a cell type and removing or altering genes, observing the effect on living cell function. In contrast, chemists are often drawn to a “bottom up” strategy, in which the smallest molecular components (such as amino acids or enzyme cofactors) are altered and examined outside the cell; this approach started as the field called biomimetic chemistry [1]. This dichotomy between biologists and chemists is also evident in the emerging discipline of synthetic biology [2], in which biological engineers design and swap pathways between different living systems, whereas chemists have been working on design of new components, building upward toward biological function. However, chemistry laboratories are increasingly adopting biological methods, and so the ability to probe/mimic the machinery of a living cell on a chemical level is being increasingly realized. A meeting in the middle, using approaches from both fields, ultimately seems likely, since the approaches and goals are often the same: altering natural components in living (or lifelike) systems, and exploiting their properties for biotechnology and medicine. At the same time, as Feynman would have recognized, we test and refine our knowledge of how biological systems work.
The biology-inspired approach has long been integral in furthering our understanding of the natural genetic molecule (DNA) and its interaction with proteins. Since Watson and Crick, chemists have been inspired to ask what it is that makes DNA so unique, and explore whether alternative architectures for transmitting biological information can exist. With advances in organic synthesis, analytical techniques and solid phase chemistry, and in combination with modern biochemical and biological methods, an increasing number of research labs have synthesized nonnatural nucleic acid base and base pair architectures designed to mimic, complement, or even replace the natural structures. The work has enabled the exploration of these fundamental questions, and application of the chemical and biological insight is leading to biotechnological advances.
Why modify DNA bases?
By designing new replacements for DNA bases, and observing their functions, we gain a better understanding of natural nucleic acids, and their crucial roles in the context of cellular function. Beyond this basic science, the knowledge gained from understanding a genetic system on a chemical level allows for design of components that may have modified or enhanced functionality, such as a means of reporting on a particular biological event, or a better design of DNA recognition scaffolds, or an important assay for detection of disease.
Although the chemical modification of DNA has long been studied, research on DNA base modifications is a newer field. The design of replacements for the sugar-phosphate backbone of DNA has proceeded for three decades and has yielded many successes, which have been reviewed elsewhere [3]. In contrast, the topic at hand here is on the chemical information encoded by the structures of DNA bases, leaving the sugar-phosphate backbone alone. In this review we outline recent accomplishments in creating nonnatural nucleic acid bases and base pairs, and describe efforts towards alternative genetic systems that contain them. We also outline both the basic scientific information and the technological benefits that have resulted from observing their function in vitro and more recently, in living systems.
Reason #1. Expanding the genetic lexicon
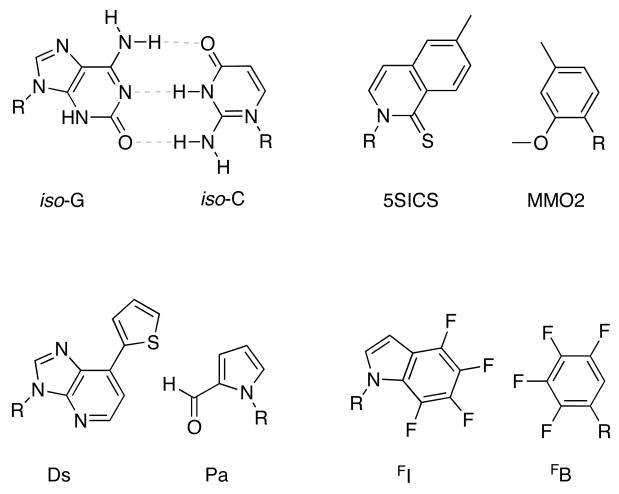
One goal of redesigning the DNA base pair architecture is to expand the information-encoding capability of DNA. To date this has been done, for the most part, in the context of DNA, with new base pairs being designed to function alongside the natural pairs (Figure 1). Such new pairs can function as selective hybridizing groups that do not pair with natural bases, or as selective encoders for incorporation of reporters into RNA, or as additions to the genetic code, leading to proteins with novel functionality. Design and synthesis of new, orthogonal base pairs capable of functioning similarly to, and in context of, the natural DNA scaffold has been a topic of extensive research since the late 1980’s.
Figure 1.
Examples of base pairs designed to augment the natural genetic set. Their dimensions are close to those of natural Watson-Crick pairs. R = deoxyribose.
In this regard, pioneering work from the Benner laboratory has yielded new base pairs in which the hydrogen bonding arrangement is different than the canonical Watson-Crick hydrogen bonding motif of the natural bases [4]. The earliest of these designs, the isoG-isoC base pair, was shown by Benner to be both replicated and translated correctly in vitro [5]. With practical applications in mind, Prudent demonstrated this as a third base pair for the polymerase chain reaction (PCR) [6], which demands high base pair fidelity. In these experiments, up to 96% fidelity per cycle was attained, and errors from mispairing during replication were found to be due to oxidation and tautomerization of the unnatural analogues [7]. To ameliorate this, Benner designed thymidine analogues which still base pair efficiently with natural adenine, but due to steric bulk of a thiocarbonyl, no longer mispair with the undesired isoG enol tautomer [8]. Recently, retaining the same shuffled hydrogen bonding pattern, Benner and colleagues have designed a new base pair between a nitropyridone (dZ) and a triaza-isoG (dP) which improves upon these problems as well [9].
The aforementioned analogues have yet to be incorporated into living cells and evaluated for their potential to expand the genetic code for in vivo translation. However, the orthogonality of such unnatural base pairs to natural DNA has been exploited elegantly in biotechnology. Currently, the isoC-isoG base pair is used as part of an amplification assay in clinics to monitor viral load of hepatitis patients [10]. The high signal-to-background of the assay relies on the design of capture probes containing unnatural bases, thereby eliminating false positives caused by accidental binding to cellular DNA, which does not recognize the unnatural bases. Prudent has also applied the isoC/isoG bases in sensitive real-time PCR quantification [11] using an endonuclease activity to report on sequence.
In contrast to the approach of rearranging the Watson-Crick hydrogen bonds, others have eliminated base pair hydrogen bonding altogether. Over a decade ago, Kool and coworkers described the concept of nonpolar nucleoside isosteres [12]; examples included difluorotoluene (dF) and 4-methylbenzimidazole (dZ), which are hydrophobic, non-hydrogen bonding base analogues of natural thymidine and adenine, respectively. These analogues were shown to form selective pairs with one another, and helped to reveal the important roles of base-base and base-solvent interactions in the stability and selectivity of pairing in DNA [13]. Results of pairing studies of nonpolar base analogues demonstrated that selective base pairing can also be achieved by shape complementarity and hydrophobic effects [14]. These nonpolar isosteres have been extensively studied both in DNA alone and in relation to DNA-protein recognition in multiple contexts, including topoisomerases [15], DNA repair enzymes [16–18], and DNA polymerases [19–25].
The roles of stacking, hydrophobic effects, solvation, and steric effects were more recently demonstrated in the Kool laboratory with pairs that were both selective and in some cases, as stable as natural Watson-Crick pairs. Examples include the “pairing” of a synthetic pyrene nucleoside and an abasic site [26], as well as super-hydrophobic fluorous pairs [14,27]. In both cases, the unnatural bases preferred pairing with their designed partners relative to any of the natural bases within the framework of DNA. Interestingly, the Klenow fragment (Exo-) of E. coli DNA Polymerase I was able to selectively and efficiently process such pairs; for example, the nucleoside triphosphate derivative of pyrene nucleoside was efficiently incorporated opposite an abasic sugar [28].
In the last decade, several laboratories have explored hydrophobic nucleobase analogues as a viable solution for expansion of the genetic alphabet. Not only can such pairs be stable, but they also commonly have the advantage that their pairing with the more polar natural bases is unfavorable as a result of strong desolvation costs. Prominent examples have come from the laboratories of Romesberg and of Hirao, both of which have made multiple successful contributions on this front. Romesberg has synthesized an impressive array of hydrophobic bases, a number of which show selective base pair formation relying on hydrophobic packing, and a few of which are exceptionally stable [29,30]. An important recent example is a methyl methoxybenzene (MMO2) - 5-methylthioisocarbostyril (5SICS) base pair, which could be efficiently inserted and extended by the Klenow fragment, making MMO2-5SICS a potentially useful hydrophobic third base pair for DNA [31].
Also utilizing the principles of shape complementarity and close hydrophobic packing, Hirao and coworkers were able to show highly efficient PCR amplification of an unnatural pair added to DNA using a 7-(2-thienyl)-imidazo(4,5-b)pyridine (Ds) and a pyrrole-2-carbaldehyde (Pa) base pair [32]. Even more impressively, Hirao designed an additional unnatural RNA nucleotide which selectively pairs with Pa, thereby allowing the amplified DNA to be transcribed. This method offers great potential for selective incorporation of unnatural bases and other functionalities into RNA. In one example, Hirao successfully incorporated a fluorescent unnatural base at specific sites into RNA as a tool for studying RNA secondary structure [33].
While the Ds-Pa base pair functions well in in vitro amplification and transcription, the modified amidotriphosphates required for optimal base pair selectivity were found to slow the PCR process. To address this, a 2-nitropyrrole base analogue (Pn) was designed, which better pairs with Ds, thereby out-competing the previously observed mispair with natural adenine. After 20 cycles of PCR amplification using Vent Exo- polymerase, less than 1% mutation rate was observed, making the Ds-Pn base pair even more promising as a third base pair for DNA [34].
Reason #2. Building tools for studying biological mechanisms
Unnatural nucleic acid bases have not only been useful in imparting novel function to DNA, but also in probing interactions within DNA itself. Unnatural nucleosides have been integral in determining the forces and mechanisms essential in base pairing, which is of course required for transfer of genetic information in a cell. Because many diseases can be traced to errors in this information transfer, it is important to understand the mechanisms of base pair recognition in this process, not only in vitro, but also in vivo.
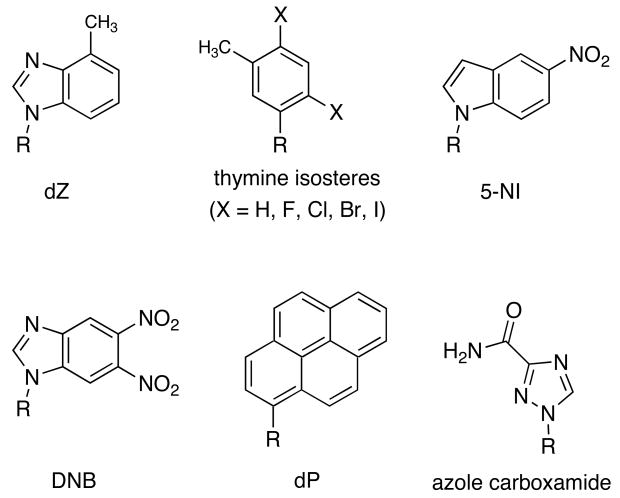
Several dozens of base analogues have been reported to date as tools for investigating these steric, shape, and electrostatic effects in both DNA and RNA (Figure 2). For example, Berdis has synthesized substituted indole analogues and found pi-surface area to be a prime factor in base pair insertion during DNA polymerization and translesion synthesis by bacteriophage T4 polymerase [35,36]. Using substituted benzimidazole analogs, Engels and Kuchta concluded that hydrophobics, electronics, and sterics cannot alone regulate polymerization of DNA by polymerase α and Klenow fragment [37]. Engels also described a number of fluorobenzenes and fluorobenzimidazoles that were used to investigate electrostatic effects of base pairing in RNA [38]. In one study, it was found that the fluorinated bases prefer to pair with themselves relative to the natural bases, similar to results seen by Kool in DNA [13, 14]. Davisson collected evidence on the roles of sterics and electrostatics in DNA synthesis using azole carboxamide base analogues [39]. McLaughlin has designed analogues lacking minor groove hydrogen bond acceptors to explore the roles of minor groove solvation in DNA stability [40].
Figure 2.
Examples of DNA base replacements that have been used for probing nucleobase recognition mechanisms.
Also in this light, ongoing work with nonpolar nucleoside analogues of varying shape and size has shown that steric effects can be critical in the biology of DNA replication. Kim and Kool reported a series of nonpolar isostere analogues of thymidine with systematically varying size, where the 2,4 carbonyl oxygens were replaced by –H, -F, -Cl, -Br, and –I [41]. Studies with this series pointed out a marked size preference during replication by Klenow fragment, with efficiency reaching a sharp maximum at the Cl analogue [42]. Sintim tested similar analogues with similar size but with varying shapes, and also found large kinetic effects of shape during DNA polymerization [43]. The studies showed that steric effects around the incipient base pair are large enough in magnitude to explain most of the selectivity that occurs during DNA replication.
While modifying the chemical parts of DNA to probe DNA structure and recognition in vitro has proven beneficial, these studies are not necessarily a good predictor for how unnatural nucleobases will be tolerated in a living cell, where multiple polymerases, repair mechanisms and biological pathways are at play. To date there are very few studies in which unnatural bases have been introduced into a living system while retaining functionality. Essigmann and Kool achieved this in 2003 when they performed replication bypass experiments in E. coli with single stranded M3 phage containing a synthetic insert having the nonpolar nucleoside isosteres dQ (a deoxyadenosine isostere) or difluorotoluene dF (thymidine isostere) [44]. By counting phage progeny, it was found that while dF and dQ are moderately to modestly bypassed by polymerases under normal conditions, and when an SOS response was induced, both isosteres were efficiently bypassed. More remarkably, a base composition assay showed that replication occurred with insertion of the correct Watson-Crick partner opposite the unnatural bases under both SOS induced and non-induced conditions. These observations show that it is possible to retain biological function with unnatural nucleobase components, and also suggested the importance of DNA base shape (in opposed to hydrogen bonding alone) as a prime determinant of fidelity in vivo.
Essigmann and Kool followed this study with an in vivo evaluation of steric effects, using the variably-sized thymidine nonpolar isostere series previously reported by Kim. Phage replication experiments in E. coli demonstrated that the cellular replicative machinery preferred the slightly-larger-than-natural the dichlorotoluene analogue, with a steep drop off in efficiency and fidelity for analogues containing substituents smaller (H, F) or larger (Br, I) than Cl. This showed a marked steric dependence for DNA replication in vivo, and also led to the suggestion that high-fidelity polymerases have more steric room than necessary, perhaps to allow for evolutionary variation [42].
Nonpolar nucleoside isosteres continue to prove useful as tools for probing biological mechanisms and interactions. More recently, two laboratories at Alnylam and at Stanford have independently used difluorobenzene ribonucleoside derivatives to test electrostatic requirements of sequence recognition in RNA interference [45,46]. Interestingly, the Watson-Crick hydrogen bonding requirement varied strongly along the sequence, and at some sites it was found that this bonding is not required to maintain activity in RNA interference in HeLa cells. The origins of selectivity and activity in RNA interference are complex and are still not well understood; thus future research involving a variety of nucleic acid analogues will be invaluable in understanding RNAi.
Reason #3. Designing alternative genetic systems
Most if not all of the unnatural bases mentioned above were designed to function within the context and geometry of the natural genetic system. Thus they maintain a general size and shape that fits well next to purine-pyrimidine pairs. However, recently an alternative strategy has begun to be explored. In this new approach, researchers are asking whether all of the natural base pairs of DNA can be replaced, giving rise to an entirely new genetic set not confined by the geometry of the natural Watson-Crick base pairs (Figure 3). Designing an alternative genetic set (and ultimately, a functioning genetic system) can be useful in exploring nature’s evolutionary arrival at the structure of DNA for storage of genetic information. This allows one to address some very basic questions: Can other base pair architectures assemble and function similarly to DNA? Do we know enough about DNA to design a similar information-storing system from novel chemical parts?
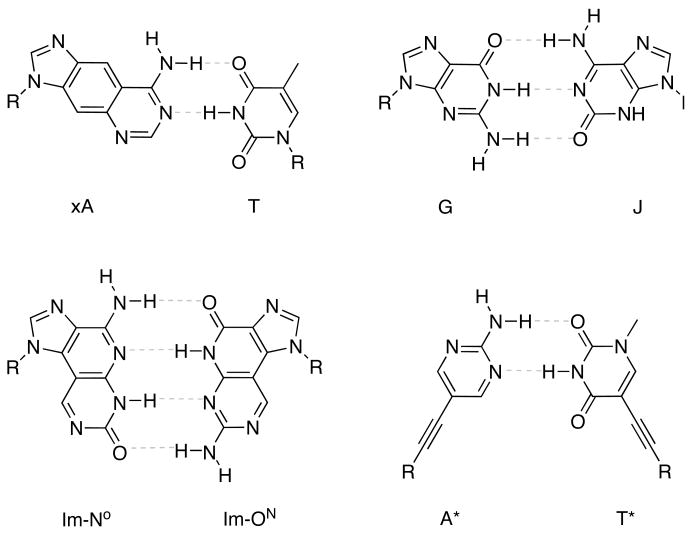
Figure 3.
Novel base pairing systems designed to form new, non-purine-pyrimidine genetic sets. All have sizes larger than Watson-Crick pairs.
In this light, Kool and coworkers have extensively explored base pairing, assembly, and structural properties of an alternative genetic set with size-expanded geometry (xDNA) [47,48]. This genetic set is comprised of eight bases with four types of pairings. Each of the pairs contains a size-expanded base that resembles a natural purine or pyrimidine, but has its Watson-Crick pairing edge shifted outward by the insertion of a benzene ring [49–51]. This gives rise to base pairs with a 2.4 Å larger C1′-C1′ backbone distance than natural DNA. In xDNA, benzopurines pair with natural purines, and benzopyrimidines pair with natural purines, making the eight-letter set orthogonal to natural DNA. Because there are eight letters, the information encoding potential of xDNA is exponentially larger than that of DNA. Kool has found while a single expanded base pair in natural DNA is destabilizing, helices composed entirely of size-expanded base pairs are more stable than natural DNA, due to more favorable base stacking [52,53]. In addition, the expanded DNA bases are brightly fluorescent [54], a property virtually absent in natural DNA.
While the xDNA genetic set is only one component of a potentially alternative genetic system (which also would require machinery for replication), the insight gained from these experiments has shed light on the tolerance of the natural phosphodiester backbone towards alternative pairing geometries that may be capable of genetic information storage. Recently, Kool has explored other expanded geometries (such as one termed “wide DNA” (yDNA)) [55, 56] and has even shown that duplex assembly is possible with base pairs containing naphtho-homologated pyrimidines paired with purines (yyDNA) [57]. Thus it appears that the natural phosphodiester backbone of DNA can be quite tolerant of different base pair sizes, as long as they are all congruent in their geometry.
Other laboratories have also experimented with altering the intermolecular backbone distance in DNA. Battersby has observed antiparallel, selective Watson-Crick-like duplex formation of oligonucleotides composed entirely of purines [58]. In this scaffold, the pairing partner for natural adenine is hypoxanthine (H), and for guanine, an N3-H tautomer of isoG (J). The demonstration of an all-purine duplex is interesting, considering all the building blocks could have potentially originated from prebiotic mixtures. This raises interesting questions about nature’s criteria for an evolving system of nucleic acids.
Taking a related expanded-size approach, Inouye has recently designed an artificial genetic set composed entirely of nonnatural C-glycosides in which added length is conferred by an ethynyl group [59]. The recognition of these bases relies on six-member heterocycles with Watson-Crick-like hydrogen-bonding faces (iG*:iC*, three hydrogen bonds, A*:T* two hydrogen bonds), linked to the C1′ of deoxyribose by the ethynyl spacer. Oligomers composed of these unnatural bases formed highly stable, antiparallel duplexes and triplexes, and displayed selectivity against mismatches as well.
Matsuda and coworkers have synthesized base pairs which not only increase the C1′-C1′ glycosidic distance relative to natural DNA, but also the number of hydrogen bonds [60,61]. It was found that the addition of one Imidazopyridopyrimidine (NN, OO, ON, NO) base pair in DNA did not add any thermal stabilization to the duplex relative to an A:T or G:C base pair. However, with the addition of consecutive imidazopyridopyrimidine base pairs (up to three), substantial duplex stabilization relative to the natural sequence could be seen, likely due to favorable aromatic stacking and relief of backbone strain [60]. Although helices composed entirely of imidazopyridopyrimidine base pairs have not yet been reported, such bases and pairs seem quite promising.
The work to date aimed at developing alternative genetic systems has clearly yielded base pairing systems that function very well in assembly, affording highly stable helices with selective base pairing. Thus they have the potential to store information (in some cases, large amounts of information). However, it is not yet clear whether this information can be read and transferred, which are clearly required for a functioning genetic system. One challenge in this regard is to find enzymes that can accept the unnatural geometries of such pairs and helices [62,63]; as it stands now, such large systems are expected to be alien to natural polymerases, which for the most part evolved to process the natural geometry and exclude molecules that do not fit.
Future outlook
While the chemical and biophysical knowledge of DNA is becoming more complete by use of nonnatural bases and pairs, the implications on the larger biological system of replacing these chemical parts is relatively new, and requires more research. In addition, the redesign of more complex biological components such as the natural polymerases is a challenging task that clearly deserves much more work. As more research is done with unnatural base pair design, alternative genetic systems, and polymerases, synthetic biologists will come closer to the goal of understanding and redesigning living systems to further benefit science and technology.
Acknowledgments
We thank the U.S. National Institutes of Health (GM63587 and GM072705) for support of work on unnatural bases and base pairs for DNA and RNA.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Breslow R. Centenary lecture. Biomimetic chemistry Chem Soc Rev. 1972;1:553–580. [Google Scholar]
- 2.(a) Rawls R. ‘Synthetic Biology’ makes its debut. Chem Eng News . 2000;78(17):49–53. [Google Scholar]; (b) Benner SA, Sismour AM. Synthetic biology: act natural. Nature. 2005;421:118. doi: 10.1038/421118a. [DOI] [PubMed] [Google Scholar]
- 3.Eschenmoser A. Chemical etiology of nucleic acid structure. Science . 1999;284:2118–2124. doi: 10.1126/science.284.5423.2118. and references therein. [DOI] [PubMed] [Google Scholar]
- 4.Switzer C, Moroney SE, Benner SA. Enzymatic incorporation of a new base pair into DNA and RNA. J Am Chem Soc. 1989;111:8322–8323. [Google Scholar]
- 5.Piccirilli JA, Krauch T, Moroney SE, Benner SA. Enzymatic incorporation of a new base pair into DNA and RNA extends the genetic alphabet. Nature. 1990;343:33–37. doi: 10.1038/343033a0. [DOI] [PubMed] [Google Scholar]
- 6.Johnson SC, Sherrill CB, Marshall DJ, Moser MJ, Prudent JR. A third base pair for the polymerase chain reaction: inserting isoC and isoG. Nucleic Acids Res. 2004;32:1937–1941. doi: 10.1093/nar/gkh522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Switzer CY, Moroney SE, Benner SA. Enzymatic recognition of the base-pair between isocytidine and isoguanosine. Biochemistry. 1993;32:10489–10496. doi: 10.1021/bi00090a027. [DOI] [PubMed] [Google Scholar]
- 8.Sismour AM, Benner SA. The use of thymidine analogs to improve the replication of an extra DNA base pair: a synthetic biological system. Nucleic Acids Res. 2005;33:5640–5646. doi: 10.1093/nar/gki873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang Z, Sismour AM, Sheng P, Puskar NL, Benner SA. Enzymatic incorporation of a third nucleobase pair. Nucleic Acids Res. 2007;35:4238–4249. doi: 10.1093/nar/gkm395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collins ML, Irvine B, Tyner D, Fine E, Zayati C, Chang C, Horn T, Ahle D, Detmer J, Shen L, et al. A branched DNA signal amplification assay for quantification of nucleic acid targets below 100 molecules/ml. Nucleic Acids Res. 1997;25:2979–2984. doi: 10.1093/nar/25.15.2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moser MJ, Marshall DJ, Grenier JK, Kieffer CD, Killeen AA, Ptacin JL, Richmond CS, Roesch EB, Scherrer CW, Sherrill CB, et al. Exploiting the enzymatic recognition of an unnatural base pair to develop a universal genetic analysis system. Clin Chem. 2003;49:407–414. doi: 10.1373/49.3.407. [DOI] [PubMed] [Google Scholar]
- 12.Schweitzer BA, Kool ET. Nonpolar aromatic nucleosides as hydrophobic isosteres of DNA nucleosides. J Org Chem. 1994;59:7238–7242. doi: 10.1021/jo00103a013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schweitzer BA, Kool ET. Hydrophobic, non-hydrogen-bonding bases and base pairs in DNA. J Am Chem Soc. 1995;117:1863–1872. doi: 10.1021/ja00112a001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lai JS, Qu J, Kool ET. Selective pairing between polyfluorinated DNA bases. J Am Chem Soc. 2004;126:3040–3041. doi: 10.1021/ja039571s. [DOI] [PubMed] [Google Scholar]
- 15.Yakovleva L, Lai JS, Kool ET, Shuman S. Nonpolar nucleobase analogs illuminate requirements for site-specific DNA cleavage by vaccinia topoisomerase. J Biol Chem. 2006;281:35914–35921. doi: 10.1074/jbc.M608349200. [DOI] [PubMed] [Google Scholar]
- 16.Chepanoske CL, Langelier CR, Chmiel NH, David SS. Recognition of the nonpolar base 4-methylindole in DNA by the DNA repair adenine glycosylase MutY. Org Lett. 2000;2:1341–1344. doi: 10.1021/ol005831o. [DOI] [PubMed] [Google Scholar]
- 17.Francis AW, Helquist SA, Kool ET, David SS. Probing the requirements for recognition and catalysis in Fpg and MutY with nonpolar adenine isosteres. J Am Chem Soc. 2003;125:16235–16242. doi: 10.1021/ja0374426. [DOI] [PubMed] [Google Scholar]
- 18.Kirby TW, DeRose EF, Beard WA, Wilson SH, London RE. A thymine isostere in the templating position disrupts assembly of the closed DNA polymerase β ternary complex. Biochemistry. 2005;44:15230–15237. doi: 10.1021/bi0511742. [DOI] [PubMed] [Google Scholar]
- 19.Moran S, Ren RXF, Rumney S, Kool ET. Difluorotoluene, a nonpolar isostere of thymine, codes specifically and efficiently for adenine in DNA replication. J Am Chem Soc. 1997;119:2056–2057. doi: 10.1021/ja963718g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morales JC, Kool ET. Efficient replication of a DNA base pair between non-hydrogen-bonded nucleoside analogues. Nat Struct Bio. 1998;5:950–954. doi: 10.1038/2925. [DOI] [PubMed] [Google Scholar]
- 21.Morales JC, Kool ET. Functional hydrogen-bonding map of the minor groove binding tracks of six DNA polymerases. Biochemistry. 2000;39:12979–12988. doi: 10.1021/bi001578o. [DOI] [PubMed] [Google Scholar]
- 22.Potapova O, Chan C, DeLucia AM, Helquist SA, Kool ET, Grindley NDF, Joyce CM. DNA polymerase catalysis in the absence of Watson-Crick hydrogen bonds: analysis by single-turnover kinetics. Biochemistry. 2006;45:890–898. doi: 10.1021/bi051792i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mizukami S, Kim TW, Helquist SA, Kool ET. Varying DNA base-pair size in subangstrom increments: evidence for a loose, not large, active site in low-fidelity Dpo4 polymerase. Biochemistry. 2006;45:2772–2778. doi: 10.1021/bi051961z. [DOI] [PubMed] [Google Scholar]
- 24.Kim TW, Brieba LG, Ellenberger T, Kool ET. Functional evidence for a small and rigid active site in a high fidelity DNA polymerase: probing T7 DNA polymerase with variably-sized base pairs. J Biol Chem. 2006;281:2289–2295. doi: 10.1074/jbc.M510744200. [DOI] [PubMed] [Google Scholar]
- 25.Lee HR, Helquist SA, Kool ET, Johnson KA. Importance of hydrogen bonding for efficiency and specificity of the human mitochondrial DNA polymerase. J Biol Chem. 2008;283:14402–14410. doi: 10.1074/jbc.M705007200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matray TJ, Kool ET. Selective and stable DNA base pairing without hydrogen bonds. J Am Chem Soc. 1998;120:6191–6192. doi: 10.1021/ja9803310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lai JS, Kool ET. Fluorous base-pairing effects in a DNA polymerase active site. Chem Eur J. 2005;11:2966–2971. doi: 10.1002/chem.200401151. [DOI] [PubMed] [Google Scholar]
- 28.Matray TJ, Kool ET. A specific partner for abasic damage in DNA. Nature. 1999;399:704–708. doi: 10.1038/21453. [DOI] [PubMed] [Google Scholar]
- 29.Leconte AM, Matsuda S, Romesberg FE. An efficiently extended class of unnatural base pairs. J Am Chem Soc. 2006;128:6780–6781. doi: 10.1021/ja060853c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matsuda S, Henry AA, Romesberg FE. Optimization of unnatural base pair packing for polymerase recognition. J Am Chem Soc. 2006;128:6369–6375. doi: 10.1021/ja057575m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leconte AM, Hwang GT, Matsuda S, Capek P, Hari Y, Romesberg FE. Discovery, characterization, and optimization of an unnatural base pair for expansion of the genetic alphabet. J Am Chem Soc. 2008;130:2336–2343. doi: 10.1021/ja078223d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hirao I, Kimoto M, Mitsui T, Fujiwara T, Kawai R, Sato A, Harada Y, Yokoyama S. An unnatural hydrophobic base pair system: site-specific incorporation of nucleotide analogs into DNA and RNA. Nat Methods. 2006;9:729–735. doi: 10.1038/nmeth915. [DOI] [PubMed] [Google Scholar]
- 33.Kimoto M, Mitsui T, Harada Y, Sato A, Yokoyama S, Hirao I. Fluorescent probing for RNA molecules by an unnatural base-pair system. Nucleic Acids Res. 2007;35:5360–5369. doi: 10.1093/nar/gkm508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kimoto M, Kawai R, Mitsui T, Yokoyama S, Hirao I. Efficient PCR amplification by an unnatural base pair system. Nucleic Acids Symp. 2008;52:469–470. doi: 10.1093/nass/nrn238. [DOI] [PubMed] [Google Scholar]
- 35.Zhang X, Lee I, Berdis AJ. The use of nonnatural nucleotides to probe the contributions of shape complementarity and pi-electron surface area during DNA polymerization. Biochemistry. 2005;44:13101–13110. doi: 10.1021/bi050585f. [DOI] [PubMed] [Google Scholar]
- 36.Zhang X, Lee I, Zhou X, Berdis AJ. Hydrophobicity, shape, and pi-electron contributions during translesion DNA synthesis. J Am Chem Soc. 2006;128:143 –149. doi: 10.1021/ja0546830. [DOI] [PubMed] [Google Scholar]
- 37.Kincaid K, Beckman J, Zivkovic A, Halcomb RL, Engels JW, Kuchta RD. Exploration of factors driving incorporation of unnatural dNTPs into DNA by Klenow fragment (DNA polymerase I) and DNA polymerase α. Nucleic Acids Res. 2005;33:2620–2628. doi: 10.1093/nar/gki563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parsch J, Engels JW. Synthesis of fluorobenzene and benzimidazole nucleic acid analogues and their influence on stability of RNA duplexes. Helv Chim Acta. 2000;83:1791–1808. [Google Scholar]
- 39.Paul N, Nashine VC, Hoops G, Zhang P, Zhou J, Bergstrom DE, Davisson VJ. DNA polymerase template interactions probed by degenerate isosteric nucleobase analogs. Chem Biol. 2003;10:815–825. doi: 10.1016/j.chembiol.2003.08.008. [DOI] [PubMed] [Google Scholar]
- 40.Woods KK, Lan T, McLaughlin LW, Williams LD. The role of minor groove functional groups in DNA hydration. Nucleic Acids Res. 2003;31:1536–1544. doi: 10.1093/nar/gkg240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim TW, Kool ET. A series of nonpolar thymine analogues of increasing size: DNA base pairing and stacking properties. J Org Chem. 2005;70:2048–2053. doi: 10.1021/jo048061t. [DOI] [PubMed] [Google Scholar]
- 42.Kim TW, Delaney JC, Essigmann JM, Kool ET. Probing the active site tightness of DNA polymerase in sub-angstrom increments. Proc Natl Acad Sci USA. 2005;102:15803–15808. doi: 10.1073/pnas.0505113102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sintim HO, Kool ET. Remarkable sensitivity to DNA base shape in the DNA polymerase active site. Angew Chem Int Ed Engl. 2006;45:1974–1979. doi: 10.1002/anie.200504296. [DOI] [PubMed] [Google Scholar]
- 44.Delaney JC, Henderson PT, Helquist SA, Morales JC, Essigmann JM, Kool ET. High-fidelity in vivo replication of DNA base shape mimics without Watson-Crick hydrogen bonds. Proc Natl Acad Sci USA. 2003;100:4469–4473. doi: 10.1073/pnas.0837277100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xia J, Noronha A, Toudjarska I, Li F, Akinc A, Braich R, Frank-Kamenetskii M, Rajeev KG, Egli M, Manoharan M. Gene silencing activity of siRNAs with a ribo-difluorotoluyl nucleotide. ACS Chem Biol. 2006;1:176–183. doi: 10.1021/cb600063p. [DOI] [PubMed] [Google Scholar]
- 46.Somoza A, Chelliserrykattil J, Kool ET. The roles of hydrogen bonding and sterics in RNA interference. Angew Chem Int Ed Engl. 2006;45:4994–4997. doi: 10.1002/anie.200601311. [DOI] [PubMed] [Google Scholar]
- 47.Liu H, Gao J, Lynch SR, Saito DY, Maynard L, Kool ET. A four-base paired genetic helix with expanded size. Science. 2003;302:868–871. doi: 10.1126/science.1088334. [DOI] [PubMed] [Google Scholar]
- 48.Krueger AT, Lu H, Lee AHF, Kool ET. Synthesis and properties of size-expanded DNAs: toward designed, functional, genetic systems. Acc Chem Res. 2007;40:141–150. doi: 10.1021/ar068200o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Leonard NJ, Sprecker MA, Morrice AG. Defined dimensional changes in enzyme substrates and cofactors. Synthesis of lin-benzoadenosine and enzymatic evaluation of derivatives of the benzopurines. J Am Chem Soc. 1976;98:3987–3994. doi: 10.1021/ja00429a040. [DOI] [PubMed] [Google Scholar]
- 50.Liu H, Gao J, Maynard L, Saito YD, Kool ET. Toward a new genetic system with expanded dimensions: size-expanded analogs of deoxyadenosine and thymidine. J Am Chem Soc. 2004;126:1102–1109. doi: 10.1021/ja038384r. [DOI] [PubMed] [Google Scholar]
- 51.Liu H, Gao J, Kool ET. Size-expanded analogues of dG and dC: synthesis and pairing properties in DNA. J Org Chem. 2005;70:639–647. doi: 10.1021/jo048357z. [DOI] [PubMed] [Google Scholar]
- 52.Liu H, Gao J, Kool ET. Helix-forming properties of size-expanded DNA, an alternative four-base genetic form. J Am Chem Soc. 2005;127:1396–1402. doi: 10.1021/ja046305l. [DOI] [PubMed] [Google Scholar]
- 53.Gao J, Liu H, Kool ET. Assembly of the complete eight-base artificial genetic helix, xDNA, and its interaction with the natural genetic system. Angew Chem Int Ed. 2005;44:3118–3122. doi: 10.1002/anie.200500069. [DOI] [PubMed] [Google Scholar]
- 54.Krueger AT, Kool ET. Fluorescence of size-expanded DNA bases: reporting on DNA sequence and structure with an unnatural genetic set. J Am Chem Soc. 2008;130:3989–3999. doi: 10.1021/ja0782347. [DOI] [PubMed] [Google Scholar]
- 55.Lu H, He K, Kool ET. yDNA: a new geometry for size-expanded base pairs. Angew Chem Int Ed. 2004;43:5834–5836. doi: 10.1002/anie.200461036. [DOI] [PubMed] [Google Scholar]
- 56.Lee AHF, Kool ET. A new four-base genetic helix, yDNA, composed of widened benzopyrimidine-purine pairs. J Am Chem Soc. 2005;127:3332–3338. doi: 10.1021/ja0430604. [DOI] [PubMed] [Google Scholar]
- 57.Lee AHF, Kool ET. Exploring the limits of DNA base pair size: naphtho-homologous DNA bases and pairs. J Am Chem Soc. 2006;128:9219–9230. doi: 10.1021/ja0619004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Battersby TR, Albalos M, Friesenhahn MJ. An unusual mode of DNA duplex association: Watson-Crick interaction of all-purine deoxyribonucleic acids. Chem Biol. 2007;14:525–531. doi: 10.1016/j.chembiol.2007.03.012. [DOI] [PubMed] [Google Scholar]
- 59.Doi Y, Chiba J, Morikawa T, Inouye M. Artificial DNA made exclusively of nonnatural C-nucleosides with four types of nonnatural bases. J Am Chem Soc. 2008;130:8762–8768. doi: 10.1021/ja801058h. [DOI] [PubMed] [Google Scholar]
- 60.Minakawa N, Kojima N, Hikishima S, Sasaki T, Kiyosue A, Atsumi N, Ueno Y, Matsuda A. New base pairing motifs. J Am Chem Soc. 2003;125:9970–82. doi: 10.1021/ja0347686. [DOI] [PubMed] [Google Scholar]
- 61.Hikishima S, Minakawa N, Kuramoto K, Fujisawa Y, Ogawa M, Matsuda A. Synthesis of 1,8-naphthyridine C-nucleosides and their base-pairing properties in oligodeoxynucleotides: thermally stable naphthyridine:imidazopyrdopyrimidine base-pairing motifs. Angew Chem Int Ed. 2005;44:596–598. doi: 10.1002/anie.200461857. [DOI] [PubMed] [Google Scholar]
- 62.Gloeckner C, Sauter KBM, Marx A. Evolving a thermostable DNA polymerase that accurately amplifies highly-damaged templates. Angew Chem Int Ed. 2007;46:3115–3117. doi: 10.1002/anie.200603987. [DOI] [PubMed] [Google Scholar]
- 63.Xia G, Chen L, Takashi S, Fa M, Schultz PG, Romesberg FE. Directed evolution of novel polymerase activities: mutation of a DNA polymerase into an efficient RNA polymerase. Proc Natl Acad Sci USA. 2002;99:6597–6602. doi: 10.1073/pnas.102577799. [DOI] [PMC free article] [PubMed] [Google Scholar]