Abstract
The Arrowsmith two-node search is a strategy that is designed to assist biomedical investigators in formulating and assessing scientific hypotheses. More generally, it allows users to identify biologically meaningful links between any two sets of articles A and C in PubMed, even when these share no articles or authors in common and represent disparate topics or disciplines. The key idea is to relate the two sets of articles via title words and phrases (B-terms) that they share. We have created a free, public web-based version of the two-node search tool (http://arrowsmith.psych.uic.edu), have described its development and implementation, and have presented analyses of individual two-node searches. In this paper, we provide an updated tutorial intended for end-users, that covers the use of the tool for a variety of potential scientific use case scenarios. For example, one can assess a recent experimental, clinical or epidemiologic finding that connects two disparate fields of inquiry -- identifying likely mechanisms to explain the finding, and choosing promising follow-up lines of investigation. Alternatively, one can assess whether the existing scientific literature lends indirect support to a hypothesis posed by the user that has not yet been investigated. One can also employ two-node searches to search for novel hypotheses. Arrowsmith provides a service that cannot be carried out feasibly via standard PubMed searches or by other available text mining tools.
Keywords: text mining, web server, hypothesis, literature-based discovery
Introduction
The Arrowsmith two-node search tool [1-5] is designed to assist biomedical investigators in formulating and assessing scientific hypotheses. More generally, it allows users to identify biologically meaningful links between any two sets of articles A and C in PubMed, even when A and C share no articles or authors in common and represent disparate topics or disciplines. This fundamental text mining strategy provides a service that cannot be carried out feasibly via standard PubMed searches. Although other ways to link disparate literatures have been studied [e.g., 6, 7], to our knowledge, no other two-node search tool has been made freely available to the scientific community.
The key idea, as shown in fig. 1, is to relate two sets of articles, or literatures (A and C) via title words and phrases (B-terms) that they share. To carry out a two-node search, the user is asked to input two separate PubMed queries that define A and C. Any articles present in both A and C are removed so that the analysis will consider only indirect linkages between the two sets of articles. Then the software identifies all words and 2- and 3-word phrases that are found in the titles of articles in both A and C. These so-called B-terms are processed and ranked according to the predicted probability that they will be relevant for some user in pointing to a meaningful link across the two literatures [5]. The interface then displays the ranked list of B-terms; clicking on a B-term opens a new window that displays the titles that contain A and B juxtaposed to the titles that contain B and C. In this fashion, the user can readily see whether, and how, the two sets of articles are related.
Fig. 1. Venn diagram illustrating the Arrowsmith data mining model.

Two disparate sets of articles (A and C) are implicitly related via title terms (Bi's) that they share. Reprinted from [5] with permission.
Ten years ago, we published a tutorial on the practical use of the two-node search tool in this journal [2]. Since then, however, the two-node search tool has undergone extensive development; its underlying algorithms have been greatly improved and its interface is now substantially more advanced [1-5]. We believe the time is right to provide an updated tutorial intended for end-users, that covers a variety of potential scientific use case scenarios.
Materials and Methods
The UIC team worked with several groups of neuroscience field testers who carried out searches during the course of their daily scientific work [4]. Their feedback led to improvements and permitted us to acquire a set of “gold standard” searches that were employed for quantitative modeling [5]. In a gold standard search, a user marked all B-terms as “relevant” that were useful in answering the question that motivated the search. We then characterized each B-term according to 8 different features [3, 5], and formulated a quantitative model that optimally separated the set of relevant B-terms from other terms. For each B-term, the model gives the estimated probability that it will be deemed as relevant by some user [5]. The model gave significantly better performance than other proposed methods both on the original set of gold standard searches and on 20 external gold standards derived from TREC Genomics Track 2006 queries [5].
A simplified version of the PubMed query box was imported into the two-node search web interface, so that users input two PubMed queries in order to define the two sets of articles (or literatures) A and C. To retrieve MEDLINE records corresponding to user queries quickly and automatically, a local customized database of MEDLINE was created and updated weekly. When a query is entered, the article ID numbers are downloaded from PubMed and the full MEDLINE records are retrieved from the local database, including a tokenized and stoplisted version of each article title. Articles not found in the local database are downloaded from PubMed as XML files, processed and stored in the local database. Note that the web interface currently processes only the most recent 50,000 articles retrieved for a given PubMed query. B-terms and their feature values are computed in a parallel fashion by processing the sets of tokenized and stoplisted titles in chunks on separate processors, and merging the results when each process is done. The baseline 2005 version of MEDLINE was processed to identify all terms (words and up to 3-word phrases) in titles. Wherever possible, B-term features were pre-computed and stored in the term database for fast look-up [5].
Results
The two-node search can contribute to at least five scientific use case scenarios:
Identifying concepts or items that have been studied in both A and C (albeit possibly from different points of view).
Assessing a recent experimental, clinical or epidemiologic finding that connects two disparate fields of inquiry: a) identifying likely mechanisms to explain the finding, and b) choosing promising follow-up lines of investigation.
Assessing a novel, but hitherto un-investigated, hypothesis, to learn if the existing scientific literature lends indirect support.
Integrating information regarding a single concept or phenomenon that has been studied in two different isolated contexts.
Searching for new hypotheses (e.g., by assessing the results of a one-node search).
1. Identifying concepts or items that have been studied in both A and C (albeit possibly from different points of view)
Perhaps the simplest task for the two-node search tool is to enumerate a list of concepts or items that have been studied in both A and C. For example, studies of microRNAs and of Xist (a noncoding RNA expressed on the X chromosome) comprise two distinct fields of research. Suppose a user wanted to identify a list of entities that have been discussed with regard to both classes of RNAs. A PubMed search on [microRNA AND Xist] finds 3 articles. However, this will only identify the set of papers that discuss both RNAs, and will miss the many papers that discuss only microRNAs or only Xist. A two-node search gives a much more complete answer: Two PubMed searches are conducted with input query A = microRNA (3582 articles) and C = Xist (576 articles). A large number of B-terms are identified (1127) of which 354 are predicted to be relevant (fig. 2). These include a heterogeneous mix of entities studied in both areas of investigation, including methods (e.g., RT-PCR), concepts (e.g., genomic organization), and types of genes (e.g., homeobox). Suppose the user is specifically interested in identifying disorders; one can select the option to restrict terms by semantic categories (using a button placed next to the B-list; fig. 2) and select the super-category of “Disorders”. The filtered list contains 78 B-terms, of which 31 are predicted to be relevant (fig. 3). Because these are listed in order of the estimated probability of relevance, the user does not necessarily need to examine all terms, but can scan down the list to peruse the most promising terms first (fig. 3). There are 4 terms with p ≥ 0.99, and 12 terms having p ≥ 0.95. Clicking on any B-term (say, germ cell tumor) opens a new window that displays the titles discussing [microRNA and germ cell tumor] next to those discussing [germ cell tumor and Xist], allowing the user to verify immediately that both microRNAs and Xist have been studied in this disorder (fig. 4). One can then explore the nature of the relationships in further detail. The interface offers a number of basic options. Clicking on the number next to any title opens a new window displaying the article's abstract with the B-term highlighted in yellow. The user also has the option to save titles to a clipboard. Options located on the B-list page allow the user to undo the last edit or all edits, and to view a text file of all B-terms (together with the full details of the query and the history of all options applied). The searches are saved temporarily (subject to server storage limitations; at present, jobs are generally stored for several months). If the user returns later to the web interface and enters the Job ID number (located at the top left of the pages; fig. 2, 3) the entire search output including edited B-list and clipboard can be viewed and analyzed without the need to repeat the search again.
Fig. 2. Screenshot of the output page for the two node search A = microRNA and C = Xist.

The ranked list of B-terms is displayed, together with other information and options for further filtering and analysis.
Fig. 3. Screenshot of the B-List page for the two node search A = microRNA and C = Xist, filtered to show only B-terms that map to the semantic super-category of “disorders”.

Fig. 4. Screenshot of the Literature page from the two-node search on A = microRNA and C = Xist.
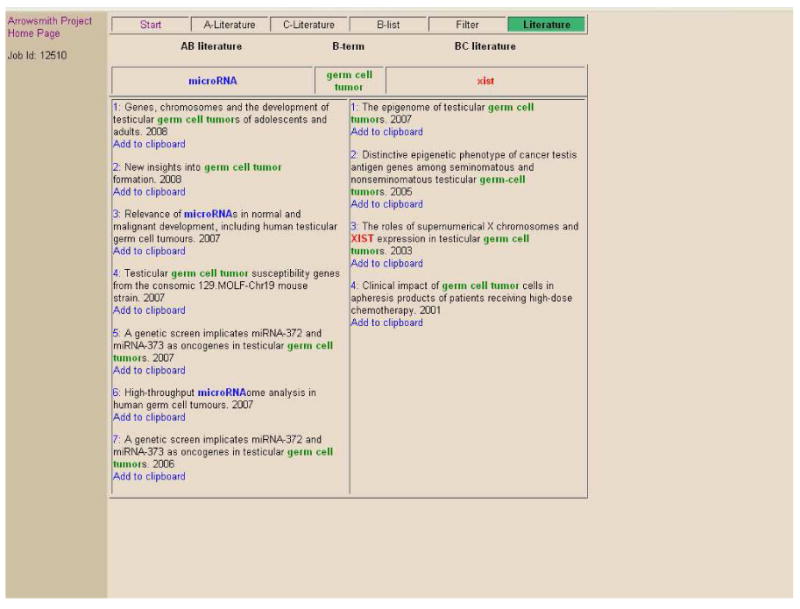
It is apparent, even from reading the titles of the listed articles, that both microRNAs and Xist have been studied in germ cell tumors. Clicking on the numbers associated with any article opens a new page that displays the abstract of the article (and links to the full-text article, if available), allowing further examination.
2. Assessing a recent experimental, clinical or epidemiologic finding that connects two disparate fields of inquiry: a) identifying likely mechanisms to explain the finding, and b) choosing promising follow-up lines of investigation
The first example is illustrative because it shows that one can identify and analyze links meaningfully across two literatures even if no hypothesis has been formulated in advance; even if the information obtained is not novel (to an expert, or even necessarily to the one doing the search); and even if the links do not point to a relationship between microRNAs and Xist. However, often the point of departure for carrying out a two-node search is a new finding reported in the literature that links two hitherto un-connected topics. For example, an early Arrowsmith analysis [8] was motivated by an (at the time) novel observation that estrogen replacement therapy appears to be protective against Alzheimer disease (AD) in older women. A two-node search on A = estrogen vs. C = Alzheimer disease generated a list of B-terms which had only been studied in contexts other than AD, that potentially could be involved in mediating this effect. For example, estrogen regulates calbindin D28k, induces cathepsin D, alters superoxide dismutase, inhibits apoE levels in plasma (and enhances its cellular uptake), enhances neuronal responses to glutamate and induces cytochrome C oxidase subunit III in rat hippocampus, and exhibits anti-oxidant activity. Furthermore, estrogen receptor polymorphism had been widely assessed as a risk factor in breast cancer but not examined at that time as possibly relevant to AD. Since this analysis was originally published in 1996 [8], many subsequent studies have appeared which investigated these concepts, confirming that investigators did, indeed, regard these as promising lines of research.
3. Assessing a novel, but un-investigated or unproven, hypothesis to learn if the existing scientific literature lends it indirect support
One can generalize the types of two-node searches further, to include not merely reported findings but original hypotheses that link two (generally disparate) topics. For example, one of us was writing a recent mini-review article on the cell biology of microRNAs [9], and wanted to assess the likelihood that phosphorylation of microRNA pathway components may regulate microRNAs in mammalian cells. A direct PubMed search on [microRNA AND phosphorylation] revealed that there is abundant evidence that microRNAs regulate kinases; a case in which phophorylation of a transcriptional coactivator (Yorkie) is required for transcription of a microRNA gene; and a case in which a RNA kinase can add 5′-phosphate groups to siRNAs. However, currently there is apparently no direct evidence that kinases (or phosphatases) regulate microRNA levels or functions.
To assess whether the existing scientific literature adds indirect support for this notion, a two-node search was carried out on A = microRNA and C = phosphorylation, and the B-list was filtered to retain terms in the category of “gene or protein names” (using a customized look-up list that was extracted from Entrez Gene). This resulted in 246 predicted relevant B-terms, of which 130 were estimated to be relevant at p ≥ 0.95. Some of these B-terms represented proteins that are targets of microRNAs (e.g., p27Kip1), and others represented proteins that regulate microRNA transcription (e.g., E2F). However, by quickly perusing the list for known microRNA pathway components, one finds 11 promising B-terms among the top 130: FMRP, hnRNP A1, RNA helicase, exportin, Ago1, RNA binding protein, RNAi, APOBEC3G, eIF6, eIF4F, and RNAse. Examining the titles corresponding to each one in turn, one can readily discern whether they are regulated by phosphorylation in the context of cellular signaling. For example, clicking on FMRP opens a new window revealing several titles that describe phosphorylation of FMRP and its relation to synaptic signaling and translation control (fig. 5). hnRNP A1, p68 RNA helicase, APOBEC3G, eIF6, and eIF4F were also identified as components that are regulated dynamically during cellular signaling by phosphorylation, in a manner that might potentially affect the way they regulate microRNAs.
Fig. 5. Screenshot of the Literature page from the two-node search on A = microRNA and C = phosphorylation.

Several articles (displayed on the right) indicate that FMRP is phosphorylated in response to mGluR signaling and that FMRP phosphorylation is correlated with translation arrest of mRNAs on polyribosomes.
The microRNA literature contains 2333 articles, and the phophorylation literature is voluminous – yet this search was conducted and analyzed in a few minutes. (Searches that are larger in scope, more complex, or that involve domains of knowledge that are less familiar to the user, may take hours to analyze.) The results of the search not only contributed to the writing of the mini-review, and provided strong indirect evidence in favor of phosphorylation as a promising mechanism for regulating microRNA pathways, but generated a short-list of specific candidate proteins that are likely to be regulated.
4. Integrating information regarding a single concept or phenomenon that has been studied in two different contexts
Protease nexin-II is a protease inhibitor that in 1989 was found to correspond to the soluble, carboxyl-truncated form of beta-amyloid precursor protein [10]. Similarly, “radial glial cells” and “neural progenitor cells” within the developing cerebral cortex have both been intensively studied for years, under the belief that they were distinct cell types. Recent studies have indicated that they are, in fact, the same cells regarded from two different perspectives (e.g., [11]). Such situations create a need to integrate information across two pre-existing, disparate literatures -- on protease nexin-II vs. amyloid precursor protein, and on radial glial cells vs. neural progenitor cells. The two-node search provides a systematic way to bring two such disparate yet complementary literatures together.
5. Searching for new hypotheses (e.g. by assessing the results of a one-node search)
The one-node search is an open-ended text mining technique in which an investigator begins with a single set of articles A representing a scientific problem, and seeks to find information published in some disparate field C (unknown to the investigator at the outset) that suggests a novel hypothesis that contributes to solving the problem [1, 12-17]. This is an active area of research [see ref. 17 for reviews] and several public web interfaces for carrying out one-node searches are available (http://kiwi.uchicago.edu/;http://www.mf.uni-lj.si/bitola/; http://litlinker.ischool.washington.edu/). The two-node search is an integral phase of the overall discovery process, since once a candidate C literature is identified in the course of a one-node search, it needs to be assessed further by carrying out a two-node search between A and C.
It is also possible to identify new hypotheses through formulating two-node searches, even without preceding one-node searches. For example, Swanson and colleagues identified a number of novel candidate viruses that were particularly amenable for future bioterror development [18], and Smalheiser identified a number of gene packaging technologies that had not been studied or discussed in the context of bioterror but might be relevant to viral weapons [19]. Other diverse examples of new biomedical hypotheses identified using the two-node search tool were presented in ref. 4. Finally, one can formulate two-node searches in a quasi-open manner by carrying out a two-node search between a small, discrete literature A representing a specific problem, and a second set C of articles that represents an entire category of articles found in MEDLINE (e.g., all articles that deal with pharmacologic agents, or all those that deal with non-drug therapies, etc.)[20].
Discussion
The Arrowsmith Project website has been running continuously since 2001, supporting the text mining analyses of our own research group and a steady number of public visitors (∼1200 unique users per month). The two-node search strategy allows one to identify information that links two disparate sets of articles in a meaningful manner. In the past, the limiting factor in using the tool was the sheer size of the list of B-terms, which can consist of hundreds or even thousands of terms. Recently, a quantitative B-term model has been created and validated, which displays a list of B-terms ranked according to the estimated probability that it will be found relevant to some user [5]. This greatly reduces the number of B-terms that a user needs to peruse. Furthermore, the user has the option to restrict the B-terms to one or more specified semantic categories such as Disease or Syndrome, Hormone, or gene/protein names. An advanced search option is also available that provides users with multiple options for manual filtering [3]. Thus, the current implementation should accommodate the needs of a wide variety of biomedical investigators and students.
Nevertheless, there is still substantial room for future improvement. Many workers in the field of text mining and natural language processing have provided new resources for terminology processing and information extraction [21-26], and we plan to integrate some of these with the Arrowsmith tool. For example, synonymous and closely related B-terms (e.g. abbreviations and their long forms) should be placed together or merged. At present, only title terms are processed, but since much of the information in a scientific article resides in the abstract and in the full-text, we plan to examine how best to analyze terms from abstracts and, eventually, from full-text articles as well. It will be necessary not merely to extract words and phrases, but to process text at the sentence level through programs which attempt to identify the Medical Subject Headings, UMLS concepts, and semantic categories for each identified term in context [21] and that parse the sentences to identify functional entities and relationships (subject/verb/object; e.g., http://www-tsujii.is.s.u-tokyo.ac.jp/medie/).
Another limitation of the current tool is that it only employs general, domain-independent information regarding B-terms, e.g., statistical information regarding the term's frequency and recency. We hope to expand the B-term model to incorporate features based on domain-specific knowledge that are derived from external knowledge sources. For example, a B-term representing a drug name could be scored according to whether the drug is FDA-approved. A B-term representing a gene could be scored according to whether the gene has been identified as a genetic cause of one or more human diseases.
The two-node search tool is only one of several tools which are available on the Arrowsmith Project website and which have been designed to allow biomedical investigators to go beyond the functionalities of the standard PubMed search interface. For example, “Author-ity” is a tool that helps disambiguate author names on Medline articles ([27]), and “Anne O'Tate” helps users summarize, drill-down and browse the results of a PubMed query [28]. We are currently studying new models of one-node searching and plan to offer one-node search functionality as part of the Arrowsmith suite of tools. Eventually, we hope to integrate all of these separate tools into a unified interface.
The two-node search admittedly involves a more advanced set of analyses than carrying out a standard PubMed search. First, the user is not simply looking up a fact or observation stated explicitly in the literature, but is seeking to identify information that is implicit and requires putting together pieces from multiple articles. Second, the user is likely to be assessing new cross-discipline scientific findings or hypotheses and trying to prioritize which of several possible new directions is most promising for further study. Such higher-level scientific strategic thinking may come naturally to principal investigators, but perhaps less readily to students or those who lack adequate domain knowledge. Nevertheless, the recent proliferation of web interfaces that carry out diverse biomedical text mining tasks [21-26] suggests that the scientific community is ready to make use of tools of increasing sophistication.
Acknowledgments
Funding: National Institutes of Health (LM007292 and LM008364 to N.S.).
We thank Marc Weeber, Don R. Swanson, and the Arrowsmith Project field testers for their many contributions to the underlying B-term model and to the web interface.
Footnotes
Conflict of interest statement. None declared.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Swanson DR, Smalheiser NR. An interactive system for finding complementary literatures: a stimulus to scientific discovery. Artificial Intelligence. 1997;91:183–203. [Google Scholar]
- 2.Smalheiser NR, Swanson DR. Using ARROWSMITH: a computer assisted approach to formulating and assessing scientific hypotheses. Computer Methods and Programs in Biomedicine. 1998;57:149–153. doi: 10.1016/s0169-2607(98)00033-9. [DOI] [PubMed] [Google Scholar]
- 3.Smalheiser NR. The Arrowsmith project: 2005 status report. In: Hoffmann A, et al., editors. Discovery Science 2005. Lecture Notes in Artificial Intelligence. Vol. 3735. Springer-Verlag Press; Berlin: 2005. pp. 26–43. [Google Scholar]
- 4.Smalheiser NR, Torvik VI, Bischoff-Grethe A, Burhans LB, Gabriel M, Homayouni R, Kashef A, Martone ME, Perkins GA, Price DL, Talk AC, West R. Collaborative development of the Arrowsmith two node search interface designed for laboratory investigators. Journal of Biomedical Discovery and Collaboration. 2006;1:e8. doi: 10.1186/1747-5333-1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Torvik VI, Smalheiser NR. A quantitative model for linking two disparate sets of articles in Medline. Bioinformatics. 2007;23:1658–1665. doi: 10.1093/bioinformatics/btm161. [DOI] [PubMed] [Google Scholar]
- 6.Wren JD. Extending the mutual information measure to rank inferred literature relationships. BMC Bioinformatics. 2004;5:145. doi: 10.1186/1471-2105-5-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Luo G, Tang C, Tian YL. Answering relationship queries on the web. Proceedings of the 16th International Conference on World Wide Web, WWW 2007; Banff, Alberta, Canada. May 8-12; 2007. pp. 561–570. [Google Scholar]
- 8.Smalheiser NR, Swanson DR. Linking estrogen to Alzheimer's Disease: an informatics approach. Neurology. 1996;47:809–810. doi: 10.1212/wnl.47.3.809. [DOI] [PubMed] [Google Scholar]
- 9.Smalheiser NR. Regulation of mammalian microRNA processing and function by cellular signaling and subcellular localization. Biochimica et Biophysica Acta (BBA) -Gene Regulatory Mechanisms. 2008 doi: 10.1016/j.bbagrm.2008.03.009. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Van Nostrand WE, Wagner SL, Suzuki M, Choi BH, Farrow JS, Geddes JW, Cotman CW, Cunningham DD. Protease nexin-II, a potent antichymotrypsin, shows identity to amyloid beta-protein precursor. Nature. 1989;341:546–549. doi: 10.1038/341546a0. [DOI] [PubMed] [Google Scholar]
- 11.Noctor SC, Flint AC, Weissman TA, Wong WS, Clinton BK, Kriegstein AR. Dividing precursor cells of the embryonic cortical ventricular zone have morphological and molecular characteristics of radial glia. Journal of Neuroscience. 2002;22:3161–3173. doi: 10.1523/JNEUROSCI.22-08-03161.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bekhuis T. Conceptual biology, hypothesis discovery, and text mining: Swanson's legacy. Biomedical Digital Librairies. 2006;3:e2. doi: 10.1186/1742-5581-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Srinivasan P. Text mining: generating hypotheses from MEDLINE. Journal of the American Society for Information Science and Technology. 2004;55:396–413. [Google Scholar]
- 14.Wren JD, Bekeredjian R, Stewart JA, Shohet RV, Garner HR. Knowledge discovery by automated identification and ranking of implicit relationships. Bioinformatics. 2004;20:389–398. doi: 10.1093/bioinformatics/btg421. [DOI] [PubMed] [Google Scholar]
- 15.Hristovski D, Peterlin B, Mitchell JA, Humphrey SM. Using literature-based discovery to identify disease candidate genes. International Journal of Medical Informatics. 2005;74:289–298. doi: 10.1016/j.ijmedinf.2004.04.024. [DOI] [PubMed] [Google Scholar]
- 16.Weeber M, Kors JA, Mons B. Online tools to support literature-based discovery in the life sciences. Briefings in Bioinformatics. 2005;6:277–286. doi: 10.1093/bib/6.3.277. [DOI] [PubMed] [Google Scholar]
- 17.Bruza Peter, Weeber Marc., editors. Literature-based Discovery. Series: Information Science and Knowledge Management. Vol. 15. Springer; Berlin Heidelberg: 2008. [Google Scholar]
- 18.Swanson DR, Smalheiser NR, Bookstein A. Information discovery from complementary literatures: categorizing viruses as potential weapons. Journal of the American Society for Information Science and Technology. 2001;52:797–812. [Google Scholar]
- 19.Smalheiser NR. Predicting emerging technologies with the aid of text-based data mining: a micro approach. Technovation. 2001;21:689–693. [Google Scholar]
- 20.Kostoff RN. Literature-Related Discovery (LRD): Introduction and background. Technological Forecasting and Social Change. 2008;75:165–185. [Google Scholar]
- 21.Aronson AR. Effective mapping of biomedical text to the UMLS Metathesaurus: the MetaMap program. Proceedings of the American Medical Informatics Association Symposium; 2001. pp. 17–21. [PMC free article] [PubMed] [Google Scholar]
- 22.Hunter L, Cohen KB. Biomedical language processing: what's beyond PubMed? Molecular Cell. 2006;21:589–594. doi: 10.1016/j.molcel.2006.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jensen LJ, Saric J, Bork P. Literature mining for the biologist: from information retrieval to biological discovery. Nature Reviews of Genetics. 2006;7:119–129. doi: 10.1038/nrg1768. [DOI] [PubMed] [Google Scholar]
- 24.Cohen AM, Hersh WR. A survey of current work in biomedical text mining. Briefings in Bioinformatics. 2005;6:57–71. doi: 10.1093/bib/6.1.57. [DOI] [PubMed] [Google Scholar]
- 25.Krallinger M, Valencia A. Text-mining and information-retrieval services for molecular biology. Genome Biology. 2005;6:e224. doi: 10.1186/gb-2005-6-7-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou W, Torvik VI, Smalheiser NR. ADAM: Another database of abbreviations in MEDLINE. Bioinformatics. 2006;22:2813–1818. doi: 10.1093/bioinformatics/btl480. [DOI] [PubMed] [Google Scholar]
- 27.Torvik VI, Weeber M, Swanson DR, Smalheiser NR. A probabilistic similarity metric for Medline records: a model for author name disambiguation. Journal of the American Society for Information Science and Technology. 2005;56:140–158. [Google Scholar]
- 28.Smalheiser NR, Zhou W, Torvik VI. Anne O'Tate: A tool to support user-driven summarization, drill-down and browsing of PubMed search results. Journal of Biomedical Discovery and Collaboration. 2008;3:2. doi: 10.1186/1747-5333-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]


