SUMMARY
We have used expression profiling and in vivo imaging to characterize C. elegans embryos as they transit from a developmentally plastic state to the onset of differentiation. Normally, this transition is accompanied by activation of developmental regulators and differentiation genes, down-regulation of early-expressed genes, and large-scale re-organization of chromatin. We find that loss of plasticity and differentiation onset depends on the Polycomb complex protein mes-2/E(Z). mes-2 mutants display prolonged developmental plasticity in response to heterologous developmental regulators. Early-expressed genes remain active, differentiation genes fail to reach wild-type levels, and chromatin retains a decompacted morphology in mes-2 mutants. By contrast, loss of developmental regulators pha-4/FoxA or end-1/GATA does not prolong plasticity. This study establishes a model to analyze developmental plasticity within an intact embryo. mes-2 orchestrates large-scale changes in chromatin organization and gene expression to promote the timely loss of developmental plasticity. Our findings indicate that loss of plasticity can be uncoupled from cell fate specification.
Keywords: PcG, FoxA, digestive tract, foregut, pluripotency, embryo, organogenesis, stem cells
INTRODUCTION
A central question in development concerns the mechanisms that underlie developmental plasticity in early embryos, and its loss during the onset of differentiation. In the mouse, for example, blastula and early gastrula cells contribute to all three germ layers, and can be cultured to establish pluripotent cell lines (Rossant, 2008). As development proceeds, embryonic cells become restricted in their cell fate potential and begin to acquire positional and cell type identities. While the molecular pathways that dictate cell fate decisions are well characterized, less is known about how the loss of developmental plasticity is coupled to cell fate establishment.
C. elegans embryogenesis is a powerful model to study developmental plasticity within the context of an intact animal. Somatic cells acquire different cellular characteristics that can be distinguished by the two-cell stage (Gonczy and Rose, 2005; Sulston et al., 1983). Traditionally, these differences were interpreted to mean that cell fates were determined very early. However, four observations suggest that somatic blastomeres are developmentally plastic until the onset of gastrulation. First, prior to gastrulation (≤2E or Endodermal stage, Figure 1A), most blastomeres contribute to diverse cell types, whereas 3 cell divisions later (8E–16E stage, ~100–200 cells), cells typically produce descendants that contribute to a single tissue or organ (Sulston et al., 1983). Second, embryonic blastomeres adopt alternative fates when C. elegans developmental transcription factors are expressed ubiquitously (Cell Fate Challenge Assay; (Fukushige and Krause, 2005; Gilleard and McGhee, 2001; Horner et al., 1998; Kiefer et al., 2007; Smith and Mango, 2007; Zhu et al., 1998)). The conversion is dramatic, such that a blastomere fated to give rise to skin, for example, can be converted into gut or muscle. This response is lost by the 8E–12E stage, and cells fail to adopt alternate fates when challenged with a heterologous regulator. Third, blastomere exchange experiments show that some cells adopt new identities when moved to new locations (Priess and Thomson, 1987; Wood, 1991). This flexibility reflects intercellular signaling, often by the Notch and wnt pathways (Priess, 2005). Thus, the reproducible cell lineage reflects, in part, reproducible cell interactions. These observations suggest that C. elegans embryonic blastomeres are developmentally plastic, and that this characteristic is lost during gastrulation.
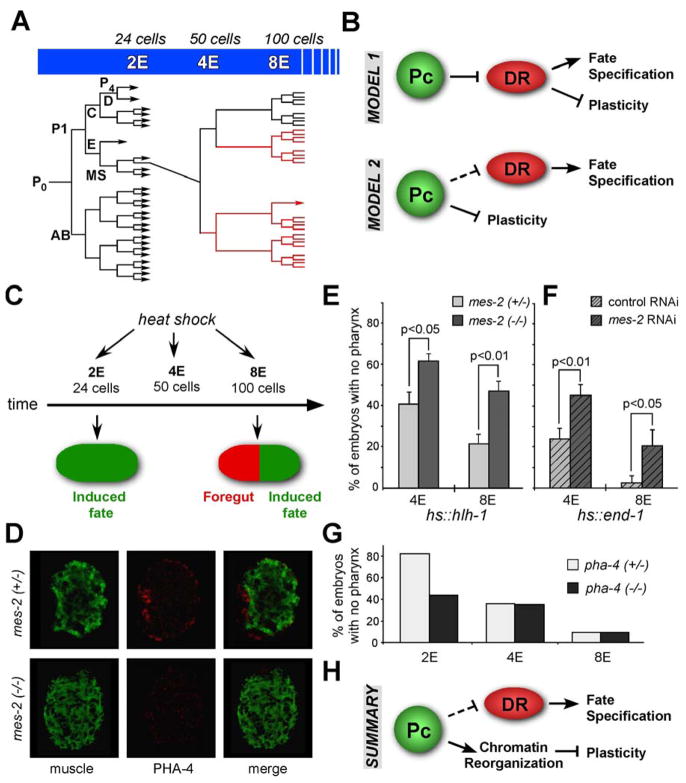
Figure 1. Loss of plasticity depends on mes-2/E(z) but not pha-4/FoxA.
A) Timeline for C. elegans embryogenesis. Embryonic cells are developmentally plastic until the 2E stage. At the ~8E stage, cells become committed to a cell fate e.g. foregut in response to pha-4/FoxA (red). B) Models for loss of developmental plasticity. Model 1: plasticity is maintained by Polycomb (Pc) repression of developmental regulators (DRs). De-repression of DRs both terminates plasticity and specifies cell fate. Model 2: cell fate specification and developmental plasticity are controlled independently. C) The Cell Fate Challenge. A heterologous DR is induced by heat-shock in 2E, 4E or 8E stage embryos. Wild-type 2E embryos adopt the induced fate (green) whereas cells from 8E embryos are resistant and assume their normal fate (e.g. red, foregut). D) and E) Embryos were challenged by HS::hlh-1 to become muscle. D) Control (mes-2(+/−)) or mes-2(−) embryos were challenged at the 8E stage and stained with anti-PHA-4 for foregut (red) and paramyosin for muscle (green). Note the PHA-4+, paramyosin- cells that were resistent to HS::hlh-1 in the mes-2(+/−) embryo. E) Percentage of control (mes-2(+/−)) or mes-2(−) embryos with widespread muscle and no foregut, indicating plasticity of the entire embryo. F) and G) Embryos were challenged to adopt intestinal fate with HS::end-1 at the indicated stages. F) Percentage of control (empty vector RNAi) or mes-2 RNAi embryos with widespread intestine and no PHA-4 staining. G) Percentage of control (pha-4(+/−)) or pha-4(−) embryos with widespread intestine staining. pha-4(−) embryos were identified by the absence of PHA-4. H) Model: Pc influences the loss of plasticity independently of cell fate regulation. Experiments in D–G were repeated ≥3x.
The discovery that mis-expression of developmental transcription factors alters cell identity implicates transcriptional regulatory mechanisms for developmental plasticity and its loss. The factors that mediate these regulatory events are unknown. One appealing candidate is the PRC2 Polycomb complex, which methylates histone H3 K27 (H3K27me) to repress transcription. Components of the PRC2 and PRC1 complexes were identified in Drosophila, where they inhibit developmental regulators to maintain cell fates (Schuettengruber et al., 2007). In ES cells, PRC2 represses developmental regulators as well, and it has been proposed that global inhibition of regulators maintains pluripotency (Figure 1B, Model 1, Niwa, 2007)). This model predicts that loss of PRC2 will derepress developmental regulators, curtail pluripotency and lead to premature differentiation. However, other studies have concluded that PRC2 components are dispensable for pluripotency, a result that is complicated by paralogous PRC components and multiple complexes (Boyer et al., 2006; Chamberlain et al., 2008; Pasini et al., 2007). Thus, the role of Polycomb for pluripotency in mammals is controversial.
C. elegans possesses a complex similar to PRC2 from other organisms. MES-2/E(z), MES-3/novel and MES-6/Esc form a complex that methylates H3K27 in vitro and in vivo (Strome, 2005). C. elegans PRC2 is required to silence the X chromosome in germline cells, which is important to produce a functional germ line (Strome, 2005). PRC2 is also highly expressed in somatic cells of the early embryo (Holdeman et al., 1998; Korf et al., 1998) and is necessary for all detectable H3K27me3 at that time (Bender et al., 2004). However, the role of H3K27me for embryogenesis has not been determined. The only known function for PRC2 in the soma occurs during post-embryonic development (Capowski et al., 1991; Ross and Zarkower, 2003; Zhang et al., 2003), and embryos bearing null mutations in mes genes are viable (Capowski et al., 1991; Strome, 2005).
Here, we characterize the molecular and cell biological features associated with early embryonic cells that are developmentally plastic, and the changes that occur as those cells transit towards differentiation. We examine how these features are altered in embryos lacking either mes factors or select developmental regulators. The data indicate that cell fate restriction and cell fate specification can be uncoupled (Figure 1B, Model 2). Our findings suggest that mes-2 influences global chromatin organization and gene expression to promote the loss of developmental plasticity in the C. elegans gastrula.
RESULTS
mes-2/E(z) inhibits cell fate plasticity
What mechanisms control developmental plasticity and its loss? According to Model 1 (Figure 1B), PRC2 represses developmental regulators to maintain plasticity in early embryos. This model predicts that without PRC2, cells will lose plasticity precociously. According to Model 2 (Figure 1B), plasticity and cell fate specification are controlled independently. To test these models, we performed the Cell Fate Challenge Assay on embryos carrying mutations in mes-2/E(z) (Figure 1C–F). Embryos were challenged to adopt a muscle fate by ectopic expression of hlh-1/MyoD (HS::hlh-1 (Fukushige and Krause, 2005)). To track resistance to hlh-1, we monitored the expression of the foregut marker PHA-4. We chose pha-4 because, in our experience, we always observed PHA-4+ cells among cells that resisted the induced fate ((Kiefer et al., 2007); J.K. and S.E.M., data not shown). Because mes-2 has a maternal effect (Capowski et al., 1991), we examined homozygous mutant embryos from homozygous mutant mothers (“mes-2 embryos”), and confirmed the genotype by staining for H3K27me3 and monitoring sterility. Embryos from mes-2(+/−) mothers served as a negative control.
When challenged by HS::hlh-1 at the 4E stage, 38% of control mes-2(+/−) embryos developed widespread muscle, based on extensive paramyosin expression and an absence of PHA-4 (Figure 1D, E). For the remaining embryos, at least a proportion of cells were no longer able to adopt a muscle fate and exhibited bright PHA-4 staining at the expense of muscle markers (Figure 1D). When challenged at the 8E stage, 19% of embryos responded to ectopic HS::hlh-1 with a complete cell-fate transformation (Figure 1E), in agreement with published results (Fukushige and Krause, 2005; Kiefer et al., 2007). This contrasts with mes-2 mutants, where twice as many embryos responded to ectopic hlh-1 compared to control embryos. 62% and 46% of mes-2 mutants adopted a muscle fate at the 4E and 8E stages, respectively, in response to HS::hlh-1 (Figure 1E). We obtained similar results when we used HS::end-1 (Zhu et al., 1998) to induce intestinal fate after mes-2 inactivation (Figure 1F). These data indicate that mes-2 embryos respond to heterologous regulators later in development than wild-type embryos, suggesting that mes-2 helps terminate developmental plasticity (Model 2, Figure 1B).
In the germ line, mes-2 silences exogenously-introduced DNA (Kelly and Fire, 1998), raising the question of whether mes-2 might alter expression of the HS::hlh-1 transgene. However, we observed no difference in HLH-1 expression in mes-2(+/−) (control) vs. mes-2(−) mothers (Figure S1).
Next, we examined the developmental regulator pha-4, which specifies foregut fate (Mango, 2007). Model 1 suggested that pha-4 might both promote foregut identity and inhibit plasticity in foregut precursor cells (Figure 1B). This hypothesis predicted that without pha-4, defective foregut precursors would retain developmental plasticity longer than normal. Alternatively, plasticity might be controlled independently of cell fate specification (Figure 1B). To distinguish between these models, we performed the Cell Fate Challenge Assay on embryos carrying mutations in pha-4/FoxA (Figure 1G). Ectopic expression of end-1 was induced in embryos from pha-4(0)/+ mothers and ectopic intestinal formation analyzed. 25% of embryos were pha-4 mutants, which could be identified by PHA-4 antibody staining. Their siblings served as controls that had been subjected to identical experimental conditions.
For the control, we observed broad, ectopic intestinal development in embryos challenged at the 2E stage, indicating cell fate plasticity. The ability to switch fate was lost by the 8E stage (Figure 1G), as observed in previous studies (Kiefer et al., 2007; Zhu et al., 1998). pha-4 mutants behaved like control embryos at the 4E and 8E stages, and we did not observe prolonged plasticity (Figure 1G). Instead, we detected a decrease in unrestricted embryos at the 2E stage, from 82% in control to 43% in pha-4 (Figure 1G). Similar results were seen with HS::hlh-1 (Figure S2). These data suggest that pha-4 does not inhibit cell fate plasticity. In fact, pha-4 may contribute to plasticity of nascent foregut cells at the 2E stage. Together, the data from pha-4 and mes-2 argue against Model 1, that PRC2 maintains plasticity by repression of developmental regulators (Figure 1B). Instead, the findings suggest that PRC2 helps terminate plasticity at the 4E – 8E stages independent of cell fate specification (Figure 1H).
Gene expression profiles for unrestricted vs. committed cells
To examine the effect of mes-2 on cellular plasticity in more detail, we developed criteria to distinguish cells with unrestricted developmental potential vs. restricted cells. We compared 2E (plastic), 4E (transition) and 8E (restricted) stage wild-type embryos. First, we examined gene expression profiles by microarray. Second, we explored chromatin morphology using artificial chromosomes assembled from exogenously-introduced DNA. Third, we probed chromatin compaction for endogenous loci using Fluorescence In Situ Hybridization (FISH). These three approaches revealed distinct molecular and morphological features for plastic vs. restricted cells in wild-type embryos.
For microarray, we prepared amplified cDNA from individual, wild-type embryos at the 2-cell, 2E, 4E and 8E stages, using previously described procedures (Robertson et al., 2004). The quality of the cDNA and staging of development were evaluated prior to microarray analysis (Supplemental Data). We identified three categories of genes: 1) “Early” genes that were expressed in 2-cell and 2E-stage embryos but downregulated to background levels by the 8E stage (Figure S3, Figure S4A, D); 2) a “Transition” group comprised of genes transiently expressed at the 2E–4E stage (Figure S3, Figure S4B, E,); 3) “Differentiation” genes that were induced at the 8E stage (Figure S3, Figure S4C, F). All positives were statistically significant, with adjusted p-values of p<0.05 (Benjamini and Hocherg, 1995). We refined the lists of Early, Transition and Differentiation genes by comparison with published microarray data (Baugh et al., 2003) and the RNA in situ database (http://nematode.lab.nig.ac.jp/db/index.html) (Figure S5). We discarded candidates if our expression data disagreed with previous reports, and compiled the remaining genes into a final list of 228 Early, 58 Transition and 50 Differentiation genes (Figure S4).
Misregulated gene expression in mes-2/E(z) mutants
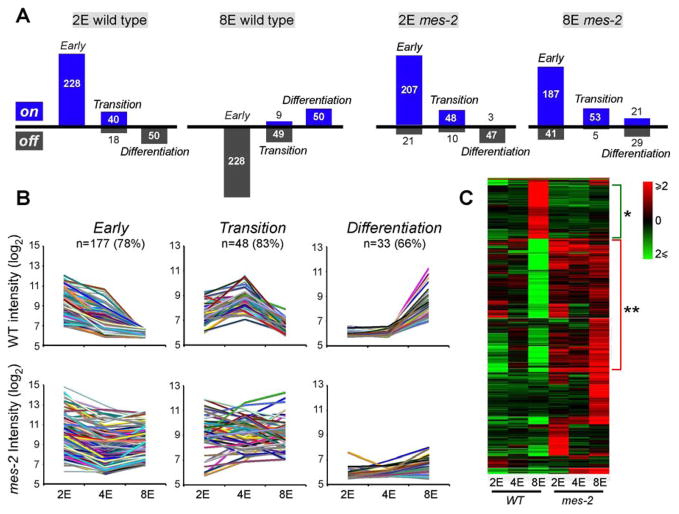
We examined the behavior of Early, Transition and Differentiation gene sets in mes-2 mutants by both microarray and RT-qPCR (Figure S6–S8). For an initial analysis, we tagged genes as ON or OFF, and compared expression in wild-type and mes-2 embryos (Figure 2A). This comparison showed that mes-2 mutants resembled wild-type embryos at the 2E stage: Early and Transition genes were expressed whereas Differentiation genes were not. At the 8E stage, however, mes-2 mutants failed to downregulate most Early or Transition genes (Figure 2A). 78% of Early genes and 83% of Transition genes showed ≥2-fold higher expression at the 8E stage in mes-2 mutants compared to wild-type embryos (q<0.05; Figure 2B). This trend was observed over the entire genome, as well as our Early and Transition genes (Figure 2C). Many genes normally down-regulated by the 8E stage in wild-type embryos were still expressed in 8E mes-2 embryos (Figure 2C, (**) group).
Figure 2. Plasticity of mes-2 mutants.
A) Early, Transition and Differentiation genes from wild-type or mes-2 mutants were designated either “on” (≥3-fold above background) or “off” (<3x background levels) based on average microarray intensity. B) 78% of Early and 83% of Transition genes were up-regulated ≥2x at the 8E stage in mes-2 mutants compared to wild-type (q<0.05). 66% of Differentiation genes were down-regulated in mes-2 embryos at the 8E stage compared to the wild type (q<0.05). C) Two-way ANOVA revealed genome-wide changes in mes-2 mutants compared to wild-type (p<0.05), including genes that failed to activate (*) or down-regulate (**) in 8E mes-2 embryos.
Interestingly, expression of some Early and Transition genes was equal or lower at the 2E or 4E stages in mes-2 mutants compared to the wild type. Thus, for at least some genes, inappropriate expression at the 8E stage reflected a selective loss of downregulation at the onset of differentiation and not a general increase at all stages (data not shown). These data indicate that mes-2 promotes the clearance of Early and Transition mRNAs at the onset of differentiation.
For Differentiation genes, 66% had defects in induction in mes-2 mutants: 29 failed to express above background levels (Figure 2A), and 33 were down-regulated at least two-fold at the 8E stage compared to wild-type 8E embryos (q<0.05; Figure 2B). 15 Differentiation genes were still expressed in mes-2 embryos, with precocious activation of three genes at the 2E and 4E stages (Figure 2A). These trends were also seen genome-wide (Figure 2C). Genes normally activated at the 8E stage in wild-type embryos failed to express in 8E mes-2 embryos (Figure 2C, (*) group). The data indicate that mes-2 is required for the timely activation of the Differentiation program. In sum, the expression studies suggest that 8E-stage mes-2 mutants retain characteristics of younger, developmentally plastic embryos.
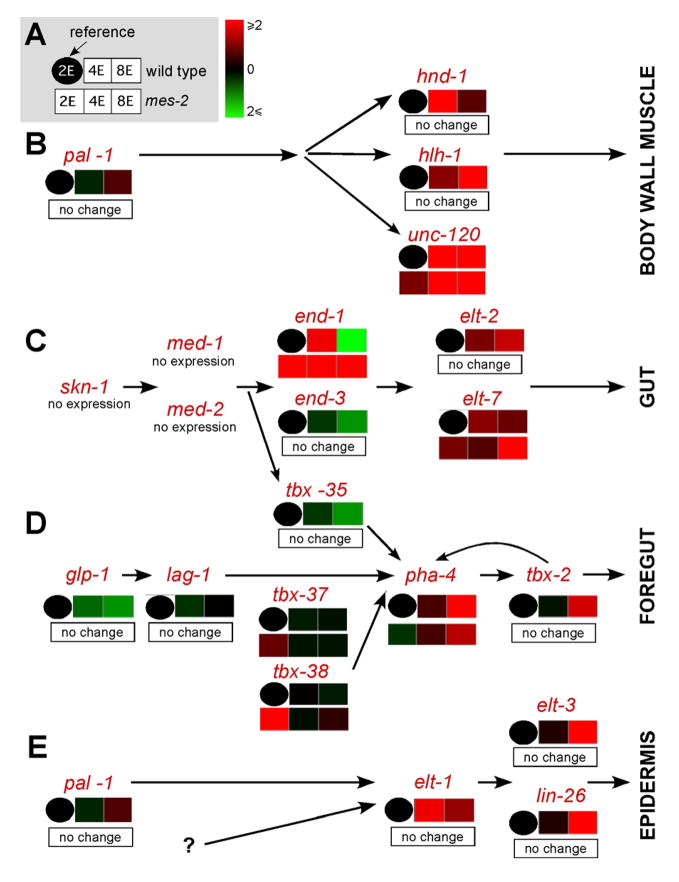
Next, we examined expression of developmental regulators in mes-2 mutants. The developmental pathways that specify skeletal muscle, intestine, foregut and epidermis are well defined in C. elegans (Okkema and Krause, 2005). We surveyed the known developmental regulators and found four that were over-expressed at the 2E stage in mes-2 mutants: the MADS-box factor unc-120/SRF for muscle development, the GATA factor end-1 for intestinal development and the Tbox proteins tbx-37 and tbx-38 for foregut development (q<0.05, Figure 3). These data show that, like mammalian ES cells, loss of C. elegans PRC2 leads to up-regulation of some developmental regulators.
Figure 3. MES-2 represses some developmental regulators.
A). Expression at the 2E, 4E and 8E stages for wild-type (upper panels) or mes-2 (lower panels) embryos relative to the reference (wild-type at the 2E stage, black circle). Down-regulation or up-regulation relative to the reference is depicted in green or red, respectively. Regulators of muscle (B), intestine (C), foregut (D) or epidermis (E) development in wild-type or mes-2 embryos, determined by microarray and graphed by GeneSifter.
To determine the outcome of increased end-1 and tbx-37/tbx-38, we examined the next tier of regulators, those that are normally activated by END-1 or TBX-37/TBX-38 at the 4E and 8E stages. elt-7 is induced by END-1 and END-3 at the 4E stage in wild-type embryos (Baugh et al., 2003; McGhee, 2007), and it began to accumulate one cell division earlier in mes-2 mutants (Figure 3C, Figure S8). At the 4E and 8E stages, elt-7 levels were variable, but not significantly different from wild-type (Figure S8). Increased tbx-37 and tbx-38 levels did not translate into increased expression of pha-4, a key downstream target (Mango 2007). pha-4 transcripts were reduced and delayed in mes-2 mutants (Figure 3D, Figure S8). Nevertheless, tbx-2/Tbox was activated appropriately by the 8E stage, suggesting recovery of foregut development in mutant embryos (Figure 3D, Figure S8). Thus, we observed inappropriate activation of some developmental regulators at the 2E stage, with relatively mild downstream consequences.
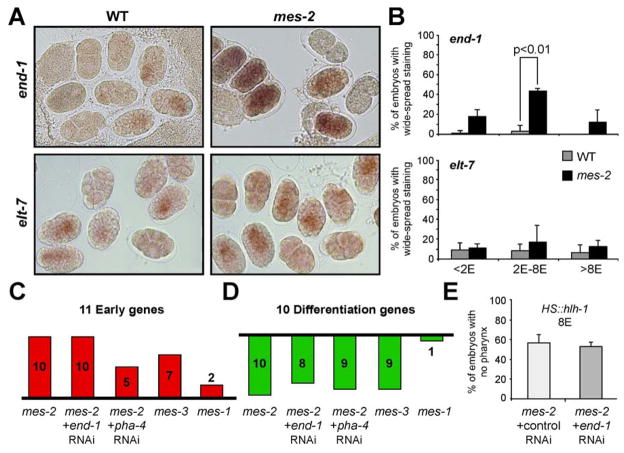
The C. elegans PRC2 complex consists of MES-3 and MES-6 in addition to MES-2 (Strome, 2005). If the role of mes-2 in early embryogenesis reflects its activity within PRC2, then loss of other PRC2 components should resemble loss of mes-2. To test this idea, we examined expression of a subset of Early or Differentiation genes in mes-3 embryos at the 8E stage. Like mes-2, clearance of Early genes and activation of Differentiation genes required mes-3 activity: 7/11 Early genes were up-regulated and 9/10 Differentiation genes were down-regulated in mes-3 mutants compared to wild-type controls (Figure 4C,D). This result implicates PRC2 for the loss of plasticity and onset of differentiation.
Figure 4. Loss of MES-2 leads to wide spread expression of end-1 but not its target elt-7.
A) RNA in situ hybridization for end-1 (upper panels) or elt-7 (lower panels) in wild-type (left) or mes-2 (right) embryos. B) Percentage of wild-type (grey) or mes-2 (black) embryos with wide-spread end-1 (upper) or elt-7 (lower) RNA. Statistically significant changes are designated with p-values. C) Expression of 11 Early genes at the 8E stage was analyzed by RT-qPCR. The number of genes up-regulated ≥2x in mes-2, mes-3 and mes-1 embryos relative to wild type is shown. Genes were considered upregulated in mes-2; end-1(RNAi) or mes-2; pha-4(RNAi) embryos if their expression level was ≥75% of mes-2; control(RNAi) embryos. D) Expression of 10 Differentiation genes at the 8E stage was analyzed by RT-qPCR. The number of genes down-regulated ≥2x in mes-2, mes-3 and mes-1 embryos relative to wild type is shown. Genes were considered down-regulated in mes-2; end-1(RNAi) or mes-2; pha-4(RNAi) embryos if their expression was ≤125% of mes-2/control(RNAi) embryos. E) mes-2 embryos with control (empty vector) or end-1 RNAi were challenged with HS::hlh-1 at the 8E stage. The percentage of embryos with widespread muscle and no PHA-4 is shown.
mes-2 phenotypes reflect somatic not germline roles
The described role for mes-2 is silencing the X chromosome in the germ line (Strome, 2005), raising the question of whether indirect effects from the maternal or embryonic germ line could account for the somatic phenotypes observed here. This scenario seems unlikely, for three reasons. First, the chromosome distribution for genes with altered expression in mes-2 mutants was random, with no enrichment for X-linked genes at either the 2E or 8E stages (Figure S9A). In contrast, previous analyses of germline genes identified a disproportionate number of X-linked genes affected by mes-4 (Bender et al., 2006). The random distribution suggests a broader role for mes-2 in the embryo compared to the germ line. Second, activation of zygotic genes, including most developmental regulators, occurred normally at the 2E and 4E stages in mes-2 mutants (Figures 2, 3). Thus, the initial progression of somatic development in pre-gastrula embryos was not altered by mes-2. Third, we examined embryos carrying mutations in the predicted receptor tyrosine kinase mes-1, which is required to form primordial germ cells (Strome 2005). No significant change in Early or Differentiation gene expression was observed in mes-1 mutants, indicating that the mes-2 phenotype did not reflect the absence of the embryonic germ line (Figure 4C, Figure S10B). Together, these observations suggest that mes-2 acts in the embryonic soma, and that this function is distinct from its role in the germ line.
mes-2 does not rely on end-1 or pha-4 to modulate plasticity
Why do mes-2 mutants display prolonged plasticity? One idea is that spurious de-repression of multiple developmental regulators within a single cell might produce a mixed identity that interfered with developmental progression. To test this idea, we used RNA in situ hybridization to determine whether increased expression of developmental regulators reflected broader, ectopic expression or higher levels within the appropriate cells. For end-1, we observed a wide-spread distribution of transcripts in many mes-2 embryos and precocious activation (Figure 4A,B). In contrast to wild-type embryos (Maduro et al., 2007), end-1 RNAs were first detected before the 1E stage in most mes-2 cells and remained in these cells up to 8E stage. The repercussions of delocalized end-1 expression appeared to be minimal, however, since the end-1 target gene elt-7 was expressed normally in mes-2 embryos (Figure 4A, B). We also surveyed tbx-37, tbx-38, hnd-1, unc-120 and elt-2. These genes either showed the wild-type pattern of expression (Figure S10A) or gave a low, undetectable signal (data not shown). Thus, only end-1 transcripts were obviously mis-localized in mes-2 mutants.
To determine if ubiquitous end-1 contributed to the mes-2 phenotype, we examined Early and Differentiation genes in embryos lacking both mes-2 and end-1. We targeted end-1 by RNAi in mes-2 mutants, which lead to a 95% decrease in end-1 transcripts (data not shown). At the 8E stage, 10/11 Early genes were still expressed in mes-2 and mes-2; end-1 embryos (Figure 4C). Conversely, 8/10 Differentiation genes failed to activate in mes-2; end-1 embryos compared to 10/10 in mes-2 single mutants (Figure 4D). Similar results were obtained with mes-2; pha-4(RNAi) embryos (Figure 4C, D). Finally, we challenged mes-2; end-1 embryos with ectopic HS::hlh-1 to track their ability to adopt a muscle fate. The mes-2; end-1 embryos responded identically to mes-2 single mutants (Figure 4E). These results demonstrate that mis-regulation of developmental regulators end-1 and pha-4 cannot account for the delayed developmental progression of mes-2 mutants.
mes-2 controls chromatin re-organization during developmental progression
A striking feature of young C. elegans embryos is the lack of obvious heterochromatin, as detected by electron microscopy (Leung et al., 1999). Given the role of Polycomb in chromatin organization, we examined global chromatin morphology in wild-type and mes-2 embryos using the Nuclear Spot Assay (Carmi et al., 1998; Fukushige et al., 1999). In previous studies, the Nuclear Spot Assay accurately reflected transcriptional regulation (Carmi et al., 1998; Fukushige et al., 1999) and provided a means to track large-scale chromatin conformation. An added advantage is the ability to examine individual cells within an embryo, for precise spatial and temporal resolution.
For the Nuclear Spot Assay, an extrachromosomal array, or pseudo-chromosome, was constructed with multiple copies of a C. elegans promoter and the Lac operator (Figure 5A). A co-selectable marker (to identify transgenic animals) and herring sperm genomic DNA (to provide sequence complexity without added C. elegans sequences (Kelly et al., 1997)) were also included. The array carried LacI::CFP, which bound the Lac operator and revealed the position and morphology of the array within the nucleus. Only interphase nuclei were monitored, to focus on changes due to developmental stage rather than those associated with mitosis. We surveyed arrays bearing promoters that were active during differentiation (myo-2 or C44H4.1 (Mango 2007)). We also examined arrays lacking any promoter (No Target), promoters that were transcriptionally silent (pax-1(mutP) or pax-1(mutA), T. Fakhouri and S.E.M., unpublished) or the active pax-1 promoter (K07C11.1 Figure S11). Arrays bearing these different promoters behaved similarly, so we describe them as a group.
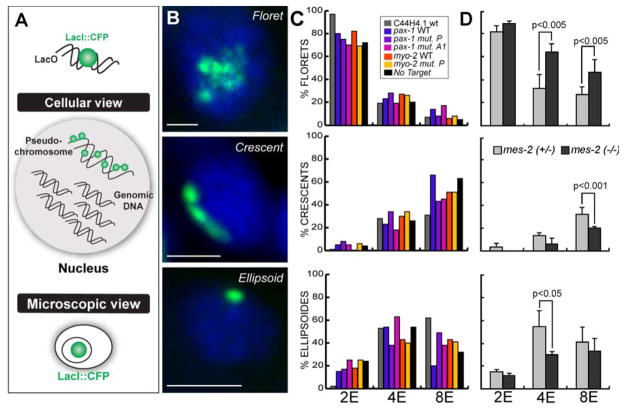
Figure 5. Decompacted chromatin in developmentally plastic embryos.
A) LacI::CFP (green) binds LacO in extrachromosomal arrays to reveal their position and morphology. B) An array carrying the myo-2 promoter forms a floret (2E stage), a crescent (8E stage) or an ellipsoid (4E stage). LacI::CFP (green), DAPI (blue). Scale bar, 3μm. C) Percentage of interphase nuclei with florets, crescents or ellipsoids for lines bearing arrays with the indicated promoters at the 2E, 4E or 8E stages. D). Percentage of interphase nuclei carrying florets, crescents or ellipsoids for No Target arrays in control (mes-2(+/−)) or mes-2(−) embryos at the indicated stages. Statistically significant changes are designated with p-values.
Using the Nuclear Spot Assay, we observed different chromatin morphologies at different stages of wild-type embryogenesis (Figure 5B, C). At the 2E stage, arrays had a distended configuration, suggesting they were globally de-compacted (Figure 5B). We named this morphology “floret,” and we observed florets in most cells of all embryos at the 2E stage (≥70% cells, Figure 5C, Table S2). Over the next two cell divisions, a growing proportion of arrays lost the floret morphology, and appeared compacted and crescent-like (Figure 5B, C, Table S2). The crescents were consistently located at the periphery of the nucleus, a location that is often associated with transcriptionally-repressed genes (Brown and Silver, 2007). Conversely, few cells at the 4E or 8E stage contained florets (Figure 5C, Table S2). The remaining arrays had an ellipsoid morphology that appeared very compact (Figure 5B, Table S2). Thus, early cells had distended arrays, and the loss of developmental plasticity was accompanied by chromatin compaction and repositioning within the nucleus. This transition was independent of transcriptional modulation of genes within the arrays, suggesting it was a general feature of plastic vs. restricted cells.
We next asked whether mes-2 was important for chromatin conformation at the 2E or 8E stages (Figure 5D). 2E stage embryos resembled the wild type, with a preponderance of florets and few crescents or ellipsoids. At the 8E stage, however, we observed more florets and fewer crescents in mes-2 mutants compared to either mes-2(+/−) or wild-type embryos. The number of florets also increased at the 4E stage, at the expense of ellipsoids. We conclude that mes-2 promotes compaction of arrays at the onset of differentiation. We note that mes-2 is not absolutely required for crescent or ellipsoid formation, since both configurations were observed in mes-2 mutant cells. An interesting possibility is that global chromatin reorganization by mes-2 contributes to the termination of plasticity and/or the onset of differentiation.
PRC2 could contribute to chromatin morphology directly, by modifying histones within arrays, or it could function indirectly, by controlling a regulator of morphology. To address this question, we examined whether H3K27me3 was associated with arrays; PRC2 is responsible for all detectable H3K27me3 in the early C. elegans embryo (Bender et al. 2004). Background levels of H3K27me3 were observed on all arrays, independent of stage or morphology (Figure 6A, B). However, it was enriched on a proportion of crescents and ellipsoids, particularly after the 4E stage. We did not observe high levels of H3K27me3 on florets at any stage. This result suggests that PRC2 directly targets nucleosomes associated with arrays. We note that indirect effects by PRC2 may also contribute towards array morphology.
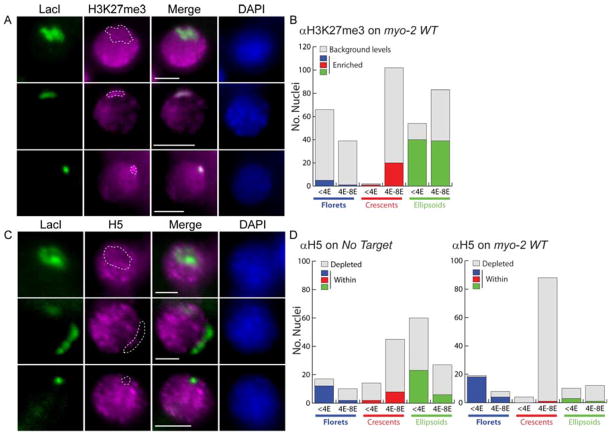
Figure 6. Histone H3K27me3 is enriched on crescents and ellipsoids.
Nuclei bearing extrachromosomal arrays stained for LacI::CFP (green), DAPI (blue) and either A) H3K27me3 or C) elongating RNA Polymerase II (H5 antibody). A) A floret with background levels of H3K27me3 (upper) vs. a crescent (middle) and ellipsoid (lower) with enriched H3K27me3. B) Number of arrays with enriched H3K27me3 (colors, “enriched”) vs. background levels (grey). C) An H5+ floret (upper) vs. a crescent (middle) and ellipsoid (lower) with H5 depleted. D) Number of arrays associated with H5 (colors, “within”) or depleted for H5 (grey). Scale bar, 3um.
The different levels of H3K27me3 on florets, crescents and ellipsoids suggested that these configurations represented different transcriptional states. To test this idea, we monitored transcriptional elongation with H5, an antibody that recognizes phosphoserine 2 within the carboxyl terminal domain of elongating RNA Polymerase II (Patturajan et al., 1998). H5 staining was depleted on crescents and ellipsoids in most nuclei between the 2E and 8E stages (Figure 6C, D). On the other hand, florets stained with H5 antibody at the <4E stage, indicating that elongating Pol II was not excluded. In 4E and 8E embryos, H5 immunoreactivty decreased, suggesting transcription was reduced on florets at later stages. These results support the idea that florets are transcriptionally active to a greater degree than are crescents or ellipsoids, particularly in pre-gastrula embryos.
Chromatin compaction of endogenous loci in wild-type vs. mes-2 mutants
We adapted techniques for FISH (Csankovszki et al. 2004) to investigate whether DNA in its natural chromosomal context underwent similar morphological changes as DNA in arrays (Figure 7). FISH was performed with probes encompassing either myo-2 or pax-1, both of which are located in gene-dense regions (Figure S12). Pairs of Cy3 and Cy5 labelled probes were located >100kb apart, and the distance between the center of each probe was used as an indicator of chromatin morphology. We focused on pre-replicative, interphase DNA (Figure S11) at the 2E or 8E stages.
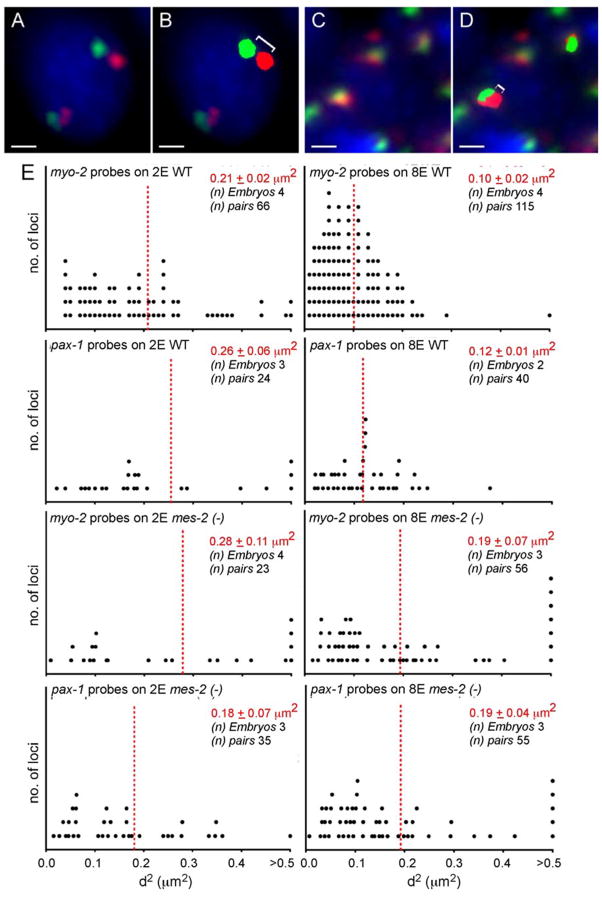
Figure 7. Reorganization of endogenous chromatin during loss of plasticity.
A) 2E and C) 8E nuclei stained with Cy3- (red) and Cy5- (green) labelled probes flanking the myo-2 λοχνσ, αυδ χονυτερσταιυεδ ωιτη ΔΑΠΙ(βλνε). Σχαλε βαρ, 1μm. B) and D) Cy3 and Cy5 signals were marked as 3D objects (solid red or green), and their X, Y and Z coordinates determined by Volocity (Experimental Procedures). E) Plots represent the distribution of 3D-distances (μm2) for myo-2 and pax-1 regions in wild-type or mes-2 embryos at the 2E or 8E stages. Averages in red.
We observed a range of distances for probes spanning either the myo-2 or pax-1 regions (Figure 7E). This variability did not reflect orientation of probes relative to the plane of focus, because distances were calculated based on optical sectioning and analysis in three dimensions (Chambeyron and Bickmore, 2004). While we cannot rule out that some variability reflected fixation conditions, the reproducibility suggested that the variability reflected biological differences.
We compared nuclei from 2E vs. 8E embryos and observed a reduction in inter-probe distances at the 8E stage. The mean-square distance for the myo-2 region decreased from 0.21 ± 0.02 μm2 (±S.D.) at the 2E stage to 0.10 ± 0.02 μm2 at the 8E stage, while the pax-1 region decreased from 0.26± 0.06 μm2 to 0.12±0.01 μm2 (Figure 7E). These changes were statistically significant (p= 5.1×10−6 and 8.2×10−5 for myo-2 and pax-1 respectively).
One potential reason for reduced probe distances at the 8E stage was a decrease in nuclear size, as C. elegans blastomeres undergo reductive divisions. To address this question, we examined nuclei of variable sizes in 2E embryos (Figure S11C). Given that 8E nuclei ranged between ~2.4 − 3.3 um in diameter, we compared inter-probe distances in 2E nuclei that were <3.3 um (n=18) or >3.3 um (n=51). Our analysis revealed no significant difference in inter-probe distances between these two groups (p>0.05; Figure S11C). Moreover, we still observed a reduction in inter-probe distances when we compared the small 2E nuclei to all 8E nuclei (n=165) (p<0.0001). Thus, compaction of endogenous chromatin during the loss of developmental plasticity is not a consequence of nuclear size reduction.
Next, we examined the effects of mes-2 on chromatin reorganization for endogenous loci. mes-2 was required to restructure the pax-1 locus at the 8E stage. Mutant 8E embryos had the same average inter-probe distances as 2E embryos, which were intermediate between wild-type 2E or 8E (Figure 7E). For myo-2, we observed a decrease in inter-probe distances at the 8E stage, but it was not as pronounced as the wild type. This compaction was not statistically significant compared to the mutant 2E stage (p=0.29), and was statistically significant compared with heterozygous 8E-stage control (p=0.031), suggesting reorganization at the 8E stage was compromised in mes-2 mutants. The remaining compaction of the myo-2 region, if it occurs in mes-2 mutants, may reflect dosage compensation since this gene is located on the X chromosome (Meyer, 2005). We note that nuclear size was not altered by mes-2 mutations (Figure S11D), and that mes-2(+/−) heterozygotes behaved like wild-type embryos, as expected (Figure S13). These data reveal that mes-2 is required to modulate chromatin morphology at the 8E stage.
DISCUSSION
This study has made three contributions towards understanding developmental plasticity in the early C. elegans embryo. First, we characterized the molecular and morphological features of developmentally plastic cells and compared them to cells that have undergone cell-fate restriction. In wild-type embryos, we observed large-scale changes in gene expression and global alterations in chromatin morphology during the transition to cell-fate restriction. Second, we examined the PRC2 component mes-2/E(z) and found that it was required for the timely onset of differentiation and not to maintain developmental plasticity. Third, we considered two possible roles for mes-2 in differentiating cells: silencing of developmental regulators vs. global reorganization of chromatin. Neither end-1 nor pha-4 was required to terminate plasticity, indicating that cell fate specification can be uncoupled from plasticity. Our findings implicate large-scale restructuring of chromatin and gene expression by MES-2 as an important facet of differentiation onset.
Developmental plasticity of pre-gastrula embryos
Studies of wild-type and perturbed C. elegans embryos have revealed that somatic blastomeres from pre-gastrula embryos are developmentally plastic (Mango, 2007). In the present study, we used gene expression profiling and chromatin analysis to characterize the plastic state, and to track the changes that accompany differentiation onset. The expression arrays revealed dramatic shifts in transcript pools from the 2E – 4E stages to the 8E stage. This observation agrees well with previous studies that noted a transition from maternal to embryonically-expressed genes at the onset of gastrulation (Baugh et al., 2003). The switch from maternal to zygotic control suggests that this stage may resemble a mid-blastula transition (MBT) similar to other animals (Heasman, 2006). Consistent with this idea, C. elegans undergo cell cycle lengthening and cell movements at the 2E stage, akin to the MBT of other animals (Edgar and McGhee, 1988; Sulston et al., 1983). These cellular behaviors and gene expression profiles suggest that the 2E to 8E period of development constitutes a major transition during embryogenesis.
At least five classes of transcription factors contribute to pluripotency in mammals: Oct4/Pou, Sox2, Nanog, Klf/Kruppel-like and c-myc (Niwa, 2007). C. elegans possess homologues of several of these factors, but their functions are not obviously linked to developmental plasticity. For example, C. elegans mep-1/Kruppel-like represses germline transcription in somatic cells, indicating that Kruppel-like factors promote a somatic or differentiated state in C. elegans (Unhavaithaya et al., 2002). mml-1 is homologous to vertebrate c-myc, but mml-1 mutants have no known embryonic phenotype (Pickett et al., 2007).
We searched genes expressed at the 2E stage for homologues of mammalian pluripotency genes, but did not identify any obvious candidates. For example, the three Pou proteins unc-6, unc-86 and ceh-18 are not expressed in the early embryo (this work; (Baugh et al., 2003)). We also examined promoters of 2E-expressed genes to determine if any were enriched for the known binding sites of these factors, but they were not (E. Johnson and S.E.M., unpublished). Thus, it is unclear if developmentally plastic cells in C. elegans depend on the same constellation of sequence-specific transcription factors as do mammalian embryos.
In addition to gene expression changes, we observed large-scale reorganization of chromatin between the 2E and 8E stages. Developmentally plastic cells contained decompacted florets, and this configuration was lost during the transition towards differentiation. Florets were associated with a marker of elongating RNA pol II and lacked repressive histone marks, suggesting an open chromatin configuration. Ellipsoids and crescents were detected at the 4E and 8E stages. These conformations were associated with a repressive histone mark and reduced elongating RNA pol II, consistent with a more closed or silenced configuration. Changes in arrays were mirrored by morphological changes near the myo-2 and pax-1 loci, indicating that chromatin reorganization is a feature of endogenous loci as well as arrays. In embryos from other species, there may also be a transition from open to compacted chromatin, based on changes in nuclear size (Ner and Travers, 1994), but this has not been investigated directly. In culture, chromatin from pluripotent ES is decondensed and becomes condensed when cells are induced to differentiate (Meshorer and Misteli, 2006). Thus, many types of cells undergo a transition in chromatin conformation as they lose developmental plasticity.
What is the underlying organization that establishes different chromatin morphologies? The florets appear relatively unstructured and diffuse within the nucleus. Ellipsoids and crescents may contain compacted and/or looped DNA that associates with the nuclear periphery. Electron microscopy studies have revealed that C. elegans nuclei at both the 2E and 8E stages lack electron-dense material, which typically characterizes heterochromatin (Leung et al., 1999). Thus, the nature of the compacted chromatin at the 4E and 8E stages is unclear.
Studies with other animals have shown a strong correlation between decompaction and the onset of transcription (Spector, 2003). What is the relationship of transcriptional activity with the chromatin configurations in early vs. late embryonic cells? The foregut promoters included in our arrays are active many hours after the 2E stage (Gaudet and Mango, 2002; Tabara et al., 1996). Moreover, arrays bearing mutated promoters or no added promoter formed Florets at the 2E stage, similar to those bearing wild-type promoters. These observations indicate that decompaction is not dependent on productive transcription that generates mature mRNAs. On the other hand, the H5 staining suggests that arrays at the 2E stage are transcriptionally active to some extent. One possibility is that early C. elegans embryos, like ES cells (Turner, 2008), are transcriptionally hyperactive, meaning DNA is transcribed promiscuously. A speculative idea is that open chromatin may be an important attribute of developmentally plastic cells that provides accessibility to the genome.
Chromatin reorganization during the transition towards differentiation may restrict transcriptional access or help partition un-needed DNA within the nucleus. This effect is unlikely to depend on the activity of specific promoters. One reason for thinking so is that arrays with and without foregut promoters underwent analogous conformational changes at the 4E and 8E stages. At endogenous loci, three genes within the myo-2 region were active in 4E–8E embryos, while at least four were silent (Baugh et al., 2003). Two genes from the pax-1 region were active in 2E–4E embryos, while at least five were not (Baugh et al., 2003). Thus, changes in chromatin conformation from the 2E to 8E developmental stages are likely distinct from the classic examples of decompaction by promoter firing. Our data suggest that chromatin reorganization can reflect different developmental states.
mes-2/E(z) promotes the transition from plasticity to differentiation
We considered two models regarding how developmental plasticity is lost (Figure 1). One appealing idea is that developmental regulators inhibit plasticity as well as promote cell fate. Conversely, silencing of developmental regulators by repressors such as the Polycomb complex might maintain developmental plasticity (Niwa, 2007). We tested this idea four ways and found no evidence for inhibition of plasticity by developmental regulators or promotion of plasticity by Polycomb. First, inactivation of the developmental regulator pha-4/FoxA failed to prolong plasticity, and loss of mes-2 failed to promote differentiation, as measured by the Cell Fate Challenge Assay. Second, expression profiling of mes-2 and mes-3 mutants revealed an inability to down-regulate Early genes or activate Differentiation genes at the 8E stage, suggesting a delayed onset of differentiation. Third, inactivation of end-1 or pha-4 in mes-2 mutants did not suppress the mes-2 phenotype. Fourth, chromatin from mes-2 mutants failed to compact at the 8E stage, and resembled chromatin at earlier stages. These data suggest that developmental regulators like pha-4 and end-1 are not key terminators of plasticity, nor that Polycomb maintains plasticity. Instead, our findings indicate that cell fate restriction can be uncoupled from cell fate specification (Figure 1H). This separation may explain how early embryos can retain developmental plasticity even as they undergo rapid changes in gene expression at the onset of embryogenesis (Heasman, 2006; Rossant, 2008). This feature of developing embryos is different from pluripotent cell lines in which the pluripotent state is associated with arrested development and a static expression landscape (Rossant, 2008).
Why is plasticity prolonged in mes-2 mutants?
What activity of mes-2 is required for the timely transition to differentiation? One possibility is that de-repression of multiple developmental regulators in mutant embryos activates diverse developmental programs within single cells. The resulting confusion might interfere with the ability of cells to terminate plasticity and differentiate in a timely fashion. This scenario seems unlikely since most developmental regulators were activated normally in mes-2 mutants. Moreover, the target genes of these regulators were also expressed normally. Thus, widespread expression of end-1 in mes-2 mutants did not generate widespread end-1 activity, as detected by elt-7 expression. We note that although de-repression of developmental regulators cannot explain the mes-2 phenotype, it is still possible that other critical genes are targeted by the MES factors.
A second appealing hypothesis is that MES factors modulate large-scale chromatin organization. In mes-2 mutants, Polycomb has been implicated in reorganization of chromatin in other contexts, including mammalian X inactivation and genomic imprinting (Erwin and Lee, 2008; Zaratiegui et al., 2007). Moreover, PRC2-Ezh1 can compact nucleosomes in vitro (Margueron et al., 2008). It will be of interest to learn whether C. elegans PRC2 shares mechanistic features with mammalian PRC2 complexes, and how these activities contribute to the loss of plasticity and the transition towards differentiation.
EXPERIMENTAL PROCEDURES
Strains are listed in Supplemental Data.
MICROARRAY ANALYSIS
Sample preparation
2-cell embryos were collected from wild-type or mes-2 mothers and incubated at 20°C for 75 min. (2E), 120 min. (4E), 3h. (8E). Comparison of mes-2 embryos with wild-type showed equivalent cell cycle timing. RNA and cDNA from individual embryos were prepared by 2 rounds of amplification and labeled with Cy5 (Robertson et al., 2004). Microarray hybridization was performed with Agilent C. elegans chips, against a mixed-stage, embryonic cDNA control labeled with Cy3. Three independent experiments were performed for each stage.
Data Processing and Normalization
microarray data were quantified with Agilent Feature Extraction (v8.5.1.1). Repeated labeling and hybridization of the same samples were used to evaluate normalization methods. Data were processed with quantile normalization and subsequent log2 transformation of the Cy5 intensity data (DNAMR software library, R language, http://www.rci.rutgers.edu/~cabrera/DNAMR/).
Data analysis
GeneSifter (www.genesifter.net) was used to analyze microarray samples. Wild-type Early, Transition and Differentiation genes were selected using the criteria outlined in Figure S3 and confirmed by comparison with data from other sources for wild-type embryos (Figure S5) and RT-qPCR for mes-2 mutants (Figures S6 – S8).
ANTIBODY STAINS
Antibody staining was performed as described previously (Kiefer et al., 2007). Antibodies used in this study are listed in Supplemental Data. Imaging was performed with the DeltaVision Core imaging system and analyzed by Softworx (Applied Precision). The number of nuclei in an embryo determined its age. Embryos containing 26–40 nuclei were considered 2E, 51–80 nuclei 4E, and 102–190 nuclei 8E. Only interphase nuclei, as determined by DAPI, were scored for array morphology.
RNA IN SITU HYBRIDIZATION
In situ hybridization was performed as described (http://www.faculty.ucr.edu/~mmaduro/). Images were acquired with SPOT Insight Digital Camera (Diagnostic Instruments, Inc).
NUCLEAR SPOT ASSAY
Nuclear spot assays were performed as described (Updike and Mango, 2006, Kiefer et al., 2007). A Student’s t-test (two-tailed) was used to test the significance of differences in array morphology between mes-2(+/−) and mes-2(−) embryos.
FLUORESCENCE IN SITU ANALYSIS (FISH)
We performed FISH (Csankovszki et al., 2004) with the following probes: myo-2 (128.3 Kb span): 5′ F17E5 and T24D3, and 3′ T24D5, pax-1 (93.3 Kb span): 5′ F20D6 and 3′ W02D7. Probes were labeled by random priming (Promega)(Csankovszki et al., 2004) or FISH Tag DNA Kits (Invitrogen #F32949, #F32947) according to the manufacturer’s protocol.
Image stacks with 0.1 or 0.15 μm Z-step were collected (DeltaVision RT Deconvolution System, SoftWoRx software). A 3D image was reconstructed and analyzed with Volocity software (Improvision). We avoided mitotic DNA by DAPI and replicating DNA (Figure S11 (Boggs and Chinault, 1997)). The XYZ coordinates of the centroid of each hybridization signal and the mean-square distance between two probes (d2) were determined according to (Chambeyron and Bickmore, 2004), where d2 = dx*dx + dy*dy + dz*dz where dx=x(cy3)– x(cy5), dy=y(cy3)–y(cy5) and dz=z(cy3)– z(cy5). A Student’s t-test (two-tailed) was used to test the significance of differences in chromatin compaction in 2E vs. 8E embryonic cells.
CELL-FATE CHALLENGE ASSAY
Two-cell embryos were collected from mes-2(+/−) or mes-2(−) mothers (Capowski et al., 1991) that carried an integrated HS::hlh-1 array (Fukushige and Krause, 2005). Embryos were incubated at 20°C for 75 min. (2E), 120 min. (4E), or 3hr. (8E). Heat shock was administered, and embryos were stained for 4C6.3 (muscle) and PHA-4 (foregut) as described (Kiefer et al., 2007). Images were acquired using an Olympus FluoView™ FV1000 confocal microscope. pha-4(+/−) mothers (Mango et al., 1994) that carried an integrated HS::end-1 array (Zhu et al., 1998) were treated as in (Kiefer et al., 2007) and stained for 1CB4 (intestine) and PHA-4 (pha-4+, high PHA-4, foregut) and intestine (pha-4+, low PHA-4). L4 HS::end-1 animals with control (empty vector) or mes-2 RNAi, and mes-2; HS::hlh-1 animals with control, end-1 or pha-4 RNAi were grown overnight. Embryos were collected the next day and subjected to heat shock as described above.
Supplementary Material
Acknowledgments
We thank A. Sanchez Alvarado, C. Murtaugh and A. Schier for critical reading of the manuscript and discussions, K. Good and J. Priess for unpublished information, J. Rand, M. Krause, J. Rothman for reagents, B. Wardell for technical help, S. Phillips and Andor Technology for use of a Spinning Disk Microscope, The Caenorhabditis Genetics Center for strains, B. Dalley, B. Milash, C. Rodesch, E. Johnson and the University of Utah Core facilities P30CA042014. T.Y. was supported by NIH F32GM077903, J.K. by NIH F32GM65728 and S.E.M. by R01 GM056264, the Huntsman Cancer Institute and the Department of Oncological Sciences.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baugh LR, Hill AA, Slonim DK, Brown EL, Hunter CP. Composition and dynamics of the Caenorhabditis elegans early embryonic transcriptome. Development. 2003;130:889–900. doi: 10.1242/dev.00302. [DOI] [PubMed] [Google Scholar]
- Bender LB, Cao R, Zhang Y, Strome S. The MES-2/MES-3/MES-6 complex and regulation of histone H3 methylation in C. elegans. Curr Biol. 2004;14:1639–1643. doi: 10.1016/j.cub.2004.08.062. [DOI] [PubMed] [Google Scholar]
- Bender LB, Suh J, Carroll CR, Fong Y, Fingerman IM, Briggs SD, Cao R, Zhang Y, Reinke V, Strome S. MES-4: an autosome-associated histone methyltransferase that participates in silencing the X chromosomes in the C. elegans germ line. Development. 2006;133:3907–3917. doi: 10.1242/dev.02584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hocherg Y. Controlling the false discovery rate—A practical and powerful approach to multiple testing. Journal of the Royal Statistical Society. 1995;57:289–300. [Google Scholar]
- Boggs BA, Chinault AC. Analysis of DNA replication by fluorescence in situ hybridization. Methods. 1997;13:259–270. doi: 10.1006/meth.1997.0525. [DOI] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- Brown CR, Silver PA. Transcriptional regulation at the nuclear pore complex. Curr Opin Genet Dev. 2007;17:100–106. doi: 10.1016/j.gde.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Capowski EE, Martin P, Garvin C, Strome S. Identification of grandchildless loci whose products are required for normal germ-line development in the nematode Caenorhabditis elegans. Genetics. 1991;129:1061–1072. doi: 10.1093/genetics/129.4.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmi I, Kopczynski JB, Meyer BJ. The nuclear hormone receptor SEX-1 is an X-chromosome signal that determines nematode sex. Nature. 1998;396:168–173. doi: 10.1038/24164. [DOI] [PubMed] [Google Scholar]
- Chamberlain SJ, Yee D, Magnuson T. Polycomb Repressive Complex 2 is Dispensable for Maintenance of Embryonic Stem Cell Pluripotency. Stem Cells. 2008 doi: 10.1634/stemcells.2008-0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambeyron S, Bickmore WA. Chromatin decondensation and nuclear reorganization of the HoxB locus upon induction of transcription. Genes Dev. 2004;18:1119–1130. doi: 10.1101/gad.292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csankovszki G, McDonel P, Meyer BJ. Recruitment and spreading of the C. elegans dosage compensation complex along X chromosomes. Science. 2004;303:1182–1185. doi: 10.1126/science.1092938. [DOI] [PubMed] [Google Scholar]
- Edgar LG, McGhee JD. DNA synthesis and the control of embryonic gene expression in C. elegans. Cell. 1988;53:589–599. doi: 10.1016/0092-8674(88)90575-2. [DOI] [PubMed] [Google Scholar]
- Erwin JA, Lee JT. New twists in X-chromosome inactivation. Curr Opin Cell Biol. 2008;20:349–355. doi: 10.1016/j.ceb.2008.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushige T, Hendzel MJ, Bazett-Jones DP, McGhee JD. Direct visualization of the elt-2 gut-specific GATA factor binding to a target promoter inside the living Caenorhabditis elegans embryo. Proc Natl Acad Sci U S A. 1999;96:11883–11888. doi: 10.1073/pnas.96.21.11883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushige T, Krause M. The myogenic potency of HLH-1 reveals wide-spread developmental plasticity in early C. elegans embryos. Development. 2005;132:1795–1805. doi: 10.1242/dev.01774. [DOI] [PubMed] [Google Scholar]
- Gaudet J, Mango SE. Regulation of organogenesis by the Caenorhabditis elegans FoxA protein PHA-4. Science. 2002;295:821–825. doi: 10.1126/science.1065175. [DOI] [PubMed] [Google Scholar]
- Gilleard JS, McGhee JD. Activation of hypodermal differentiation in the Caenorhabditis elegans embryo by GATA transcription factors ELT-1 and ELT-3. Mol Cell Biol. 2001;21:2533–2544. doi: 10.1128/MCB.21.7.2533-2544.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonczy P, Rose LS. Asymmetric cell division and axis formation in the embryo. WormBook. 2005:1–20. doi: 10.1895/wormbook.1.30.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heasman J. Patterning the early Xenopus embryo. Development. 2006;133:1205–1217. doi: 10.1242/dev.02304. [DOI] [PubMed] [Google Scholar]
- Holdeman R, Nehrt S, Strome S. MES-2, a maternal protein essential for viability of the germline in Caenorhabditis elegans, is homologous to a Drosophila Polycomb group protein. Development. 1998;125:2457–2467. doi: 10.1242/dev.125.13.2457. [DOI] [PubMed] [Google Scholar]
- Horner MA, Quintin S, Domeier ME, Kimble J, Labouesse M, Mango SE. pha-4, an HNF-3 homologue, specifies pharyngeal organ identity in Caenorhabditis elegans. Genes Dev. 1998;12:1947–1952. doi: 10.1101/gad.12.13.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly WG, Fire A. Chromatin silencing and the maintenance of a functional germline in Caenorhabditis elegans. Development. 1998;125:2451–2456. doi: 10.1242/dev.125.13.2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly WG, Xu S, Montgomery MK, Fire A. Distinct requirements for somatic and germline expression of a generally expressed Caernorhabditis elegans gene. Genetics. 1997;146:227–238. doi: 10.1093/genetics/146.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiefer JC, Smith PA, Mango SE. PHA-4/FoxA cooperates with TAM-1/TRIM to regulate cell fate restriction in the C. elegans foregut. Dev Biol. 2007;303:611–624. doi: 10.1016/j.ydbio.2006.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korf I, Fan Y, Strome S. The Polycomb group in Caenorhabditis elegans and maternal control of germline development. Development. 1998;125:2469–2478. doi: 10.1242/dev.125.13.2469. [DOI] [PubMed] [Google Scholar]
- Leung B, Hermann GJ, Priess JR. Organogenesis of the Caenorhabditis elegans intestine. Dev Biol. 1999;216:114–134. doi: 10.1006/dbio.1999.9471. [DOI] [PubMed] [Google Scholar]
- Mango SE. The C. elegans pharynx: a model for organogenesis. WormBook. 2007:1–26. doi: 10.1895/wormbook.1.129.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Li G, Sarma K, Blais A, Zavadil J, Woodcock CL, Dynlacht BD, Reinberg D. Ezh1 and Ezh2 maintain repressive chromatin through different mechanisms. Mol Cell. 2008;32:503–518. doi: 10.1016/j.molcel.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGhee JD. The C. elegans intestine. WormBook. 2007:1–36. doi: 10.1895/wormbook.1.133.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meshorer E, Misteli T. Chromatin in pluripotent embryonic stem cells and differentiation. Nat Rev Mol Cell Biol. 2006 doi: 10.1038/nrm1938. [DOI] [PubMed] [Google Scholar]
- Meyer BJ. X-Chromosome dosage compensation. WormBook. 2005:1–14. doi: 10.1895/wormbook.1.8.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H. How is pluripotency determined and maintained? Development. 2007;134:635–646. doi: 10.1242/dev.02787. [DOI] [PubMed] [Google Scholar]
- Okkema PG, Krause M. Transcriptional regulation. WormBook. 2005:1–40. doi: 10.1895/wormbook.1.45.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini D, Bracken AP, Hansen JB, Capillo M, Helin K. The polycomb group protein Suz12 is required for embryonic stem cell differentiation. Mol Cell Biol. 2007;27:3769–3779. doi: 10.1128/MCB.01432-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patturajan M, Schulte RJ, Sefton BM, Berezney R, Vincent M, Bensaude O, Warren SL, Corden JL. Growth-related changes in phosphorylation of yeast RNA polymerase II. J Biol Chem. 1998;273:4689–4694. doi: 10.1074/jbc.273.8.4689. [DOI] [PubMed] [Google Scholar]
- Pickett CL, Breen KT, Ayer DE. A C. elegans Myc-like network cooperates with semaphorin and Wnt signaling pathways to control cell migration. Dev Biol. 2007;310:226–239. doi: 10.1016/j.ydbio.2007.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priess JR. Notch signaling in the C. elegans embryo. WormBook. 2005:1–16. doi: 10.1895/wormbook.1.4.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priess JR, Thomson JN. Cellular interactions in early C. elegans embryos. Cell . 1987;48:241–250. doi: 10.1016/0092-8674(87)90427-2. [DOI] [PubMed] [Google Scholar]
- Robertson SM, Shetty P, Lin R. Identification of lineage-specific zygotic transcripts in early Caenorhabditis elegans embryos. Dev Biol. 2004;276:493–507. doi: 10.1016/j.ydbio.2004.09.015. [DOI] [PubMed] [Google Scholar]
- Ross JM, Zarkower D. Polycomb group regulation of Hox gene expression in C. elegans. Dev Cell. 2003;4:891–901. doi: 10.1016/s1534-5807(03)00135-7. [DOI] [PubMed] [Google Scholar]
- Rossant J. Stem cells and early lineage development. Cell. 2008;132:527–531. doi: 10.1016/j.cell.2008.01.039. [DOI] [PubMed] [Google Scholar]
- Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell. 2007;128:735–745. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- Smith PA, Mango SE. Role of T-box gene tbx-2 for anterior foregut muscle development in C. elegans. Dev Biol. 2007;302:25–39. doi: 10.1016/j.ydbio.2006.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector DL. The dynamics of chromosome organization and gene regulation. Annu Rev Biochem. 2003;72:573–608. doi: 10.1146/annurev.biochem.72.121801.161724. [DOI] [PubMed] [Google Scholar]
- Strome S. Specification of the germ line. WormBook. 2005:1–10. doi: 10.1895/wormbook.1.9.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston JE, Schierenberg E, White JG, Thomson JN. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev Biol. 1983;100:64–119. doi: 10.1016/0012-1606(83)90201-4. [DOI] [PubMed] [Google Scholar]
- Tabara H, Motohashi T, Kohara Y. A multi-well version of in situ hybridization on whole mount embryos of Caenorhabditis elegans. Nucleic Acids Res. 1996;24:2119–2124. doi: 10.1093/nar/24.11.2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner BM. Open chromatin and hypertranscription in embryonic stem cells. Cell Stem Cell. 2008;2:408–410. doi: 10.1016/j.stem.2008.04.006. [DOI] [PubMed] [Google Scholar]
- Unhavaithaya Y, Shin TH, Miliaras N, Lee J, Oyama T, Mello CC. MEP-1 and a homolog of the NURD complex component Mi-2 act together to maintain germline-soma distinctions in C. elegans. Cell. 2002;111:991–1002. doi: 10.1016/s0092-8674(02)01202-3. [DOI] [PubMed] [Google Scholar]
- Wood WB. Evidence from reversal of handedness in C. elegans embryos for early cell interactions determining cell fates. Nature. 1991;349:536–538. doi: 10.1038/349536a0. [DOI] [PubMed] [Google Scholar]
- Zaratiegui M, Irvine DV, Martienssen RA. Noncoding RNAs and gene silencing. Cell. 2007;128:763–776. doi: 10.1016/j.cell.2007.02.016. [DOI] [PubMed] [Google Scholar]
- Zhang H, Azevedo RB, Lints R, Doyle C, Teng Y, Haber D, Emmons SW. Global regulation of Hox gene expression in C. elegans by a SAM domain protein. Dev Cell. 2003;4:903–915. doi: 10.1016/s1534-5807(03)00136-9. [DOI] [PubMed] [Google Scholar]
- Zhu J, Fukushige T, McGhee JD, Rothman JH. Reprogramming of early embryonic blastomeres into endodermal progenitors by a Caenorhabditis elegans GATA factor. Genes Dev. 1998;12:3809–3814. doi: 10.1101/gad.12.24.3809. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.