Abstract
The G65R mutation in the N-lobe of human transferrin was created to mimic a naturally occurring variant (G394R) found in the homologous C-lobe. Because Gly65 is hydrogen-bonded to the iron-binding ligand Asp63, it comprises part of the second shell hydrogen bond network surrounding the iron within the metal binding cleft of the protein. Substitution with an arginine residue at this position disrupts the network, resulting in much more facile removal of iron from the G65R mutant. As shown by UV-vis and EPR spectroscopy, and by kinetic assays measuring the release of iron, the G65R mutant can exist in three forms. Two of the forms (yellow and pink in color) are inter-convertible. The yellow form predominates in 1 M bicarbonate; the pink form is generated from the yellow form upon exchange into 1 M HEPES buffer, pH 7.4. The third form (also pink in color) is produced by the addition of Fe3+-(nitrilotriacetate)2 to apo-G65R. Hydrogen/deuterium exchange experiments are consistent with all forms of the G65R mutant assuming a more open conformation. Additionally, mass spectroscopic analysis reveals the presence of nitrilotriacetate in the third form. The inability to obtain crystals of the G65R mutant, led to development of a novel crystallization strategy in which the double mutation G65R/K206E stabilizes a single closed pink conformer and captures Arg65 in a single position. Collectively, these studies highlight the importance of the hydrogen bond network in the cleft, as well as the inherent flexibility of the N-lobe which although able to adapt to accommodate the large arginine substitution exists in multiple conformations.
The transferrins are a family of iron-binding and transport proteins that can be isolated from various biological fluids. The major functions of serum transferrin (TF)1 are to bind ferric iron in a stable complex (preventing its hydrolysis and maintaining its solubility in vivo) and to transport iron to cells by a mechanism involving receptor-mediated endocytosis (see reviews (1, 2)). TF is comprised of a single polypeptide chain containing homologous N- and C-lobes connected by a short bridging region. Each lobe contains an iron-binding site situated at the bottom of a deep cleft made up of two subdomains (the NI and NII subdomain in the N-lobe, and the CI and CII subdomain in the C-lobe) that provide four amino acid ligands that bind the iron (3–5). Coordination of the iron is completed by two oxygen atoms from a synergistic anion (carbonate) crucial to high affinity iron binding. Release of iron from the human TF N-lobe (hTF N-lobe) is accompanied by a large conformational change in which the cleft opens 63° by a rigid body rotation of the two subdomains around a hinge comprised of two anti-parallel β-strands at the bottom of the cleft (6).
The X-ray crystal structure of the isolated hTF N-lobe, produced by a recombinant approach, reveals that the iron-binding ligands consist of the side chains of Asp63, Tyr95, Tyr188 and His249 (3). In addition, there is a second shell comprised of residues that are hydrogen-bonded to the liganding residues (3, 6). Not surprisingly, mutation of any of the metal binding ligands drastically alters the spectroscopic, kinetic and iron-binding properties of the N-lobe (7–10). Mutation of the second-shell residues also has a considerable effect on the metal-binding properties of the N-lobe due to their proximity to the iron center and the disturbance of the hydrogen-bonding network within the cleft (11–15).
A naturally occurring variant was identified in a woman in England that affects the function of human TF (hTF) (16). In this variant, an arginine residue substituted for the second shell glycine residue at position 394 in the C-lobe results in weaker iron binding as indicated by electrophoresis on 6 M urea gels (17). It has been well established that either monoferric hTF binds to the specific TF receptor (TFR) with a lower affinity than diferric hTF (18, 19). Cell binding experiments with the G394R variant are consistent with the C-lobe of full-length hTF adopting an open (apo) conformation (20, 21). To mimic the G394R mutant in the hTF C-lobe, the equivalent conserved residue, Gly65, has been mutated to an arginine creating the G65R mutant in the isolated hTF N-lobe (22). This approach was taken because production of recombinant C-lobe is poor and placement of the mutation into either lobe of full length hTF complicates the interpretation of its effect on the structure and function.
The unusual behavior of this G65R N-lobe mutant (no C-lobe) with regard to its iron binding, thermodynamic stability and EPR spectra has been described previously (11, 22, 23); however the reasons for the abnormal metal-binding properties of the mutant were not completely understood. Therefore we have revisited this mutant using a combination of absorption and EPR spectroscopies, kinetics, and mass spectrometry. Additionally, an X-ray structure of the G65R/K206E mutant is provided. Since we were unable to crystallize the G65R mutant per se, a new strategy was developed in which the G65R mutation was placed in the background of a K206E N-lobe mutant. The substitution of lysine by glutamic acid results in an N-lobe which is locked in the closed conformation by a salt bridge between the glutamic acid in the NI subdomain and Lys296 in the NII subdomain (15). Ordinarily Lys206 and Lys296 comprise a pH sensitive trigger which is critical to the release of iron from the N-lobe in a physiologically relevant time frame. What makes this approach reasonable is that, to date, virtually all of the single point N-lobe mutations show a very localized effect, i.e. they produce no significant changes except at the site of the mutation (where a water molecule often takes the place of the missing side chain atoms) (24). We now provide a more comprehensive assessment of the effect of changing the glycine at position 65 to arginine on the metal binding and spectroscopic behavior of the protein and offer structural information to aid in understanding the unusual behavior of the G65R mutant and, by inference, of the naturally occurring G394R variant of hTF.
MATERIALS AND METHODS
Materials
All chemicals were of reagent grade. Stock solutions of TRIS, HEPES, MES, and other buffers were prepared by dissolving the anhydrous salts in Milli-Q (Millipore) purified water, and adjusting the pH to desired values with 1 N NaOH or HCl. A standard solution of Fe(II) (1,000 μg/ml) in 5% HNO3 was obtained from Johnson Matthey, Ward Hill, MA. Ethylenediamine tetraacetate (EDTA) was obtained from Mann Research Laboratories, Inc., nitrilotriacetate (NTA) from Sigma, DMEM-F12 medium from Invitrogen, and the spiral cartridge concentrator and Centricon 10 microconcentrators were from Amicon. The anion exchange resins Poros 50 HQ and Poros QE/M (quaternized polyethyleneimine) were from Applied Biosystems.
Mutagenesis, Production and Purification
The DNA manipulations used to generate the G65R and K206E mutants have been reported previously (15, 22). The double mutant was made by placing the G65R mutation into the plasmid containing the K206E mutant. Expression and purification of the hTF N-lobe and hTF N-lobe mutants was carried out as described (25). Briefly, the recombinant hTF N-lobe was isolated from the culture medium of adherent BHK cells attached to the surface of expanded surface roller bottles. Four to five batches of medium were pooled, concentrated, exchanged and applied to a Poros 50 HQ anion exchange column to remove phenol red and most of the albumin present in the medium. Following a single step elution with 160 mM TRIS buffer, pH 8.0, the samples were concentrated and purified to homogeneity by gel filtration chromatography on a Sephacryl S-200HR (26/60) column; polyacrylamide gel electrophoresis (10%) in the presence of SDS was used to verify purity.
Upon isolation from the S-200HR column (equilibrated and developed in 100 mM ammonium bicarbonate), the G65R mutant is yellow. To convert the yellow sample to one of the pink forms, it was placed into a YM-10 microconcentrator and exchanged (~2 mL, 4 times) into 1 M HEPES buffer, pH 7.4. To remove iron, samples were placed in YM-10 microconcentrators and treated in a step-wise manner with aliquots of ~2 mL of: “iron removal buffer” (0.5 M sodium acetate, pH 4.9, containing 1 mM NTA and 1 mM EDTA), followed by 100 mM KCl (once), 100 mM sodium perchlorate (once) and 100 mM KCl (4 times). Fe3+-loaded proteins were prepared by addition of a slight excess of Fe3+(NTA)2 (pH 3.5) to the apo-proteins in 50 mM HEPES, pH 7.4 buffer (or to samples which were not completely iron saturated as indicated by the ratio of A280 divided by the Amax ) (7). NTA is commonly used to keep the Fe3+ in solution. Apo and iron loaded samples were then exchanged into 50 mM HEPES, pH 7.4 using YM-10 microconcentrators.
Electronic Spectra
UV-vis spectra were recorded on a Cary 100 spectrophotometer at 25°C. The appropriate buffer (pH 7.4) served as the reference for full-range spectra from 650 to 300 nm. Difference spectra were generated by storing the spectrum of the apo-N-lobe as the baseline and subtracting it from the sample spectra.
EPR Spectroscopy
Frozen solution EPR spectra of iron loaded hTF N-lobe were obtained at 90 K using a flow of cold N2 gas and a laboratory constructed X-band (9.4 GHz) EPR spectrometer described elsewhere (8, 26). Typical spectrometer settings were microwave frequency = 9.386 GHz, microwave power = 20 mW, amplitude = 10.0 G and scan rate = 4 G/s and time constant = 1.0 s.
Kinetics of Iron Release
The kinetics of iron release from the N-lobe and mutants of the N-lobe were carried out at 25°C on an Applied Photophysics SX.18MV stopped-flow spectrofluorometer fitted with a 20 μL observation cell with a 2 mm light path and a dead time of ~1.1 ms for monitoring fast release (27). One syringe contained protein (375 nM in Milli-Q water) and the other syringe contained MES buffer (200 mM, pH 5.6) and EDTA (8 mM). The samples were excited at 280 nm (wavelength selection from a monochromator) and fluorescence emission was monitored using a high-pass 320 nm cut-on filter. For measuring the change in absorbance at 280 nm, we used the absorbance detector on the same stopped-flow instrument. For these experiments, one syringe contained protein (6.0 μM in Milli-Q water) and the other syringe contained MES buffer (200 mM, pH 5.6) and EDTA (8 mM). Rate constants were determined by fitting the change in absorbance or fluorescence intensity-versus-time using Origin software (version 7.5) with either a single-exponential (y = A1*exp(−x/t1) + y0), a double-exponential (y = A1*exp(−x/t1) + A2*exp(−x/t2) + y0) function or a triple-exponential function (y = A1*exp(−x/t1) + A2*exp(−x/t2) + A3*exp(−x/t3) + y0) as required for each mutant.
Direct ESI MS measurements
The composition of the protein-metal-synergistic anion complex was determined using direct mass measurements of protein ions produced by electrospray ionization mass spectrometry (ESI MS). The measurements were carried out with a hybrid quadrupole/time-of-flight mass spectrometer (QStar XL, Sciex/Applied Biosystems, Toronto, Canada) equipped with a Turbospray™ ESI source and collisional cooling of ions in the interface region. Mild ion desolvation conditions were achieved by maintaining a low declustering potential (<75 V), in comparison to enhanced desolvation conditions (165 V). Protein solutions were prepared for ESI MS analysis by diluting to a final concentration of 10 μM in 20 mM ammonium acetate.
Hydrogen/deuterium exchange
Hydrogen/deuterium exchange (HDX) measurements were carried out using a slightly modified procedure described in detail elsewhere (28). Briefly, HDX of intact proteins was initiated by diluting the protein stock solutions (125 μM) 20-fold with exchange buffer (25 mM potassium phosphate pH 7.4 in D2O) equilibrated at 37°C. After 15 minutes of exchange, the samples were placed in an ice bath for 30 seconds and then an equal volume of pre-chilled quench solution (4.0% formic acid) was added and rapidly mixed, yielding a pH of 2.5. The reaction was rapidly desalted using a protein micro trap cartridge (Michrom Bioresources, Inc.) on ice with a mobile phase of 5% acetonitrile, 0.1% formic acid driven by an HPLC (Agilent). The protein was eluted from the trap directly into the mass spectrometer using an isocratic flow of 70% acetonitrile, 0.1% formic acid at a flow rate of 150 μL/min. Fully exchanged samples (which served as the endpoint) were prepared by performing the HDX incubation in 2.0% formic acid (in lieu of buffer) for 6 hrs at 37 °C. The extent of deuterium incorporation for each protein following the exchange was determined as a protein mass change (measured with QStar XL, vide supra). Since these measurements were carried out under denaturing conditions, any observed mass change is ascribed to the increase of deuterium content, as opposed to the presence of ferric ion and/or synergistic anion.
Crystallization and cryoprotection
Crystals of the G65R/K206E N-lobe were grown using a vapor diffusion sitting drop method. The double mutant was crystallized at 20°C from a solution of 200 mM potassium acetate buffer (pH 7.7) containing 10 mM KCl and 18% polyethylene glycol 3350. The initial concentration of the mutant in the sitting drop was 17.5 mg/mL. Crystals appeared in about a week following streak seeding with wild-type N-lobe. Crystals were cryoprotected by a 1 hour soak in mother liquor containing 35% polyethylene glycol 3350 prior to flash freezing in liquid nitrogen.
Data collection, Structure determination, and Refinement
X-ray diffraction data on the frozen crystal was collected using a Mar345 image plate detector on a Rigaku RU-300 generator with a copper rotating anode. All data sets were indexed using DENZO (29) and scaled and merged using SCALEPACK (29) (Table 1). Phasing information was generated using molecular replacement, MOLREP (30), with the wild-type iron containing N-lobe (PDB 1A8E). The atoms representing the iron, carbonate and all water molecules were removed from the search model to eliminate bias. The model was refined using successive rounds of simulated annealing, occupancy, and B-factor refinement using CNS (31). Throughout the refinement, revision of the model based upon map interpretation with the graphics program O was performed (32, 33). The final model includes residues 3–331 of the polypeptide chain, iron, carbonate and 269 water molecules. Coordinates have been deposited in the Protein Data Bank (PDB code 3FGS).
Table 1.
Data Collection and Model Refinement Statistics
| K206E/G65R | |
|---|---|
| space group | P212121 |
| unit cell (Å) | a = 43.35 |
| b = 57.06 | |
| c = 133.74 | |
| resolution limits (Å) | 30 – 1.80 |
| Rsym (%)a | 4.9 (11.3) |
| I/σa | 22.25 (11.51) |
| completeness (%)a | 93.3 (87.2) |
| molecules in asymmetric unit | 1 |
| no. of unique reflectionsa | 29637 (2701) |
| R-factor (Rfree) (%) | 21.6 (24.6) |
| model details | |
| no. of protein atoms | 2557 |
| no. of solvent atoms | 269 |
| Ions | Fe3+, CO32− |
| average B-factor (Å2) | |
| protein main chain atoms | 13.76 |
| protein side chain atoms | 15.59 |
| solvent molecules | 18.32 |
| rms deviation from ideal geometry | |
| bond distances (Å) | 0.006 |
| bond angles (deg) | 1.3 |
| Ramachandran plot statistics % (Gly and Pro excluded) | |
| most favored regions | 86.4 (248 residues) |
| allowed regions | 12.5 (36 residues) |
| generously allowed regions | 0.7 (2 residues) |
| disallowed regions | 0.3 (1 residue) |
Data in parentheses are for the outermost shell (1.86 – 1.80Å)
RESULTS
Absorption Spectra
In the presence of excess bicarbonate, pH ~8.1, the iron-containing G65R N-lobe mutant is yellow. Multiple exchanges of the yellow form into1 M HEPES buffer, pH 7.4 on a microconcentrator produces a pink form. The spectral parameters for the G65R mutant in bicarbonate and in HEPES buffer are compared to those for the wild-type N-lobe (Table 2). Additionally, the parameters for the D63A and H249A mutants which each lack a ligand are included in Table 2. We note that the spectral characteristics of the yellow form of the G65R mutant are similar to both the D63A and the H249A N-lobe mutants; all are significantly blue-shifted and yellow in color. In contrast the pink form in HEPES is more similar to the wild-type N-lobe spectrum but still has a slight red shift. When the G65R mutant from which iron has been removed is loaded with Fe3+(NTA)2 (in the presence of ~10 mM bicarbonate) a pink product with spectral characteristics close to those of the wild-type N-lobe is observed (Table 2). The spectra of the two pink forms, while having similar band maxima of 474 and 469 nm, differ somewhat in their A280/Amax with ratios of 28.2 and 22.0, respectively, (with Amax defined as the absorbance maximum in the visible range). The spectral parameters of the G65R/K206E double mutant and those of the single K206E mutant are included in Table 2; both show slight blue shifts and a somewhat higher ratio of A280/Amax ratios relative to the wild-type N-lobe. This ratio is commonly measured as an indicator of the degree of iron saturation/coordination.
Table 2.
Summary of the spectral characteristics for recombinant wild-type and various mutants of the hTF N-lobe.
EPR Spectra
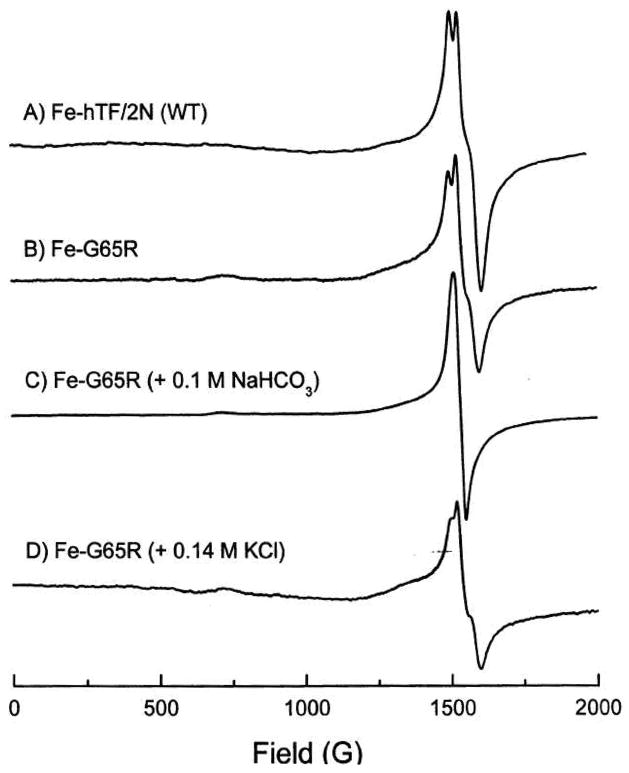
The Fe3+ EPR spectrum of the G65R mutant demonstrates the existence of at least two forms (Figure 1). In 100 mM HEPES buffer, pH 7.4, the protein (pink) displayed a near rhombic EPR spectrum similar to that of wild-type N-lobe with the three component signal near g′ = 4.3 (compare spectra A and B) (12). Addition of 100 mM NaHCO3 to this sample results in a color change from pink to yellow and an EPR signal that is purely rhombic (spectrum C), similar to spectra observed for all other mutants containing only three amino acid ligands (including D63A, Y95F and H249A) (8, 9, 11). Addition of 140 mM KCl to the pink G65R sample of spectrum B causes spectral changes (spectrum D) similar to the mixture of the yellow and pink forms previously reported (11). Additionally, a shoulder at ~1360 G is observed and attributed to the well known salt effect on the EPR spectra of transferrins (reviewed in (11)). The shoulder is thought to arise from conformational change(s) in the protein induced by a combination of Cl− binding and ionic strength (11), but strain in the zero field parameters D and E of the Fe3+ center may also contribute to this feature (34).
Figure 1.
X-band (9.38 GHz) EPR spectra of frozen samples of iron saturated wild-type N-lobe and the G65R mutant. Protein samples (~ 0.3 mM), HEPES (~ 100 mM), pH 7.5, temperature 90 K.
The EPR spectrum for the G65R/K206E mutant is presented in Figure 2 (spectrum A). In the presence of 100 mM HEPES the spectrum of the double mutant is very similar to that of the wild-type N-lobe with little change induced by addition of bicarbonate (Figure 2B); i.e., the K206E mutation abrogates the destabilizing effect of bicarbonate on the spectrum. The presence of 140 mM KCl leads to a spectrum (Figure 2C) which again resembles the wild-type N-lobe with the appearance of a shoulder at ~ 1325 G to the low field side of the g′ = 4.3 signal, due to the salt effect (11). The appearance of the shoulder indicates that some “axial character” has been introduced in the ligand field of the high-spin Fe3+center (11).
Figure 2.
X-band (9.38 GHz) EPR spectra of frozen samples of the iron saturated G65R/K206E mutant. Protein samples (~ 0.3 mM), HEPES (~ 100 mM), pH 7.5, temperature 90 K.
Kinetics of Iron Release
The apparent first order rate constants for iron-release from wild-type N-lobe and the various forms of the G65R mutant at pH 5.6 (the putative pH of the endosome) monitored by the increase in the fluorescent signal are given in Table 3, and the kinetic traces are provided in Figure 3A. The two observed rate constants for the wild-type N-lobe have been definitively attributed to the release of iron (the faster rate constant) and a conformational change in two of the three Trp residues in the N-lobe associated with cleft opening (27). These assignments are based on the ability to monitor iron release directly by the decrease in absorbance at 470 nm. In the current study, the decrease in absorbance at 280 nm for the wild-type N-lobe yields a very similar rate constant, 8.0 ± 0.9 min−1 versus 9.1 ± 0.7 min−1 measured previously by absorbance at 470 nm (26). Since the change in absorbance at 280 nm provides a signal that is considerably stronger than that at 470 nm (data not shown), it is preferable to monitor the kinetics at the shorter wavelength. As noted previously (26), these values compare very well to kobsN1 of 8.9 ± 0.8 min−1 determined by fluorescence (Table 3).
Table 3.
Rate constants kobs (min−1) of iron release for iron-containing N-lobe and N-lobe mutants monitored by the increase in fluorescence (Figure 3) or by the decrease in absorbance at 280 nm (kinetic traces not shown). Measurements were made in 100 mM MES, pH 5.6 and 4 mM EDTA,
| Protein | Fluorescence kobsN (min−1) | I1/I2/I3 (%)b | Absorbance kobsN (min−1) | I1/I2 (%)c |
|---|---|---|---|---|
| N-lobe (wild-type)a | 8.9 ± 0.8 1.3 ± 0.3 |
64 36 |
8.0 ± 0.9 | 100 |
| G65R (HCO3−)a | 322.7 ± 23 13.6 ± 3 |
95 5 |
411.6 ± 30 | 100 |
| G65R (HEPES)a | 370.4 ± 10 58.9 ± 3 3.7 ± 0.1 |
83 9 8 |
427.2 ± 26 36.7 ± 3 |
83 17 |
| G65R (Fe3+(NTA)2)a | 318.0 ± 2 13.7 ± 0.6 |
92 8 |
305.4 ± 31 | 100 |
| G65R/K206E | 1.0 ± 0.01 | 100 | ||
| K206E | Too slow to measure accurately |
Kinetic data was fit to a sum of exponentials (see Material and Methods).
I1 I2 and I3 refer to the % change in fluorescent intensity corresponding to kobsN1 kobsN2, and kobsN3, respectively.
I1 and I2 refer to the % change in absorbance intensity corresponding to kobsN1 and kobsN2, respectively.
Figure 3.
Figure 3A. Kinetic traces monitoring the time-based change in fluorescence of each form of the G65R mutant as indicated. Inset shows the curves for the first 2 sec to highlight the difference in the kinetic curves for the three samples. Figure 3B. Kinetic traces of the time-based change in fluorescence when iron is removed from the wild-type N-lobe and the G65R/K206E mutant. All samples are in 100 mM MES, pH 5.6, containing 4 mM EDTA. Excitation was at 280 nm, and emission was recorded with a 320 nm cut-on filter.
It is clear that all of the forms of the G65R mutant release iron significantly faster (a 35–50 fold increase) compared to the fastest rate constant for the wild-type N-lobe. As indicated by the rate constants from the absorbance data, the three forms all feature a fast event which correlates with actual iron removal (compare Table 3). Interestingly, the carbonate complex and the NTA complex show similar kinetics with a fast event accounting for most of the change in the intensity of the signal (95 and 92% of the intensity) and a slower event accounting for the remaining change. When the change in absorbance is monitored, a single rate constant due to iron release is found for the wild-type N-lobe and two of the mutants, indicating that iron has been completely removed (Table 3). When monitored by fluorescence, the HEPES form of G65R has a more complex kinetic profile with three rate constants. Additionally, this form yields two rate constants from absorbance measurement (see Discussion). It is significant that for the G65R/K206E mutant we observe iron release that is considerably slower than both the wild-type N-lobe and the G65R samples (Table 3). Since no iron release is found for the K206E mutant alone, the result indicates that the G65R mutation counterbalances the K206E mutation, i.e. the arginine at position 65 is more destabilizing than the salt bridge between Glu206 and Lys296 is stabilizing. This statement is supported by analysis on 6 M urea gels; all of the iron is removed from the G65R/K206E mutant following a 15 minute incubation in 100 mM MES buffer, pH 5.6 containing 4 mM EDTA, while none is removed from the K206E mutant alone (data not shown). Of interest is that the change in fluorescence intensity observed for the K206E/G65R double mutant is considerably less than that found for the wild-type N-lobe (Figure 3B).
ESI MS measurements
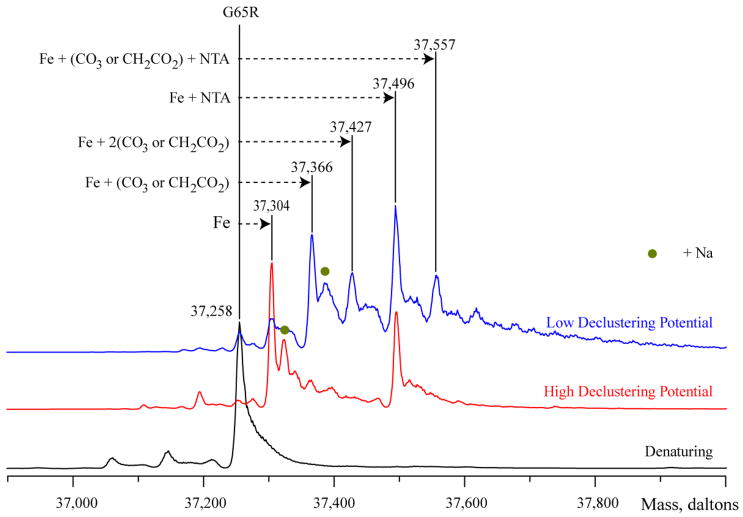
Direct ESI MS measurements of the Fe3+(NTA)2 form of the G65R mutant were carried out to establish the composition and stoichiometry of the protein-metal complex. Previously we demonstrated that it is possible to preserve the protein-metal-synergistic anion complex in the gas phase by selecting mild ion desolvation conditions in the ESI MS interface, while enhanced desolvation of ions in the ESI source results in the loss of the synergistic anion (35–37). The major ionic species observed in the mass spectrum of the Fe3+(NTA)2 form of the G65R mutant acquired under mild (near-native) conditions is a protein-metal complex (Figure 4). Another prominent peak in this mass spectrum corresponds to a ternary protein/metal/NTA complex (1:1:1 stoichiometry). This species becomes predominant in the mass spectrum acquired under mild desolvation conditions in the ESI MS interface, with the second most abundant species corresponding to a ternary complex where NTA is replaced with either carbonate or acetate (a distinction between these two anions could only be made if the accuracy of mass measurements exceeds 0.005%).
Figure 4.
ESI mass spectra of iron, synergistic anion, and NTA binding to the G65R mutant. Spectra recorded for the G65R-Fe-NTA sample illustrate the dependence of the observed ternary complex on the source conditions. Binding of NTA and Fe is observed with a high declustering potential (red trace), whereas more mild conditions (blue trace) were required to observe anion binding. The spectrum collected for denatured protein serves as a ligand-free control (black trace). The presence of a Na+ ion is indicated by a green dot.
Hydrogen deuterium exchange
HDX MS of the apo- and iron bound-forms of the hTF N-lobe reveals a noticeably higher degree of amide protection in the presence of iron (Table 4), consistent with our earlier results (38). The higher extent of deuterium incorporation in the apo-protein is indicative of the lower protection from solvent due to the open conformation of the hTF N-lobe. Though this difference in protection between apo and holo hTF remains evident at longer time points, a single 15 minute exchange time was selected to evaluate the G65R mutants. The extent of exchange observed for all forms of the G65R mutant is indistinguishable from that of the apo-wild-type N-lobe, clearly suggesting that the mutant remains in the open conformation regardless of the presence (or absence) of iron. Interestingly, the extent of exchange displayed by the double mutant G65R/K206E lies in the middle of the range defined by the apo- and holo-forms of the wild-type N-lobe (Table 4).
Table 4.
Hydrogen-deuterium exchange reactions after 15 min at 37°C
| Protein | Increase in Mavga | % of Exchangea |
|---|---|---|
| Fe N-lobe (wild-type) | 112 ± 2 | 52 ± 1 |
| Apo N-lobe (wild-type) | 124 ± 2 | 59 ± 1 |
| G65R (HCO3−) | 129 ± 2 | 61 ± 1 |
| G65R (HEPES) | 126 ± 2 | 58 ± 1 |
| G65R (Fe3+(NTA)2) | 127 ± 2 | 58 ± 1 |
| G65R/K206E | 119 ± 2 | 55 ± 1 |
Measured value ± standard deviation
Crystal structure of the G65R/K206E mutant
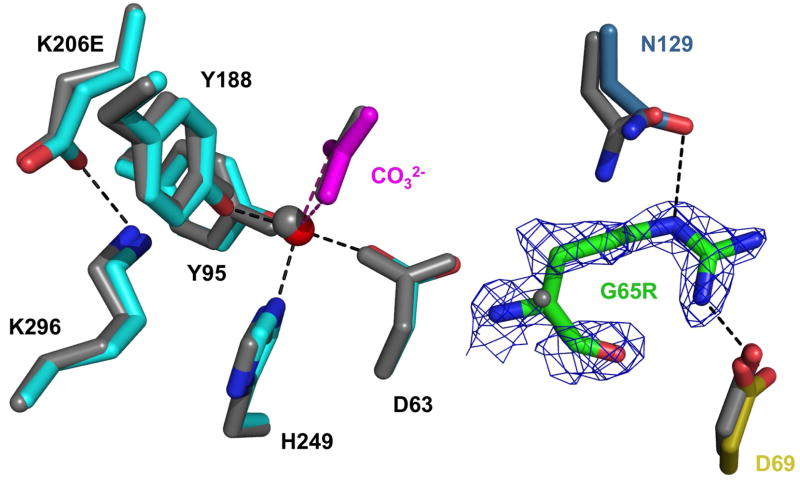
We were unable to obtain crystals of the G65R mutant, most likely because of both the flexibility of the arginine and the instability and heterogeneity of the metal complex. By placing the G65R mutation into a stable environment (the K206E N-lobe mutant) we were able to crystallize the double mutant and to identify at least one conformation available to Arg65. The iron coordination in the wild-type N-lobe, the K206E mutant, and the G65R/K206E double mutant is shown in Table 5. In comparison to the parent K206E (RMSD for 1312 atoms is 0.48 Å), the major difference in the structure of the G65R/K206E mutant is that the Arg65 is within hydrogen bonding distance of both Asp69 and Asn129 (Figure 5). In both the wild-type N-lobe and the K206E mutant, Asp69 contacts Arg327; in the G65R/K206E double mutant structure, Arg327 resides in a water hydrogen bonding network and no longer contacts Asp69. In the wild-type N-lobe and the K206E mutant, Asn129 contacts Tyr319 which is in a hydrogen network comprised of Ser125 and the ligand Asp63. In the double mutant, Asn129 only makes contact with the substituted Arg65. The substituted glutamic acid forms a salt bond with Lys296 accounting for inability to remove the iron from the K206E mutant and the decreased rate constant for iron release from the double mutant.
Table 5.
Bond lengths (Å) for the iron coordination in wild-type N-lobe and the two mutants, K206E and G65R/K206E
| Residue | N-lobe (wild-type) (1A8E) | K206E (2O84) | G65R/K206E (3FGS) |
|---|---|---|---|
| Asp63 OD1 | 2.03 | 2.20 | 2.04 |
| Tyr95 OH | 1.97 | 2.03 | 2.04 |
| Tyr188 OH | 1.80 | 2.31 | 2.20 |
| His249 NE2 | 2.04 | 2.58 | 2.23 |
| Arg124 carbonate | 2 conformations | 1 conformation | 1 conformation |
| Carbonate ligands NE NH2 |
2.81 2.57 |
2.93 2.90 |
2.67 2.64 |
Figure 5.
Overlay of the G65R/K206E (multicolor) structure on the K206E structure (gray). Iron (red) with its amino acid ligands (cyan) and synergistic anion (carbonate, magenta) are shown for orientation. A simulated annealing omit map (1.5s) is drawn for Arg65. Note that Arg65 is within hydrogen bonding distance of both Asp69 and Asn129 (•••••).
DISCUSSION
The current work clearly shows that the G65R mutant is extremely unstable kinetically and exists in multiple forms in a buffer induced manner. Previous EPR studies of the mutant showed that the spectrum could be simulated by summing the spectra of the fully open and fully closed conformations (11). The present work demonstrates that the situation is more complex. Although the absorbance maxima and ratios of both of the pink forms (HEPES and Fe3+(NTA)2) are similar to each other and to the iron bound wild-type N-lobe (Table 2), neither of these samples is in the ‘closed’ conformation as indicated by the kinetic data and the HDX experiments. Based on the blue shifted absorbance spectrum (Table 2) and the EPR spectrum (Figure 2, spectrum C), the yellow carbonate form of the G65R mutant is clearly missing a ligand. Surprisingly, this does not seem to make it any more labile than the other two forms of the mutant as indicated by the kinetic rate constants.
In the complex with NTA, the possibility that the chelator might remain bound to the iron and stabilize the metal-binding site must be considered. Precedence for this idea comes from crystallographic studies of ovotransferrin (oTF) (39) in which colorless apo-oTF N-lobe crystals were soaked in a solution of 3 mM Fe3+(NTA)2 for 4 hours, yielding red crystals. The x-ray structure revealed that the binding cleft remained open, with the NTA providing four ligands to the iron with the two tyrosines completing the six coordinate complex and giving rise to the color. Reminiscent of the G65R mutant, this oTF ternary protein-iron-NTA complex could only be displaced by a high concentration of carbonate. Nevertheless, the experimental conditions in the oTF work would preclude the dynamic conformational change(s) which ordinarily results in elimination of the NTA. In solutions of wild-type N-lobe of either oTF or hTF the amino acid ligands from the protein easily displace the NTA from the iron due to the higher affinity with which they bind. In the specific case of the G65R mutant iron loaded with Fe3+(NTA)2, the weak binding of ferric iron (as inferred from the kinetic data in Table 3) means that it might exist as a complex with NTA. Indeed, the mass spectrometry analysis reveals the presence of NTA in the complex (Figure 4). The presence of the smaller synergistic anions (carbonate and bicarbonate) in some portion of the sample further indicates that there is a mixture of forms. We note that mass spectrometry is a powerful (and relatively straightforward) method to access the precise nature of the ternary complex.
Unfortunately, the sample in nonvolatile HEPES buffer is inaccessible to direct ESI MS analysis so its precise makeup could not be determined by this method. However, as described previously (40), the negatively charged ethanesulfonic acid group of HEPES can act as an anion although the binding is very weak, consistent with the fact that a concentration of 1 M HEPES is required to “convert” the sample to this form. The complexity of the sample is underscored by the fact that one additional rate constant is required to fit the kinetic data from both the fluorescence and absorbance experiments compared to the wild-type N-lobe (Table 3). The “weakness” introduced by the presence of the large arginine residue in place of the glycine clearly allows access to several conformations with unique kinetic properties. The observation of two rate constants in the absorbance data (Table 3) indicates that the iron is released from two distinct forms with differing iron binding affinities.
X-ray crystal structure
The X-ray structure of the G65R mutant in the K206E background reveals the precise structural changes that can occur. Since the structure obtained is expected to represent a single low energy conformation, it is essential to look at the changes due directly to the K206E mutation compared to those that can be attributed to the G65R mutation. Contributing to the stability of both the K206E mutant and the G65R/K206E double mutant is the finding that Arg124 and the carbonate occupy only a single position in both the K206E and the G65R/K206E mutants (Table 5). A unique feature of the wild-type N-lobe of hTF is the observation from the original crystal structure that Arg124, which stabilizes the synergistic carbonate anion, exists in two different conformations (termed “near” and “far”) leading to the suggestion that protonation of the synergistic carbonate anion is the first step of iron release. This hypothesis has been validated by studies in which the position of Arg124 was forced into the near or the far conformation by strategic mutagenesis (41). This work showed that the position of Arg124 correlates perfectly with the rate of iron release. Further support for the role of protonation of the synergistic carbonate anion comes from substitution of carbonate by the larger anion oxalate which results in extremely sluggish iron release (42).
Interestingly, the double mutant G65R/K206E is considerably less stable than the K206E mutant as indicated by the fact that it releases iron at pH 5.6 in the presence of EDTA, whereas the K206E mutant does not (Table 3). Of interest, the HDX experiment with G65R/K206E shows a level of protection which is intermediate between fully open and fully closed conformations (Table 4). Thus, as noted earlier, the destabilization due to the Arg substitution overcomes the stabilization provided by the K206E mutation. The significant reduction in the fluorescence intensity when iron is released from the double mutant relative to the wild-type N-lobe (Figure 3B) is very curious. Combined with the HDX data and the slow rate constant for iron release (Table 3), we suggest that the G65R mutation loosens the cleft sufficiently for iron to be captured by the chelator at pH 5.6, but that the cleft without iron remains more closed than open due to the presence of the K206E mutation. As recently reported (26), only two of the three Trp residues (Trp128 and Trp264) make a substantive contribution to the change in the fluorescent signal. Iron removal accounts for ~60% of the increase in the signal and conformational changes in the local environment of Trp128 and Trp264 account for the remaining ~40%. In the G65R/K206E mutant the local environment of these two Trp residues undoubtedly differs from that found in the fully apo conformation and apparently mute the fluorescent signal. Steady state fluorescence data on the iron containing and apo conformations of the G65R/K206E mutant indicates that the net change due to iron removal is ~ 30% less than the change observed for the wild-type N-lobe (data not shown). Taken together, these differences provide an explanation for the significant reduction in the fluorescence intensity when iron is released from the double mutant relative to the wild-type N-lobe.
In summary, the G65R mutant exists in at least three conformations, none of which is very stable. The yellow form exists only in the presence of excess bicarbonate and is largely open. A second form (pink in color) is the complex formed with NTA, which remains bound to the metal. In this form the iron is undoubtedly also bound to the two tyrosine ligands and possibly is stabilized by interaction with other amino acids in the cleft (as observed for oTF). Again the cleft is more open than closed. The third form induced by HEPES buffer is the most labile and complex, as indicated by the kinetic analysis. None of the forms is very stable and all are probably sampling other conformations. Obviously the least labile conformation has been captured in the crystal structure of the double mutant. This conformation is closed and is stabilized by the formation of a hydrogen bond between Arg65 and Asn129 residing on opposite sides of the binding cleft. Due to the dynamic nature of the N-lobe it seems likely that all of the conformations sample this form occasionally affording them some stability.
Acknowledgments
This work was supported by USPHS grant R01-DK-21739 from the National Institute of Diabetes, and Digestive and Kidney Diseases (ABM), grant R37-GM-20194 from the National Institute of General Medical Sciences (to NDC) and grant R01 GM061666 (to IAK). Support for NGJ and SLB came from Hemostasis and Thrombosis Training Grant (5T32HL007594) issued to Dr. KG Mann at The University of Vermont by the National Heart, Lung and Blood Institute.
We wish to thank Ted and Heather Baker for providing us with the coordinates for the hTF N-lobe K206E structure in advance of its deposition to the PDB.
Footnotes
Abbreviations: TF, serum transferrin; hTF, human serum transferrin; hTF N-lobe, recombinant N-lobe of human transferrin comprising residues 1–337, referred to as wild-type N-lobe; the G65R designation indicates that the glycine at position 65 has been mutated to arginine; oTF, ovotransferrin from chicken egg white; TFR, the specific transferrin receptor; BHK, baby hamster kidney cells; EPR, electron paramagnetic resonance; NTA, nitrilotriacetate; EDTA, ethylenediamine tetraacetate; DMEM-F12, Dulbecco’s Modified Eagle Medium supplemented with Ham’s F12 nutrient mixture; HDX, hydrogen/deuterium exchange; ESI MS, electrospray ionization mass spectrometry.
References
- 1.Wessling-Resnick M. Biochemistry of iron uptake. Crit Rev Biochem Mol Biol. 1999;34:285–314. doi: 10.1080/10409239991209318. [DOI] [PubMed] [Google Scholar]
- 2.Drakesmith H, Prentice A. Viral infection and iron metabolism. Nature Reviews. 2008;6:541–552. doi: 10.1038/nrmicro1930. [DOI] [PubMed] [Google Scholar]
- 3.MacGillivray RTA, Moore SA, Chen J, Anderson BF, Baker H, Luo YG, Bewley M, Smith CA, Murphy ME, Wang Y, Mason AB, Woodworth RC, Brayer GD, Baker EN. Two high-resolution crystal structures of the recombinant N-lobe of human transferrin reveal a structural change implicated in iron release. Biochemistry. 1998;37:7919–7928. doi: 10.1021/bi980355j. [DOI] [PubMed] [Google Scholar]
- 4.Hall DR, Hadden JM, Leonard GA, Bailey S, Neu M, Winn M, Lindley PF. The crystal and molecular structures of diferric porcine and rabbit serum transferrins at resolutions of 2.15 and 2.60 Å, respectively. Acta Crystallogr D Biol Crystallogr. 2002;58:70–80. doi: 10.1107/s0907444901017309. [DOI] [PubMed] [Google Scholar]
- 5.Wally J, Halbrooks PJ, Vonrhein C, Rould MA, Everse SJ, Mason AB, Buchanan SK. The crystal structure of iron-free human serum transferrin provides insight into inter-lobe communication and receptor binding. J Biol Chem. 2006;281:24934–24944. doi: 10.1074/jbc.M604592200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jeffrey PD, Bewley MC, MacGillivray RTA, Mason AB, Woodworth RC, Baker EN. Ligand-induced conformational change in transferrins: crystal structure of the open form of the N-terminal half-molecule of human transferrin. Biochemistry. 1998;37:13978–13986. doi: 10.1021/bi9812064. [DOI] [PubMed] [Google Scholar]
- 7.He QY, Mason AB, Woodworth RC, Tam BM, Wadsworth T, MacGillivray RTA. Effects of mutations of aspartic acid 63 on the metal-binding properties of the recombinant N-lobe of human serum transferrin. Biochemistry. 1997;36:5522–5528. doi: 10.1021/bi963028p. [DOI] [PubMed] [Google Scholar]
- 8.He QY, Mason AB, Woodworth RC, Tam BM, MacGillivray RTA, Grady JK, Chasteen ND. Inequivalence of the two tyrosine ligands in the N-lobe of human serum transferrin. Biochemistry. 1997;36:14853–14860. doi: 10.1021/bi9719556. [DOI] [PubMed] [Google Scholar]
- 9.He QY, Mason AB, Pakdaman R, Chasteen ND, Dixon BL, Tam BM, Nguyen V, MacGillivray RTA, Woodworth RC. Mutations at the histidine 249 ligand profoundly alter the spectral and iron-binding properties of human serum transferrin N-lobe. Biochemistry. 2000;39:1205–1210. doi: 10.1021/bi9915216. [DOI] [PubMed] [Google Scholar]
- 10.MacGillivray RTA, Bewley MC, Smith CA, He QY, Mason AB, Woodworth RC, Baker EN. Mutation of the iron ligand his 249 to Glu in the N-lobe of human transferrin abolishes the dilysine "trigger" but does not significantly affect iron release. Biochemistry. 2000;39:1211–1216. doi: 10.1021/bi991522y. [DOI] [PubMed] [Google Scholar]
- 11.Grady JK, Mason AB, Woodworth RC, Chasteen ND. The effect of salt and site-directed mutations on the iron(III)-binding site of human serum transferrin as probed by EPR spectroscopy. Biochem J. 1995;309:403–410. doi: 10.1042/bj3090403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He QY, Mason AB, Woodworth RC, Tam BM, MacGillivray RTA, Grady JK, Chasteen ND. Mutations at nonliganding residues Tyr-85 and Glu-83 in the N-lobe of human serum transferrin - Functional second shell effects. J Biol Chem. 1998;273:17018–17024. doi: 10.1074/jbc.273.27.17018. [DOI] [PubMed] [Google Scholar]
- 13.Li YJ, Harris WR, Maxwell A, MacGillivray RTA, Brown T. Kinetic studies on the removal of iron and aluminum from recombinant and site-directed mutant N-lobe half transferrins. Biochemistry. 1998;37:14157–14166. doi: 10.1021/bi9810454. [DOI] [PubMed] [Google Scholar]
- 14.Steinlein LM, Ligman CM, Kessler S, Ikeda RA. Iron release is reduced by mutations of lysine 206 and 296 in recombinant N-terminal half-transferrin. Biochemistry. 1998;37:13696–13703. doi: 10.1021/bi980318s. [DOI] [PubMed] [Google Scholar]
- 15.He QY, Mason AB, Tam BM, MacGillivray RTA, Woodworth RC. Dual role of Lys206-Lys296 interaction in human transferrin N-lobe: Iron-release trigger and anion-binding site. Biochemistry. 1999;38:9704–9711. doi: 10.1021/bi990134t. [DOI] [PubMed] [Google Scholar]
- 16.Evans RW, Williams J, Moreton K. A variant of human transferrin with abnormal properties. Biochem J. 1982;201:19–26. doi: 10.1042/bj2010019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Evans RW, Meilak A, Aitken A, Patel KJ, Wong C, Garratt RC, Chitnavis B. Characterization of the amino acid change in a transferrin variant. Biochem Soc Trans. 1988;16:834–835. [Google Scholar]
- 18.Young SP, Bomford A, Williams R. The effect of the iron saturation of transferrin on its binding and uptake by rabbit reticulocytes. Biochem J. 1984;219:505–510. doi: 10.1042/bj2190505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mason AB, Halbrooks PJ, James NG, Connolly SA, Larouche JR, Smith VC, MacGillivray RTA, Chasteen ND. Mutational analysis of C-lobe ligands of human serum transferrin: insights into the mechanism of iron release. Biochemistry. 2005;44:8013–8021. doi: 10.1021/bi050015f. [DOI] [PubMed] [Google Scholar]
- 20.Young SP, Bomford A, Madden AD, Garratt RC, Williams R, Evans RW. Abnormal in vitro function of a variant human transferrin. Br J Haematol. 1991;56:581–587. doi: 10.1111/j.1365-2141.1984.tb02183.x. [DOI] [PubMed] [Google Scholar]
- 21.Evans RW, Crawley JB, Garratt RC, Grossmann JG, Neu M, Aitken A, Patel KJ, Meilak A, Wong C, Singh J, Bomford A, Hasnain SS. Characterization and structural analysis of a functional human serum transferrin variant and implications for receptor recognition. Biochemistry. 1994;33:12512–12520. doi: 10.1021/bi00207a019. [DOI] [PubMed] [Google Scholar]
- 22.Woodworth RC, Mason AB, Funk WD, MacGillivray RTA. Expression and initial characterization of five site-directed mutants of the N-terminal half-molecule of human transferrin. Biochemistry. 1991;30:10824–10829. doi: 10.1021/bi00109a002. [DOI] [PubMed] [Google Scholar]
- 23.Lin LN, Mason AB, Woodworth RC, Brandts JF. Calorimetric studies of the N-terminal half-molecule of transferrin and mutant forms modified near the Fe3+-binding site. Biochem J. 1993;293:517–522. doi: 10.1042/bj2930517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nurizzo D, Baker HM, He QY, MacGillivray RTA, Mason AB, Woodworth RC, Baker EN. Crystal structures and iron release properties of mutants (K206A and K296A) that abolish the dilysine interaction in the N-lobe of human transferrin. Biochemistry. 2001;40:1616–1623. doi: 10.1021/bi002050m. [DOI] [PubMed] [Google Scholar]
- 25.Mason AB, Funk WD, MacGillivray RTA, Woodworth RC. Efficient production and isolation of recombinant amino-terminal half-molecule of human serum transferrin from baby hamster kidney cells. Protein Expr Purif. 1991;2:214–220. doi: 10.1016/1046-5928(91)90074-s. [DOI] [PubMed] [Google Scholar]
- 26.Yang X, Chasteen ND. Molecular diffusion into horse spleen ferritin: a nitroxide radical spin probe study. Biophys J. 1996;71:1587–1595. doi: 10.1016/S0006-3495(96)79361-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.James NG, Berger CL, Byrne SL, Smith VC, MacGillivray RTA, Mason AB. Intrinsic fluorescence reports a global conformational change in the N-lobe of human serum transferrin following iron release. Biochemistry. 2007;46:10603–10611. doi: 10.1021/bi602425c. [DOI] [PubMed] [Google Scholar]
- 28.Bobst CE, Abzalimov RR, Houde D, Kloczewiak M, Mhatre R, Berkowitz SA, Kaltashov IA. Detection and characterization of altered conformations of protein pharmaceuticals using complementary mass spectrometry-based approaches. Anal Chem. 2008;80:7473–7481. doi: 10.1021/ac801214x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Otwinowski Z, Minor W, Carter CW, Sweet RM. Methods in Enzymology, Part A. Academic Press; San Diego: 1997. Processing of x-ray diffraction data collected in oscillation mode; pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 30.Vagin A, Teplyakov A. MOLREP: an Automated Program for Molecular Replacement. J Appl Crystallogr. 1997;30:1022–1025. [Google Scholar]
- 31.Brunger AT. Slow-cooling protocols for crystallographic refinement by simulated annealing. Acta Crystallogr. 1990;A46:585–593. doi: 10.1107/s0108767390002355. [DOI] [PubMed] [Google Scholar]
- 32.Jones TA. A graphics model building and refinement system for macromolecules. J Appl Crystallogr. 1978;11:268–272. [Google Scholar]
- 33.Kleywegt GJ, Jones TA, Carter CW, Sweet RM. Methods in Enzymology, Part B. Academic Press; San Diego: 1997. Model building and refinement practice; pp. 208–230. [DOI] [PubMed] [Google Scholar]
- 34.Weisser JT, Nilges MJ, Sever MJ, Wilker JJ. EPR investigation and spectral simulations of iron-catecholate complexes and iron-peptide models of marine adhesive cross-links. Inorg Chem. 2006;45:7736–7747. doi: 10.1021/ic060685p. [DOI] [PubMed] [Google Scholar]
- 35.Gumerov DR, Kaltashov IA. Dynamics of iron release from transferrin N-lobe studied by electrospray ionization mass spectrometry. Anal Chem. 2001;73:2565–2570. doi: 10.1021/ac0015164. [DOI] [PubMed] [Google Scholar]
- 36.Gumerov DR, Mason AB, Kaltashov IA. Interlobe communication in human serum transferrin: Metal binding and conformational dynamics investigated by electrospray ionization mass spectrometry. Biochemistry. 2003;42:5421–5428. doi: 10.1021/bi020660b. [DOI] [PubMed] [Google Scholar]
- 37.Zhang M, Gumerov DR, Mason AB, Kaltashov IA. Indirect detection of protein-metal binding: interaction of serum transferrin with In3+ and Bi3+ J Am Soc Mass Spectrom. 2004;15:1658–1664. doi: 10.1016/j.jasms.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 38.Kaltashov IA, Zhang M, Eyles SJ, Abzalimov RR. Investigation of structure, dynamics and function of metalloproteins with electrospray ionization mass spectrometry. Anal Bioanal Chem. 2006;386:472–481. doi: 10.1007/s00216-006-0636-6. [DOI] [PubMed] [Google Scholar]
- 39.Mizutani K, Yamashita H, Kurokawa H, Mikami B, Hirose M. Alternative structural state of transferrin - The crystallographic analysis of iron-loaded but domain-opened ovotransferrin N-lobe. J Biol Chem. 1999;274:10190–10194. doi: 10.1074/jbc.274.15.10190. [DOI] [PubMed] [Google Scholar]
- 40.Cheng YG, Mason AB, Woodworth RC. pH dependence of specific divalent anion binding to the N-lobe of recombinant human transferrin. Biochemistry. 1995;34:14879–14884. doi: 10.1021/bi00045a032. [DOI] [PubMed] [Google Scholar]
- 41.Adams TE, Mason AB, He QY, Halbrooks PJ, Briggs SK, Smith VC, MacGillivray RT, Everse SJ. The position of arginine 124 controls the rate of iron release from the N-lobe of human serum transferrin. A structural study. J Biol Chem. 2003;278:6027–6033. doi: 10.1074/jbc.M210349200. [DOI] [PubMed] [Google Scholar]
- 42.Halbrooks PJ, Mason AB, Adams TE, Briggs SK, Everse SJ. The oxalate effect on release of iron from human serum transferrin explained. J Mol Biol. 2004;339:217–226. doi: 10.1016/j.jmb.2004.03.049. [DOI] [PubMed] [Google Scholar]