Abstract
Ionotropic glutamate receptors mediate the majority of vertebrate excitatory synaptic transmission. The development of selective antagonists for glutamate receptor subtypes is of interest in the treatment of a variety of neurological disorders. This study presents the structure of the binding domain of GluR2 bound to two antagonists (UBP277 and UBP282) that are derivatives of the natural product, willardiine. The antagonists bind to one lobe of the protein with interactions similar to agonists. Interaction with the second lobe differs between the two antagonists, resulting in a different position of the uracil ring and different orientations of the bilobed structure. UBP277 binding produces a stable lobe orientation that is similar to the apo state, but the binding of UBP282 produces the largest hyperextension of the lobes yet reported for an AMPA receptor. The carboxyethyl (UBP277) and carboxybenzyl (UBP282) substituents in the N3 position keep the lobes separated by a “foot-in-the-door” mechanism and the internal dynamics are minimal compared to the CNQX-bound form of the protein (which makes minimal contacts with one of the two lobes). In contrast to the antagonists CNQX and DNQX, UBP277 and UBP282 produce a complex with higher thermal stability, but the affinity of these compounds is more than 100-fold lower. These structures support the idea that antagonism is associated with the overall orientation of the lobes rather than with specific interactions, and antagonism can rise either from specific interactions with both lobes (“foot-in-the-door” mechanism) or from the lack of extensive interactions with one of the two lobes.
Ionotropic glutamate receptors (iGluRs) mediate the majority of excitatory synaptic transmission in the central nervous system of higher vertebrates (1) and play important roles in the formation of synaptic plasticity underlying higher-order processes such as learning and memory as well as in neuronal development (2). In addition, iGluRs have been implicated in various neurodegenerative disorders such as Parkinson's and Alzheimer's diseases, Huntington's chorea, and neurologic disorders including epilepsy and ischemic brain damage. Antagonists of glutamate receptors have been shown to limit tumor growth in a variety of human tumors and to inhibit tumor cell migration (3). In recent years, many advances in characterizing the relationship between iGluR structure and function have been made. iGluRs are membrane-bound receptor ion channels composed of four subunits surrounding a central ion channel in which each subunit contributes to pore formation. Individual subunits are categorized by pharmacological properties, sequence, functionality, and biological role into those that are sensitive: (1) to the synthetic agonist N-methyl-D-aspartic acid or glycine (NMDA; NR1, NR2A-D, NR3A-B); (2) to the synthetic agonist α-amino-3-hydroxy-5-methyl-4-isoxazole-propionic acid (AMPA; GluR1-4); and (3) to the naturally occurring neurotoxin kainate (GluR5-7, KA1,2).
The three-dimensional structures of the binding domain (S1S2) of a number of glutamate receptors are known from X-ray crystallography (reviewed by 4). In particular, the structures of the AMPA subunit, GluR2, bound to a wide variety of agonists, partial agonists and antagonists have provided compelling clues to the structural basis of channel activation and desensitization (5, 6). The binding domain consists of two weakly interacting lobes with the agonist-binding site between the lobes. In the case of full agonists, binding of ligands results in a closure of the lobes that is essentially complete. Partial agonists differ in that the lobes close to a lesser extent in some cases. In the case of a series of partial agonists based on the willardiine backbone, at least some of the partial agonists show a variable degree of lobe closure (7, 8) and considerable internal dynamics (9, 10). The partial agonist, kainate, has a stable degree of lobe closure (10, 11) and minimal internal dynamics (12). Kainate seems to lock the lobes in a partially closed state through what has been described as a “foot-in-the-door” mechanism by which the isoprenyl group clashes with L650 (13). Mutation of L650 to T results in increased activation of the channel by kainate and a greater degree of lobe closure (13, 14). Antagonists, on the other hand, show little or no lobe closure (15-17) and, in some cases, a further opening of the binding cleft (18). Previous NMR (12) studies suggested that both the apo form and the CNQX-bound (antagonist) forms are very dynamic, probably exhibiting a variable degree of lobe closure or, possibly, extension. Molecular dynamics measurements suggested that the lobe orientation of the DNQX-bound form was more variable than the glutamate-bound for m (19). We report here the structure, thermodynamics, and preliminary analysis of the dynamics of GluR2 S1S2 bound to two antagonists, UBP277 and UBP282, which behave like antagonist versions of kainate. These antagonists are derivatives of the natural product, willardiine, with substitutions in the N3 position. This is in contrast to the 5-position substituents that function as partial agonists. The carboxyethyl (UBP277) and carboxybenzyl (UBP282) substituents in the N3 position keep the lobes separated by a “foot-in-the-door” mechanism and the internal dynamics are minimal compared to the CNQX-bound form of the protein.
Experimental Procedures
Materials
The willardiine compounds were synthesized as described previously (20, 21). DNQX and CNQX were purchased from Tocris (Ellisville, MO) and Ascent Scientific (Princeton, NJ). 5-Fluorotryptophan was obtained from Sigma and Fisher. The GluR2 S1S2J construct was obtained from Eric Gouaux (Vollum Institute; 15).
Protein Preparation and Purification
S1S2 consists of residues N392 - K506 and P632 - S775 of the full rat GluR2-flop subunit (22), a `GA' segment at the N-terminus, and a `GT' linker connecting K506 and P632 (15). For 1H,15N-NMR experiments, poly-histidine-tagged S1S2 was over-expressed at 37°C as inclusion bodies in BL-21(DE3) E. coli, refolded, partially digested with trypsin for histidine-tag removal, and purified, essentially according to protocols described elsewhere (23, 24). Typically, 1.25L of isotopically enriched, fully deuterated, vitamin supplemented, minimal media (25) yielded 3-4 samples of 0.25 - 0.35 mM purified protein, as measured using UV absorbance at 280 nm and assuming a molar absorption coefficient of 41,500 M-1cm-1 (26). Nearly complete deuteration at non-labile positions was achieved by dissolving lyophilized media components in pure D2O and culturing with deuterated glucose. Exchange of deuterons for protons at labile sites occurred naturally after bacterial lysis in aqueous refolding and purification buffers. The NMR buffer containing S1S2 and ligand was an aqueous solution of 25 mM sodium acetate, 25 mM sodium chloride, 3 mM sodium azide, 12% D2O, pH 5.0. Protein was prepared for ITC experiments using a similar protocol, but the medium was not isotopically labeled.
For crystallography and thermal denaturation studies, pET-22b(+) plasmids were transformed in E. coli strain Origami B (DE3) cells and were grown at 37°C to OD600 of 0.9 to 1.0 in LB medium supplemented with the antibiotics (ampicillin and kanamycin). The cultures were cooled to 20°C for 20 min. and isopropyl-β-D-thiogalactoside (IPTG) was added to a final concentration of 0.5 mM. Cultures were allowed to grow at 20°C for 20 h. The cells were then pelleted and the S1S2 protein purified using a Ni-NTA column, followed by a sizing column (Superose 12, XK 26/100), and finally an HT-SP-ion exchange-Sepharose column (Amersham Pharmacia). Glutamate (1 mM) was maintained in all buffers throughout purification. After the last column, the protein was concentrated and stored in 20 mM sodium acetate, 1 mM sodium azide, and 10 mM glutamate at pH 5.5. For 19F NMR experiments, the procedure was modified to incorporate 5-fluorotryptophan (27) as described previously (12).
Crystallography
For crystallization trials, the proteins were concentrated to 0.2 to 0.4 mM using a Centricon 10 centrifugal filter (Millipore, Bedford, MA). UBP277 and UBP282 were exchanged into the sample by successive concentration [Amicon Ultra-4 (10K) filter] and dilution using buffer only in the first few steps followed by 5 mM ligand. The final protein concentration was approximately 0.2 mM. The final ligand concentrations were 5 mM for the antagonists. All crystals were grown at 4°C using the hanging drop technique, and the drops contained a 1:1 (v/v) ratio of protein solution to reservoir solution. The reservoir solution in case of GluR2-UBP277 was 14-16% polyethylene glycol (PEG) 8K, 0.1 M sodium cacodylate, 0.25 M ammonium sulfate, and 0.1-0.15 M zinc acetate (pH 6.5). In case of GluR2-UBP282, the reservoir solution contained 15-17% PEG 8K, 0.2 M sodium cacodylate (pH 6.5) and 0.2 M sodium acetate.
Data were collected at the Cornell High Energy Synchrotron Source beam line A1 using a Quantum-210 Area Detector Systems charge-coupled device detector. Data sets were indexed and scaled with HKL-2000 (28). Structures were solved with molecular replacement using Phenix (29). Refinement was performed with CNS 1.2 (30), and Coot 0.5 (31) was used for model building.
Radioligand binding
The binding of [3H]AMPA (40 Ci/mmol; Amersham) to GluR2 S1S2 was determined as described by Chen et al. (32). The binding buffer was 30 mM Tris-HCl, 100 mM KSCN, 2.5 mM CaCl2, 10% glycerol, pH 7.2, maintained at all times at 4°C. Glutamate was removed from the S1S2 proteins by successive concentration and dilution as described above, and diluted to a final concentration of 70 nM. After the addition of the tested ligand, [3H]AMPA (10 nM) was added, the reaction (200 μl) proceeded for 1 hour followed by filtration through Millipore GSWP filters and two 2 ml washes with binding buffer. All analysis was done using Kaleidagraph (Synergy Software).
ITC
Protein bound to 1 mM glutamate was exchanged into ITC buffer (20 mM sodium phosphate buffer, 50 mM NaCl, 1 mM azide, pH 7.3) by successive concentration [Amicon Ultra-4 (10K) filter] until the glutamate concentration was 2 μM. The resulting solution was then exchanged into either 2 μM glutamate, 190 μM UBP277, or 250 μM UBP282. The final protein concentration ranged from 15 to 30 μM, as determined from the absorbance at 280 nm using a molar extinction coefficient of 1.0 ml/mg (28600 M-1). The sample was stirred at 300 rpm in a Microcal VP-ITC calorimeter, while competing ligand was injected (40 injections at 6 μL per injection, spaced by 180 seconds). All experiments were performed at 10°C. For measurements involving CNQX and DNQX, the competing ligand was the antagonist (CNQX or DNQX in the injection syringe at 350-400 μM), and the protein in the sample cell was prepared with 2 μM glutamate. Because UBP277 and UBP282 are of much lower affinity than glutamate, measurements involving UBP277 and UBP282 were reversed, so that the competing ligand was the agonist (glutamate or IW in the injection syringe at 500 μM), and the protein in the sample cell was prepared with antagonist (190 μM UBP277 or 250 μM UBP282).
The thermograms were analyzed with the competitive binding approach described by Sigurskjold et al. (33) using Origin software. In all cases, the value of ΔH for binding of ligand B (the ligand in the sample cell) to apo protein was taken to be zero as a reference. For competitive binding experiments using IW, CNQX, or DNQX as ligand A and glutamate as ligand B, data were fit using an equilibrium association constant for binding of glutamate of 1.0 × 106. For competitive binding experiments using IW or glutamate as ligand A and UBP282 or UBP277 as ligand B, data was fit by iteratively varying the equilibrium association constant of ligand B until the association constant of ligand A was 1.0 × 106 (glutamate) or 3.0 × 106 (IW). After an initial fit to the data, the protein concentration was adjusted and the data re-analyzed using an iterative process until the fit yielded a binding stoichiometry of 1. The value of the association constant for ligand B was then varied separately until the fit converged.
Thermal denaturation
Thermal unfolding was monitored by intrinsic tryptophan fluorescence with a Varian Cary Eclipse spectrofluorimeter, using an excitation wavelength of 280 nm and an emission wavelength of 336 nm. The temperature was monitored in the cuvettes and adjusted at a rate on 0.06°C/min between 15 and 65°C. The protein concentrations were 1-2 μM in NMR buffer (25 mM sodium acetate pH 6.0, 25 mM sodium chloride, and 1 mM sodium azide). The concentrations of CNQX, DNQX, UBP277 and UBP282 were 10 μM, 10 μM, 1 mM, and 1 mM, respectively. The data were analyzed using Kaleidagraph (Synergy Software) as described by Madden et al. (34).
NMR Spectroscopy
1H and 15N measurements were made on a Varian Inova 500 spectrometer equipped with a triple resonance, z-gradient cryogenic probe. 19F NMR spectra were made using a broadband probe on a Varian Inova 500 spectrometer with the proton channel downtuned to fluorine (470 MHz). Spectra were processed using NMRPipe v. 1.6 (35). The data were apodized with a mixed exponential-Gauss window function and zero-filled to double the original number of data points before Fourier transform.
Results
Structure of UBP277 bound to GluR2 S1S2
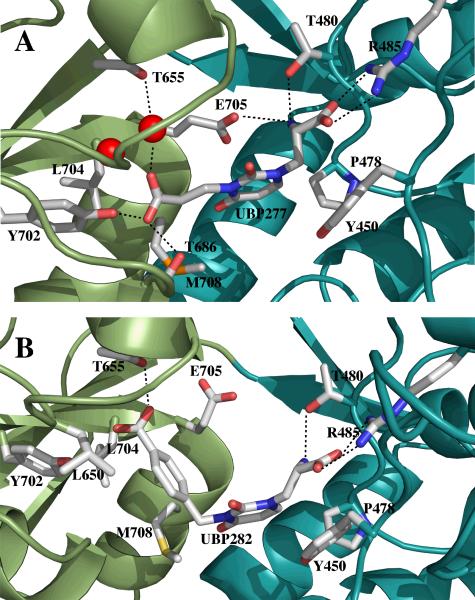
The structure of DNQX bound to GluR2 S1S2 (15) was used as the search probe for the molecular replacement solution of UBP277 bound to GluR2 S1S2. The refinement statistics are given in Table 1. The resolution was 1.9 Å, and two unique copies were found in the unit cell. Continuous density for the backbone was observed from K393 to C773 (including the GT linker that replaces the ion channel domain), and the ligand was well resolved. Because of the variability of the apo structures, we report here the opening of the lobe relative to the S1S2-glutamate structure (C protomer; 15). The UBP277-S1S2 structures are slightly more open than the corresponding S1S2-DNQX structures and not significantly different from the apo structures (15). The α-carboxyl and α-amino groups are positioned in the binding site in exactly the same manner as glutamate in its corresponding structure (Figure 1A), which has been described in detail by Armstrong and Gouaux (1ftj, 15). This involves interaction of the α-carboxyl and primarily R485, with additional stabilization from the backbone amides of T480 and S654. Likewise, the α-amino of UBP277 is stabilized by the backbone carbonyl of P478, the γ-carboxylate of E705, and the hydroxyl of T480. These interactions are common to most agonists and antagonists that bind to AMPA receptors.
Table 1.
Structural Statistics
| Structure | GluR2-UBP277 | GluR2-UBP282 |
|---|---|---|
| Space Group | P21 | P1 |
| Unit Cell (Å) | a=98.9 b=57.7 c=105.4 β=114.9 | a=91.0 b=90.9 c=92.5 |
| α=85.6 β=85.5 γ=72.4 | ||
| X-ray source | CHESS (A1) | CHESS (A1) |
| Wavelength (Å) | 0.977 | 0.977 |
| Resolution (Å) | 50-1.90 (1.98-1.90) | 50-2.8 (2.9-2.8) |
| Measured reflections (#) | 314093 | 162249 |
| Unique reflections (#) | 100335 | 69322 |
| Data redundancy | 3.6 (3.5) | 2.4 (2.2) |
| Completeness (%) | 99.0 (99.4) | 96.7 (79.0) |
| Rsym(%) | 20.5 (86.1) | 10.2 (37.2) |
| I/σi | 10.2 (2.95) | 7.42 (2.79) |
| PDB ID | 3H03 | 3H06 |
| Current Model Refinement Statistics | ||
| Phasing | MR | MR |
| Molecules/AU | 4 | 1 |
| Rwork/Rfree (%) | 22.4/25.8 | 23.0/29.6 |
| Free R test set size (#/%) | 6935 (10.0) | 6536 (9.4) |
| Number of protein atoms | 8036 | 16080 |
| Number of heteroatoms | 76 | 192 |
| Rmsd bond length (Å) | .0045 | .0079 |
| Rmsd bond angles (°) | 1.1 | 1.3 |
| Rmsd B factors (Å2, main/side) | 1.17/2.22 | 1.20/1.66 |
Figure 1.
Structures of the binding sites for UBP277 (A) and UBP282 (B) on GluR2 S1S2. In each case, Lobe 1 is shown in cyan and Lobe 2 in green. Interactions between the ligand and protein sidechains are indicated. Additional interactions with the backbone are also made with Lobe 1.
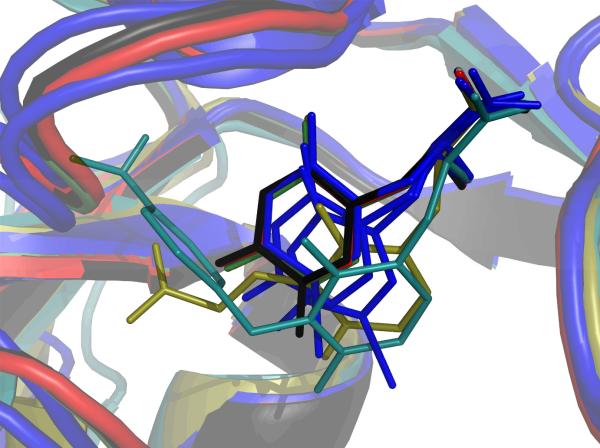
The position of the uracil-like ring, however, differs relative to the position of willardiine partial agonists in the S1S2 binding site (Figure 2). The χ1 angle determines the position of the uracil ring. For willardiines with small substitutents (e.g., willardiine, fluorowillardiine), the 2-carbonyl oxygen interacts with the sidechains of S654 and T655. The larger iodowillardiine is apparently more mobile in the binding site, showing at least three different positions in various structures (7). In these alternative positions, the ring is rotated deeper into the cleft away from S654 and T655. This is accommodated by a rotation of the sidechain of M708 and variable degrees of lobe opening, depending upon the crystallization conditions. For UBP277, χ1 angle is rotated by ~22° relative to willardiine, moving the ring even deeper into the cleft and further away from S654 and T655. Compared with willardiine partial agonists, the 5′ carboxyethyl group is relatively large and is accommodated by a large-scale rotation of lobe 2 relative to lobe 1. This consists of an opening of the lobes relative to the glutamate-bound structure of 19.8 ± 0.6° (1ftj, C protomer), which is very similar to the orientation in which the apo structure crystallized (15) and is similar to the orientation of the lobes measured in solution (10). The carboxyl group then inserts into a relatively hydrophilic pocket consisting of the sidechains of T686, Y702, and T655. A water molecule is coordinated between the hydroxyl of T655 and the γ-carboxylate of UBP277.
Figure 2.
The orientation of different willardiines in the GluR2 S1S2 binding site is illustrated. The residues in Lobe 1 were aligned. The positions of HW (red; 1mqj), BrW (green, 1mqh) and FW (black, 1mqi) are superimposable. However, the positions of IW (blue, 1mqg) varies within the same crystal. IW is the lowest efficacy of the four partial agonists. UBP277 (yellow) and UBP282 (cyan) move even further down the binding pocket than IW and make contact with Lobe 2 through the substituents in the 2 position.
Structure of UBP282 bound to GluR2 S1S2
The structure of UBP277 bound to GluR2 S1S2 was used as the search probe for the molecular replacement solution of UBP282 bound to GluR2 S1S2. The resolution was 2.8 Å, with P1 symmetry (eight copies in the asymmetric unit). Continuous density for the backbone was observed from K393 to C773 in two of the structures; whereas, the density was poorly resolved between F659 and I664 in four protomers and between M674 and S680 in two protomers. The ligand was clearly resolved, but not as well as the higher resolution structure of UBP277. The orientation in the binding site was similar to that of UBP277 (Figure 1B), with the α-carboxylate and α-amino groups interacting with the same residues in Lobe 1. One exception is E705, which is rotated away from the α-amino of UBP282 to make room for the carboxylbenzyl group. The position of E705 is similar to that in the apo state (15) and for N3-substituted willardiines (UBP310 and UBP302) bound to GluR5 (36). The larger carboxybenzyl group results in a small rotation of the uracil ring further into the cleft relative to the UBP277-bound structure, and a ~33° rotation of the χ1 angle relative to FW and HW. Because of the size of the substituent, the lobe is opened further (26.8 ± 1.8°) relative to the glutamate-bound structure (1ftj, protomer C). This is greater than the reported opening of the apo structure (15) and is the largest hyperextension of an AMPA receptor yet reported. The rotation of the N3-substituent places the sidechain carboxyl in a position different from that in the bound UBP277 structure. In the willardiine and fluorowillardiine structure, the 2-carbonyl on the ring interacts with S654 and T655. In this case it is the carboxyl group that forms a similar interaction with T655 as well as an interaction with Y702. Thus, lobe 2 is tethered to the antagonist in a position somewhat similar to that of a partial agonist, but with the lobes rotated apart by more than 20°. The aromatic ring of the carboxybenzyl group is positioned in a hydrophobic patch made up of L650, L704, and the methylene groups of E705.
Binding affinity
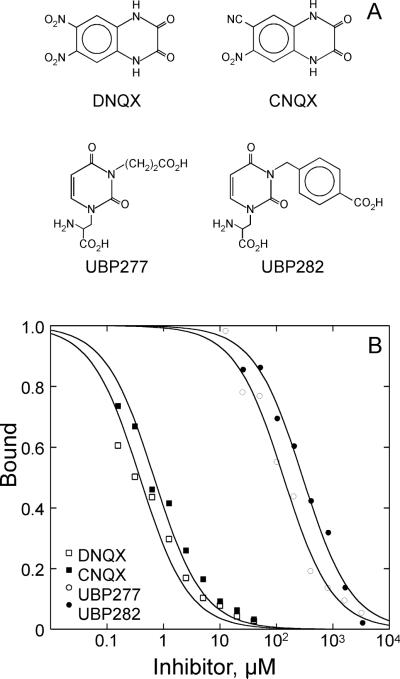
The binding of UBP277 (37) and UBP282 were compared with that of CNQX and DNQX using the inhibition of [3H]AMPA binding to soluble GluR2 S1S2 (Figure 3). Both CNQX (IC50 = 0.68 ± 0.06 μM) and DNQX (IC50 = 0.38 ± 0.04 μM) bound with submicromolar affinity. In contrast, both UBP277 (37; IC50 = 135 ± 12 μM) and UBP282 (IC50 = 292 ± 26 μM) bound with significantly lower affinity than CNQX and DNQX.
Figure 3.
(A) Structures of the antagonists used in the binding study. (B) Inhibition of the binding of [3H]AMPA to GluR2 S1S2 by DNQX, CNQX, UBP277, and UBP282. The IC50 values were: 0.38 ± 0.04 μM, 0.68 ± 0.06 μM, 135 ± 12 μM, 292 ± 26 μM, respectively.
Dynamics of antagonist-bound complexes
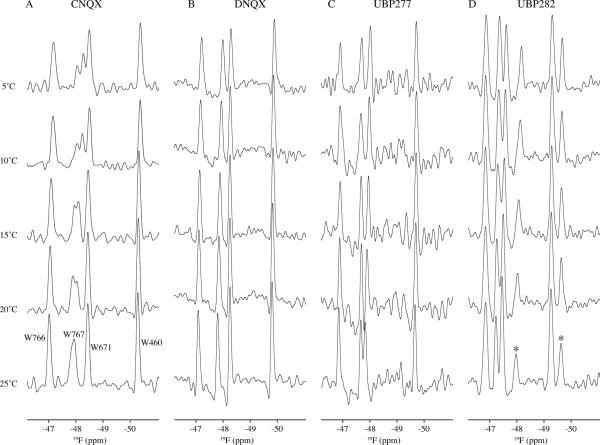
19F NMR
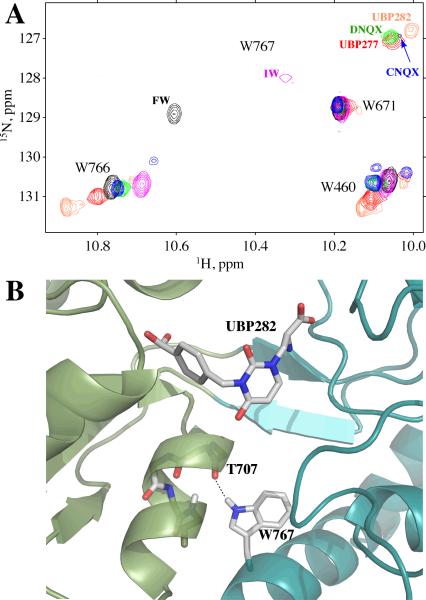
Previous studies with 19F-tryptophan-labeled S1S2 showed that 19F-W767 exhibited clear two-site chemical exchange (kex of approximately 4 ms) when bound to CNQX (12). No exchange was observed in any of the tryptophans for 19F-S1S2 bound to UBP282, UBP277 or DNQX (Figure 4). In all cases, the linewidths were approximately equal and no excessive linebroadening was observed as the temperature was decreased from 25°C to 5°C. The indole amide proton of W767 is hydrogen-bonded to the carbonyl of T707, presumably making this tryptophan an indicator of large-scale relative motions of the lobes (Figure 5B). One interpretation of the exchange dynamics seen in S1S2-CNQX is that it is exchanging between at least two different lobe orientations (12).
Figure 4.
19F NMR spectra of antagonists bound to 5-19F-tryptophan-labeled GluR2 S1S2 as a function of temperature: (A) CNQX, (B) DNQX, (C) UBP277, and (D) UBP282. The signal for W767 is in chemical exchange for the CNQX-bound form but apparently not for S1S2 bound to the other three antagonists. Two additional peaks (marked with asterisks) were consistently seen in spectra of the UBP282-bound form and are consistent with chemical shifts observed in the apo form.
Figure 5.
(A) A portion of the 1H,15N-HSQC (TROSY) spectrum at 10°C showing the sidechain tryptophan amides for GluR2 S1S2 bound to a variety of willardiine agonists and antagonists as well as CNQX and DNQX. The signal for W767 varies with the orientation of the two lobes, moving upfield as the lobe opening increases. The amide proton makes a hydrogen bond across the lobe interface with the carbonyl of T707, shown here in the UBP277-S1S2 structure (B). The peak for this correlation is dramatically broadened at this temperature for IW and CNQX, but not the other compounds shown. This broadening is consistent with exchange between multiple relative orientations of the two lobes (10).
1H,15N-HSQC
The 1H,15N-HSQC(TROSY) spectra for each of the four antagonists bound to S1S2 was determined at 10°C (Figure 5A). In each case, the spectra are well resolved, and indicate a structured protein similar to that reported for agonists (24, 38, 39). As noted above, the W767 sidechain NH is diagnostic of the orientation of the lobes and the large-scale dynamics of the lobes (10; Figure 5B). It is near the axis of rotation so that the hydrogen bond is largely conserved but seems to vary in strength for different relative rotations of the two lobes (Figure 8A). With different ligands in the binding site, the peak for W767 moves progressively upfield (Figure 5A) with an increasing separation of the lobes, which is consistent with a weaker hydrogen bond (40). In the case of CNQX, however, the W767 sidechain NH peak is very weak, consistent with the chemical exchange observed in the 19F spectra described by Ahmed et al. (12) and discussed above. Also shown in Figure 5B are spectra for S1S2 bound to the partial agonists, FW and IW. Like CNQX-S1S2, the W767 NH peak for IW-S1S2 is exchange broadened and indicative of chemical exchange between different lobe orientations (10). In the case of S1S2-CNQX, the protein unfolded and became partially degraded for long-term experiments (>1 hr) at or above 20°C; whereas for the other antagonists, the protein remained stable for several days at 25°C (data not shown).
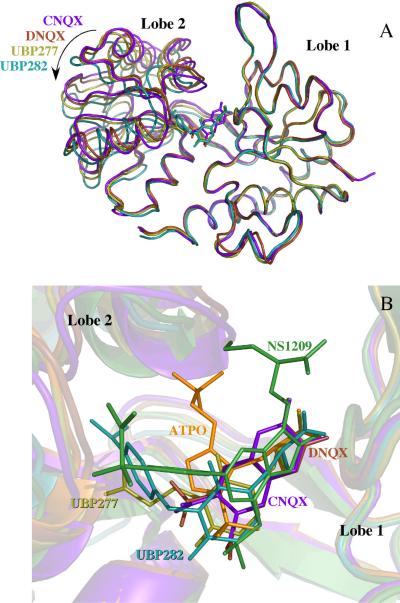
Figure 8.
(A) Ribbon structures of GluR2-S1S2 bound to CNQX (purple, 3b7d, 17), DNQX (brown, 1ftl, 15), UBP277 (yellow), and UBP282 (cyan). The residues in Lobe 1 are aligned to illustrate the different lobe orientations. Using DynDom (41), the lobe opening relative to the glutamate-bound form (1ftj, C protomer, 15) is 15.7° ± 0.5° for CNQX, 16.5° ± 1.1° for DNQX, 19.8° ± 0.6° for UBP277, and 26.7° ± 1.8° for UBP282. (B) Overlay of the binding site for six antagonists showing the multiple sites of interaction. Shown are CNQX, DNQX, UBP277, UBP282 (as in part A), ATPO (orange, 1n0t, 16), and NS1209 (green, 2cmo, 18). As in part A, the residues in Lobe 1 were aligned.
Thermodynamics
Isothermal titration calorimetry
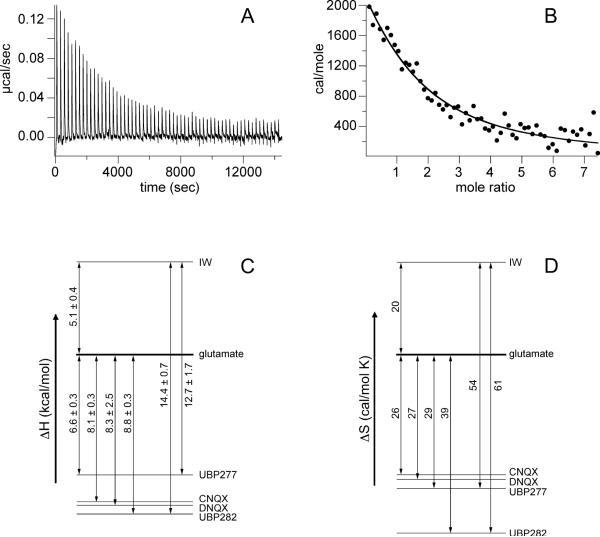
Titration of glutamate into a GluR2-S1S2 sample which had been dialyzed extensively against buffer in the absence of glutamateric ligands showed little signal at 10°C. This is consistent with previous work on GluR4, which would predict only a small enthalphic change at this temperature (34). While the signal would be expected to be larger at 25°C, the protein is unstable at higher temperatures in the apo state (12) and the removal of glutamate is sometimes incomplete (18), resulting in low signal-to-noise and large variability in direct binding studies. However, competition between antagonist and glutamate or other ligands was reliably measured. In this case, glutamate-bound S1S2 was used as the reference state. The thermodynamics of binding of DNQX and CNQX was determined by titrating DNQX or CNQX into protein bound to glutamate or IW. Because UBP277 and UBP282 have lower affinity for S1S2, ITC competition experiments were performed by exchanging the antagonist into the binding site and titrating with IW or glutamate (Figure 6A). IW was used in some cases because IW binding is endothermic relative to glutamate (ΔΔH = 5.2 ± 0.4 kcal/mol), and thus competition with antagonists provides a higher signal-to-noise. For DNQX and CNQX at 10°C, the binding was exothermic relative to glutamate binding, with a ΔΔH of approximately to -8 kcal/mole (Table 2). Relative to glutamate, the entropic cost of binding was relatively high in that ΔΔS was approximately -25 cal/mole•K for both antagonists. Similarly, binding of UBP277 and UBP282 are exothermic relative to glutamate (Figure 6A). Although the affinities of UBP277 and UBP282 for GluR2 S1S2 are more than 100-fold lower than CNQX and DNQX, the enthalpy of binding is favorable relative to glutamate (ΔΔH = -6.6 ± 0.3 and -8.8 ± 0.3 kcal/mole, respectively). However, there is an even more unfavorable entropic cost of binding (-29 and -39 cal/mole•K, respectively). In contrast, the willardiine partial agonist, IW, has a generally unfavorable enthalpy of binding relative to glutamate, and a more favorable entropy term (Table 2 and Figure 6C and D). As expected, the titration of UBP277- and UBP282-bound S1S2 with IW (Figure 6C and D) resulted in larger values of ΔΔH (-14.4 and -12.7 kcal/mol, respectively) and a larger ΔΔS (-51 and -54 cal/mole•K). As shown in Figure 6C and 6D, the differences in ΔΔH and ΔΔS for titrating UBP277 or UBP282 with IW vs. glutamate are consistent with the direct competition between IW and glutamate.
Figure 6.
(A & B) Sample data from ITC competitive binding studies: 500 μM UBP277 titrated into 12 μM S1S2 bound to glutamate at 10°C. The data were fit using Origin 7 and the equations described by Sigurskjold et al. (33) for competition binding. (C & D) Schematic of the relative enthalpies and entropies of binding of various ligands to S1S2 as determined by competitive binding ITC experiments.
Table 2.
Thermodynamics of antagonist binding Isothermal titration calorimetry (ITC) data from competitive binding experiments at 10°C. All values are reported relative to the binding of glutamate.
| Ligand | Binding Constant (Ka, M-1) | ΔΔH (kcal/mol) | ΔΔS (cal/mol•K) |
|---|---|---|---|
| IW | 3.0 × 106 | +5.1± 0.4 | +20 |
| Glutamate | 1.0 × 106 | 0 | 0 |
| UBP277 | 5.1 × 104 | -6.6± 0.3 | -29 |
| UBP282 | 2.2 × 104 | -8.8± 0.3 | -39 |
| DNQX | 3.4 × 106 | -8.3± 2.5 | -27 |
| CNQX | 3.0 × 106 | -8.1± 0.3 | -26 |
Thermal denaturation
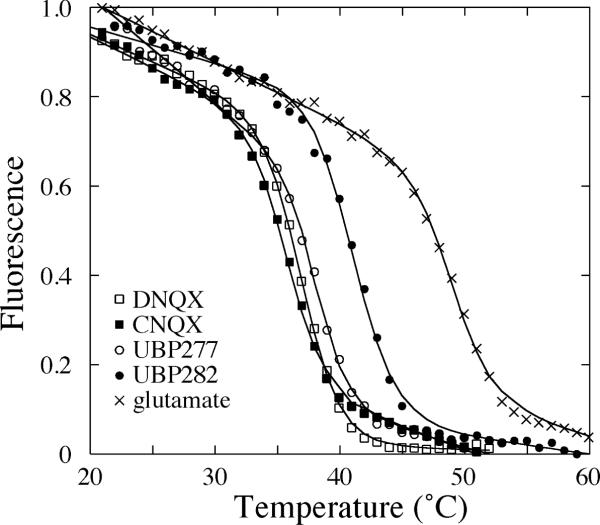
Intrinsic tryptophan fluorescence was used to monitor the thermal unfolding of the S1S2 domain in the presence of various antagonists. The temperature was increased at 0.06 °C/min over the range of 15°C to 70°C, which resulted in a linear decrease in fluorescence overlayed with a clear transition associated with protein unfolding (Figure 7). Complexes with CNQX, DNQX, UBP277, and UBP282 are all significantly less stable than the glutamate-bound form, with transition temperatures of 35.8°C, 37.0°C, 38.1°C and 41.2°C, respectively, relative to the Tm of the glutamate-bound form, which was 49.1°C.
Figure 7.
Thermal denaturation of GluR2-S1S2 bound to glutamate, CNQX, DNQX, UBP277 and UBP282. The temperature was increased at a rate of 0.06 min/°C. The data were fit as described by Madden et al. (34).
Discussion
Antagonists bind to GluR2 and, in the intact protein, block the binding of agonist but do not produce channel activation. Whereas the binding of agonist to the isolated binding domain leads to a closure of the two lobes, antagonists bind to the same site on Lobe 1, but do not promote closure. In fact, for some antagonists, contacts with Lobe 2 prevent lobe closure or lead to hyperextension of the two lobes. Hyperextension has also been observed in GluR5 using two N3 substituted willardiine antagonists (UBP310 and UBP302; 36) and for NS1209 bound to GluR2 (18). The structures of GluR2 S1S2 bound to two antagonists are reported here. In both cases, the antagonists make contacts with both Lobes 1 and 2, maintaining a separation between the two lobes that is equal or greater than observed in the apo crystal structure. Although some interactions (e.g., with T655) are similar to the 5-substituted willardiine partial agonists, the larger substituents in the N3 position and their more extensive interactions with lobe 2 are responsible for the large lobe separations.
The structures of GluR2 S1S2 bound to four antagonists have been reported (CNQX, 17; DNQX, 15; ATPO, 16; NS1209, 18) in addition to the two reported here. The contacts with Lobe 1 are relatively constant, including interactions with the side chain of R485, the carbonyl of P478, and the backbone amide proton and side chain hydroxyl of T480. Contacts with Lobe 2, however, are variable (Figure 8B). The interactions of ATPO, NS1209, UBP277 and UBP282 with lobe 2 are extensive, whereas a direct interaction with Lobe 2 is limited to T686 in the CNQX-bound structure. For the DNQX-bound structures, one of the two protomers makes extensive interactions with Lobe 2 though a sulfate ion, whereas the other protomer is similar to the CNQX-bound structures. As proposed previously (16), the variable interactions with Lobe 2 suggest that it is the separation of the lobes rather than specific interactions that account for their action as antagonists.
In the case of partial agonists, the lobes reorient by closing around the ligand but not to the same extent as full agonists. This is a result of steric hindrance as is the case for the foot-in-the-door mechanism described for the partial agonist kainate (13) or a weaker interaction with Lobe 2 that leads to multiple orientations of the two lobes, typified by iodowillardiine (10). The action of ATPO, NS1209, UBP277 and UBP282 would be more similar to the foot-in-the-door mechanism of kainate, however, with a greater lobe separation. CNQX, and to a lesser extent DNQX, seem to function as antagonists more by virtue of minimal interactions with Lobe 2. In fact, CNQX can function as a partial agonist when GluR2 is expressed in mammalian cells with TARPs (17). The 2,3-dione of CNQX and DNQX make canonical interactions with Lobe 1 of GluR2. The nitro group of CNQX and one of the nitro groups of DNQX interacts with T686 of Lobe 2, and the other nitro group of DNQX can interact with Y732 and T707 in Lobe 2. This weak interaction of the cyano and nitro groups of CNQX with the protein is consistent with markers for lobe dynamics that suggest multiple lobe orientations. This comes from two lines of evidence. The first is 19F-NMR (12) of W767, a residue that spans the lobe interface and is clearly in slow chemical exchange between two environments (kex of ~4 ms). Likewise, the hydrogen bond across the lobe interface made by the sidechain amide proton of W767 is exchanged broadened in the CNQX-bound form but none of the other antagonist-bound forms. The most likely interpretation is that the more extensive interactions of UBP277 and UBP282 with Lobe 2 result in a more fixed lobe orientation than with CNQX. Consistent with this is the finding that the orientation of the lobes in the solution form of UBP277-S1S2 complex is similar to that found in the crystal (10). Thus, like partial agonists, the mechanism of antagonism can consist either of a foot-in-the-door mechanism or a weak interaction with Lobe 2 that results in multiple lobe orientations.
The thermodynamics of antagonist binding to GluR2 S1S2 has been studied previously for ATPO, CNQX and NS1209 and found to be exothermic relative to glutamate (18). At least for GluR4, the change in heat capacity is nonzero, so that the relative importance of entropy and enthalpy is different depending upon the temperature (34). At 10°C, the binding of glutamate to GluR4 is almost exclusively driven by a favorable entropy change (34). One contribution to this is the exclusion of ordered water from the surface area on the protein that forms the lobe interface. Another significant contribution to the entropy of binding may arise from the conformational entropy associated with the internal packing of the protein. Since the conformational entropy decreases with increases in efficacy of the bound ligand (Maltsev & Oswald, unpublished observations), this contribution would likely be unfavorable for the glutamate-bound relative to the apo state. Likewise, the loss of flexibility of glutamate upon binding would be an unfavorable entropic contribution. Measurements of thermodynamics of competition between glutamate and antagonist shows that at 10°C, all antagonists are enthalpically favored relative to glutamate and entropically unfavored. If one assumes that ΔH for glutamate binding is close to zero at 10°C as is the case for GluR4 (34), the favorable ΔS for glutamate binding is similar to the ΔΔS for the competition between glutamate and CNQX/DNQX (unfavorable for CNQX/DNQX relative to glutamate), suggesting that the entropic change for CNQX or DNQX binding to the apo form is small. This is consistent with the idea that exclusion of water is the driving force for glutamate binding at this temperature, because the water is not excluded to the same extent in the CNQX/DNQX structures as it is in the glutamate structures. Also, CNQX and DNQX are relatively rigid molecules in solution so that binding would be associated with less of an entropic cost. UBP277 and UBP282 bind with considerably lower affinity to S1S2 than CNQX/DNQX. At least in the case of UBP282, the entropic cost is the predominant factor that decreases binding affinity. The exclusion of water from the binding site is unlikely to be less than that seen for CNQX/DNQX, so the additional entropic factors must arise from the interaction of the ligand with the protein. This could include the stabilization of the multiple orientations of the two lobes, the loss of flexibility of the ligand upon binding, and an increase in packing of the structure that would decrease the conformational entropy. The stability of the antagonist-bound structures, measured by thermal unfolding, is clearly lower than that of the glutamate-bound form. Yet, despite the significantly lower binding affinity, the UBP277- and UBP282-bound forms are more stable than the bound forms of CNQX and DNQX, with the UBP282-bound form more stable than the UBP277-bound form. Thus, the more rigid structures are more stable despite the fact that the lobes are in a more open conformation. Paradoxically, the increased contacts of UBP282 with S1S2 and increased thermal stability may in fact lead to a lower binding affinity due to the secondary effects on the entropy of the system.
Conclusions
Competitive antagonism of GluR2 requires a ligand that makes the canonical binding interactions on Lobe 1, but actively prevents the lobes from closing. Alternatively, antagonists can bind in a similar manner to Lobe 1 but contain a bulky group with only minor interactions with Lobe 2, leading to considerable flexibility but little or no lobe closure. The actual details of the interactions with Lobe 2 are likely to be unimportant in terms of efficacy, but can affect the affinity of binding dramatically. Particularly in the case of UBP282, the binding affinity is surprisingly low despite a number of favorable interactions with S1S2. The reason for the low binding affinity is not apparent from the crystal structure and arises from unfavorable entropic considerations. This can be contrasted with CNQX and DNQX, which make fewer direct contacts with lobe 2 but bind with a lower entropic cost.
Acknowledgement
We thank Prof. Eric Gouaux (Vollum Institute) for the GluR2 S1S2J construct, and Prof. Linda Nowak (Cornell), Prof. Gregory A. Weiland (Cornell), Alex Maltsev (Cornell) and Dr. Chris Ptak (Cornell) provided useful discussions and valuable advice. We are grateful to Drs. Colin Parrish and Christian Nelson (Baker Institute, Cornell) for the use of the Varian Cary Elipse fluorescence spectrometer, and to Dr. William Horne (Cornell) for use of the Microcal VP-ITC calorimeter.
This work was supported by a grant from the National Institutes of Health (R01-GM068935). Travel funds were provided by the Abraham and Henrietta Brettschneider Oxford and Cornell Exchange Fund. The chemical synthesis of willardiine derivatives was supported by a grant from the MRC and BBSRC, UK. This work is based upon research conducted at the Cornell High Energy Synchrotron Source (CHESS), which is supported by the National Science Foundation under award DMR 0225180, using the Macromolecular Diffraction at the CHESS (MacCHESS) facility, which is supported by award RR-01646 from the National Institutes of Health, through its National Center for Research Resources.
Abbreviations
- AMPA
α-amino-3-hydroxy-5-methyl-4-isoxazole-propionic acid
- CNQX
6-cyano-7-nitroquinoxaline-2,3-dione
- BrW
(S)-5-bromowillardiine
- CPMG
Carr, Purcell, Meiboom, Gill (an NMR pulse sequence element)
- CSA
chemical shift anisotropy
- DNQX
6,7-Dinitroquinoxaline-2,3-dione
- F W
(S)-5-fluorowillardiine
- HW
(S)-willardiine
- iGluR
ionotropic glutamate receptor
- IPTG
isopropyl-β-D-thiogalactoside
- IW
(S)-5-iodowillardiine
- KSCN
potassium thiocyanate
- NMDA
N-methyl-D-aspartic acid
- S1S2
extracellular ligand-binding domain of GluR2
- UBP277
(S)-3-(2-carboxyethyl)willardiine
- UBP282
(S)-3-(4-carboxybenzyl)willardiine
References
- 1.Dingledine R, Borges K, Bowie D, Traynelis S. The glutamate receptor ion channels. Pharmacol. Rev. 1999;51:7–61. [PubMed] [Google Scholar]
- 2.Asztely F, Gustafsson B. Ionotropic glutamate receptors. Their possible role in the expression of hippocampal synaptic plasticity. Mol Neurobiol. 1996;12:1–11. doi: 10.1007/BF02740744. [DOI] [PubMed] [Google Scholar]
- 3.Rzeski W, Turski L, Ikonomidou C. Glutamate antagonists limit tumor growth. Proc Natl Acad Sci U S A. 2001;98:6372–6377. doi: 10.1073/pnas.091113598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mayer ML. Glutamate receptors at atomic resolution. Nature. 2006;440:456–462. doi: 10.1038/nature04709. [DOI] [PubMed] [Google Scholar]
- 5.Jin R, Horning M, Mayer ML, Gouaux E. Mechanism of activation and selectivity in a ligand-gated ion channel: structural and functional studies of GluR2 and quisqualate. Biochemistry. 2002;41:15635–15643. doi: 10.1021/bi020583k. [DOI] [PubMed] [Google Scholar]
- 6.Sun Y, Olson R, Horning M, Armstrong N, Mayer M, Gouaux E. Mechanism of glutamate receptor desensitization. Nature. 2002;417:245–253. doi: 10.1038/417245a. [DOI] [PubMed] [Google Scholar]
- 7.Jin R, Banke TG, Mayer ML, Traynelis SF, Gouaux E. Structural basis for partial agonist action at ionotropic glutamate receptors. Nat Neurosci. 2003;6:803–810. doi: 10.1038/nn1091. [DOI] [PubMed] [Google Scholar]
- 8.Jin R, Gouaux E. Probing the function, conformational plasticity, and dimer-dimer contacts of the GluR2 ligand-binding core: Studies of 5-substituted willardiines and GluR2 S1S2 in the crystal. Biochemistry. 2003;42:5201–5213. doi: 10.1021/bi020632t. [DOI] [PubMed] [Google Scholar]
- 9.Fenwick MK, Oswald RE. NMR spectroscopy of the ligand-binding core of ionotropic glutamate receptor 2 bound to 5-substituted willardiine partial agonists. J. Mol. Biol. 2008;378:673–685. doi: 10.1016/j.jmb.2008.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maltsev AS, Ahmed AH, Fenwick MK, Jane DE, Oswald RE. Mechanism of partial agonism at the GluR2 AMPA receptor: Measurements of lobe orientation in solution. Biochemistry. 2008;47:10600–10610. doi: 10.1021/bi800843c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Armstrong N, Sun Y, Chen GQ, Gouaux E. Structure of a glutamate-receptor ligand-binding core in complex with kainate. Nature. 1998;395:913–917. doi: 10.1038/27692. [DOI] [PubMed] [Google Scholar]
- 12.Ahmed AH, Loh AP, Jane DE, Oswald RE. Dynamics of the S1S2 glutamate binding domain of GluR2 measured using 19F NMR spectroscopy. J Biol Chem. 2007;282:12773–12784. doi: 10.1074/jbc.M610077200. [DOI] [PubMed] [Google Scholar]
- 13.Armstrong N, Mayer M, Gouaux E. Tuning activation of the AMPA-sensitive GluR2 ion channel by genetic adjustment of agonist-induced conformational changes. Proc Natl Acad Sci U S A. 2003;100:5736–5741. doi: 10.1073/pnas.1037393100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ramanoudjame G, Du M, Mankiewicz KA, Jayaraman V. Allosteric mechanism in AMPA receptors: a FRET-based investigation of conformational changes. Proc Natl Acad Sci U S A. 2006;103:10473–10478. doi: 10.1073/pnas.0603225103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Armstrong N, Gouaux E. Mechanisms for activation and antagonism of an AMPA-sensitive glutamate receptor: crystal structures of the GluR2 ligand binding core. Neuron. 2000;28:165–181. doi: 10.1016/s0896-6273(00)00094-5. [DOI] [PubMed] [Google Scholar]
- 16.Hogner A, Greenwood JR, Liljefors T, Lunn ML, Egebjerg J, Larsen IK, Gouaux E, Kastrup JS. Competitive antagonism of AMPA receptors by ligands of different classes: crystal structure of ATPO bound to the GluR2 ligand-binding core, in comparison with DNQX. J Med Chem. 2003;46:214–221. doi: 10.1021/jm020989v. [DOI] [PubMed] [Google Scholar]
- 17.Menuz K, Stroud RM, Nicoll RA, Hays FA. TARP auxiliary subunits switch AMPA receptor antagonists into partial agonists. Science. 2007;318:815–817. doi: 10.1126/science.1146317. [DOI] [PubMed] [Google Scholar]
- 18.Kasper C, Pickering DS, Mirza O, Olsen L, Kristensen AS, Greenwood JR, Liljefors T, Schousboe A, Watjen F, Gajhede M, Sigurskjold BW, Kastrup JS. The structure of a mixed GluR2 ligand-binding core dimer in complex with (S)-glutamate and the antagonist (S)-NS1209. J Mol Biol. 2006;357:1184–1201. doi: 10.1016/j.jmb.2006.01.024. [DOI] [PubMed] [Google Scholar]
- 19.Lau AY, Roux B. The free energy landscapes governing conformational changes in a glutamate receptor ligand-binding domain. Structure. 2007;15:1203–1214. doi: 10.1016/j.str.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jane DE, Hoo K, Kamboj R, Deverill M, Bleakman D, Mandelzys A. Synthesis of willardiine and 6-azawillardiine analogs: pharmacological characterization on cloned homomeric human AMPA and kainate receptor subtypes. J Med Chem. 1997;40:3645–3650. doi: 10.1021/jm9702387. [DOI] [PubMed] [Google Scholar]
- 21.Dolman NP, Troop HM, More JC, Alt A, Knauss JL, Nistico R, Jack S, Morley RM, Bortolotto ZA, Roberts PJ, Bleakman D, Collingridge GL, Jane DE. Synthesis and pharmacology of willardiine derivatives acting as antagonists of kainate receptors. J Med Chem. 2005;48:7867–7881. doi: 10.1021/jm050584l. [DOI] [PubMed] [Google Scholar]
- 22.Hollmann M, Heinemann S. Cloned glutamate receptors. Annu. Rev. Neurosci. 1994;17:31–108. doi: 10.1146/annurev.ne.17.030194.000335. [DOI] [PubMed] [Google Scholar]
- 23.Chen GQ, Sun Y, Jin R, Gouaux E. Probing the ligand binding domain of the GluR2 receptor by proteolysis and deletion mutagenesis defines domain boundaries and yields a crystallizable construct. Protein Sci. 1998;7:2623–2630. doi: 10.1002/pro.5560071216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McFeeters RL, Oswald RE. Structural mobility of the extracellular ligand-binding core of an ionotropic glutamate receptor. Analysis of NMR relaxation dynamics. Biochemistry. 2002;41:10472–10481. doi: 10.1021/bi026010p. [DOI] [PubMed] [Google Scholar]
- 25.Maniatis T, Fritsch EF, Sambrook J. Molecular Cloning. Cold Spring Harbor Laboratory; New York: 1982. [Google Scholar]
- 26.Pace CN, Vajdos F, Fee L, Grimsley G, Gray T. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 1995;4:2411–2423. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salopek-Sondi B, Luck LA. 19F NMR study of the leucine-specific binding protein of Escherichia coli: mutagenesis and assignment of the 5-fluorotryptophan-labeled residues. Protein Eng. 2002;15:855–859. doi: 10.1093/protein/15.11.855. [DOI] [PubMed] [Google Scholar]
- 28.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. In: Carter CW, Sweet RM, editors. Methods in Enzymology, Vol. 276, Macromolecular Crystallography, part A. Academic Press; New York: 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 29.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 30.Brunger AT. Version 1.2 of the Crystallography and NMR system. Nature protocols. 2007;2:2728–2733. doi: 10.1038/nprot.2007.406. [DOI] [PubMed] [Google Scholar]
- 31.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 32.Chen GQ, Gouaux E. Overexpression of a glutamate receptor (GluR2) ligand binding domain in Escherichia coli: application of a novel protein folding screen. Proc Natl Acad Sci U S A. 1997;94:13431–13436. doi: 10.1073/pnas.94.25.13431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sigurskjold BW. Exact analysis of competition ligand binding by displacement isothermal titration calorimetry. Anal Biochem. 2000;277:260–266. doi: 10.1006/abio.1999.4402. [DOI] [PubMed] [Google Scholar]
- 34.Madden DR, Abele R, Andersson A, Keinanen K. Large-scale expression and thermodynamic characterization of a glutamate receptor agonistbinding domain. Eur J Biochem. 2000;267:4281–4289. doi: 10.1046/j.1432-1033.2000.01481.x. [DOI] [PubMed] [Google Scholar]
- 35.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 36.Mayer ML, Ghosal A, Dolman NP, Jane DE. Crystal structures of the kainate receptor GluR5 ligand binding core dimer with novel GluR5-selective antagonists. J Neurosci. 2006;26:2852–2861. doi: 10.1523/JNEUROSCI.0123-06.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ahmed AH, Wang Q, Sondermann H, Oswald RE. Structure of the S1S2 glutamate binding domain of GluR3. Proteins: Structure, Function, and Bioinformatics. 2009 doi: 10.1002/prot.22274. DOI: 10.1002/prot.22274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Valentine ER, Palmer AG., 3rd Microsecond-to-millisecond conformational dynamics demarcate the GluR2 glutamate receptor bound to agonists glutamate, quisqualate, and AMPA. Biochemistry. 2005;44:3410–3417. doi: 10.1021/bi047984f. [DOI] [PubMed] [Google Scholar]
- 39.Zeng L, Lu L, Muller M, Gouaux E, Zhou MM. Structure-based functional design of chemical ligands for AMPA-subtype glutamate receptors. J Mol Neurosci. 2002;19:113–116. doi: 10.1007/s12031-002-0020-8. [DOI] [PubMed] [Google Scholar]
- 40.Wagner G, Pardi A, Wuthrich K. Hydrogen-bond length and H-1-NMR chemical-shifts in proteins. Journal of the American Chemical Society. 1983;105:5948–5949. [Google Scholar]
- 41.Hayward S, Lee RA. Improvements in the analysis of domain motions in proteins from conformational change: DynDom version 1.50. J Mol Graph Model. 2002;21:181–183. doi: 10.1016/s1093-3263(02)00140-7. [DOI] [PubMed] [Google Scholar]