Abstract
Benzene is an established cause of leukemia, and possibly lymphoma, in humans, but the underlying molecular pathways remain largely undetermined. We used two microarray platforms to identify global gene expression changes associated with well-characterized occupational benzene exposure in the peripheral blood mononuclear cells (PBMC) of a population of shoefactory workers. Differential expression of 2692 genes (Affymetrix) and 1828 genes (Illumina) was found and the concordance was 50 % (based on an average fold-change ≥ 1.3 from the two platforms), with similar expression ratios among the concordant genes. Four genes (CXCL16, ZNF331, JUN and PF4), which we previously identified by microarray and confirmed by real-time PCR, were among the top 100 genes identified by both platforms in the current study. Gene Ontology analysis showed overrepresentation of genes involved in apoptosis among the concordant genes while pathway analysis identified pathways related to lipid metabolism. The two-platform approach allows for robust changes in the PBMC transcriptome of benzene-exposed individuals to be identified.
Keywords: benzene exposure, gene expression, human blood, toxicogenomics
INTRODUCTION
Benzene is an established cause of leukemia and a possible cause of lymphoma in humans [1]. A possible mechanism underlying these pathologies is the induction by benzene of genetic changes leading to chromosome aberrations, translocations, aneuploidy and long-arm deletions [2,3] along with alterations in cell differentiation and immune surveillance. Benzene is hematotoxic, causing a decrease in total white blood cells, granulocytes and lymphocytes even among workers with relatively low-level exposure to benzene [4]. Benzene is thought to lower blood cell counts via metabolite effects on hematopoietic progenitor cells [4,5]. Depression of the mitogenic response of B and T lymphocytes, as well as impairment of macrophage activity, also result from benzene exposure [6]. Damage to the bone marrow stromal microenvironment is another aspect of benzene-associated hematoxicity [7,8]. Individual susceptibility to the genotoxic and hematotoxic effects of benzene is mediated through polymorphisms in DNA-repair genes [9], cytokine and cell adhesion genes [10], and genes involved in benzene metabolism [4,11].
While pathological outcome and susceptibility studies have generated some understanding of the mechanisms of action of benzene, global gene expression studies have the ability to inform on a more detailed level the involvement of specific genes and molecular pathways. The p53-dependent nature of benzene toxicity and carcinogenesis was revealed by examination of gene expression changes in mouse bone marrow (BM) in response to a 2-week exposure to inhaled benzene at 300 ppm [12]. Gene expression in mouse hematopoietic stem cells (HSC) exposed to inhaled benzene (100 ppm) implicated a number of response pathways including apoptosis, growth control of damaged HSC, repair of damaged DNA, and HSC growth arrest [13]. We previously identified several genes (ZNF331, CXCL16, JUN, and PF4) altered by benzene in peripheral blood mononuclear cells (PBMC) from benzene-exposed (>10 ppm) workers compared with unexposed controls. The genes were identified by the application of highthroughput microarray analysis to discover potential biomarkers and relatively low-throughput real-time PCR for confirmation [3].
In order to confirm previous findings and to discover more differentially expressed genes associated with benzene exposure, in the current study we analyzed more samples using the Affymetrix microarray platform, and have expanded the study to include a second microarray platform (Illumina). Recent reports have shown good inter-platform reproducibility of gene expression measurements between these two platforms [14]. The approach integrates highthroughput confirmation with discovery, helping to further elucidate genetic pathways and mechanisms underlying hematoxicity induced by benzene exposure.
RESULTS
The PBMC transcriptome of 8 individuals occupationally exposed to benzene compared with 8 matched controls was examined on two microarray platforms. Distinct processing protocols appropriate to each platform, from labeling through hybridization and detection, were applied. Data was analyzed using a novel Quantile Transformation approach [15].
Cross-Comparison of Genes Associated with Benzene Exposure by Affymetrix and Illumina Microarray Platforms
On the Affymetrix platform, 2692 genes (represented by 3549 probes) were differentially expressed (raw p ≤ 0.05). Considering genes with expression levels altered by 1.5-fold or greater, 65 genes were down-regulated while 180 genes were up-regulated. On the Illumina platform, 1828 genes (1856 probes) were differentially expressed (raw p ≤ 005) Modification of expression levels by 1.5-fold and higher occurred in 171 genes (88 down-regulated and 83 up-regulated). Supplementary Tables S1 and S2 contain lists of all genes identified as significant by Affymetrix and Illumina platforms, respectively, and show multiple test correction values. Among the Affymetrix data, 15 genes remained significant after multiple testing using the Quantile Transformation (QT) approach while 1 gene among the Illumina data remained significant.
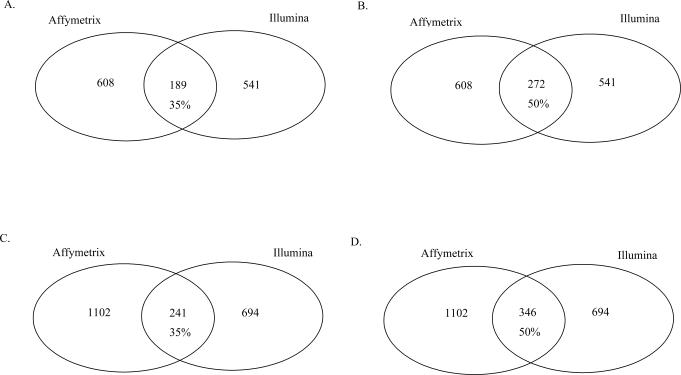
The subset of genes that was identified as differentially expressed (based on raw p-values) in common by both platforms was determined using two approaches. First, only those genes identified by each platform which were common to a stringent platform comparison reference file (described in Material and Methods), were analyzed by the same approach used to analyze the full complement of genes from each platform. This approach yielded 1345 significant genes by Affymetrix and 1275 genes by Illumina, which were directly comparable based on the reference file (supplementary Tables S3 and S4, respectively). We determined the number of genes with a fold-change ≥ 1.3 (up or down) on both platforms and found a concordance of 35 % (189 genes), Figure 1A. We also determined the number of common genes with an average fold-change ≥ 1.3 (from both platforms) and found a concordance of 50 % (272 genes), Figure 1B.
Figure 1.
Concordance between platforms in the identification of genes induced by benzene exposure. The numbers of significant genes (P ≤ 0.05) from the analysis of differential expression performed by t-test/quantile transformation (QT) are shown. A. Genes with fold-change ≥ 1.3-fold on both platforms, based on the stringent comparison data set. B. Genes with an average fold-change ≥ 1.3-fold, based on the stringent comparison data set. C. Genes with a fold-change ≥ 1.3-fold on both platforms, based on the complete data set. D. Genes with an average fold-change ≥ 1.3-fold, based on the complete data set.
In the second, less stringent approach, all of the significant genes identified by each platform (2692 by Affymetrix and 1828 by Illumina) were compared by gene symbol. We determined the number of genes with a fold-change ≥ 1.3 on both platforms in this less stringent data set and found a concordance of 35 % (241 genes), Figure 1C. As above, we also determined the number of common genes with an average fold-change ≥ 1.3 (from both platforms) and found a concordance of 50 % (346 genes) similar to that obtained in the stringent comparison dataset, Figure 1D.
All concordant genes are listed in supplementary Table S5. Expression ratios were similar among the concordant genes (mean difference in expression ratio = 0.13, standard deviation = 0.12). Among the common genes, 57 genes were down-regulated while 66 genes were up-regulated, by 1.5-fold or greater. Four genes (CXCL16, ZNF331, JUN and PF4), which we previously identified by microarray and confirmed by real-time PCR, were identified by both platforms in the current study. JUN and ZNF331 are among the top 20 common genes (ranked by p-value) which are listed in Table 1. Both of these genes remained significant after correction for multiple testing (Quantile Transformation) of the Affy data set (JUN QT-p = 0.044; ZNF331 QT-p = 0.042), while ZNF331 remained significant (QT-p = 0.039) and JUN (QT-p = 0.072) approached significance upon correction of the Illumina data set. Other genes of note among the top 20 are HSPA1A and HSPA1B, members of the heat-shock 70 (HSP70) multigene family.
Table 1.
Top 20 genes associated with benzene exposure cross-validated by Affymetrix and Illumina microarray platforms
| Affymetrix |
RefSeq ID | Illumina |
||||||
|---|---|---|---|---|---|---|---|---|
| ID | p-value* | Ratio† | Gene Title | Symbol | ID | p-value* | Ratio | |
| Downregulated (N=6) | ||||||||
| 202581_at | 3.98E-05 | 0.18 | heat shock 70kDa protein 1B | HSPA1B | NM_005346 | GI_26787974-S | 3.74E-05 | 0.19 |
| 200799_at | 6.57E-05 | 0.34 | heat shock 70kDa protein 1A | HSPA1A | NM_005345 | GI_26787973-S | 1.48E-04 | 0.32 |
| 201466_s_at | 1.47E-05 | 0.39 | v-jun sarcoma virus 17 oncogene homolog (avian) | JUN | NM_002228 | GI_44890066-S | 2.43E-05 | 0.33 |
| 208960_s_at | 1.45E-02 | 0.55 | Kruppel-like factor 6 | KLF6 | NM_001008490 | GI_37655156-S | 1.10E-04 | 0.41 |
| 229054_at | 4.37E-07 | 0.60 | FLJ39779 protein | FLJ39779 | NM_207442 | GI_42660305-S | 1.92E-04 | 0.51 |
| 202014_at | 1.33E-03 | 0.62 | protein phosphatase 1,regulatory (inhibitor) subunit 15A | PPP1R15A | NM_014330 | GI_9790902-S | 4.12E-05 | 0.55 |
| Upregulated (N=14) | ||||||||
| 201939_at | 1.05E-03 | 1.82 | polo-like kinase 2 (Drosophila) | PLK2 | NM_006622 | GI_5730054-S | 1.45E-04 | 1.62 |
| 227613_at | 8.78E-03 | 1.82 | zinc finger protein 331 | ZNF331 | NM_018555 | GI_20127571-S | 6.38E-06 | 1.62 |
| 235568_at | 5.09E-04 | 1.81 | mast cell-expressed membrane protein 1 | MCEMP1 | NM_174918 | GI_27885012-S | 9.11E-05 | 1.95 |
| 202856_s_at | 5.84E-04 | 1.76 | solute carrier family 16 member 3 | SLC16A3 | NM_004207 | GI_4759111-S | 3.99E-04 | 1.69 |
| 216248_s_at | 8.94E-03 | 1.74 | nuclear receptor subfamily 4, group A, member 2 | NR4A2 | NM_173173.1 | GI_27894352 | 3.94E-04 | 1.47 |
| 200768_s_at | 2.47E-03 | 1.64 | methionine adenosyltransferase II, alpha | MAT2A | NM_005911 | GI_34147493-S | 1.04E-04 | 1.98 |
| 218421_at | 6.52E-04 | 1.51 | ceramide kinase | CERK | NM_022766 | GI_32967302-A | 3.86E-04 | 1.40 |
| 209272_at | 2.24E-02 | 1.44 | NGFI-A binding protein 1 (EGR1 binding protein 1) | NAB1 | NM_005966 | GI_19923347-S | 2.46E-04 | 1.47 |
| 219862_s_at | 5.74E-04 | 1.38 | nuclear prelamin A recognition factor | NARF | NM_001038618 | GI_14165459-A | 1.84E-05 | 1.35 |
| 217964_at | 1.81E-02 | 1.31 | tetratricopeptide repeat domain 19 | TTC19 | NM_017775 | GI_22547158-S | 2.05E-04 | 1.37 |
| 223093_at | 2.27E-02 | 1.26 | ankylosis, progressive homolog (mouse) | ANKH | NM_054027 | GI_34452701-S | 3.99E-04 | 1.32 |
| 36554_at | 5.28E-03 | 1.25 | acetylserotonin O-methyltransferase-like | ASMTL | NM 004192 | GI_ 4757793-S | 1.02E-04 | 1.29 |
| 223740_at | 2.28E-03 | 1.24 | chromosome 6 open reading frame 59 | C6orf59 | NM_024929.1 | GI_13376403 | 3.16E-04 | 1.55 |
| 205791_x_at | 8.96E-03 | 1.23 | zinc finger protein 155, transcript variant 1 | ZNF155 | NM_198089 | GI_37655172-A | 2.63E-04 | 1.29 |
The central colums list gene title symbol, symbol and RefSeq ID, while platform-specific IDs, p-values, and differential expression ratios (relative to control) are detailed on the left (Affymetrix) and right (Illumina).
Raw p-values i.e. not adjusted for multiple testing are shown.
Differential Expression ratio.
Classification of Genes by Gene Ontology and Pathway Analyses
Several GO categories were identified by GOstat as enriched among both the Affymetrix and Illumina datasets (genes with ≥ 1.5-fold up- or down-regulation). These GO categories, as well as their associated genes, are listed in Table 2 and include immune response, defense response, and response to stress, suggesting concordance between the two platforms at the functional level. Genes involved in apoptosis were significant among the Affymetrix dataset but not the Illumina dataset. Among the genes identified by both platforms, GOstat analysis showed significant association with the GO term apoptosis (GO:0006915; p-value of 0.0113), but only when all genes were included regardless of the magnitude of the fold-change.
Table 2.
Functional classification of genes modified by benzene exposure.
| GO Term | Genes† | Illumina | Affymetrix | ||
|---|---|---|---|---|---|
| p-value‡ | No. Genes | p-value‡ | No. Genes | ||
| Apoptosis | RHOB; BCL2; SPP1; CIAS1; CUL4A; TNFSF14; NFKB1; ANXA1; STK17B; PPP1R15A; TRAF3; IFI6; CTSB; MAEA; BID; TLR2; HSPA1A; MX1 | - | - | 0.0208 | 18 |
| Immune response | BCL2, CLEC5A, CXCL16, IFI6, IFNG, IL1R2, IL21R, ISG15, KLF6, MX1, PLA2G7, TNFSF14 (Overlap N = 12) | 2.58E-05 | 21 | 4.04E-14 | 37 |
| Defense response | BCL2, CD69, CLEC5A, CXCL16, HIST2H2BE, IFI6, IFNG, IL1R2, IL21R, ISG15, KLF6, MX1, PLA2G7, TNFSF14 (Overlap N = 14) | 8.23E-07 | 25 | 1.218E-13 | 39 |
| Response to stress | BCL2, CLEC5A, CXCL16, DNAJB1, DUSP1, HIST2H2BE, HSPA1A, IFI6, IFNG, ISG15, MX1, PLA2G7, PPP1R15A, SRXN1 (Overlap N = 14) | 7.26E-06 | 26 | 2.17E-06 | 35 |
| Inflammatory Response | NFKB1, CCR5, ANXA1, PLA2G7, SPP1, CIAS1, FPRL1, CHST2, CXCL16, C3AR1, TLR2, IL1R1, CD93 | - | - | 0.00121 | 13 |
| Chromatin Assembly | H2BFS; HIST1H1C; HIST2H2AC; HIST1H2AC; HIST2H2BE; HIST1H2BH; HIST2H2AA3 | 0.000819 | 7 | - | - |
*GOStat was used to assess for enrichment of GO terms among the genes with significant differential expression ≥ 1.5-fold from both the Affymetrix and Illumina analyses.
Genes associated with significant GO terms are listed; in the case of a GO term being associated with both platforms common genes are listed.
A χ2 test is used to generate a p-value and adjustment for multiple comparisons is based on False Discovery Rate (FDR).
Ingenuity canonical pathway analysis identified significant pathways among the common genes as well as among the significant genes from both platforms. Significant pathways and associated genes are shown in Table 3. Lipid metabolism was a key theme among the common genes with involvement of ganglioside biosynthesis, glycerolipid metabolism, glycerphospholipid metabolism and sterol biosynthesis pathways. From the Affymetrix dataset protein ubiquitination was strongly impacted with 23 genes up-regulated and 4 genes down-regulated. Among the Illumina dataset multiple pathways were involved as shown in Table 3.
Table 3.
Canonical pathways impacted by benzene exposure.
| Pathway* | Genes | Affymetrix | Illumina | Common | |||
|---|---|---|---|---|---|---|---|
| p-value† | No. Genes | p-value | No. Genes | p-value | No. Genes | ||
| Death receptor Signaling | BCL2, CFLAR, CRADD, FASLG, IKBKE, NFKB2, TNFRSF1A | - | - | 0.016 | 7 | - | - |
| ERK/MAPK signaling | DUSP1, DUSP4, FOS, MYC, PIK3R1, PPARG, PPP1CB, PPP1R10, PRKAR2B, RPS6KA1, SRC, STAT3 | - | - | 0.024 | 12 | - | - |
| Ganglioside biosynthesis | ST3GAL1, ST3GAL4, ST3GAL5, ST6GALNAC2, B3GALT4, ST8SIA4 | 0.046 | 4 | 0.00034 | 5 | 0.043 | 2 |
| Globoside metabolism | B3GALT3, HEXA, ST3GAL1, ST8STA4 | - | - | 0.022 | 4 | - | - |
| Glycerolipid metabolism | AGPAT4, AGPAT6, CERK, DHRS9, DGAT2, GK, LAC89944, LPL, PPAP2B | - | - | 0.026 | 9 | 0.019 | 6 |
| Glycerophospholipid metabolism | CERK, PPAP2B, HMOX1, PAFAH1B1,PLAG2G7 | - | - | - | - | 0.017 | 5 |
| Il-10 signaling | CCR5, FOS, HMOX1, IKBKE, IL1R2, JUN, NFKB2, RELB, STAT3 | - | - | 0.0031 | 9 | 0.004 | 6 |
| IL-6 signaling | ABCB1, FOS, IKBKE, IL1R2, JUN, MAPKAPK2, NFKB2, STAT3, TNFRSF1A | - | - | 0.0081 | 9 | - | - |
| PDGF signaling | ABL1, FOS, JUN, MYC, PIK3R1, SRC, STAT3 | - | - | 0.021 | 7 | - | - |
| PPAR signaling | FOS, IKBKE, IL1R2, JUN, NCOA1, NFKB2, PPARG, RXRA, STAT5A, TNFRSF1A | - | - | 0.0017 | 10 | - | - |
| Protein ubiquitination | ANAPC1, BAP1, BIRC4, BTRC, CUL1, IFNG, PSMA3, PSMB4, PSMC2, PSMC4, PSMD2, PSMD4, PSMD11, PSMD12, SMURF2, UBC, UBE2I, UBE2Q1, USP3, USP18, USP24, USP28, USP33, USP36, USP39, USP47, USP9X | 0.0017 | 27 | - | - | - | - |
| Sterol biosynthesis | FDFT1, HMGCR, MVD, SC5DL | - | - | 0.025 | 4 | 0.019 | 3 |
| Toll-like receptor signaling | FOS, JUN, NFKB2, RELB, TLR2, TOLLIP | - | - | 0.031 | 6 | - | - |
Derived from Ingenuity® Pathway Analysis of Affymetrix, Illumina and common datasets.
Fischer's exact test is used to calculate the p-value.
Evaluation of Transcripts Discordant between Platforms
As described above the overall concordance between the two platforms was 35-50 %. The most significant genes (Top 50 by p-value) identified by either platform were more likely to be ranked as significant by the other platform. Thus of the top 50 genes identified by Illumina, 31 genes were ranked as significant by Affymetrix, raising the concordance to 62 %. Similarly, 27 of the top 50 genes identified by Affymetrix also appeared in the Illumina dataset (concordance 59 %).
Some genes, which were significant on one platform, approached significance on the other platform. For example, C3AR1, which was up-regulated 2-fold on Affymetrix, approached significance on the Illumina platform (p-value = 0.056, ratio 1.38). In the case of FYN, which was up-regulated significantly (1.4-fold) by Affymetrix, the Illumina platform also detected up-regulation (1.2-fold) but only approached significance (p = 0.07) and therefore did not appear in the list of common genes. As both of these genes were present in the stringent comparison file, this suggests that the two platforms may differ in their ability to detect small changes in expression for specific probes.
DISCUSSION
We identified robust changes in gene expression in response to benzene exposure in 8 occupationally exposed individuals compared with 8 unexposed controls, by cross-comparison using two microarray platforms (Affymetrix and Illumina). This approach enabled identification of a greater number of robust biomarkers than our previous approach of single-platform array analysis in conjunction with quantitative PCR confirmation.
A total of 346 genes were cross-validated by our two-platform approach. Further validation was provided by the fact that four genes (CXCL16, ZNF331, JUN and PF4), which we previously showed to be highly significantly associated with benzene exposure [3], were present in the cross-validated dataset from the current study. JUN was previously shown to be down-regulated by benzene exposure in mouse HSC [13]. Expression of FOSB expression was also down-regulated (~1.6-fold) by both platforms in the current study. JUN and FOS are basic regionleucine zipper (bZIP) members of the AP-1 transcription complex [16], which modulates the decision of a cell to proliferate, differentiate, or die by apoptosis [17]. Since JUN promotes proliferation of many cell types [17], reduced levels of JUN could indicate that the PBMCs of benzene-exposed individuals are not proliferating or progressing through the cell cycle as quickly as those of non-exposed individuals. Platelet Factor 4 (PF4), a chemokine secreted from activated platelets [18], activated T cells and mast cells [19], is a chemoattractant for neutrophils and fibroblasts and plays a role in inflammation and wound repair. PF4 was down-regulated in the current study in agreement with previous observations [3,20].
We used gene ontology and pathway analyses to discern potential underlying biology from the data. GO analysis showed enrichment in genes involved in apoptosis among the 346 common genes, while pathway analysis identified an impact on lipid metabolism. Plasma cholesterol (and phospholipids) were found previously to be slightly elevated in rat liver following a 28-day oral benzene exposure [21]. Lipid levels have been shown to be altered in hematological disorders including acute leukemia and non-Hodgkin lymphoma [22], CLL [23] and ALL [24] and might represent a novel therapeutic target [23]. It is unclear whether changes in lipid metabolism are causal in malignancy or arise as a consequence of the disease process. It is known that changes in lipid metabolism occur during infection and that TNF and other cytokines are capable of altering lipid metabolism in a variety of tissues leading to hypertriglyceridemia [25,26]. The potential link between benzene exposure, lipid metabolism and leukemogenesis is unclear. One possibility is that maintenance of the phopsholipid membrane may be compromised during oxidative stress arising from benzene exposure.
The fact that PBMC profiling reflects liver gene expression is not surprising in view of a recent study showing that the peripheral blood transcriptome dynamically reflects systems wide biology with 83% of liver genes also expressed in blood [27]. Genes involved in the GO categories of immune response, stress response and defense response were enriched in the separate platform datasets, with overlap of genes between platforms. Therefore, despite the fact that concordance between the microarray platforms was 35-50 %, several mechanisms (concordance at the pathway level) underlying benzene effects in human PBMC are in close agreement and fit well with the phenotypic effects of benzene including decreased blood cell counts [5], and depression of the immune system [6].
Chromatin assembly was identified as an overrepresented GO category in the Illumina data only. If confirmed, it could represent a potential mechanism by which benzene causes leukemia as histone proteins are involved in the regulation of DNA transcription, replication, repair and recombination and post-translational modifications on histone tails epigenetically regulate the genome-wide transcriptome. Reduction of histone levels such as by the Human T Lymphotropic Virus Type 1 protein, Tax [28], ionizing radiation (IR) [29], adriamycin [29], and if confirmed, benzene, may directly induce chromosomal instability and deregulate gene expression, leading to cancer. Reduced histone levels have been proposed to have similar effects to those observed with loss of imprinting (LOI) through DNA hypomethylation, including aberrant chromosome rearrangements, deregulation of cellular gene expression and activation of latent viral genomes [28]. Chromatin structure, nucleosome remodeling and histone tail modifications influence double-strand DNA break repair [30-33] and defects in chromatin assembly have been shown to impair double-strand break repair and activate S phase arrest [34,35].
In the current study the Bax:Bcl2 ratio, an indicator of the degree of apoptosis was 1:0.5, suggesting a shift towards apoptosis. Down-regulation of two anti-apoptotic hsp70-encoding genes [36] was also observed in the current study. Increased apoptosis is a mechanism that could potentially underlie benzene-associated leukemia, one theory being that cells escape from apoptosis with DNA breaks which can result in chromosomal translocation [37-39], and induction of apoptosis in hematopoietic progenitor cells [40,41] and cell lines [42] by benzene metabolites has been previously demonstrated. Removal of cells predestined to die by apoptosis is facilitated, at least in some tissues by macrophages [43,44], and dysfunction of macrophages may lead to survival of cells that would otherwise have been removed. Poisoning of the BM stromal environment [7], particularly macrophages [8], is a hematotoxic effect of benzene.
While overall concordance between the two microarray platforms was 35-50 %, the most significant genes identified by either array were much more likely to be ranked as significant by the other platform. While very high concordance levels have been reported (~90%) between the Affymetrix and Illumina platforms, these were based on extremely different biological samples with large fold changes in expression [14]. Smaller concordance levels were seen when comparing less biologically similar samples [14] or analyzing rat toxicogenomic data [45]. Our study was based on occupationally-exposed individuals with inherent inter-individual variability in baseline expression, determined by factors such as blood count, blood type, genotype, presence of subclinical infection [46], which are less easy to control for by study design as are factors such as age and gender. Other explanations of microarray data discordance have been discussed [14,47]. Our inter-platform concordance is higher than that of two studies using a similar approach to ours, which showed average concordances of 22.8% [48] and ~10%) [49].
As well as concordance among individual genes, similar mechanisms of benzene effect such as response to stress and immune response were found in our study. However the results potentially caution against use of a single platform to identify biomarkers/pathways in human exposure studies, in which subtle perturbations may be detected. Our data also suggest the need to be very stringent in the selection of potential biomarkers based on a single platform as platform concordance was much higher among the most highly significant genes.
Challenges are inherent to this type of molecular epidemiology study. While many potential biomarkers of benzene exposure were generated, few genes remained significant after multiple testing (QT p-value ≤ 0.05). Another challenge is the biological relevance of small fold-changes in gene expression. Increasing the number of individuals studied is one way to increase the power to select true biomarkers. Cross-comparison by two platforms increases the chances that the genes identified in our study represent true potential biomarkers, but validation of biomarkers in a larger population is also necessary, at both the RNA and protein levels. While limited sample material precluded the validation of the microarray findings by QPCR in the current study, use of two microarray platforms offers a type of inherent validation in that distinct processing protocols appropriate to each platform, from labeling through hybridization and detection, were applied.
The question of the appropriateness of PBMC as a cell target in which to examine benzene's hematotoxic effects must also be addressed. While some of benzene's immunotoxic effects are thought to involve damage to BM stromal cells [7] and early progenitor cells [4] the knock-on effects of damage to these cells might be expected to be manifest in the transcriptome of their downstream cell targets. As discussed above, the peripheral blood transcriptome dynamically reflects system wide biology [27]. However, relevant changes may be masked by looking at heterogenous populations of cells e.g. BM compared with HSC [13]. Many of the genes identified in this study are expressed in several cell types and have pleiotropic effects, making it challenging to induce function and mechanism when examining PBMC. Further, as benzene exposure has been shown to decrease all types of white blood cells and platelets, by 16% (lymphocytes) to 35.8% (B cells) [4], some of the observed changes in expression could reflect these altered cell populations. However, PBMC are convenient for molecular epidemiology research studies and pathways and mechanisms identified as potentially impacted by benzene exposure such as apoptosis, can be further tested in in vitro studies using targeted cell subsets.
In conclusion, we have demonstrated robust changes in the PBMC transcriptome of benzene-exposed individuals, using a two-platform approach. The genes identified contribute to further understanding of the mechanisms underlying benzene-induced hematoxicity and leukemia.
MATERIALS AND METHODS
Study Subjects
Eight highly exposed workers (mean air benzene level ± SD = 39.0 ± 25.5 ppm) and eight unexposed controls (<0.04 ppm) who were frequency-matched to these subjects on the basis of age and gender, were chosen from a large molecular epidemiology study [50] investigating occupational exposure to benzene. Six subject pairs (twelve individuals) were included in a previous study [3] and another pair was chosen to give a better balance among the subjects for the confounders of age and gender. The study was approved by institutional review boards at all institutions. Participation was voluntary, written informed consent was obtained, and the participation rate was approximately 95%.
Four pairs were male and the other four female. Mean age was 33.5 ± 7.0 years for the eight exposed workers and 35.4 ± 7.0 years for the controls. Four of the exposed workers and one of the control workers were smokers. Mean white blood cell count (mean ± SD cells/μL blood) was 4812.5 ± 974.6 for the exposed workers compared with 5762.5 ± 1785.6 for controls (normal range for Chinese population 4000 - 10,000). Similarly, in the exposed workers granulocyte (2862.5 ± 652.3) and platelet (160750 ± 36405.5) counts were lower than in the control workers (granulocyte 3562.5 ± 1261.5, platelet 209875 ± 74299.5), but still within the normal range for the Chinese population (2000 - 8000, 100,000 - 361,500, respectively). Hemoglobin (g/L) was 139.9 ± 15.5 in the exposed workers and 145.8 ± 19.5 in the controls (normal range 110 - 170). Among all 390 subjects (140 controls and 250 benzene-exposed) in the overall molecular epidemiology study, all types of white blood cells (WBCs) measured and platelets were significantly decreased in workers exposed to >10 ppm benzene (and also in those exposed to doses as low as <1ppm benzene) compared to controls [4]. Exposure assessment, biologic sample collection and RNA isolation were described previously [3,4]. Workers had been employed an average of 6.1 ± 2.9 years and individual benzene exposure was monitored repeatedly up to 16 months before phlebotomy. A single RNA isolation was performed from each individual and stored in aliquots.
Affymetrix microarray analysis
The Affymetrix Human U133 GeneChip set containing ~44,000 probes targeting >39,000 unique transcripts derived from approximately 33,000 well-substantiated genes, are included in this chip set. The complete protocol used for Affymetrix microarray analysis was described previously [3].
Illumina Microarray analysis
RNA samples, with A260:A280 ratios between 1.7 and 2.1, and with integrity confirmed by denaturing agarose gel electrophoresis, were labeled using the Illumina® RNA Amplification kit (Ambion, Austin, TX). Samples were reverse transcribed in 20μL reactions comprising 200 ng sample RNA, 1X First Strand Buffer, dNTPs, RNase inhibitor, and ArrayScript enzyme. Reactions were incubated at 37°C for 2 hr after which components of the second strand synthesis reaction including 10X Second Strand Buffer, dNTP mix, DNA polymerase, and RNase H were added to yield a final reaction volume of 100μL. Reactions were incubated at 16°C for 2hr and the resulting cDNAs were purified. cDNA binding buffer (250μL) was added to each reaction which was then mixed and passed through a cDNA filter cartridge by centrifugation at 10,000 × g for 1 min. Filters were washed with wash Buffer (500μL) and dried by centrifugation for an additional minute. cDNA was eluted using 2 × 10μL aliquots of Nuclease-free Water at 55°C. The purified cDNA was dried to completion in a vacuum centrifuge concentrator set to medium heat and resuspended in 10μL in vitro transcription (IVT) reaction mix comprising 1X reaction buffer, dNTP mix, biotin labeled UTP (10 mM; Roche Applied Science, Indianapolis, IN), and T7 enzyme. Reactions were incubated at 37°C for 14 hr after which volumes were adjusted to 100μL by addition of Nuclease-free water. cRNA Binding Buffer (350μL) and 100% ethanol (250μL) were added and mixed by pipetting before passing through a cRNA filter cartridge under centrifugation at 10,000 × g for 1 min. Filters were washed with wash Buffer (650μL) and dried by centrifugation for an additional minute. cDNA was eluted using 100μL of Nuclease-free Water at 55°C. cRNA was quantified using nce-based assay the RiboGreen® fluorescence-based assay (Invitrogen, Carlsbad, CA).
Hybridization, washing and detection were performed using the Illumina Gene Expression System Buffer Kit for HumanRef-8 BeadChips (Illumina, San Diego, CA) according to the manufacturer's protocol. An aliquot containing 850 ng cRNA was transferred to a new tube and adjusted to a volume of 11.3 μL. Hybridization Mix was prepared by mixing Hyb E1 buffer (125 μL), which had been prewarmed in a 55°C oven for 10 min, to formamide (75μL). Hybridization Mix (22.7 μL) was added to each cRNA sample. Following sample incubation at 65°C for 5 min, 34 μL was dispensed onto the center of each HumanRef-8 BeadChip onto array. BeadChips were assembled onto Hybridization cartridges, mixed by shaking to ensure bubbles moved freely, and then placed on the BeadChip Hyb Wheel and incubated for 16 hr at 55°c rotation.
Following hybridization, a manual washing procedure was followed in which BeadChips were placed in a slide rack and washed in supplied solutions in glass staining dishes. Slide racks were plunged in and out of the appropriate solution 5-10 times and then mixing was performed on an orbital shaker (Thermolyne Roto Mix, Type 50800, Barnstead International, Dubuque, IA) at highest possible speed or on a rocker shaker (Rocker II 260350, Boekel Scientific, Feasterville, PA) at medium-high speed, for the times indicated. Following hybridization, slides were washed successively in wash E1BC solution (250 mL), 100% ethanol (250 mL), and fresh Wash E1BC solution (250 mL), with 15 min, 10 min and 2 min orbital shaking, respectively. In order to block the slides, BeadChips were placed face-up in a wash tray (supplied) containing Block E1 buffer (4 mL) and rocked for 10 min. For detection BeadChips were transferred to a fresh wash tray containing Block E1 buffer (2 mL) containing streptavidin-Cy3 (1μg/mL; Amersham Biosciences, Piscataway, NJ) and rocked for 10 min. Slides were then placed in a staining rack and washed in Wash E1BC solution (250 mL) with 5 minutes of orbital shaking. The slides were then dried by centrifugation at 275 × g for 4 min at 20°C in a Jouan CR4.22 centrifuge (Thermo Electron Corporation, Waltham, MA) and stored in the dark until scanned. Scanning was performed using a BeadArray Reader and BeadScan software (Illumina).
Data Analysis
Raw data files for each microarray experiment have been deposited at GEO, accession number Series GSE9569 (GSM241938 through GSM243811) and access is available at: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=dhovtgesoygsyng&acc=GSE9569. Data was normalized by quantile normalization using Bioconductor (Affymetrix data by RMA and Illumina data by “Affy” package) and two-sample Welch t-statistics (unequal variance) were calculated. We used a multiple testing procedure that controls familywise error rates (FWER) and has been shown to provide sharp control (for instance relative to the standard Bonferroni procedure) by accounting for the strong correlation of gene expression measures typically observed in microarray studies [51]. It works using a re-sampling based technique (bootstrapping) to generate a sample of the test statistics from the empirical joint distribution of the data and then quantile transforms these random samples into the appropriate marginal null distribution (in this case, the t-distribution). Then, the observed test statistics are compared to the maximum of each random draw from this null distribution to derive the estimated FWER for various cut-offs in a list ordered by statistical significance. In the supplementary tables, we also provide the more traditional (and typically conservative) methods for reporting FWER.
Comparison of array platforms
In order to determine comparable targets from the Affymetrix (Human U133 GeneChip set; ~45,000 probe sets targeting 39,000 transcripts from 33,000 well-substantiated genes) and Illumina (HumanRef-8 BeadChip; > 23,000 RefSeq-curated gene targets) platforms, probe sequences from each platform and transcript sequences from RefSeq Release 13 (http://www.ncbi.nlm.nih.gov/RefSeq/) were compared. For both platforms, probes that were not valid were filtered out. A probe was defined as valid if it perfectly matched a transcript sequence and did not perfectly match any other transcript sequences with a different gene symbol. If a transcript sequence contained multiple valid probes, the one closest to the 3' end of the transcript was selected. As cDNA synthesis is primed by olgodT primers from the polyA tails of the mRNAs, this minimizes the effects of RNA degradation on cDNA integrity. Based on these criteria 14,708 targets were included in the cross-platform analysis.
In a second approach, all the significant genes identified by each platform (2692 by Affymetrix and 1828 by Illumina) were subjected to an ID conversion program called Gene Expression Pattern Analysis Suite v3.1 (http://www.gepas.org) [52] and significant gene lists were then compared by gene symbol.
Pathway Analysis
Gene RefSeq accession numbers were imported into Ingenuity Pathway Analysis software (Ingenuity® Systems, Redwood City, CA, (www.ingenuity.com) a web-based application, which queries the Ingenuity Pathway Knowledge Base (IPKB) for genetic interactions. To evaluate the significance of the association of a particular gene set with the relevant canonical pathway within Ingenuity, a ratio of the number of genes from the data set that map to the pathway divided by the total number of genes that map to the canonical pathway is displayed and Fischer's exact test is used to calculate the corresponding p-value.
Gene Ontology Analysis
A publicly available tool was applied to assess enrichment of Gene Ontology (GO) terms over that which would be expected by chance alone. In GOstat [50], a χ2 test is used to generate a p-value. Adjustment for multiple comparisons is based on False Discovery Rate (FDR).
Supplementary Material
ACKNOWLEDGEMENTS
We thank the participants for taking part in this study. Supported by NIH grants RO1ES06721 and P42ES04705 (to Martyn T. Smith), and P42ES05948 and P30ES10126 (to Stephen M. Rappaport) from the National Institute of Environmental Health Sciences and Intramural Funds from the National Cancer Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Hayes RB, Songnian Y, Dosemeci M, Linet M. Benzene and lymphohematopoietic malignancies in humans. Am J Ind Med. 2001;40:117–26. doi: 10.1002/ajim.1078. [DOI] [PubMed] [Google Scholar]
- [2].Smith MT, Zhang L, Wang Y, Hayes RB, Li G, Wiemels J, Dosemeci M, Titenko-Holland N, Xi L, Kolachana P, Yin S, Rothman N. Increased translocations and aneusomy in chromosomes 8 and 21 among workers exposed to benzene. Cancer Res. 1998;58:2176–81. [PubMed] [Google Scholar]
- [3].Forrest MS, Lan Q, Hubbard AE, Zhang L, Vermeulen R, Zhao X, Li G, Wu YY, Shen M, Yin S, Chanock SJ, Rothman N, Smith MT. Discovery of novel biomarkers by microarray analysis of peripheral blood mononuclear cell gene expression in benzene-exposed workers. Environ Health Perspect. 2005;113:801–7. doi: 10.1289/ehp.7635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Lan Q, Zhang L, Li G, Vermeulen R, Weinberg RS, Dosemeci M, Rappaport SM, Shen M, Alter BP, Wu Y, Kopp W, Waidyanatha S, Rabkin C, Guo W, Chanock S, Hayes RB, Linet M, Kim S, Yin S, Rothman N, Smith MT. Hematotoxicity in workers exposed to low levels of benzene. Science. 2004;306:1774–6. doi: 10.1126/science.1102443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Yoon BI, Hirabayashi Y, Kawasaki Y, Kodama Y, Kaneko T, Kim DY, Inoue T. Mechanism of action of benzene toxicity: cell cycle suppression in hemopoietic progenitor cells (CFU-GM) Exp Hematol. 2001;29:278–85. doi: 10.1016/s0301-472x(00)00671-8. [DOI] [PubMed] [Google Scholar]
- [6].Snyder R. Overview of the toxicology of benzene. J Toxicol Environ Health A. 2000;61:339–46. doi: 10.1080/00984100050166334. [DOI] [PubMed] [Google Scholar]
- [7].Garnett HM, Cronkite EP, Drew RT. Effect of in vivo exposure to benzene on the characteristics of bone marrow adherent cells. Leuk Res. 1983;7:803–10. doi: 10.1016/0145-2126(83)90074-7. [DOI] [PubMed] [Google Scholar]
- [8].Thomas DJ, Sadler A, Subrahmanyam VV, Siegel D, Reasor MJ, Wierda D, Ross D. Bone marrow stromal cell bioactivation and detoxification of the benzene metabolite hydroquinone: comparison of macrophages and fibroblastoid cells. Mol Pharmacol. 1990;37:255–62. [PubMed] [Google Scholar]
- [9].Shen M, Lan Q, Zhang L, Chanock S, Li G, Vermeulen R, Rappaport SM, Guo W, Hayes RB, Linet M, Yin S, Yeager M, Welch R, Forrest MS, Rothman N, Smith MT. Polymorphisms in genes involved in DNA double-strand break repair pathway and susceptibility to benzene-induced hematotoxicity. Carcinogenesis. 2006;27:2083–9. doi: 10.1093/carcin/bgl061. [DOI] [PubMed] [Google Scholar]
- [10].Lan Q, Zhang L, Shen M, Smith MT, Li G, Vermeulen R, Rappaport SM, Forrest MS, Hayes RB, Linet M, Dosemeci M, Alter BP, Weinberg RS, Yin S, Yeager M, Welch R, Waidyanatha S, Kim S, Chanock S, Rothman N. Polymorphisms in cytokine and cellular adhesion molecule genes and susceptibility to hematotoxicity among workers exposed to benzene. Cancer Res. 2005;65:9574–81. doi: 10.1158/0008-5472.CAN-05-1419. [DOI] [PubMed] [Google Scholar]
- [11].Rothman N, Smith MT, Hayes RB, Traver RD, Hoener B, Campleman S, Li GL, Dosemeci M, Linet M, Zhang L, Xi L, Wacholder S, Lu W, Meyer KB, Titenko-Holland N, Stewart JT, Yin S, Ross D. Benzene poisoning, a risk factor for hematological malignancy, is associated with the NQO1 609C-->T mutation and rapid fractional excretion of chlorzoxazone. Cancer Res. 1997;57:2839–42. [PubMed] [Google Scholar]
- [12].Yoon BI, Li GX, Kitada K, Kawasaki Y, Igarashi K, Kodama Y, Inoue T, Kobayashi K, Kanno J, Kim DY, Inoue T, Hirabayashi Y. Mechanisms of benzene-induced hematotoxicity and leukemogenicity: cDNA microarray analyses using mouse bone marrow tissue. Environ Health Perspect. 2003;111:1411–20. doi: 10.1289/ehp.6164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Faiola B, Fuller ES, Wong VA, Recio L. Gene expression profile in bone marrow and hematopoietic stem cells in mice exposed to inhaled benzene. Mutat Res. 2004;549:195–212. doi: 10.1016/j.mrfmmm.2003.12.022. [DOI] [PubMed] [Google Scholar]
- [14].Shi L, Reid LH, Jones WD, Shippy R, Warrington JA, Baker SC, Collins PJ, de Longueville F, Kawasaki ES, Lee KY, Luo Y, Sun YA, Willey JM, Setterquist RA, Fischer GM, Tong W, Dragan YP, Dix DJ, Frueh FW, Goodsaid FM, Herman D, Jensen RV, Johnson CD, Lobenhofer EK, Puri RK, Schrf U, Thierry-Mieg J, Wang C, Wilson M, Wolber PK, Zhang L, Slikker W, Jr., Shi L, Reid LH. The MicroArray Quality Control (MAQC) project shows inter- and intraplatform reproducibility of gene expression measurements. Nat Biotechnol. 2006;24:1151–61. doi: 10.1038/nbt1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].van der Laan MJ, Hubbard AE. Quantile-function based null distribution in resampling based multiple testing. Stat Appl Genet Mol Biol. 2006;5 doi: 10.2202/1544-6115.1199. Article14. [DOI] [PubMed] [Google Scholar]
- [16].Shaulian E, Karin M. AP-1 as a regulator of cell life and death. Nat Cell Biol. 2002;4:E131–6. doi: 10.1038/ncb0502-e131. [DOI] [PubMed] [Google Scholar]
- [17].Hess J, Angel P, Schorpp-Kistner M. AP-1 subunits: quarrel and harmony among siblings. J Cell Sci. 2004;117:5965–73. doi: 10.1242/jcs.01589. [DOI] [PubMed] [Google Scholar]
- [18].Brandt E, Ludwig A, Petersen F, Flad HD. Platelet-derived CXC chemokines: old players in new games. Immunol Rev. 2000;177:204–16. doi: 10.1034/j.1600-065x.2000.17705.x. [DOI] [PubMed] [Google Scholar]
- [19].Boehlen F, Clemetson KJ. Platelet chemokines and their receptors: what is their relevance to platelet storage and transfusion practice? Transfus Med. 2001;11:403–17. doi: 10.1046/j.1365-3148.2001.00340.x. [DOI] [PubMed] [Google Scholar]
- [20].Vermeulen R, Lan Q, Zhang L, Gunn L, McCarthy D, Woodbury RL, McGuire M, Podust VN, Li G, Chatterjee N, Mu R, Yin S, Rothman N, Smith MT. Decreased levels of CXC-chemokines in serum of benzene-exposed workers identified by array-based proteomics. Proc Natl Acad Sci U S A. 2005;102:17041–6. doi: 10.1073/pnas.0508573102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Heijne WH, Jonker D, Stierum RH, van Ommen B, Groten JP. Toxicogenomic analysis of gene expression changes in rat liver after a 28-day oral benzene exposure. Mutat Res. 2005;575:85–101. doi: 10.1016/j.mrfmmm.2005.02.003. [DOI] [PubMed] [Google Scholar]
- [22].Kuliszkiewicz-Janus M, Malecki R, Mohamed AS. Lipid changes occuring in the course of hematological cancers. Cell Mol Biol Lett. 2008;13:465–74. doi: 10.2478/s11658-008-0014-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Pallasch CP, Schwamb J, Konigs S, Schulz A, Debey S, Kofler D, Schultze JL, Hallek M, Ultsch A, Wendtner CM. Targeting lipid metabolism by the lipoprotein lipase inhibitor orlistat results in apoptosis of B-cell chronic lymphocytic leukemia cells. Leukemia. 2008;22:585–92. doi: 10.1038/sj.leu.2405058. [DOI] [PubMed] [Google Scholar]
- [24].Moschovi M, Trimis G, Apostolakou F, Papassotiriou I, Tzortzatou-Stathopoulou F. Serum lipid alterations in acute lymphoblastic leukemia of childhood. J Pediatr Hematol Oncol. 2004;26:289–93. doi: 10.1097/00043426-200405000-00006. [DOI] [PubMed] [Google Scholar]
- [25].Grunfeld C, Feingold KR. The metabolic effects of tumor necrosis factor and other cytokines. Biotherapy. 1991;3:143–58. doi: 10.1007/BF02172087. [DOI] [PubMed] [Google Scholar]
- [26].Marshall MK, Doerrler W, Feingold KR, Grunfeld C. Leukemia inhibitory factor induces changes in lipid metabolism in cultured adipocytes. Endocrinology. 1994;135:141–7. doi: 10.1210/endo.135.1.8013346. [DOI] [PubMed] [Google Scholar]
- [27].Liew CC, Ma J, Tang HC, Zheng R, Dempsey AA. The peripheral blood transcriptome dynamically reflects system wide biology: a potential diagnostic tool. J Lab Clin Med. 2006;147:126–32. doi: 10.1016/j.lab.2005.10.005. [DOI] [PubMed] [Google Scholar]
- [28].Bogenberger JM, Laybourn PJ. Human T Lymphotropic Virus Type 1 protein Tax reduces histone levels. Retrovirology. 2008;5:9. doi: 10.1186/1742-4690-5-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Su C, Gao G, Schneider S, Helt C, Weiss C, O'Reilly MA, Bohmann D, Zhao J. DNA damage induces downregulation of histone gene expression through the G1 checkpoint pathway. Embo J. 2004;23:1133–43. doi: 10.1038/sj.emboj.7600120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Bao Y, Shen X. Chromatin remodeling in DNA double-strand break repair. Curr Opin Genet Dev. 2007;17:126–31. doi: 10.1016/j.gde.2007.02.010. [DOI] [PubMed] [Google Scholar]
- [31].Fillingham J, Keogh MC, Krogan NJ. GammaH2AX and its role in DNA double-strand break repair. Biochem Cell Biol. 2006;84:568–77. doi: 10.1139/o06-072. [DOI] [PubMed] [Google Scholar]
- [32].Karagiannis TC, El-Osta A. Epigenetic changes activate widespread signals in response to double-strand breaks. Cancer Biol Ther. 2004;3:617–23. doi: 10.4161/cbt.3.7.917. [DOI] [PubMed] [Google Scholar]
- [33].Osley MA, Tsukuda T, Nickoloff JA. ATP-dependent chromatin remodeling factors and DNA damage repair. Mutat Res. 2007;618:65–80. doi: 10.1016/j.mrfmmm.2006.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Lewis LK, Karthikeyan G, Cassiano J, Resnick MA. Reduction of nucleosome assembly during new DNA synthesis impairs both major pathways of double-strand break repair. Nucleic Acids Res. 2005;33:4928–39. doi: 10.1093/nar/gki806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Ye X, Franco AA, Santos H, Nelson DM, Kaufman PD, Adams PD. Defective S phase chromatin assembly causes DNA damage, activation of the S phase checkpoint, and S phase arrest. Mol Cell. 2003;11:341–51. doi: 10.1016/s1097-2765(03)00037-6. [DOI] [PubMed] [Google Scholar]
- [36].Garrido C, Gurbuxani S, Ravagnan L, Kroemer G. Heat shock proteins: endogenous modulators of apoptotic cell death. Biochem Biophys Res Commun. 2001;286:433–42. doi: 10.1006/bbrc.2001.5427. [DOI] [PubMed] [Google Scholar]
- [37].Khodarev NN, Sokolova IA, Vaughan AT. Abortive apoptosis as an initiator of chromosomal translocations. Med Hypotheses. 1999;52:373–6. doi: 10.1054/mehy.1997.0672. [DOI] [PubMed] [Google Scholar]
- [38].McHale CM, Smith MT. Prenatal origin of chromosomal translocations in acute childhood leukemia: implications and future directions. Am J Hematol. 2004;75:254–7. doi: 10.1002/ajh.20030. [DOI] [PubMed] [Google Scholar]
- [39].Vaughan AT, Betti CJ, Villalobos MJ, Premkumar K, Cline E, Jiang Q, Diaz MO. Surviving apoptosis: a possible mechanism of benzene-induced leukemia. Chem Biol Interact. 2005;153-154:179–85. doi: 10.1016/j.cbi.2005.03.022. [DOI] [PubMed] [Google Scholar]
- [40].Abernethy DJ, Kleymenova EV, Rose J, Recio L, Faiola B. Human CD34+ hematopoietic progenitor cells are sensitive targets for toxicity induced by 1,4-benzoquinone. Toxicol Sci. 2004;79:82–9. doi: 10.1093/toxsci/kfh095. [DOI] [PubMed] [Google Scholar]
- [41].Moran JL, Siegel D, Sun XM, Ross D. Induction of apoptosis by benzene metabolites in HL60 and CD34+ human bone marrow progenitor cells. Mol Pharmacol. 1996;50:610–5. [PubMed] [Google Scholar]
- [42].Inayat-Hussain SH, Ross D. Intrinsic pathway of hydroquinone induced apoptosis occurs via both caspase-dependent and caspase-independent mechanisms. Chem Res Toxicol. 2005;18:420–7. doi: 10.1021/tx049762o. [DOI] [PubMed] [Google Scholar]
- [43].Lang RA, Bishop JM. Macrophages are required for cell death and tissue remodeling in the developing mouse eye. Cell. 1993;74:453–62. doi: 10.1016/0092-8674(93)80047-i. [DOI] [PubMed] [Google Scholar]
- [44].Diez-Roux G, Lang RA. Macrophages induce apoptosis in normal cells in vivo. Development. 1997;124:3633–8. doi: 10.1242/dev.124.18.3633. [DOI] [PubMed] [Google Scholar]
- [45].Guo L, Lobenhofer EK, Wang C, Shippy R, Harris SC, Zhang L, Mei N, Chen T, Herman D, Goodsaid FM, Hurban P, Phillips KL, Xu J, Deng X, Sun YA, Tong W, Dragan YP, Shi L. Rat toxicogenomic study reveals analytical consistency across microarray platforms. Nat Biotechnol. 2006;24:1162–9. doi: 10.1038/nbt1238. [DOI] [PubMed] [Google Scholar]
- [46].Whitney AR, Diehn M, Popper SJ, Alizadeh AA, Boldrick JC, Relman DA, Brown PO. Individuality and variation in gene expression patterns in human blood. Proc Natl Acad Sci U S A. 2003;100:1896–901. doi: 10.1073/pnas.252784499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Canales RD, Luo Y, Willey JC, Austermiller B, Barbacioru CC, Boysen C, Hunkapiller K, Jensen RV, Knight CR, Lee KY, Ma Y, Maqsodi B, Papallo A, Peters EH, Poulter K, Ruppel PL, Samaha RR, Shi L, Yang W, Zhang L, Goodsaid FM. Evaluation of DNA microarray results with quantitative gene expression platforms. Nat Biotechnol. 2006;24:1115–22. doi: 10.1038/nbt1236. [DOI] [PubMed] [Google Scholar]
- [48].Cheadle C, Becker KG, Cho-Chung YS, Nesterova M, Watkins T, Wood W, 3rd, Prabhu V, Barnes KC. A rapid method for microarray cross platform comparisons using gene expression signatures. Mol Cell Probes. 2006 doi: 10.1016/j.mcp.2006.07.004. [DOI] [PubMed] [Google Scholar]
- [49].Elo LL, Katajamaa M, Lund R, Oresic M, Lahesmaa R, Aittokallio T. Improving identification of differentially expressed genes by integrative analysis of Affymetrix and Illumina arrays. Omics. 2006;10:369–80. doi: 10.1089/omi.2006.10.369. [DOI] [PubMed] [Google Scholar]
- [50].Beissbarth T, Speed TP. GOstat: find statistically overrepresented Gene Ontologies within a group of genes. Bioinformatics. 2004;20:1464–5. doi: 10.1093/bioinformatics/bth088. [DOI] [PubMed] [Google Scholar]
- [51].Chen J, van der Laan MJ, Smith MT, Hubbard AE. A comparison of methods to control type I errors in microarray studies. Stat Appl Genet Mol Biol. 2007;6 doi: 10.2202/1544-6115.1310. Article28. [DOI] [PubMed] [Google Scholar]
- [52].Herrero J, Diaz-Uriarte R, Dopazo J. Gene expression data preprocessing. Bioinformatics. 2003;19:655–6. doi: 10.1093/bioinformatics/btg040. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.