Abstract
Estrogen synthesis evolved in chordates to control reproduction. The terminal enzyme in the cascade directly responsible for estrogen synthesis is aromatase cytochrome P450 (P450arom) encoded by the CYP19 gene. Mammals typically have a single CYP19 gene but pigs, peccaries and other Suiformes have two or more resulting from duplication in a common ancestor. Duplication of CYP genes in the steroid synthetic cascade has occurred for only one other enzyme, also terminal, 11β-hydroxylase P450 (P450c11). P450arom and P450c11 share common substrates and even physiological functions as possible remnants from a common P450 progenitor, perhaps an ancestral P450arom, which is supported by phylogenetic analysis. Conserved tissue-specific expression patterns of P450arom paralogs in placenta and gonads of pigs and peccaries suggest how functional adaptation may have proceeded divergently and influenced adopted reproductive strategies including ovulation rate and litter size. Data suggest that the porcine placental paralog evolved catalytically to protect female conceptuses from testosterone produced by male siblings; the gonadal paralog to synthesize a novel, nonaromatizable testosterone metabolite (1OH-testosterone) that may increase ovulation rate. This would represent a coevolution facilitating litter bearing as pigs diverged from peccaries. Evidence of convergence between the peccary CYP19 genes and lower tissue expression may therefore represent initiation of loss of the functional paralogs. Studies on the Suiforme aromatases provide insights into the evolution of the steroidogenic cascade and metabolic pathways in general, how it translates into physiological adaptations (altered reproductive strategies for instance), and how duplicated genes become stabilized or disappear from genomes.
Aromatase cytochrome P450 (P450arom) is the terminal enzyme in the metabolic pathway that leads to the synthesis of estrogens (Fig. 1). As far as is known, it is expressed in the gonads (and brain or nervous system) of all vertebrates, even the protochordate amphioxus (Callard et al., ’84; Mizuta and Kubokawa, 2007), regulating estrogen production and reproductive function (Conley and Hinshelwood, 2001; Lange et al., 2002). The absolute dependence of fertility on estrogen synthesis from androgens by P450arom has no doubt contributed to conservation of the peptide sequence and the catalytic function of the enzyme. However, P450arom is just one of many proteins involved in the synthesis and metabolism of sex steroids, corticoids and a variety of other sterols, and how it evolved from or along with these other steroid hydroxylating P450s is unknown (Nebert et al., ’89). Eukaryotic P450s evolved from soluble, prokaryotic enzymes (Werck-Reichhart and Feyereisen, 2000) to become anchored in the endoplasmic reticulum, diversifying subsequently to mitochondrial forms (Omura, 2006). Aromatase, and other key P450s including 17α-hydroxylase/17,20-lyase (P450c17; androgen synthesis) and 21-hydroxylase (P450c21; corticoid synthesis) are anchored in the outer microsomal membrane where they couple with a redox partner, cytochrome P450 oxido-reductase (CPR; Hanukoglu, ’92; Miller, 2005). Others including cholesterol side chain cleavage (P450scc; pregnane synthesis) and 11β-hydroxylase (P450c11; corticoid synthesis) are embedded in the inner mitochondrial membrane, therefore requiring and utilizing a completely different system of redox partner proteins (Miller, 2005; Omura, 2006). Again, how or why the P450s involved in sterol synthesis and transformation diverged and expanded to utilize mitochondrial, as well as microsomal, subcellular compartmentalization is unknown. Hypotheses on the potential origins of these proteins and the evolutionary forces that may have influenced the assembly of P450-based metabolic cascades are likely to be refined as sequences from more species are cloned, and their functional characteristics are explored and better defined.
Fig. 1.
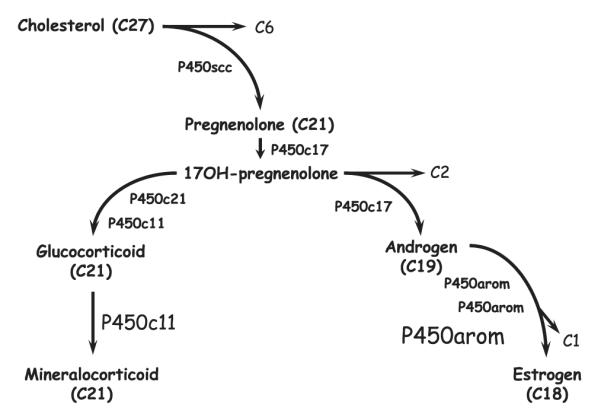
Schematic representation of the steroidogenic cascade involving the steroid hydroxylating cytochromes P450 (P450) that catalyze cleavage and/or hydroxylation of sequential substrates from cholesterol through pregnenolone, androstane and estrane (synthesis). The enzymes catalyzing each step are shown; cholesterol side chain cleavage (P450scc) cleaves the 6 carbon side chain (C6) from the 27 carbon skeleton (C27) releasing pregnenolone (C21). Subsequent metabolism by 17α-hydroxylase/17,20-lyase (P450c17) produces 17-hydroxypregnanes (C21) that are intermediates in the production of either corticoids or sex steroids. Metabolism of 17-hydroxypregnanes by 21-hydroxylase (P450c21) and 11β-hydroxylase (P450c11) directs the pathway toward corticoid production. Alternatively, cleavage of the 17-hydroxypregnanes in a second oxidation catalyzed by P450c17 directs the pathway to androgen (C19) synthesis and then, after three oxidations culminating in cleavage of a methyl, to estrogens (C18). Note that P450c11 and P450arom represent the terminal enzymes in the steroid synthetic cascade.
It is well known that gene duplication has helped to shape the evolution of genomes (Hughes, ’94, ’99) and the biochemical and physiological processes they control. The discovery that genes encoding some of the steroid hydroxylating P450s have duplicated relatively recently provides an unusual opportunity to gain insight into the process leading to the evolution of those P450s forming the biochemical backbone of the steroidogenic pathway. Two examples of gene duplication events leading to the expression of true paralogues of P450 enzymes involved in steroid synthesis (one microsomal and one mitochondrial) have been discovered in vertebrates. The one most recently discovered involves the CYP19 gene encoding P450arom (Callard and Tchoudakova, ’97; Choi et al., ’97b; Conley et al., ’97; Graddy et al., 2000; Conley and Hinshelwood, 2001; Gaucher et al., 2004), which duplicated in fish long ago and more recently among species of at least one mammalian suborder, the Suiformes. In fish (Tchoudakova and Callard, ’98; Tchoudakova et al., 2001) and suiforms (Corbin et al., ’95, 2007; Hinshelwood et al., ’95; Conley et al., ’96, ’97), the duplicated CYP19 genes exhibit strong tissue specific expression, which may well have facilitated gene survival with functional adaptation (Hughes, 2005), a theme that will be explored in greater detail below. Full-length sequences for aromatases cloned from over a dozen other mammals representing seven orders including the artiodactyla suggest the existence of a single functional CYP19 gene in these species. The second known example of gene duplication among the steroid hydroxylase genes is that of CYP11B, which appears to have duplicated independently in humans and several rodent species. In species with a single CYP11B gene, the P450c11 enzyme catalyzes the synthesis of corticosterone, by 11β-hydroxylation of 11-deoxycorticosterone. This same enzyme also catalyzes the synthesis of aldosterone (in much lower yield) by subsequent 18-hydroxylation and 18-oxidation of corticosterone. This presumably represents the enzymatic function of the ancestral enzyme (Bulow and Bernhardt, 2002). The duplication of CYP11B in rats, mice, guinea pigs, hamsters and humans (Mornet et al., ’89; Lehoux et al., ’94; Bulow and Bernhardt, 2002; Okamoto et al., 2005), as well as some fish (Jiang et al., ’96; Kusakabe et al., 2002), has allowed for some measure of functional adaptation. The resultant enzyme paralogs exhibit properties that favor corticosterone (P450c11, 11β-hydroxylase) or aldosterone (P450aldo, aldosynthase) production. These enzymes exhibit expression that is specific to the outer glomerulosa and fasciculata zones of the mammalian adrenal cortex that promotes differential regulation (Rainey, ’99), and therefore gene survival, despite their lack of sequence divergence in some cases (the human paralogs are 93% identical). Perhaps it is not coincidental that both enzyme systems catalyze the formation of terminal products of the sex steroid (P450arom, estrogen; P450c11, 11keto-testosterone) and mineralo-corticoid (P450c11 or P450aldo, aldosterone) pathways (Fig. 1). The acquisition of adaptive functions of more proximal enzymes may be limited by the strict requirements of intermediates required for terminal product formation. If true, this implies that gene duplication and the expansion of metabolic function from established pathways is more likely to involve the terminal and therefore more ancient of the component enzymes.
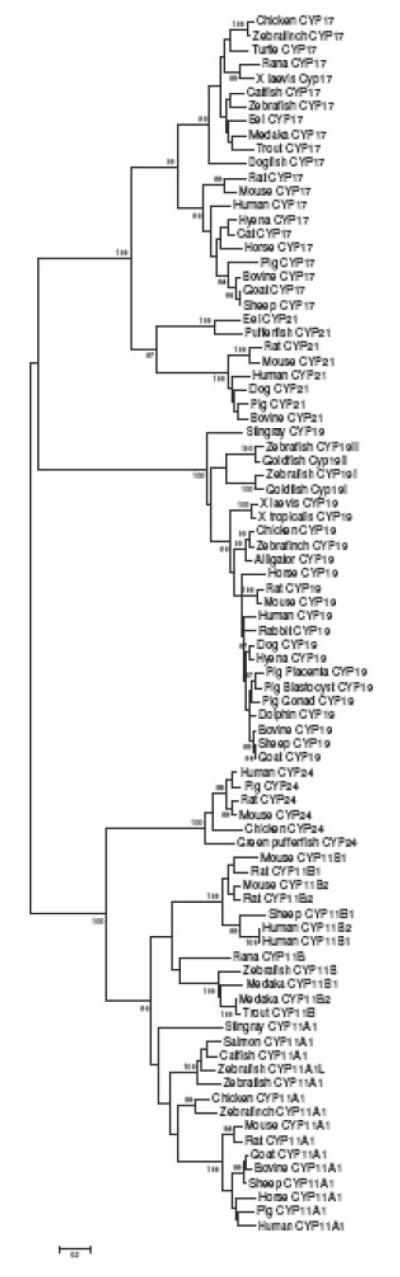
How the enzymatic cascade leading to steroid synthesis became established in vertebrates is unknown, but phylogenetic analyses may provide insights into the process, which may be equally relevant to the evolution of other integrated metabolic pathways. The expanding database of sequences of the genes encoding all the steroid hydroxylases in numerous species provides ever increasing power for studies of this kind. The results of past analyses suggest that CYP19 is an ancient gene (Aoyama et al., ’96; Nelson, 2003), but that it is not easily resolved from the mitochondrial clan (Fig. 2). This suggests a close evolutionary relationship, remnants of which may still exist from functional analyses of the P450arom and mitochondrial enzymes such as P450c11. For instance, P450c11 metabolizes both androstenedione (Rainey et al., ’94) and testosterone (Kusakabe et al., 2002) as substrates utilized in common with P450arom. Importantly, like P450arom (Kautsky and Hagerman, ’80), P450c11 is an active 19-hydroxylase (Veronneau et al., ’96) and is the only other enzyme shown to be capable of aromatizing an androgen (19OH-androstenedione) to estrogen (estrone; Suhara et al., ’88). The steric proximity of the C-19 and C-11 positions on the steroid nucleus further suggests that substrate orientation is also conserved (Fig. 3). As 19OH-androstenedione is potently hypertensive in mammals (Sekihara, ’82), amplifying aldosterone action (Sekihara et al., ’93) and 11keto-testosterone is a major sex steroid in fish (Borg, ’94; Lokman et al., 2002), the physiological role of P450arom and P450c11 also overlap potentially. In addition to sharing common substrates, enzyme inhibitors that are generally considered specific show surprisingly similar binding characteristics. The form of P450arom expressed in the porcine gonads is sensitive to inhibition by etomidate (Corbin et al., ’95; Conley et al., ’96, 2002), a compound well known to exhibit specificity for P450c11 (Engelhardt and Weber, ’94), though it is not capable of metabolizing either 11-deoxycortisol or 11-deoxycorticosterone (unpublished data). Conversely, the imidazole fadrozole is a very selective inhibitor of P450arom (Njar and Brodie, ’99) but has notable inhibitory potency toward P450c11 and the synthesis of aldosterone and cortisol (Lamberts et al., ’89, Table 1). However, other fundamental properties of these enzymes help resolve the ancestry of the CYP19 and CYP11 genes. As mineralocorticoids appeared with the first terrestrial migration (Colombo et al., 2006), it is unlikely that CYP11 would have predated CYP19 and the regulation of reproductive function by estrogen in vertebrate evolution (Lange et al., 2002). The most likely ancestral characteristic with respect to subcellular localization further supports this notion. Lanosterol demethylase (encoded by CYP51) is the only P450 enzyme for which homologues have been found in all biological kingdoms (Yoshida et al., 2000), and it is a microsomal enzyme in eukaryotes (Mitropoulos et al., ’76). This enzyme is essential in the synthesis of cholesterol, a function important in the evolution of eukaryotic membranes (Bloch, ’79; Yeagle, ’85; Arora et al., 2004). The fact that lanosterol demethylase is a microsomal P450, suggests that microsomal localization was an ancestral condition for eukaryotic P450s, and that mitochondrial forms evolved later, hence CYP11 (and other mitochondrial P450s) from CYP19. Perhaps on a subcellular level, the strict microsomal expression of the ancestral P450arom, and mitochondrial distribution of P450c11 and related CYPs was as critical as tissue-specificity in promoting survival of these gene clades after ancestral duplication. Compartmentalization in different organelles is likely to have facilitated their functional adaptation and divergence. Collectively, available evidence suggests that the steroidogenic pathway evolved in a retrograde fashion from CYP19, from the terminal enzyme in the sex steroid synthetic cascade. Retrograde evolution of other metabolic pathways has been hypothesized (Lazcano and Miller, ’99; Roy, ’99; Rison and Thornton, 2002; Schmidt et al., 2003). If true, then the retrograde “assembly” of enzymes comprising the steroid synthetic pathway also coevolved with their cognate nuclear receptors from an ancestral protein resembling an estrogen receptor (Thornton, 2001; Thornton et al., 2003; Bridgham et al., 2006). Metabolic redundancy or catalytic promiscuity, such as that demonstrated for P450scc (Guryev et al., 2003; Slominski et al., 2005a,b, 2006), would facilitate assembly of a metabolic cascade from preexisting elements. Few alternative substrates for P450arom enzymes have been investigated to date but might provide evidence supporting such a hypothesis.
Fig. 2.
Phylogenetic tree based on amino acid sequence of vertebrate P450s (represented here by the names of their respective CYP genes) involved in sterol synthesis and metabolism analyzed using the JTT distance (Jones et al., ’92). Sequences in the analysis included the enzymes 17α-hydroxylase/17,20-lyase (CYP17), 21-hydroxylase (CYP21), aromatase (CYP19), 24-hydroxylase (CYP24), 11β-hydroxylase (CYP11B1), aldosynthase (CYP11B2) and cholesterol side chain cleavage (CYP11A1). One thousand bootstrap pseudo-samples were used.
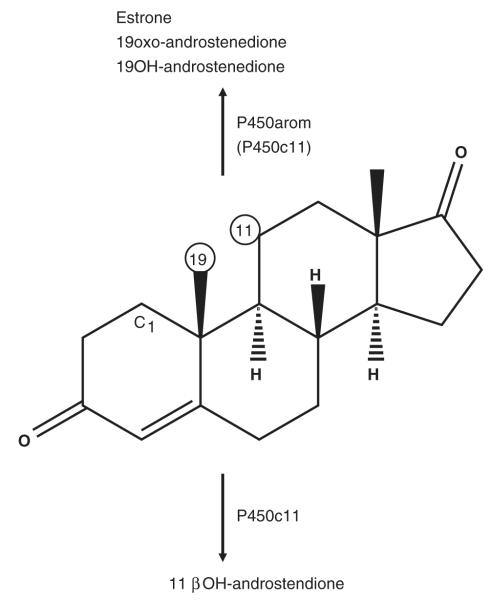
Fig. 3.
The structure of the substrate androstenedione, showing proximity of the C-19 methyl and C-11 carbon residues that are sites of oxidation by aromatase (P450arom) and 11β-hydroxylase (P450c11) cytochromes P450 as shown. Testosterone is utilized also by both enzymes for the formation of estradiol by P450arom in all vertebrates and 11keto-testosterone by P450c11 in certain teleosts. The site of 1-hydroxylation (C1) of testosterone by the porcine gonadal P450arom is also shown.
TABLE 1.
Estimated 50% inhibitory concentrations (IC50: nM) for human 11β-hydroxylase and porcine gonadal aromatase (P450arom) cytochromes P450 for the compounds etomidate and fadrozole, presumed specific for P450c11 and P450arom, respectively. Data were taken from the sources indicated
| Human P450c11 | Porcine gonadal P450arom | |
|---|---|---|
| etomidate | 1–10 | 100 |
| fadrozole | 10–30 | 2 |
As for CYP11B, duplication of CYP19 genes encoding P450arom(s) provides an opportunity for functional diversification and/or specialization (Lynch and Force, 2000; Lynch and Katju, 2004). Although this has occurred in numerous species, data on functional properties and possible physiological adaptation are sparse among the fish P450arom enzymes (Tong et al., 2001). However, the functional properties of paralogs of P450arom have been better studied in pigs (Corbin et al., ’95, ’99, 2001, 2003, 2004; Conley et al., ’96, 2002). The porcine genome contains three separate and complete copies of CYP19 (Graddy et al., 2000) spanning approximately one megabase of the porcine genome (Conley et al., ’97). The sites of expression of each gene are distinct, giving rise to gonadal (expression in both ovaries: Corbin et al., ’95; testes: Conley et al., ’96), placental (Corbin et al., ’95) and embryonic (Choi et al., ’96) P450arom enzymes. Analysis indicates that the embryonic (blastocyst) form arose from the most recent duplication of the placental CYP19 and has evolved most rapidly since (Gaucher et al., 2004). Few analyses of the function of this particular paralogue have been conducted, though it is highly expressed in the trophoblast (Conley et al., ’92, ’94), which very actively synthesizes estrogens (Geisert et al., ’90). Interestingly, it appears that the gonadal paralog of porcine P450arom is expressed in the hypothalamus, based on sequence analysis and sensitivity of enzyme activity to inhibition by etomidate (Conley et al., 2005), a distinct biochemical characteristic of this protein (Conley et al., 2002). Therefore, unlike fishes, duplication of porcine CYP19 did not give rise to brain- and gonad-specific paralogs of P450arom, but paralogs that are expressed (tissue-specifically) in the gonads and placenta, and subsequently the preimplantation trophoblast, and the same paralog previously referred to as the gonadal P450arom is also expressed in the brain. Catalytic adaptations of these paralogs are therefore likely to pertain to functions specific to the establishment and success of mammalian pregnancy.
Studies on the catalytic characteristics of the paralogs of porcine P450arom have provided clues as to the biochemical adaptations and physiological consequences that have influenced the evolutionary divergence of these enzymes. Specifically, extensive kinetic and functional comparisons have been conducted on the porcine gonadal and placental paralogs of P450arom (Corbin et al., ’95, ’99, 2001; Conley et al., 2004), along with the human and bovine enzymes (Corbin et al., 2003). These studies have shown that substantial catalytic differences exist at least between the gonadal and placental paralogs, and the results are consistent with the likelihood of functional adaptation. Early experiments examining substrate affinities indicated that the placental paralog has a higher affinity than the gonadal enzyme (Corbin et al., ’99). This would support efficient androgen metabolism at lower substrate concentrations by trophoblast than would be supported by the gonadal enzyme. A careful analysis of substrate turnover further emphasizes their functional divergence; the porcine gonadal P450arom catalyzes the aromatization of androstenedione at one third the rate of the placental enzyme, and the catalytic efficiency is one fifth (Corbin et al., 2003). Collectively, we hypothesize that the greater catalytic efficiency of androgen metabolism by the placental P450arom would provide female fetuses with protection from the masculinizing influence of their male siblings in utero (Corbin et al., ’99). There are well-recognized positional effects of male fetal androgens on female fetal development and postnatal reproductive behavior (vom Saal, ’81; Ryan and Vandenbergh, 2002). Similarly, a deficiency of human placental P450arom results in profound virilization of female infants at birth (Grumbach and Auchus, ’99). Thus, the catalytic evolution of this enzyme and its expression in the placenta might well represent an adaptation facilitating normal reproductive development and fertility of female offspring in pigs having evolved multiple ovulation and litter bearing ability.
The possible adaptation of the porcine placental P450arom does not preclude, and on the basis of the likelihood of gene survival may even dictate, a parallel adaptation emerging by evolution of the gonadal form. In fact, given the absolute dependence of normal reproductive development and function on estrogen (Federman, ’94), it seems counterintuitive that a gonadal paralog might not efficiently catalyze estrogen formation in comparison with the porcine placental, bovine or human P450arom enzymes (Corbin et al., 2003). The much higher levels of protein expression found in porcine ovarian follicular tissues than in placenta (Fig. 4), must at least partially mitigate any potential deficiencies in estrogen synthesis. However, investigation of the metabolic products of purified recombinant enzyme reconstituted in vitro suggested that catalytic characteristics of the gonadal paralog differed from those of the placental enzyme and of any other known P450arom in fact (Corbin et al., 2004). Specifically, the porcine gonadal P450arom metabolized testosterone but oxidized it in a way that resulted hydroxylations at two separate sites on the A ring. Oxidation was directed toward the C10 methyl group forming 19OH-testosterone, 19oxo-testosterone and subsequently estradiol, as with other vertebrate aromatases. However, oxidation was also directed at C1 position (Fig. 3) and resulted in accumulation of 1OH-testosterone as an additional metabolite. This product accumulated because it could not be metabolized further by either the porcine gonadal or the placental paralogs or the bovine or human P450arom enzymes. In other words, 1OH-testosterone appears to be a nonaromatizable androgen. Results of these studies (Corbin et al., 2004) also indicated that 1OH-testosterone activated the androgen receptor in a prostate cell line. No 1-hydroxylation was observed when androstenedione was metabolized to estrone by the porcine gonadal P450arom or with testosterone or androstenedione metabolism by the placental paralog (Corbin et al., 2004). Therefore, the unique catalytic property was paralog and substrate specific. Ovarian follicular granulosa cells and testicular microsomes were also shown to catalyze 1OH-testosterone synthesis that was inhibited by a specific P450arom inhibitor providing evidence that this unusual steroid is likely synthesized in vivo. Therefore, we hypothesized that these unusual, possibly unique, biochemical attributes of the gonadal paralog of porcine P450arom compensated for the inefficiency of estrogen synthesis and promoted the survival of the gene and protein in its present form (Corbin et al., 2004).
Fig. 4.
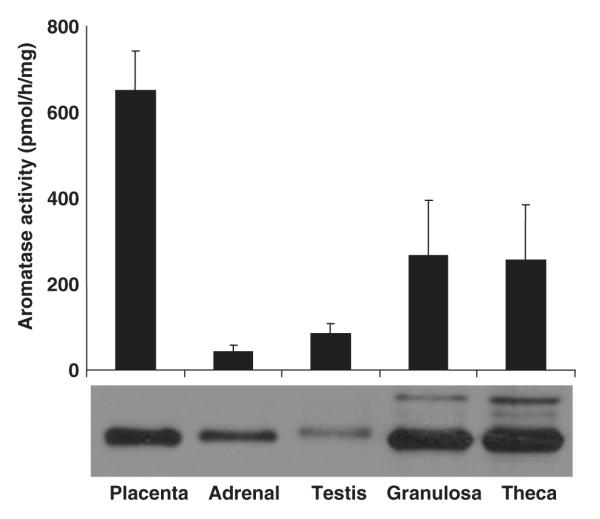
Aromatase enzyme activity and immuno-detectable levels of enzyme protein in porcine tissues. Aromatase activity (pmol/h/mg microsomal protein) is shown for placenta, adrenal gland, testis, and for preovulatory follicular granulosa and theca cell microsomal proteins. Equal amounts (20 μg/lane) from the same protein samples were subjected to western immunoblot analysis as shown below. Note that placental microsomes exhibit twice the enzyme activity of the ovarian (granulosa and theca) tissues but, by comparison, clearly expresses much lower levels of enzyme per microgram of crude protein. This suggests that the enzyme paralog expressed in the placenta has a significantly higher turnover rate, as was shown subsequently to be the case in kinetic studies with purified recombinant protein (Corbin et al., 2003). The differential inhibition of aromatase activity by etomidate in gonadal (follicular and testicular) versus placental tissues (Corbin et al., ’95; Conley et al., ’96) further suggests that expression of the gonadal and placental paralogs is very tissue-specific.
At the present time, the physiological significance and potential gain of function represented by the unique catalytic capacity of the porcine gonadal P450arom to synthesize 1OH-testosterone as well as estradiol is only speculation. However, some evidence supports the suggestion that functional evolution of the gonadal form may well have been reproductively advantageous and perhaps even synergistic with the catalytic evolution of the placental enzyme. Studies in pigs have shown that administration of testosterone markedly increases ovulation rate in gilts (Cardenas and Pope, ’94). More importantly, dihydrostestosterone, a nonaromatizable androgen has the same effects (Cardenas et al., 2002), suggesting that it is mediated not through conversion to estradiol in the follicle but through activation of androgen receptors, known to be expressed in porcine granulosa cells (Garrett and Guthrie, ’96). We propose that 1OH-testosterone synthesized in granulosa cells of porcine preovulatory follicles is an endogenously synthesized nonaromatizable androgen that promotes increased ovulation rates in pigs (Corbin et al., 2004). Moreover, we hypothesize that the development of this unique catalytic activity was an important element in the evolution of litter bearing as a reproductive strategy of pigs. Increasing the capacity of placental tissues to metabolize testosterone to estradiol would facilitate this strategy by minimizing potentially harmful effects of male androgens on female reproductive development (Corbin et al., ’99). Thus, we propose that the appearance of testosterone 1-hydroxylase activity by the gonadal paralog of porcine P450arom, coincidentally and together with increased testosterone metabolic efficiency by the placental paralog, promoted the success of litters as a reproductive strategy in the evolution of pigs.
In contrast to the placental and gonadal paraologs of porcine aromatase, very little functional data exist on the third porcine blastocyst paralog, though it is believed to be critical for the establishment of pregnancy (Bazer and Thatcher, ’77). Estradiol stimulates ovarian (luteal) progesterone production (Conley and Ford, ’89), decreases uterine prostaglandin release (Frank et al., ’77; Conley et al., ’89) and plays an important role in the spacing of embryos within the lumen of the uterine horns before attachment (Pope et al., ’86). Perhaps embryo spacing, facilitated by the expression of the blastocyst form of aromatase, promotes maximum litter size by reducing embryo mortality (Pope, ’99). Aromatase expression peaks around the time of blastocyst elongation (Geisert et al., ’82) on day 11 or 12 of pregnancy, based on western immunoblot (Conley et al., ’92), immunohistochemistry (Conley et al., ’94), transcript (Ko et al., ’94; Choi et al., ’97a), enzyme activity (van der et al., ’89) and estrogen levels in utero and in uterine venous blood (Ford et al., ’82). Despite substantial physiological evidence of estrogen synthesis by blastocysts, the sole attempt at characterization of catalytic function of the blastocyst paralog provided very little evidence of aromatization of testosterone to estradiol (Kao et al., 2000). However, these studies were conducted using transiently transfected CHO cells. Data utilizing purified recombinant enzyme to determine valid kinetic parameters and turnover rates were not reported nor were experiments performed using blastocysts as a source of the wildtype enzyme. No evidence has been presented that blastocysts express either the placental or the gonadal CYP19 genes, at least none was reported from the cloning or expression studies. The blastocyst form is believed to have arisen from the placental form and to have undergone extremely rapid evolution since (Gaucher et al., 2004), but more careful biochemical studies are required to define any functional adaptations that might have resulted. Hence, the question remains, was it “necessary” or advantageous to reduplicate CYP19 to achieve expression in preimplantation trophoblast? Blastocyst expression of either preexisting CYP19 gene might have been achieved by development of a tissue-specific untranslated first exon, a strategy utilized by the placental trophoblast of other mammals (Hinshelwood et al., ’95). Differential expression of the single CYP19 gene of other species (Simpson et al., ’94, ’97; Conley and Hinshelwood, 2001; Vanselow et al., 2001) is controlled by multiple, tissue-specific, untranslated first exons and is a strategy equally evident in pigs despite CYP19 gene duplication. Specifically, a different promoter controls expression of the gonadal paralog in the ovary than the one used in testis and adrenal expression for instance (Conley et al., ’96, ’97). As the gonadal gene was likely the ancestral form, alternative, tissue-specific promoters may have predated duplication of the ancestral porcine CYP gene, which is consistent with the occurrence of multiple promoters in all mammals in which CYP19 gene expression has been examined at this level. The fact that this strategy was not utilized to promote expression in porcine blastocysts is an additional reason to speculate that this enzyme paralog may well be adapted catalytically in some way also.
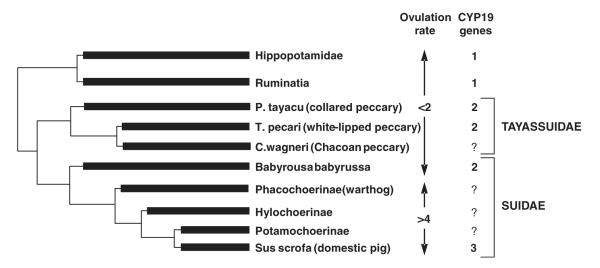
The development of litter bearing as a reproductive strategy in pigs assumes that their ancestral reproduction favored lower ovulations rates, but is this likely, and what of the CYP19 gene(s) in related and ancestral suiforms? An examination of reproductive patterns among close relatives supports the view that one or two ovulations were more likely in the ancestral suiform (Fig. 5). Peccaries are distant relatives of the pig, within the Suiforme suborder of artiodactyla (Ducrocq, ’95). There are three extant species of peccary that have been studied, the collared (Pecari tayassu), the white-lipped (Tayassu pecari) and the Chacoan (Catagonus wagnerii) (Sowls, ’97). All give birth to only two young on average which, at least in the case of the collared peccary, occupy different uterine horns (unpublished observations). Other artiodactyls including cattle, most breeds of sheep and hippopotami, also ovulate one or two follicles (Hayssen et al., ’93). Only one functional CYP19 gene exists in the bovine and ovine genomes (Vanselow et al., 2001), as in the human (Means et al., ’89; Bulun et al., 2004) and in the equine and canine genomes (data not shown). However, Gaucher et al. (2004) provided evidence for two CYP19 genes in white-lipped peccaries and babirusa, suggesting that duplication predated divergence of these species from domestic pigs. To further address issues relating to the ancestry of the CYP19 duplication, and of the conservation or divergence of function since, similar studies were initiated in other related species. Specifically, genomic DNA was obtained from collared peccaries and compared with pigs, white-lipped peccaries as well hippopotamus by Southern analysis. The presence of multiple bands hybridizing to a porcine P450arom probe in lanes containing collared and white-lipped peccary genomic DNA was consistent with duplication of CYP19 as previously reported (Gaucher et al., 2004). Subsequent analyses provided evidence of two CYP19 genes in the genome of collared peccaries, and confirmed the probable duplication in white-lipped peccaries. A single amplicon was also isolated from hippopotamus genomic DNA. Amplicons (spanning exons IV and V and an intervening intron) representing both CYP19 genes in the collared peccary and the single gene in hippopotamus were cloned, and sequenced to confirm their identity and to provide sequence information for phylogenetic analysis (Corbin et al., 2007). This analysis provided evidence supporting the hypothesis that the CYP19 gene duplicated in the ancestor of pigs, peccaries and babirusa but after divergence of the hippopotomus.
Fig. 5.
Diagram showing presumed divergence of species and reproductive strategies (ovulation rate/litter size) among members of the Suiforme suborder together with the Hippopotamidae and Ruminatia. Ovulation rates (<2 or >4 follicles/oocytes) and the number of genes encoding for aromatase cytochrome P450 (CYP19) are also shown for all species of the Tayassuidae and Suidae families where known. To date, full-length sequences for aromatases cloned from over a dozen other mammals representing seven orders including the artiodactyla suggest the existence of a single functional CYP19 gene in these species.
The existence of two genes encoding CYP19 in both collared and white-lipped peccaries helps establish the phylogeny of CYP19 duplication in the Suiformes (Fig. 5), but what of the evolution of function in these species? The phylogenetic analysis of genomic clones provided statistical support for a clustering of the pig gonadal P450arom with one of the peccary amplicons. The sequence obtained for the other peccary CYP19 gene could not be resolved, in part because of apparent gene conversion event with the gonadal paralog (Corbin et al., 2007). Therefore, studies were initiated to investigate the functional conservation of peccary P450arom enzymes using tissues from the collared species. Gonads were collected from mature males and placentas from females at different stages of pregnancy (Corbin et al., 2007). Transcript was isolated and partial sequences encoding peccary P450arom paralogs were obtained from amplicons using the same exon IV and exon V primers as for the genomic analysis. These data demonstrated that the form of the peccary gene shown to cluster with the porcine gonadal CYP19 was expressed in the testis of the collared peccary. Even though not resolved by phylogenetic analysis, the other peccary CYP19 gene was expressed in peccary placenta. Thus, both genes appear to have conserved their gonad and placenta-specific expression (Corbin et al., 2007). The data on relative levels of expression were preliminary but indicated that expression levels in peccary tissues were low. This was suggested for the placental paralog of the collared peccary, based on the low efficiency of amplification with peccary specific primers. The same was true of the testis expression, which was 100-fold higher in the testis of the domestic boar than in peccary based on analysis of enzyme activity and western immunoblot data (Corbin et al., 2007). Thus, it appears that collared peccaries at least have a gonadal and placental CYP19 gene and that the tissue-specific expression of these genes has been conserved as this species diverged from pigs. A preliminary examination of peccary testicular enzyme also failed to find evidence of the production of 1OH-testosterone. Our data suggest that the functional properties of CYP19 genes have evolved differently in pigs and peccaries with respect to levels of expression and perhaps catalytic characteristics that may serve their differing reproductive strategies.
Despite conservation of tissue-specific expression patterns of the gonadal and placental CYP19 genes in the collared peccary, the levels of expression appear to be substantially lower than those in pigs. Although levels of gene expression do not necessarily reflect functional relevance, they do provide another facet of adaptive evolution (Liao and Zhang, 2006a,b). Recent studies in our laboratory demonstrate in pigs that mature testis size and sperm production capacity are influenced by P450arom expression in neonatal life (At-Taras et al., 2006). In as much as an increase in levels of expression may signal physiological adaptation, loss of expression may also suggest a loss of functional significance. The apparently low level of expression in the peccary placenta may simply reflect the ancestral state, but the observed gene conversion may also represent the early stage of loss of the CYP19 gene duplication in this species. The identification of a CYP19 pseudogene expressed in the bovine placenta (Furbass and Vanselow, ’95) may be the evidence supporting this possibility. Certainly, there are many examples of gene conversion among CYP genes (Werck-Reichhart and Feyereisen, 2000; Goldstone and Stegeman, 2006), and conversion is a mechanism whereby gene redundancy can be eliminated from the genome (Hughes and Friedman, 2005). Therefore, it is possible that the duplication of suiform CYP19 genes not only provides insight into the evolution and fixation of characters but their disappearance as well. There are relatively few biological systems where gene duplication and loss has been examined in relation to biochemical and presumed physiological fitness (Zhang et al., 2002; Piatigorsky, 2003), fewer yet involving reproductive traits. Other members of the Suiforme suborder must be examined to properly explore the potential value of studies on CYP19 for further understanding the forces influencing genome evolution.
In conclusion, studies of suiform CYP19 genes and the P450arom enzyme paralogs encoded by them offer considerable promise of advancing understanding of the evolution of steroid hydroxylases. CYP19 may well have been the progenitor of the steroid hydroxylases that constitute the backbone of the steroidogenic cascade, from which the clade of mitochondrial enzymes first diverged. Evidence of close evolutionary ties may exist between CYP19 and the mitochondrial P450s, P450c11 in particular provides support for this view. Both are terminal enzymes in the sex steroid and corticoid synthetic pathways, respectively, and both have experienced gene duplication in certain species with the survival of functionally adapted paralogs. Unlike most P450s, both P450arom and P450c11 are capable of catalyzing up to three concerted oxygenation reactions. In addition, they share common substrates and exhibit comparable sensitivities to imidazoles that are thought to be otherwise specific, suggesting structural similarities in the active site. It appears likely also that the combination of phylogenetic, biochemical and physiological studies of the porcine CYP19 gene cluster will provide valuable information on the evolution of reproductive strategies, such as litter bearing, and possibly of reproductive or social behavior, based on the expression of P450arom in the brain. Therefore, some of the fundamental processes shaping metabolic pathways, reproductive and ecological fitness and genome evolution, may all be enlightened by investigations into the suiform aromatase system.
ACKNOWLEDGMENTS
This study was presented as part of a mini-symposium on Vertebrate Aromatases at the International Congress of Comparative Physiology and Biochemistry, Salvador, Brazil, August 11–16th, 2007. The mini-symposium was made possible by a grant from the United States Department of Agriculture-National Research Initiative, and funding from the United States Department of Agriculture-Agricultural Research Service.
Grant sponsors: United States Department of Agriculture-National Research Initiative; United States Department of Agriculture-Agricultural Research Service.
LITERATURE CITED
- Aoyama Y, Noshiro M, Gotoh O, Imaoka S, Funae Y, Kurosawa N, Horiuchi T, Yoshida Y. Sterol 14-demethylase P450 (P45014DM*) is one of the most ancient and conserved P450 species. J Biochem (Tokyo) 1996;119:926–933. doi: 10.1093/oxfordjournals.jbchem.a021331. [DOI] [PubMed] [Google Scholar]
- Arora A, Raghuraman H, Chattopadhyay A. Influence of cholesterol and ergosterol on membrane dynamics: a fluorescence approach. Biochem Biophys Res Commun. 2004;318:920–926. doi: 10.1016/j.bbrc.2004.04.118. [DOI] [PubMed] [Google Scholar]
- At-Taras EE, Berger T, McCarthy MJ, Conley AJ, Nitta-Oda BJ, Roser JF. Reducing estrogen synthesis in developing boars increases testis size and total sperm production. J Androl. 2006;27:552–559. doi: 10.2164/jandrol.05195. [DOI] [PubMed] [Google Scholar]
- Bazer FW, Thatcher WW. Theory of maternal recognition of pregnancy in swine based on estrogen controlled endocrine versus exocrine secretion of prostaglandin F2alpha by the uterine endometrium. Prostaglandins. 1977;14:397–400. doi: 10.1016/0090-6980(77)90185-x. [DOI] [PubMed] [Google Scholar]
- Bloch KE. Speculations on the evolution of sterol structure and function. CRC Crit Rev Biochem. 1979;7:1–5. doi: 10.3109/10409237909102566. [DOI] [PubMed] [Google Scholar]
- Borg B. Androgens in teleost fishes. Comp Biochem Physiol C-Pharmacol Toxicol Endocrinol. 1994;109:219–245. [Google Scholar]
- Bridgham JT, Carroll SM, Thornton JW. Evolution of hormone-receptor complexity by molecular exploitation. Science. 2006;312:97–101. doi: 10.1126/science.1123348. [DOI] [PubMed] [Google Scholar]
- Bulow HE, Bernhardt R. Analyses of the CYP11B gene family in the guinea pig suggest the existence of a primordial CYP11B gene with aldosterone synthase activity. Eur J Biochem. 2002;269:3838–3846. doi: 10.1046/j.1432-1033.2002.03076.x. [DOI] [PubMed] [Google Scholar]
- Bulun SE, Takayama K, Suzuki T, Sasano H, Yilmaz B, Sebastian S. Organization of the human aromatase p450 (CYP19) gene. Semin Reprod Med. 2004;22:5–9. doi: 10.1055/s-2004-823022. [DOI] [PubMed] [Google Scholar]
- Callard GV, Tchoudakova A. Evolutionary and functional significance of two CYP19 genes differentially expressed in brain and ovary of goldfish. J Steroid Biochem Mol Biol. 1997;61:387–392. doi: 10.1016/s0960-0760(97)80037-4. [DOI] [PubMed] [Google Scholar]
- Callard GV, Pudney JA, Kendall SL, Reinboth R. In vitro conversion of androgen to estrogen in amphioxus gonadal tissues. Gen Comp Endocrinol. 1984;56:53–58. doi: 10.1016/0016-6480(84)90060-1. [DOI] [PubMed] [Google Scholar]
- Cardenas H, Pope WF. Administration of testosterone during the follicular phase increased the number of corpora lutea in gilts. J Anim Sci. 1994;72:2930–2935. doi: 10.2527/1994.72112930x. [DOI] [PubMed] [Google Scholar]
- Cardenas H, Herrick JR, Pope WF. Increased ovulation rate in gilts treated with dihydrotestosterone. Reproduction. 2002;123:527–533. doi: 10.1530/rep.0.1230527. [DOI] [PubMed] [Google Scholar]
- Choi I, Simmen RC, Simmen FA. Molecular cloning of cytochrome P450 aromatase complementary deoxyribonucleic acid from periimplantation porcine and equine blastocysts identifies multiple novel 5′-untranslated exons expressed in embryos, endometrium, and placenta. Endocrinology. 1996;137:1457–1467. doi: 10.1210/endo.137.4.8625924. [DOI] [PubMed] [Google Scholar]
- Choi I, Collante WR, Simmen RC, Simmen FA. A developmental switch in expression from blastocyst to endometrial/placental-type cytochrome P450 aromatase genes in the pig and horse. Biol Reprod. 1997a;56:688–696. doi: 10.1095/biolreprod56.3.688. [DOI] [PubMed] [Google Scholar]
- Choi I, Troyer DL, Cornwell DL, Kirby-Dobbels KR, Collante WR, Simmen FA. Closely related genes encode developmental and tissue isoforms of porcine cytochrome P450 aromatase. DNA Cell Biol. 1997b;16:769–777. doi: 10.1089/dna.1997.16.769. [DOI] [PubMed] [Google Scholar]
- Colombo L, Dalla VL, Fiore C, Armanini D, Belvedere P. Aldosterone and the conquest of land. J Endocrinol Invest. 2006;29:373–379. doi: 10.1007/BF03344112. [DOI] [PubMed] [Google Scholar]
- Conley A, Hinshelwood M. Mammalian aromatases. Reproduction. 2001;121:685–695. doi: 10.1530/rep.0.1210685. [DOI] [PubMed] [Google Scholar]
- Conley A, Corbin J, Smith T, Hinshelwood M, Liu Z, Simpson E. Porcine aromatases: studies on tissue-specific, functionally distinct isozymes from a single gene? J Steroid Biochem Mol Biol. 1997;61:407–413. [PubMed] [Google Scholar]
- Conley A, Mapes S, Corbin CJ, Greger D, Graham S. Structural determinants of aromatase cytochrome P450 inhibition in substrate recognition site-1. Mol Endocrinol. 2002;16:1456–1468. doi: 10.1210/mend.16.7.0876. [DOI] [PubMed] [Google Scholar]
- Conley AJ, Ford SP. Direct luteotrophic effect of oestradiol-17 beta on pig corpora lutea. J Reprod Fertil. 1989;87:125–131. doi: 10.1530/jrf.0.0870125. [DOI] [PubMed] [Google Scholar]
- Conley AJ, Pusateri AE, Van Orden DE, Ford SP. Effect of intraluteal estradiol-17beta implants on weight and progesterone secretion of porcine corpora lutea. Anim Reprod Sci. 1989;20:221–230. [Google Scholar]
- Conley AJ, Christenson RK, Ford SP, Geisert RD, Mason JI. Steroidogenic enzyme expression in porcine conceptuses during and after elongation. Endocrinology. 1992;131:896–902. doi: 10.1210/endo.131.2.1379167. [DOI] [PubMed] [Google Scholar]
- Conley AJ, Christenson LK, Ford SP, Christenson RK. Immunocytochemical localization of cytochromes P450 17 alpha-hydroxylase and aromatase in embryonic cell layers of elongating porcine blastocysts. Endocrinology. 1994;135:2248–2254. doi: 10.1210/endo.135.5.7956948. [DOI] [PubMed] [Google Scholar]
- Conley AJ, Corbin CJ, Hinshelwood MM, Liu Z, Simpson ER, Ford JJ, Harada N. Functional aromatase expression in porcine adrenal gland and testis. Biol Reprod. 1996;54:497–505. doi: 10.1095/biolreprod54.2.497. [DOI] [PubMed] [Google Scholar]
- Conley AJ, Corbin CJ, Mapes SM, Heffelfinger JR. Pigs, peccaries and the evolution of aromatase. Biol Reprod. 2004:102. (abstract) [Google Scholar]
- Conley AJ, Corbin CJ, Ford JJ. Brain aromatase in male and female pigs: hypothalamic levels and the identification of the tissue-specific form. Biol Reprod. 2005:113. (abstract) [Google Scholar]
- Corbin CJ, Khalil MW, Conley AJ. Functional ovarian and placental isoforms of porcine aromatase. Mol Cell Endocrinol. 1995;113:29–37. doi: 10.1016/0303-7207(95)03607-9. [DOI] [PubMed] [Google Scholar]
- Corbin CJ, Trant JM, Walters KW, Conley AJ. Changes in testosterone metabolism associated with the evolution of placental and gonadal isozymes of porcine aromatase cytochrome P450. Endocrinology. 1999;140:5202–5210. doi: 10.1210/endo.140.11.7140. [DOI] [PubMed] [Google Scholar]
- Corbin CJ, Trant JM, Conley AJ. Porcine gonadal and placental isozymes of aromatase cytochrome P450: subcellular distribution and support by NADPH-cytochrome P450 reductase. Mol Cell Endocrinol. 2001;172:115–124. doi: 10.1016/s0303-7207(00)00373-7. [DOI] [PubMed] [Google Scholar]
- Corbin CJ, Mapes SM, Lee YM, Conley AJ. Structural and functional differences among purified recombinant mammalian aromatases: glycosylation, N-terminal sequence and kinetic analysis of human, bovine and the porcine placental and gonadal isozymes. Mol Cell Endocrinol. 2003;206:147–157. doi: 10.1016/s0303-7207(02)00422-7. [DOI] [PubMed] [Google Scholar]
- Corbin CJ, Mapes SM, Marcos J, Shackleton CH, Morrow D, Safe S, Wise T, Ford JJ, Conley AJ. Paralogues of porcine aromatase cytochrome P450: a novel hydroxylase activity is associated with the survival of a duplicated gene. Endocrinology. 2004;145:2157–2164. doi: 10.1210/en.2003-1595. [DOI] [PubMed] [Google Scholar]
- Corbin CJ, Hughes AL, Heffelfinger JR, Berger T, Waltzek T, Roser JF, Santos TC, Miglino MA, Oliveira M, Braga F, Mereilles F, Conley AJ. Evolution of suiform aromatases: ancestral duplication with conservation of tissue-specific expression in the collared paccary (Pecari tayassu) J Mol Evol. 2007;65:403–412. doi: 10.1007/s00239-007-9021-0. [DOI] [PubMed] [Google Scholar]
- Ducrocq S. An Eocene peccary from Thailand and the biogeographical origins of the artiodactyl family Tayassuidae. Palaeontology. 1995;37:765–779. [Google Scholar]
- Engelhardt D, Weber MM. Therapy of Cushing’s syndrome with steroid biosynthesis inhibitors. J Steroid Biochem Mol Biol. 1994;49:261–267. doi: 10.1016/0960-0760(94)90267-4. [DOI] [PubMed] [Google Scholar]
- Federman DD. Life without estrogen. N Engl J Med. 1994;331:1088–1089. doi: 10.1056/NEJM199410203311611. [DOI] [PubMed] [Google Scholar]
- Ford SP, Christenson RK, Ford JJ. Uterine blood flow and uterine arterial, venous and luminal concentrations of oestrogens on days 11, 13 and 15 after oestrus in pregnant and non-pregnant sows. J Reprod Fertil. 1982;64:185–190. doi: 10.1530/jrf.0.0640185. [DOI] [PubMed] [Google Scholar]
- Frank M, Bazer FW, Thatcher WW, Wilcox CJ. A study of prostaglandin F2alpha as the luteolysin in swine: III effects of estradiol valerate on prostaglandin F, progestins, estrone and estradiol concentrations in the utero-ovarian vein of nonpregnant gilts. Prostaglandins. 1977;14:1183–1196. doi: 10.1016/0090-6980(77)90295-7. [DOI] [PubMed] [Google Scholar]
- Furbass R, Vanselow J. An aromatase pseudogene is transcribed in the bovine placenta. Gene. 1995;154:287–291. doi: 10.1016/0378-1119(94)00754-g. [DOI] [PubMed] [Google Scholar]
- Garrett WM, Guthrie HD. Expression of androgen receptors and steroidogenic enzymes in relation to follicular growth and atresia following ovulation in pigs. Biol Reprod. 1996;55:949–955. doi: 10.1095/biolreprod55.5.949. [DOI] [PubMed] [Google Scholar]
- Gaucher EA, Graddy LG, Li T, Simmen RC, Simmen FA, Schreiber DR, Liberles DA, Janis CM, Benner SA. The planetary biology of cytochrome P450 aromatases. BMC Biol. 2004;2:19. doi: 10.1186/1741-7007-2-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisert RD, Brookbank JW, Roberts RM, Bazer FW. Establishment of pregnancy in the pig: II. Cellular remodeling of the porcine blastocyst during elongation on day 12 of pregnancy. Biol Reprod. 1982;27:941–955. doi: 10.1095/biolreprod27.4.941. [DOI] [PubMed] [Google Scholar]
- Geisert RD, Zavy MT, Moffatt RJ, Blair RM, Yellin T. Embryonic steroids and the establishment of pregnancy in pigs. J Reprod Fertil Suppl. 1990;40:293–305. [PubMed] [Google Scholar]
- Goldstone HM, Stegeman JJ. A revised evolutionary history of the CYP1A subfamily: gene duplication, gene conversion, and positive selection. J Mol Evol. 2006;62:708–717. doi: 10.1007/s00239-005-0134-z. [DOI] [PubMed] [Google Scholar]
- Graddy LG, Kowalski AA, Simmen FA, Davis SL, Baumgartner WW, Simmen RC. Multiple isoforms of porcine aromatase are encoded by three distinct genes. J Steroid Biochem Mol Biol. 2000;73:49–57. doi: 10.1016/s0960-0760(00)00054-6. [DOI] [PubMed] [Google Scholar]
- Grumbach MM, Auchus RJ. Estrogen: consequences and implications of human mutations in synthesis and action. J Clin Endocrinol Metab. 1999;84:4677–4694. doi: 10.1210/jcem.84.12.6290. [DOI] [PubMed] [Google Scholar]
- Guryev O, Carvalho RA, Usanov S, Gilep A, Estabrook RW. A pathway for the metabolism of vitamin D3: unique hydroxylated metabolites formed during catalysis with cytochrome P450scc (CYP11A1) Proc Natl Acad Sci USA. 2003;100:14754–14759. doi: 10.1073/pnas.2336107100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanukoglu I. Steroidogenic enzymes: structure, function, and role in regulation of steroid hormone biosynthesis. J Steroid Biochem Mol Biol. 1992;43:779–804. doi: 10.1016/0960-0760(92)90307-5. [DOI] [PubMed] [Google Scholar]
- Hayssen V, van Tienhoven A, van Tienhoven A. Asdell’s patterns of mammalian reproduction. Comstock Publishing Associates; Ithaca: 1993. [Google Scholar]
- Hinshelwood MM, Liu Z, Conley AJ, Simpson ER. Demonstration of tissue-specific promoters in nonprimate species that express aromatase P450 in placentae. Biol Reprod. 1995;53:1151–1159. doi: 10.1095/biolreprod53.5.1151. [DOI] [PubMed] [Google Scholar]
- Hughes AL. The evolution of functionally novel proteins after gene duplication. Proc R Soc Lond B Biol Sci. 1994;256:119–124. doi: 10.1098/rspb.1994.0058. [DOI] [PubMed] [Google Scholar]
- Hughes AL. Adaptive evolution of genes and genomes. Oxford University Press; New York: 1999. [Google Scholar]
- Hughes AL. Gene duplication and the origin of novel proteins. Proc Natl Acad Sci USA. 2005;102:8791–8792. doi: 10.1073/pnas.0503922102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AL, Friedman R. Loss of ancestral genes in the genomic evolution of Ciona intestinalis. Evol Dev. 2005;7:196–200. doi: 10.1111/j.1525-142X.2005.05022.x. [DOI] [PubMed] [Google Scholar]
- Jiang JQ, Kobayashi T, Ge W, Kobayashi H, Tanaka M, Okamoto M, Nonaka Y, Nagahama Y. Fish testicular 11beta-hydroxylase: cDNA cloning and mRNA expression during spermatogenesis. FEBS Lett. 1996;397:250–252. doi: 10.1016/s0014-5793(96)01187-8. [DOI] [PubMed] [Google Scholar]
- Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- Kao YC, Higashiyama T, Sun X, Okubo T, Yarborough C, Choi I, Osawa Y, Simmen FA, Chen S. Catalytic differences between porcine blastocyst and placental aromatase isozymes. Eur J Biochem. 2000;267:6134–6139. doi: 10.1046/j.1432-1327.2000.01705.x. [DOI] [PubMed] [Google Scholar]
- Kautsky MP, Hagerman DD. Kinetic properties of steroid 19-hydroxylase and estrogen synthetase from porcine ovary microsomes. J Steroid Biochem. 1980;13:1283–1290. doi: 10.1016/0022-4731(80)90088-6. [DOI] [PubMed] [Google Scholar]
- Ko Y, Choi I, Green ML, Simmen FA, Simmen RC. Transient expression of the cytochrome P450 aromatase gene in elongating porcine blastocysts is correlated with uterine insulin-like growth factor levels during peri-implantation development. Mol Reprod Dev. 1994;37:1–11. doi: 10.1002/mrd.1080370102. [DOI] [PubMed] [Google Scholar]
- Kusakabe M, Kobayashi T, Todo T, Mark LP, Nagahama Y, Young G. Molecular cloning and expression during spermatogenesis of a cDNA encoding testicular 11beta-hydroxylase (P45011beta) in rainbow trout (Oncorhynchus mykiss) Mol Reprod Dev. 2002;62:456–469. doi: 10.1002/mrd.10145. [DOI] [PubMed] [Google Scholar]
- Lamberts SW, Bruining HA, Marzouk H, Zuiderwijk J, Uitterlinden P, Blijd JJ, Hackeng WH, de Jong FH. The new aromatase inhibitor CGS-16949A suppresses aldosterone and cortisol production by human adrenal cells in vitro. J Clin Endocrinol Metab. 1989;69:896–901. doi: 10.1210/jcem-69-4-896. [DOI] [PubMed] [Google Scholar]
- Lange IG, Hartel A, Meyer HH. Evolution of oestrogen functions in vertebrates. J Steroid Biochem Mol Biol. 2002;83:219–226. doi: 10.1016/s0960-0760(02)00225-x. [DOI] [PubMed] [Google Scholar]
- Lazcano A, Miller SL. On the origin of metabolic pathways. J Mol Evol. 1999;49:424–431. [PubMed] [Google Scholar]
- Lehoux JG, Mason JI, Bernard H, Ducharme L, LeHoux J, Veronneau S, Lefebvre A. The presence of two cytochrome P450 aldosterone synthase mRNAs in the hamster adrenal. J Steroid Biochem Mol Biol. 1994;49:131–137. doi: 10.1016/0960-0760(94)90003-5. [DOI] [PubMed] [Google Scholar]
- Liao BY, Zhang J. Evolutionary conservation of expression profiles between human and mouse orthologous genes. Mol Biol Evol. 2006a;23:530–540. doi: 10.1093/molbev/msj054. [DOI] [PubMed] [Google Scholar]
- Liao BY, Zhang J. Low rates of expression profile divergence in highly expressed genes and tissue-specific genes during Mammalian evolution. Mol Biol Evol. 2006b;23:1119–1128. doi: 10.1093/molbev/msj119. [DOI] [PubMed] [Google Scholar]
- Lokman PM, Harris B, Kusakabe M, Kime DE, Schulz RW, Adachi S, Young G. 11-Oxygenated androgens in female teleosts: prevalence, abundance, and life history implications. Gen Comp Endocrinol. 2002;129:1–12. doi: 10.1016/s0016-6480(02)00562-2. [DOI] [PubMed] [Google Scholar]
- Lynch M, Force A. The probability of duplicate gene preservation by subfunctionalization. Genetics. 2000;154:459–473. doi: 10.1093/genetics/154.1.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Katju V. The altered evolutionary trajectories of gene duplicates. Trends Genet. 2004;20:544–549. doi: 10.1016/j.tig.2004.09.001. [DOI] [PubMed] [Google Scholar]
- Means GD, Mahendroo MS, Corbin CJ, Mathis JM, Powell FE, Mendelson CR, Simpson ER. Structural analysis of the gene encoding human aromatase cytochrome P-450, the enzyme responsible for estrogen biosynthesis. J Biol Chem. 1989;264:19385–19391. [PubMed] [Google Scholar]
- Miller WL. Minireview: regulation of steroidogenesis by electron transfer. Endocrinology. 2005;146:2544–2550. doi: 10.1210/en.2005-0096. [DOI] [PubMed] [Google Scholar]
- Mitropoulos KA, Gibbons GF, Reeves BE. Lanosterol 14alpha-demethylase. Similarity of the enzyme system from yeast and rat liver. Steroids. 1976;27:821–829. doi: 10.1016/0039-128x(76)90141-0. [DOI] [PubMed] [Google Scholar]
- Mizuta T, Kubokawa K. Presence of sex steroids and cytochrome P450 genes in amphioxus. Endocrinology. 2007;148:3554–3565. doi: 10.1210/en.2007-0109. [DOI] [PubMed] [Google Scholar]
- Mornet E, Dupont J, Vitek A, White PC. Characterization of two genes encoding human steroid 11 beta-hydroxylase (P-450(11) beta) J Biol Chem. 1989;264:20961–20967. [PubMed] [Google Scholar]
- Nebert DW, Nelson DR, Feyereisen R. Evolution of the cytochrome P450 genes. Xenobiotica. 1989;19:1149–1160. doi: 10.3109/00498258909043167. [DOI] [PubMed] [Google Scholar]
- Nelson DR. Comparison of P450s from human and fugu: 420 million years of vertebrate P450 evolution. Arch Biochem Biophys. 2003;409:18–24. doi: 10.1016/s0003-9861(02)00553-2. [DOI] [PubMed] [Google Scholar]
- Njar VC, Brodie AM. Comprehensive pharmacology and clinical efficacy of aromatase inhibitors. Drugs. 1999;58:233–255. doi: 10.2165/00003495-199958020-00003. [DOI] [PubMed] [Google Scholar]
- Okamoto M, Nonaka Y, Takemori H, Doi J. Molecular identity and gene expression of aldosterone synthase cytochrome P450. Biochem Biophys Res Commun. 2005;338:325–330. doi: 10.1016/j.bbrc.2005.07.187. [DOI] [PubMed] [Google Scholar]
- Omura T. Mitochondrial P450 s. Chem Biol Interact. 2006;163:86–93. doi: 10.1016/j.cbi.2006.06.008. [DOI] [PubMed] [Google Scholar]
- Piatigorsky J. Crystallin genes: specialization by changes in gene regulation may precede gene duplication. J Struct Funct Genomics. 2003;3:131–137. [PubMed] [Google Scholar]
- Pope WF. Embryonic mortality in swine. In: Zavy MT, Geisert RD, editors. Embryonic mortality in domestic species. CRC Press; Boca Raton, FL: 1999. pp. 53–77. [Google Scholar]
- Pope WF, Lawyer MS, First NL. Intrauterine migration of the porcine embryo: coordination of bead migration with estradiol. J Anim Sci. 1986;63:848–853. doi: 10.2527/jas1986.633848x. [DOI] [PubMed] [Google Scholar]
- Rainey WE. Adrenal zonation: clues from 11beta-hydroxylase and aldosterone synthase. Mol Cell Endocrinol. 1999;151:151–160. doi: 10.1016/s0303-7207(99)00051-9. [DOI] [PubMed] [Google Scholar]
- Rainey WE, Bird IM, Mason JI. The NCI-H295 cell line: a pluripotent model for human adrenocortical studies. Mol Cell Endocrinol. 1994;100:45–50. doi: 10.1016/0303-7207(94)90277-1. [DOI] [PubMed] [Google Scholar]
- Rison SC, Thornton JM. Pathway evolution, structurally speaking. Curr Opin Struct Biol. 2002;12:374–382. doi: 10.1016/s0959-440x(02)00331-7. [DOI] [PubMed] [Google Scholar]
- Roy S. Multifunctional enzymes and evolution of biosynthetic pathways: retro-evolution by jumps. Proteins. 1999;37:303–309. doi: 10.1002/(sici)1097-0134(19991101)37:2<303::aid-prot15>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- Ryan BC, Vandenbergh JG. Intrauterine position effects. Neurosci Biobehav Rev. 2002;26:665–678. doi: 10.1016/s0149-7634(02)00038-6. [DOI] [PubMed] [Google Scholar]
- Schmidt S, Sunyaev S, Bork P, Dandekar T. Metabolites: a helping hand for pathway evolution? Trends Biochem Sci. 2003;28:336–341. doi: 10.1016/S0968-0004(03)00114-2. [DOI] [PubMed] [Google Scholar]
- Sekihara H. 19-hydroxyandrostenedione as a new hypertensinogenic agent. J Steroid Biochem. 1982;16:329–331. doi: 10.1016/0022-4731(82)90185-6. [DOI] [PubMed] [Google Scholar]
- Sekihara H, Yazaki Y, Kojima T. 19-Hydroxyandrostenedione amplifies the hypertensive action of mineralocorticoids in rats. J Endocrinol. 1993;138:31–40. doi: 10.1677/joe.0.1380031. [DOI] [PubMed] [Google Scholar]
- Simpson ER, Mahendroo MS, Means GD, Kilgore MW, Hinshelwood MM, Graham-Lorence S, Amarneh B, Ito Y, Fisher CR, Michael MD. Aromatase cytochrome P450, the enzyme responsible for estrogen biosynthesis. Endocr Rev. 1994;15:342–355. doi: 10.1210/edrv-15-3-342. [DOI] [PubMed] [Google Scholar]
- Simpson ER, Zhao Y, Agarwal VR, Michael MD, Bulun SE, Hinshelwood MM, Graham-Lorence S, Sun T, Fisher CR, Qin K, Mendelson CR. Aromatase expression in health and disease. Recent Prog Horm Res. 1997;52:185–213. [PubMed] [Google Scholar]
- Slominski A, Semak I, Zjawiony J, Wortsman J, Gandy MN, Li J, Zbytek B, Li W, Tuckey RC. Enzymatic metabolism of ergosterol by cytochrome p450scc to biologically active 17alpha,24-dihydroxyergosterol. Chem Biol. 2005a;12:931–939. doi: 10.1016/j.chembiol.2005.06.010. [DOI] [PubMed] [Google Scholar]
- Slominski A, Semak I, Zjawiony J, Wortsman J, Li W, Szczesniewski A, Tuckey RC. The cytochrome P450scc system opens an alternate pathway of vitamin D3 metabolism. FEBS J. 2005b;272:4080–4090. doi: 10.1111/j.1742-4658.2005.04819.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slominski A, Semak I, Wortsman J, Zjawiony J, Li W, Zbytek B, Tuckey RC. An alternative pathway of vitamin D metabolism. Cytochrome P450scc (CYP11A1)-mediated conversion to 20-hydroxyvitamin D2 and 17,20-dihydroxyvitamin D2. FEBS J. 2006;273:2891–2901. doi: 10.1111/j.1742-4658.2006.05302.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowls LK. Javelinas and other peccaries: their biology, management, and use. Texas A & M University Press; College Station: 1997. [Google Scholar]
- Suhara K, Ohashi K, Takahashi K, Katagiri M. Aromatase and nonaromatizing 10-demethylase activity of adrenal cortex mitochondrial P-450(11)beta. Arch Biochem Biophys. 1988;267:31–37. doi: 10.1016/0003-9861(88)90004-5. [DOI] [PubMed] [Google Scholar]
- Tchoudakova A, Callard GV. Identification of multiple CYP19 genes encoding different cytochrome P450 aromatase isozymes in brain and ovary. Endocrinology. 1998;139:2179–2189. doi: 10.1210/endo.139.4.5899. [DOI] [PubMed] [Google Scholar]
- Tchoudakova A, Kishida M, Wood E, Callard GV. Promoter characteristics of two cyp19 genes differentially expressed in the brain and ovary of teleost fish. J Steroid Biochem Mol Biol. 2001;78:427–439. doi: 10.1016/s0960-0760(01)00120-0. [DOI] [PubMed] [Google Scholar]
- Thornton JW. Evolution of vertebrate steroid receptors from an ancestral estrogen receptor by ligand exploitation and serial genome expansions. Proc Natl Acad Sci USA. 2001;98:5671–5676. doi: 10.1073/pnas.091553298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton JW, Need E, Crews D. Resurrecting the ancestral steroid receptor: ancient origin of estrogen signaling. Science. 2003;301:1714–1717. doi: 10.1126/science.1086185. [DOI] [PubMed] [Google Scholar]
- Tong SK, Chiang EF, Hsiao PH, Chung B. Phylogeny, expression and enzyme activity of zebrafish cyp19 (P450 aromatase) genes. J Steroid Biochem Mol Biol. 2001;79:299–303. doi: 10.1016/s0960-0760(01)00146-7. [DOI] [PubMed] [Google Scholar]
- van der MJ, te KG, van Deursen R, Geelen J. Aromatase activity in individual day-11 pig blastocysts. J Reprod Fertil. 1989;87:783–788. doi: 10.1530/jrf.0.0870783. [DOI] [PubMed] [Google Scholar]
- Vanselow J, Furbass R, Zsolnai A, Kalbe C, Said HM, Schwerin M. Expression of the aromatase cytochrome P450 encoding gene in cattle and sheep. J Steroid Biochem Mol Biol. 2001;79:279–288. doi: 10.1016/s0960-0760(01)00144-3. [DOI] [PubMed] [Google Scholar]
- Veronneau S, Bernard H, Cloutier M, Courtemanche J, Ducharme L, Lefebvre A, Mason JI, Lehoux JG. The hamster adrenal cytochrome P450C11 has equipotent 11beta-hydroxylase and 19-hydroxylase activities, but no aldosterone synthase activity. J Steroid Biochem Mol Biol. 1996;57:125–139. doi: 10.1016/0960-0760(95)00249-9. [DOI] [PubMed] [Google Scholar]
- vom Saal FS. Variation in phenotype due to random intrauterine positioning of male and female fetuses in rodents. J Reprod Fertil. 1981;62:633–650. doi: 10.1530/jrf.0.0620633. [DOI] [PubMed] [Google Scholar]
- Werck-Reichhart D, Feyereisen R. Cytochromes P450: a success story. Genome Biol. 2000;1 doi: 10.1186/gb-2000-1-6-reviews3003. REVIEWS3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeagle PL. Cholesterol and the cell membrane. Biochim Biophys Acta. 1985;822:267–287. doi: 10.1016/0304-4157(85)90011-5. [DOI] [PubMed] [Google Scholar]
- Yoshida Y, Aoyama Y, Noshiro M, Gotoh O. Sterol 14-demethylase P450 (CYP51) provides a breakthrough for the discussion on the evolution of cytochrome P450 gene superfamily. Biochem Biophys Res Commun. 2000;273:799–804. doi: 10.1006/bbrc.2000.3030. [DOI] [PubMed] [Google Scholar]
- Zhang J, Zhang YP, Rosenberg HF. Adaptive evolution of a duplicated pancreatic ribonuclease gene in a leaf-eating monkey. Nat Genet. 2002;30:411–415. doi: 10.1038/ng852. [DOI] [PubMed] [Google Scholar]