Abstract
Plasma glutathione peroxidase (GPx-3) is a selenocysteine-containing extracellular antioxidant protein that catalyzes the reduction of hydrogen peroxide and lipid hydroperoxides. Selenoprotein expression involves the alternate recognition of a UGA codon as a selenocysteine codon and requires signals in the 3′-untranslated region (UTR), including a selenocysteine insertion sequence (SECIS), as well as specific translational cofactors. To ascertain regulatory determinants of GPx-3 expression and function, we generated recombinant GPx-3 (rGPX-3) constructs with various 3′-UTR, as well as a Sec73Cys mutant. In transfected Cos7 cells, the Sec73Cys mutant was expressed at higher levels than the wild type rGPx-3, although the wild type rGPx-3 had higher specific activity, similar to plasma purified GPx-3. A 3′-UTR with only the SECIS was insufficient for wild type rGPx-3 protein expression. Selenocompound supplementation and co-transfection with SECIS binding protein 2, increased wild type rGPx-3 expression. These results demonstrate the importance of translational mechanisms in GPx-3 expression.
Keywords: antioxidant, selenocysteine, 3′-untranslated region, regulation
Introduction
Glutathione peroxidases (GPx) comprise a family of enzymes that scavenge peroxides. Eight known isoforms exist [1, 2]; however, only five isoforms [1, 3] contain selenocysteine and are capable of catalyzing the reduction of hydrogen peroxide and lipid hydroperoxides using glutathione (GSH) as a reducing cofactor. Of these, plasma glutathione peroxidase (GPx-3) is the only known selenocysteine-containing extracellular antioxidant isoform; GPx-5 is extracellular, but does not contain a selenocysteine [4].
The human GPx-3 gene is approximately 10 kb in length spanning 5 exons on chromosome 5q32 with the first exon coding for the signal peptide and the second exon containing the selenocysteine (Sec) codon (UGA) [5]. GPx-3 mRNA expression is tissue-specific; it has been detected in the heart, lung, gastrointestinal tract, liver, brain, kidney, adipose tissue, breast, and placenta [6–11]; and is translated into a secreted protein found in extracellular fluids, such as ascitic fluid in rats, epithelial lining fluid, alveolar fluid, plasma, milk, and aqueous humor [6, 12–15]. The kidney has the greatest level of GPx-3 mRNA expression, which is specifically localized to the epithelial cell of the proximal tubule [15, 16]. The kidney was also demonstrated to be the main source of GPx-3 in plasma, as anephric patients show significantly lower levels of plasma GPx-3 activity compared to healthy controls [17] and plasma GPx-3 activity correlates with the adequacy of renal function.
As a secreted protein, GPx-3 is proposed to be a major scavenger of reactive oxygen species (ROS) in the extracellular space and within the vasculature [18] preserving nitric oxide (NO) bioavailability and promoting an antithrombotic environment. Our group previously showed that a deficiency of GPx-3 leads to enhanced platelet activation and an increased risk of thrombotic stroke [19, 20]. In addition, we identified a promoter haplotype that decreases GPx-3 transcriptional activity and gene expression and, in genetic studies, this haplotype correlates with an increased stroke risk in children and young adults [21], further supporting a role for GPx-3 in maintaining vascular homeostasis. GPx-3 has also been suggested to play a role in preventing plasma LDL oxidation, vascular inflammation, and atherogenesis [22].
The regulation of GPx-3 expression is thought to occur at the transcriptional, translational, and, perhaps, post-translational levels, although studies have been limited compared with other selenoproteins. Regulation of GPx-3 expression has been shown for hypoxia [23] at the transcriptional level [10], and for selenium and oxidative stress at the transcriptional and translational levels [23, 24]. In general, selenoprotein expression is known to be regulated by selenium, which is incorporated into the protein as selenocysteine (Sec). Several cis-acting elements, such as a UGA codon (coding for Sec) and the selenocysteine insertion sequence (SECIS), and trans-acting elements, such as a Sec-elongation factor (eEFSec), SECIS binding protein 2 (SBP2), and selenocysteine-specific tRNA (tRNAsec) [25], are important for incorporation of Sec in the polypeptide. The SECIS element is a stem loop structure found in the 3′ UTR of eukaryotic selenoprotein mRNA that serves to direct Sec incorporation into the growing polypeptide chain [26]. Eukaryotic SECIS binds SBP2 [27] presumably by bringing together cofactors such as tRNAsec, and an elongation factor, eEFsec [28], on the ribosome for efficient translation of the selenoprotein.
In order to understand in more detail the translational regulation of GPx-3, we generated recombinant human GPx-3 and compared its size, quaternary structure, and activity with those of GPx-3 purified from human plasma. We assessed: 1) the effect of the length of the 3′-UTR; 2) the effect of a Sec73Cys mutation; and 3) the roles of selenium, SBP2, eEFsec, and tRNAsec on protein expression. In addition, we examined the relationship between quaternary structure and specific enzymatic activity.
Materials and Methods
Cell Culture
All cell media, Dulbecco’s Phosphate Buffered Saline, and trypsin-EDTA (0.05%) were purchased from Invitrogen, Carlsbad, CA, except for Eagles’s Minimum Essential Medium, which was purchased from Invitrogen, Carlsbad, CA. All cell lines were obtained from ATCC, Manassas, VA. Fetal bovine serum (FBS) was purchased from Mediatech, Herndon, VA, and cells were maintained at 37° C in 95% O2 and 5% CO2. Sodium selenite, seleno-L-methionine, and Se-methylselenocysteine hydrochloride were purchased from Sigma, St. Louis, MO. African green monkey kidney fibroblast (Cos7) cells were grown in Dulbecco’s Modified Eagle Medium with 4.5 g/L D-glucose, 4 mM L-glutamine, 110 mg/L sodium pyruvate, and 10% FBS. Human renal proximal tubule (Caki-2) cells were grown in McCoy’s 5A medium with L-glutamine, 25 mM HEPES (Gibco), and 10% FBS. Human hepatoma (HepG2) cells were grown in Eagle’s Minimum Essential Medium containing non-essential amino acids, sodium pyruvate, and 10% FBS. Unless otherwise specified, all media were supplemented with 10 ng/ml sodium selenite.
Construction of Recombinant C-terminal His6-tagged GPx-3 (rGPx-3)
GPx-3 cDNA was amplified from a GPx-3 cDNA [23] using primers, GPx-3 cDNA NheI Forward, containing a Nhe I restriction site on the 5′ end of the GPx-3 cDNA, and GPx-3 cDNA XhoI Reverse, containing a Xho I restriction site on the 3′end of the GPx-3 cDNA (Table 1); the Nhe I/Xho I fragment was inserted into the pcDNA 4/V5 His C vector upstream of the V5/His-tag where expression of rGPx-3 is driven by a CMV promoter. A C-terminal His-tag vector was chosen to avoid the possibility of loss of an N-terminal His-tag during cleavage of N-terminal signal sequences in the secreted protein.
Table 1.
Summary of primers detecting rGPx-3 mRNA and endogenous GPx-3.
| Primer Name | Sequence | Position based on Accession # AY310878 | Size |
|---|---|---|---|
| GPx-3 cDNA | |||
| GPx-3 cDNA NheI Forward | 5′ ATA TGC TAG C CC ATG GCC CGG CTG CTG CAG G 3′ | 2181 | 693 bp |
| GPx-3 cDNA XhoI Reverse | 5′ CCT ACT CGA G CC TTC CTC TTG ACC CCC AGG GCT 3′ | 9656 | |
| 100 bp 3′ UTR SECIS-containing fragment | |||
| GPx-3 SECIS_100 PmeI Forward | 5′ ATA TGT TTA AAC GGA CCC CAT GGC AGG GGT 3′ | 10296 | 125 bp |
| GPx-3 SECIS_100 PmeI Reverse | 5′ CCT AGT TTA AAC TTA GCC TGA ATG CAC TAA 3′ | 10379 | |
| 500 bp 3′ UTR SECIS-containing fragment | |||
| GPx-3 SECIS_500 PmeI Forward | 5′ ATAT GT TTA AAC T GCC TAC AGG TAT GCG TGA TTG 3′ | 9959 | 529 bp |
| GPx-3 SECIS_500 PmeI Reverse | 5′ CCT A GT TTA AAC AAG CCA GTG GAC CAG TGA GGG GTG A 3′ | 10476 | |
| Full Length 3′ UTR SECIS-containing fragment | |||
| GPx-3 SECIS Full Length PmeI Forward | 5′ ATA TGT TTA AAC CTG AAG GCC GTC TCA TCC 3′ | 9660 | 872 bp |
| GPx-3 SECIS Full Length PmeI Reverse | 5′ CCT AGT TTA AAC CTG CAG AAA GGC TTT TAC 3′ | 10515 | |
| GPx-3 cDNA Selenocysteine Mutant | |||
| SeCysMut Forward | 5′ CAA CGT GGC CAG CTA CTG TGG CCT GAC GGG CCA G 3′ | 6982 | Sec→Cys |
| SeCysMut Reverse | 5′ CTG GCC CGT CAG GCC ACA GTA GCT GGC CAC GTT G 3′ | 7015 | |
| Recombinant GPx-3 Transfects Detection | |||
| GPx3 His-tag Forward | 5′ GGT CAT CAT CAC CAT CAC CAT TGA 3′ | pcDNA 4/V5 His C vector 1053 | 95 bp (p100), 428 bp (p500), |
| SECIS Reverse | 5′ AGG TCC GCC CAC AAG GGC TTT G 3′ | 10358 | 731 bp (p800) |
| Recombiant GPx-3 Transfectants Detection with Real Time PCR | |||
| Real Time GPx-3 Forward | 5′ GCA ACG TCA AGA TGG ACA TCC T 3′ | 9592 | 112 bp |
| Real Time GPx-3 Reverse | 5′ GGA GAG GGT TAG GGA TAG GCT TAC 3′ | pcDNA 4/V5 His C vector 1029 | |
| TaqMan GPx-3 Probe | 5′ AGG TCA CCC ATT CGA A 3′ | pcDNA 4/V5 His C vector 989 | |
| Endogenous GPx-3 Detection | |||
| Endogenous 5′UTR GPx-3 Forward | 5′ ATA TCA CCC CGC CAT GGC CCG GCT G 3′ | Position 56 based on Bierl et al JBC 2004 | 688 bp |
| Endogenous GPx-3 cDNA Reverse | 5′ CC TTC CTC TTG ACC CCC AGG GCT 3′ | 9656 |
Restriction enzymes sites for NheI, XhoI, and PmeI are italicized preceded by 4 bp overhangs.
Endogenous 5′UTR GPx-3 Forward also contains 4 bp overhangs.
The Cys codon in the rGPx-3 Cys mutant is highlighted in bold and underlined.
To insert the SECIS element, the 3′-UTR was amplified using various primers containing a Pme I restriction site inserted downstream of the V5/His-tag of the construct described above. The full length 3′-UTR was amplified as a fragment approximately 800 bp in length. In addition, fragments were amplified that contained the SECIS and the 3′-terminal 500 bp of the 3′-UTR; and another fragment containing only the SECIS element, approximately 100 bp in length, was also amplified. The final constructs are denoted by the size of the 3′-UTR as p800 WT (rGPx-3 WT), p500, and p100, respectively. pcDNA 4/V5 LacZ and pcDNA 4/V5 empty vectors were also used as controls. All constructs were confirmed by sequencing (Dana-Farber/Harvard Cancer Center DNA Resource Core). All plasmids and restriction enzymes were purchased from Invitrogen, Carlsbad, CA, and New England Biolabs, Ipswich, MA, respectively. Primers are listed in Table 1.
Site Directed Mutagenesis
Site-directed mutagenesis was performed according to the manufacturer’s protocol (Stratagene, La Jolla, CA) using primers, SeCysMut Forward and SeCysMut Reverse (Table 1), to introduce a Cys in place of a Sec (TGA→TGT) at position 73 (Sec73Cys) in the p800 construct. This construct is identified as p800 Cys (rGPx-3 Cys).
Transfection of rGPx-3 Constructs
Cos7, Caki-2, and HepG2 cells were grown to 70–90% confluence on 100 mm plates, and then transfected according the manufacturer’s protocol with Superfect Reagent (Qiagen, Valencia, CA). Both lysate and/or media were examined by Western blot and/or ELISA, while mRNA from all transiently transfected constructs, selenium supplementation, and cofactor studies were analyzed by quantitative real time PCR (qRT-PCR).
For selenium supplementation experiments, Cos7 cells were first selenium-starved for 5 days in 0.1% FBS with no supplemental selenium added. After 5 days, sodium selenite, seleno-L-methionine, or Se-methylselenocysteine hydrochloride was added at 0, 29, 58, 145, 289 and 578 nM in 0.1% FBS to p800 WT transiently transfected cells. Lysate and media were collected after 48 hours.
For the translational cofactor experiments, cDNAs were transiently transfected at 2.5 μg each along with 15 μg p800 WT rGPx-3 construct. The p800 WT was also transfected with 7.5 μg, 5 μg, and 2.5 μg empty vector as transfection controls. Both lysate and media were collected after 48 hours. Human selenophosphate synthetase D (SelD) [29] (Genebank accession # U34044), Xenopus Laevis selenocysteine-specific tRNA (tRNAsec) [30] (Genebank accession #M34507), and rat (full-length) SECIS-binding protein 2 (SBP2) [31] (Genebank accession # NM_024002) cDNAs were kind gifts from Drs. Marla Berry, Dolph Hatfield, and Paul Copeland, respectively.
Stable Cell Line Generation
Cos7 cells were transfected as described above. Stable cell lines were established according to the manufacturer’s protocol (Qiagen, Valencia, CA). The cells were plated at several dilutions for selection with 400 μg/ml Zeocin (Invitrogen, Carlsbad, CA) for 1–2 months or until colony formation appeared, and were maintained with 200 μg/ml Zeocin.
Reverse-Transcriptase PCR (RT-PCR)
Endogenous and recombinant GPx-3 mRNA expression was monitored by RT-PCR. RNA was isolated from 100 mm plates using the RNeasy Mini Kit according to the manufacturer’s protocol (Qiagen, Valencia, CA). RNA was quantitated by UV absorbance using a DU 800 spectrophotometer (Beckman Coulter, Fullerton, CA). Approximately 1 μg RNA was used to generate cDNA using the RT-for-PCR kit (Clontech Mountain View, CA). Primers were generated to detect only rGPx-3 transcripts by locating a forward primer, GPx3 His-tag Forward, that binds to the His-tag sequence and a reverse primer, GPx-3 SECIS Reverse, that binds to the SECIS element. Glyceraldehyde-3-phosphate dehydrogenase (G3PDH) was used as an endogenous control. PCR was performed according to the conditions described above using the Titanium Taq PCR Kit according to the manufacturer’s protocol (Clontech, Mountain View, CA). Samples were visualized by agarose gel electrophoresis and ethidium bromide staining (American Bioanalytical, Natick, MA).
Quantitative Real Time PCR (qRT-PCR)
RNA was isolated, quantitated, and cDNA generated as described above. The TaqMan detection system was used to monitor synthesis of recombinant GPx-3. Primers and the TaqMan probe used are listed in Table 1. Recombinant GPx-3 was normalized to rhesus G3PDH (AB Biosystems, Foster City, CA) as the endogenous control and values were then compared to those of p800 WT to provide relative levels. Samples were run on a 7900HT Fast Real Time PCR System (AB Biosystems, Foster City, CA).
Purification of rGPx-3
rGPx-3 from transiently and stably transfected Cos7 cell media was purified using His-Gravi Trap immobilized metal ion affinity chromatography columns (IMAC) for large-scale preparations (GE Healthcare, Chalfont St. Giles, UK), while small-scale purification of rGPx-3 from transiently transfected Cos7 cells involved the use His-Spin Trap columns (GE Healthcare, Chalfont St. Giles, UK) according to the manufacturer’s protocols Binding buffer contained 20 mM sodium phosphate, 500 mM NaCl, and 30 mM imidazole, pH 7.4, and wash buffer contained 20 mM sodium phosphate, 30 mM imidazole, pH 7.4, with 0.5% Tween 20. Protein was eluted with 20 mM sodium phosphate, 500 mM imidazole, pH 7.4, with 0.5% Tween 20. For some experiments, cell lysates were prepared in parallel using RIPA buffer (Sigma, St. Louis, MO). Both cell lysate and media were processed in the presence of inhibitors of aspartic, cysteine, and serine aminopeptidases (Calbiochem, San Diego, CA). Following purification, some samples were concentrated with a YM-10 centrifugal device (Millipore, Billerica, MA). Protein concentrations were quantitated using the DC Protein Assay (Biorad, Hercules, CA).
Purification of Human Plasma Glutathione Peroxidase
Human plasma was obtained from Brigham and Women’s Hospital Blood Bank with institutional approval. Human GPx-3 was purified from 300 ml plasma according to the method of Maddipati and Marnett [32] with the following modifications. Purification was performed using a FPLC system (GE Healthcare, Chalfont St. Giles, UK). The enzyme was loaded onto DEAE Sephadex column and eluted with a linear gradient of 0–1 M NaCl in buffer Q (20 mM Tris, 5mM EDTA, and 1mM glutathione, pH 8.0). Following gel filtration on a Superdex 200 column, the protein sample was applied to a MonoQ column (GE Healthcare, Chalfont St. Giles, UK) and washed with a 40 ml bed volume of buffer Q. The sample was eluted with a linear gradient of 0–1M NaCl in buffer Q.
SDS-PAGE and Western Blot
Cell media or purified samples were prepared in reducing/denaturing gel loading buffer containing 250 mM Tris-HCl, pH 6.8, 8% sodium dodecyl sulfate (SDS), 8% β-mercaptoethanol (βME), glycerol, and 0.02% bromphenol blue, and heated at 100°C for 10 minutes. To prepare non-reduced samples, βME was omitted from the loading buffer and samples were not heat-denatured prior to electrophoresis. Samples were separated on 4–15% gradient sodium dodecyl sulfate polyacrylamide (SDS-PAGE) gels (Biorad, Hercules, CA), then transferred to methanol-treated polyvinylidene fluoride (PVDF) membranes (Biorad, Hercules, CA) using a semi-dry blotting system. Membranes were blocked with 5% milk in Tris-Buffered Saline Tween-20 (TBST) (Biorad, Hercules, CA). Recombinant proteins or plasma purified GPx-3 was detected using a 1:250 dilution of the primary anti-V5 antibody or a 1:500 dilution of the primary monoclonal GPx-3 antibody (Abcam, Cambridge, MA) followed by a 1:1000 dilution of a secondary anti-mouse IgG HRP antibody (Sigma, St. Louis, MO). Actin was detected using an anti-actin antibody (Sigma, St. Louis, MO) at a 1:4000 dilution and a secondary anti-rabbit IgG HRP secondary antibody at a 1:2000 dilution (Cell Signaling, Danvers, MA). Positope was used as an anti-V5 primary antibody control (Invitrogen, Carlsbad, CA), while partially purified human plasma GPx-3 served as a positive control for the monoclonal anti-GPx-3 antibody (Abcam, Cambridge, MA).
Primary antibody incubation was performed overnight at 4° C, and secondary antibody incubation was performed for 1–1.5 hours at room temperature. An ECL Plus Western Blotting Detection System was used for protein detection (GE Healthcare, Chalfont St. Giles, UK), membranes were exposed to Amersham Hyperfilm HCl (GE Healthcare, Chalfont St. Giles, UK), and the film was developed with an X-OMAT 2000A Processor (Kodak, Rochester, NY). Densitometry was performed with the VersaDoc 4000 Imaging System (Biorad, Hercules, CA).
Enzyme-linked ImmunoSorbent Assay (ELISA)
To develop an ELISA method for rGPx-3 detection, we used a His-tag antibody plate (Novagen, San Diego, CA) that contains an immobilized His-tag monoclonal antibody. In order to generate a standard for the His-tagged protein, the pcDNA 4/V5 Lac Z vector was transiently transfected in Cos7 cells, and recombinant Lac Z (rLacZ) in cell extracts was quantified by comparing recombinant β-galactosidase activity to that of a LacZ (Sigma, St. Louis, MO) standard. Since rLacZ contains an identical His-tag and V5 epitope as the rGPx-3, known quantities of rLacZ were used to standardize the molar quantity of rGPx-3. Briefly, rGPx-3 and rLacZ standards were added to the ELISA plate at 4°C overnight or at room temperature for 3 hr. After hybridization, the plate was washed and a primary anti-V5-HRP antibody at a 1:500 dilution (Invitrogen, Carlsbad, CA) was added. After 1 hr of antibody incubation at room temperature, the plate was washed and next incubated with 3, 3′ 5, 5′-tetramethylbenzidine (TMB) (Calbiochem, San Diego, CA) to detect HRP activity. The reaction was stopped with 250 mM HCl and absorbance was measured at 450 nm wavelength using a Spectro Max 190 plate reader (Molecular Devices, Sunnyvale, CA).
Mass Spectrometry
Purified rGPx-3 samples (p800 WT and p800 Cys mutant) were separated on 4–15% gradient SDS-PAGE gels, and fragments migrating at approximately 30 kDa were excised from SDS-PAGE gels stained with Biosafe Coomassie Blue (Biorad, Hercules, CA) or with silver stain (Invitrogen, Carlsbad, CA). Excised samples were subjected to tryptic digestion, and the proteins identified by matrix-assisted laser desorption/ionization (MALDI) mass spectrometry (Dana Farber Mass Spectrometry Facility).
GPx Activity Assay
GPx-3 activity was also determined using a commercially available kit for GPx-1 activity (Calbiochem, San Diego, CA). Activity was indirectly monitored spectrophotometrically by the reduction of glutathione disulfide (GSSG) recycled to reduced glutathione (GSH) by glutathione reductase using NADPH as a reducing agent. Enzymatic activity is quantitated by the change in NADPH absorbance at 340 nm over time.
In-gel Activity Assay
The current method of the in-gel activity assay uses reduced glutathione (GSH) and hydrogen peroxide to visualize a band of GPx activity in a polyacrylamide gel as described by Lin and colleagues [33]. GPx-1 standards from bovine erythrocytes along with the sample of interest were run on a 4–15% SDS-PAGE gel. The gel was then rinsed with 25% isopropanol in 10 mM Tris-HCl buffer (pH 7.9) for 10 min and then equilibrated in 50 mM Tris-HCl buffer (pH 7.9) for 15 min. The gel was then soaked in substrate solution containing 50 mM Tris-HCl buffer (pH 7.9), 13 mM GSH, 0.007% t-butyl peroxide (t-BuOOH), for 15 min with gentle shaking and next rinsed with water. The gel was developed by gently shaking in the dark at room temperature for 10 min with a developing solution composed of 1.2 mM 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide and 1.6 mM phenazine methosulfate. The gel was then rinsed with water until clear zones were observed on a purple background representing GPx activity. Activity was measured based on the density of the reversed image obtained from the Biorad, Hercules, CA, Versadoc scanner.
Amplex Red GPx-3 Endpoint Activity Assay
The Amplex Red/GPx fluorometric endpoint assay is a variation of the GPx1 spectrophotometric activity assay (Calbiochem, San Diego, CA) and modified from the Amplex Red Catalase Assay Kit (Molecular Probes, Invitrogen, Carlsbad, CA). In this modification, GPx3 first reduces H2O2 (H2O2 + 2GSH→GSSG + 2H2O), then Amplex Red in the presence of horseradish peroxidase reacts with un-reacted H2O2 to produce a fluorescent resorufin product. GPx-1 standards at 0, 3.13, 6.25, 12.5, 25, and 50 mU in assay buffer (50 mM Tris HCl pH 7.6, 5 mM EDTA) were used as controls. To initiate H2O2 reduction by GPx, 35 μl 2 mM GSH/well and 35 μl 0.007% t-BuOOH/well were added to 42 μl standard or sample per well in a 96-well plate. Reaction mixes were incubated at room temperature for 5 min. After the initial incubation, 100 μl Amplex Red (10 mM) and 40 μl HRP (100 U/ml) in 1X reaction buffer were added to each well, and the plate was incubated in the dark at 37° C for 30 min. Fluorescence was measured at 530 λ (excitation) and 590 λ (emission) with the Spectro Max Gemini XPS fluorescent plate reader (Molecular Devices, Sunnyvale, CA). Fluorescence is inversely proportional to enzyme activity.
Statistics
Statistical analyses were performed using SigmaStat 3.01 (SPSS Inc, Chicago, IL). An unpaired, two-tailed Student’s t-test was performed to compare two groups or individual treatment groups to their respective controls. One-way ANOVA followed by pair-wise analysis with the Student Newman-Keuls test was used to determine significant differences in experiments with multiple treatments.
Results
rGPx-3 Transient and Stable Cell Line Generation
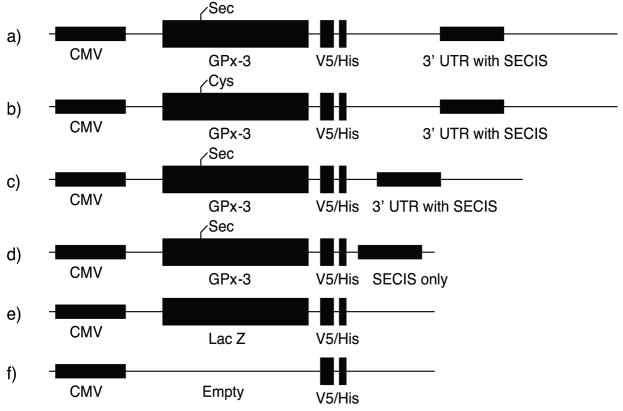
The low level of expression of selenoproteins and the unique translational mechanism render regulation of their expression complex. Thus, we first sought to overexpress recombinant GPx-3 by cloning the human GPx-3 cDNA and its full length 3′-UTR containing the SECIS element into a pcDNA 4/V5 His C vector, the resulting construct denoted p800 WT rGPx-3. In addition, past studies on GPx-3 overexpression in cultured cells demonstrated GPx-3 detection with [75Se] based only on transient transfection of HEK293 cells [23]. For more quantitative detection of transient and permanent transfectants, we used a vector that contains a unique C-terminal 6xHis-tag and a V5 epitope. Various lengths of the 3′-UTR containing the SECIS element were also cloned into the GPx-3 construct and denoted by the approximate length (in base pairs) of the 3′-UTR (p100, p500). As Sec insertion at a UGA codon involves several translational cofactors, we also constructed a Sec73Cys mutant to compare the translational efficiency of Sec-containing vs. Cys-containing constructs in this expression system. Table 1 summarizes the primers used for rGPx-3 generation, and Fig. 1 illustrates all constructs used.
Fig. 1. Summary of Recombinant GPx-3 Constructs.
Recombinant GPx-3 contains a human GPx-3 cDNA driven by a CMV promoter through a V5 epitope and His-tag. The constructs include: a) rGPx-3 with full length 3′-UTR; b) a Cys mutant (Sec73Cys) with a full length 3′-UTR; c) rGPx-3 with a 500 bp 3′-UTR; d) rGPx-3 with a 100 bp SECIS-only 3′-UTR fragment; e) rLacZ control; and f) an empty vector control.
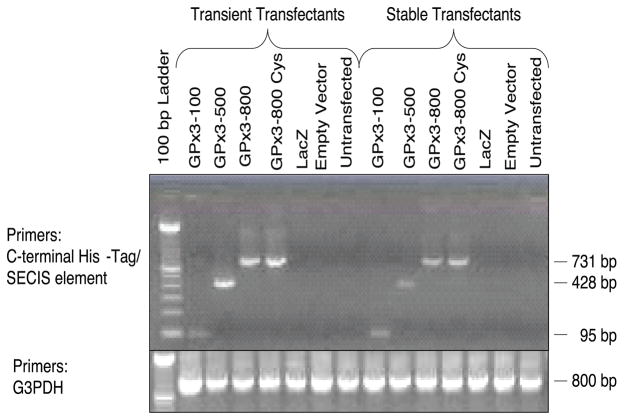
In order to understand better the requirements for selenoprotein synthesis and selenocysteine incorporation in GPx-3, we generated stable cell lines for each of the constructs for large scale purification and characterization. Avissar and colleagues showed that Caki-2, a human proximal tubule cell line, had greater levels of endogenous GPx-3 expression than several other cell types tested [16]. In our preliminary studies, stable cell lines established at first using Caki-2 cells showed very low rGPx-3 expression (data not shown). Transient transfection in Caki-2 and Hep62 resulted in detectable levels of rGPx-3 following His-Spin column purification. We found a consistent higher yield of rGPx-3 from the p800 Cys construct compared with the p800 WT construct in these cells. In order to enhance protein expression, Cos7 cells were used to overexpress the various recombinant GPx-3 proteins. We achieved successful transcription of rGPx-3 mRNA in both transiently and stably transfected Cos7 cells for all of the constructs used as detected by reverse transcriptase PCR (Fig. 2).
Fig. 2. rGPx-3 mRNA Expression in Transient and Stable Cos7 cells.
Reverse transcriptase-PCR detecting rGPx-3 in transiently and stably transfected cell lines.
Effects of 3′-UTR on rGPx-3 Protein Expression
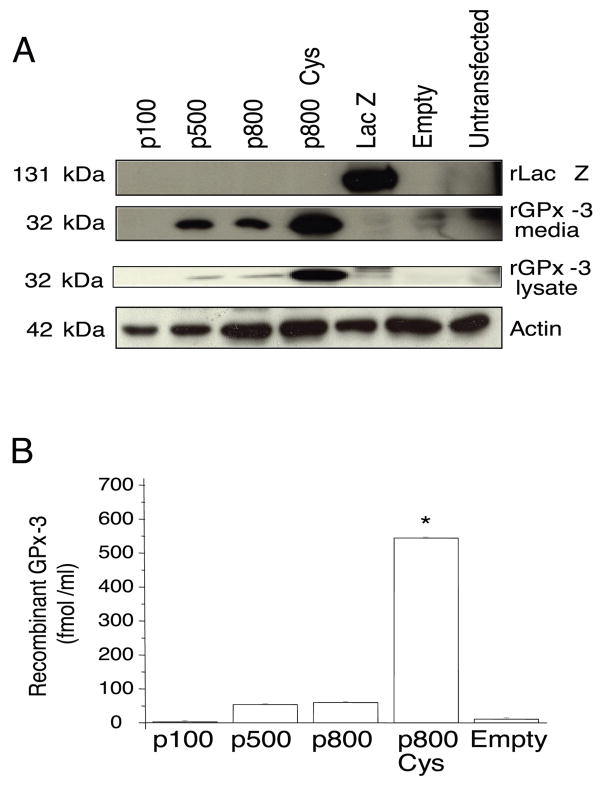
To understand the role the 3′-UTR has on rGPx-3 protein expression and secretion, Cos7 cells were transiently transfected with p100, p500, p800 WT, and p800 Cys mutants. Western blots using anti-V5 antibody detected rGPx-3 protein in media and lysates when Cos7 cells were transfected with the p500, p800 WT, and p800 Cys constructs (Fig. 3a), and in media in stably transfected cells (data not shown). The truncated p500 construct allowed for expression of rGPx-3 at levels similar to the p800 WT; however, no rGPx-3 protein expression was observed when Cos7 cells were transfected with the p100 mutant (Fig. 3a). These findings are consistent with previous studies in our laboratory showing that the truncated 500 bp GPx-3 3′-UTR is sufficient to direct selenium-dependent translation (using a GPx-1 promoter-driven luciferase expression “read-through” assay) [34].
Fig. 3. Effect of 3′ UTR on rGPx-3 Protein Expression.
a) SDS-PAGE and immunoblot of purified rGPx-3 constructs from the media and lysates of transiently transfected Cos7 cells detected by anti-V5 antibody. rLac Z was used as a His-Spin purification control, and actin was used as a loading control. The Western blot is representative of three separate experiments. b) ELISA was used to quantitate protein expression directly for each non-purified secreted rGPx-3 form (p<0.03). Each bar represents the average of three experiments performed in duplicate, and values are given as mean +/− SD.
His-tag ELISA detection confirmed these observations using anti-V5-HRP. The p100 construct did not produce any detectable rGPx-3 from Cos7 cells, while a similar amount of rGPx-3 was detected intracellularly and in the media of transient transfectants using the p500 and p800 WT constructs. The p800 Cys mutant, in contrast, was expressed at levels nearly 10-fold higher (Fig. 3b) than any Sec-containing species.
To determine whether the increased recovery of rGPx-3 with the Cys mutation was due to alterations in mRNA levels or improved translational efficiency, we developed a TaqMan-based assay to quantitate rGPx-3 transcripts from Cos7 cells and compared levels of rGPx-3 relative to p800 WT after normalization to endogenous G3PDH mRNA. Basal levels of rGPx-3 transcripts were similar in p800 WT and p800 Cys permanent transfectants (1.02 ± 0.11 vs. 1.06 ± 0.22) and also in transiently transfected cells (1.00 ± 0.5 for p800 WT vs. 1.27 ± 0.17 for p800 Cys). To determine whether there was a difference in mRNA stability between p800 WT and p800 Cys transcripts, we treated transfectants with actinomycin D and measured rGPx-3 mRNA over 8 hours. There was no difference in the half-lives (approximately 7 hours) of the p800 WT and p800 Cys rGPx-3 transcripts (data not shown). These findings indicate that translational mechanisms account for the differences in rGPx-3 between p800 WT and p800 Cys transfectants as the amounts and stability of the rGPx-3 transcripts are the same in cells transfected with either of these constructs.
To confirm that the protein we had isolated was GPx-3, protein products of appropriate molecular size were excised from silver and Coomassie-stained SDS-PAGE gels and examined by mass spectrometry. Table 2 shows the specific peptides detected. Consistent with the greater amount of rGPx-3 isolated from p800 Cys than p800 WT transfectants, more peptides were identified by mass spectrometric analysis in the former than the latter. Similarly, more peptides were detected from the purified plasma GPx-3 owing to its greater abundance within the gel run and possibly a reflection of its comparatively high abundance in plasma. Peptide coverage was approximately 9, 22, and 50% for p800 WT, p800 Cys, and partially purified GPx-3 from plasma, respectively. These data confirm that the proteins we isolated are GPx-3.
Table 2.
Summary of p800 WT, p800 Cys, and Partially Purified Plasma GPx-3 by MALDI-TOF Analysis
| Sample Name | m/z submitted | MH+ Matched | Modifications | Start | End | Sequence |
|---|---|---|---|---|---|---|
| rGPx-3 p800 WildType Media | 1851.907 | 1851.9932 | 209 | 224 | (K)MDILSYMRRQAALGVK(R) | |
| 1867.8704 | 1867.9881 | 1Met-ox | 209 | 224 | (K)MDILSYMRRQAALGVK(R) | |
| 1945.8593 | 1945.9834 | 2Met-ox | 202 | 217 | (R)TTVSNVKMDILSYMRR(Q) | |
| rGPx-3 p800 Cys Mutant Media | 1203.6006 | 1203.6408 | 145 | 153 | (K)EQKFYTFLK(N) | |
| 1314.6949 | 1314.7239 | 186 | 197 | (K)FLVGPDGIPIMR(W) | ||
| 1537.5616 | 1537.7744 | pyroGlu | 107 | 120 | (K)QEPGENSEILPTLK(Y) | |
| 1851.8746 | 1851.9932 | 209 | 224 | (K)MDILSYMRRQAALGVK(R) | ||
| 1867.7609 | 1867.9881 | 1Met-ox | 209 | 224 | (K)MDILSYMRRQAALGVK(R) | |
| 2581.0235 | 2581.3953 | 202 | 224 | (R)TTVSNVKMDILSYMRRQAALGVK(R) | ||
| Partially Purified Plasma GPx-3 | 1314.6752 | 1314.7239 | 186 | 197 | (K)FLVGPDGIPIMR(W) | |
| 1537.723 | 1537.7744 | pyroGlu | 107 | 120 | (K)QEPGENSEILPTLK(Y) | |
| 1554.7397 | 1554.801 | 107 | 120 | (K)QEPGENSEILPTLK(Y) | ||
| 1570.7498 | 1570.8199 | 169 | 180 | (R)LFWEPMKVHDIR(W) | ||
| 1633.6906 | 1633.7486 | 154 | 168 | (K)NSCPPTSELLGTSDR(L) | ||
| 1930.9224 | 1931.0221 | 186 | 201 | (K)FLVGPDGIPIMRWHHR(T) | ||
| 1954.9465 | 1955.0174 | 121 | 137 | (K)YVRPGGGFVPNFQLFEK(G) | ||
| 2581.1133 | 2581.3953 | 202 | 224 | (R)TTVSNVKMDILSYMRRQAALGVK(R) | ||
| 2654.2266 | 2654.3362 | 121 | 144 | (K)YVRPGGGFVPNFQLFEKGDVNGEK(E) | ||
| 3039.4189 | 3039.5323 | 121 | 147 | (K)YVRPGGGFVPNFQLFEKGDVNGEKEQK(F) |
Effects of Selenium on rGPx-3 Expression
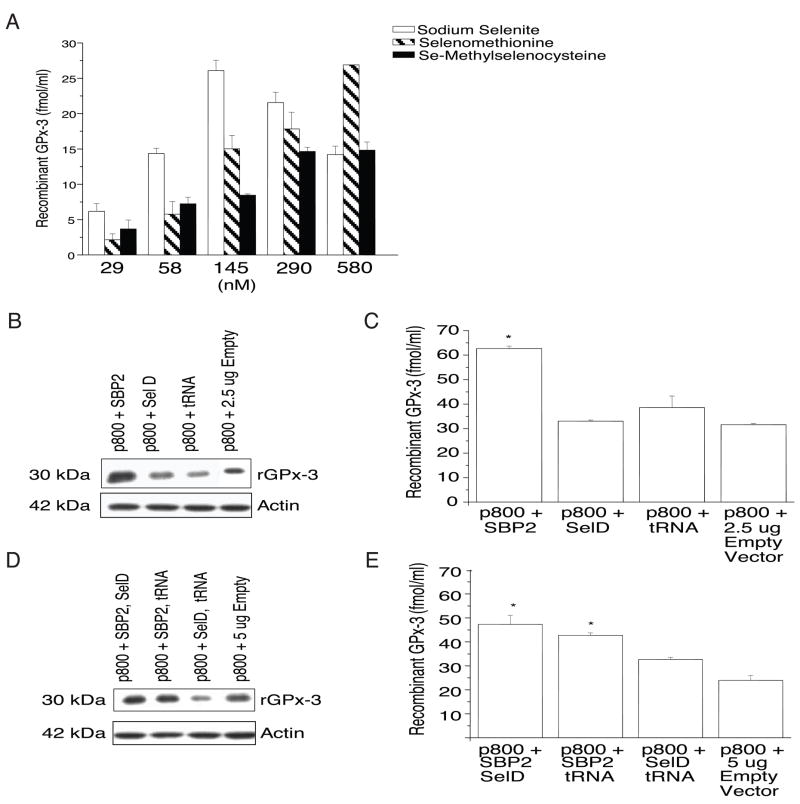
To study further the determinants of rGPx-3 expression, we transiently transfected Cos7 cells with the p800 WT construct to examine the effects of selenium supplementation. First, cells were selenium-starved for 5 days, followed by addition of various concentrations of inorganic selenium (sodium selenite), or organified selenium (as seleno-L-methionine or Se-methylselenocysteine hydrochloride) at the time of transfection. qRT-PCR revealed no change in mRNA expression by selenium treatment with any of the compounds used (data not shown), suggesting GPx-3 mRNA stability is not influenced by selenium levels in this system and that transcript stability is distinctly different for GPx-3 than GPx-1, the stability of which is reduced by selenium restriction, and similar to GPx4, which is not affected by selenium restriction [35]. ELISA quantitation showed that p800 WT rGPx-3 expression peaked at ~ 145 nM sodium selenite, decreasing at higher concentrations (Fig. 4a). Supplementation with either seleno-L-methionine or Se-methylselenocysteine also increased rGPx-3 expression; however, rGPx-3 protein expression appeared to plateau at ~ 290 nM with Se-methylselenocysteine treatment, but showed a dose-dependent increase over the full range of concentrations used for seleno-L-methionine.
Fig. 4. Effect of Selenium and Translational Cofactor Treatment on p800 WT rGPx-3 Expression.
a) Sodium selenite, seleno-L-methionine, or Se-Methylselenocysteine hydrochloride was added to transiently transfected p800 WT for 48 hr and media examined by His-tag ELISA (p<0.05). Each bar represents the average of three experiments performed in duplicate, and values are given as mean +/− SD. b-e) Addition of 2.5 μg of SBP2, SelD, and tRNAsec to transiently transfected p800 WT Cos7 cells, after which the media and lysate were examined (after 48 hours) by Western blotting using an anti-V5 antibody; (b, d) and by His-tag ELISA (c, e) with one (b, c) cofactor co-transfection (p<0.05), or two (d, e) cofactor co-transfection (p<0.04). Each bar represents the average of five experiments performed in duplicate, and values are given as mean +/− SD.
Effects of Cofactors on rGPx-3 Expression
To determine effects of translational cofactors on p800 WT rGPx-3 expression, transient transfection of the p800 WT was performed together with transfection with cDNAs for the translational cofactors, SBP2, SelD, and tRNAsec, in Cos7 cells. Recombinant mRNA was examined by qRT-PCR (data not shown), and cell media was examined by immunobloting (Fig. 4b, d) and His-tag ELISA (Fig. 4c, e). SBP2 co-transfection increased rGPx-3 expression 2.5-fold compared to the control vector (Fig. 4c). Co-transfection with two cofactors also increased rGPx-3 expression in this system (Fig. 4e).
When three cofactor cDNAs were transfected, there was no difference in rGPx-3 expression compared to no cofactor treatment (data not shown). Co-transfection of equivalent amounts of vector showed decreasing rGPx-3 with increasing amounts of empty vector, suggesting transfection competition among vectors. qRT-PCR showed no effect on GPx-3 mRNA expression with any of the translational cofactors tested (data not shown).
Non-reduced and Reduced rGPx-3 Forms
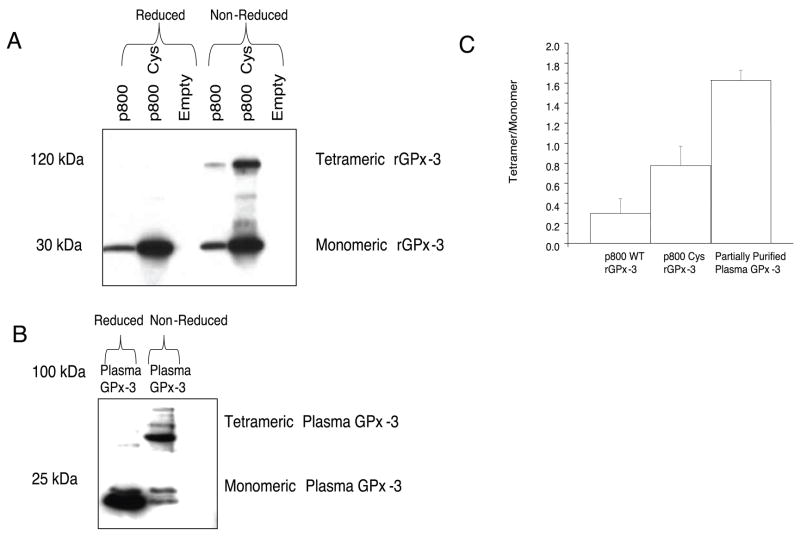
In order to ascertain the quaternary structure of rGPx-3, we examined the migration of non-denatured, non-reduced p800 WT and p800 Cys mutant from His-Gravi Trap large-scale purified stable cell lines on SDS-PAGE gels. In these samples, Western blot analysis showed an approximate 112 kDa band and a 28 kDa band, presumably the homotetrameric and monomeric rGPx-3 forms, respectively, as detected with the anti-V5 antibody (Fig. 5a). Heat-denaturation in the presence of βME eliminated quaternary structure, resulting only in bands of 28 kDa. In the partially purified plasma GPx-3, both the tetrameric (approximately 88 kDa) and monomeric (approximately 22 kDa) forms were detected in non-reduced samples (Fig. 5b). The differences in the size of rGPx-3 compared to GPx-3 from plasma reflect the approximate size of both the V5 epitope and His-tag. Densitometry showed that there was an approximate 2.5-fold increase in the p800 Cys homotetrameric form compared to the p800 WT homotetrameric form when comparing the ratio of tetramer-to-monomer (Fig. 5c). Additional bands may correspond to dimeric and trimeric forms in the non-denatured p800 Cys mutant preparation (Fig. 5a) [36].
Fig. 5. rGPx-3 and Plasma GPx-3 Forms.
Western blot of His-Gravi Trap purified rGPx-3 in Cos7 media were run on a SDS-PAGE gel under reducing and non-reducing conditions and compared to partially purified GPx-3 from human plasma. a) rGPx-3 was detected using an anti-V5 antibody; b) GPx-3 from human plasma was detected by using a monoclonal GPx-3 antibody; and c) densitometric ratio of tetrameric-to-monomeric forms of rGPx-3 and partially purified GPx-3 from plasma. The Western blot is representative of three separate experiments. Each bar represents the average densitometry of three experiments, with values given as mean +/− SD.
GPx-3 Enzyme Activity
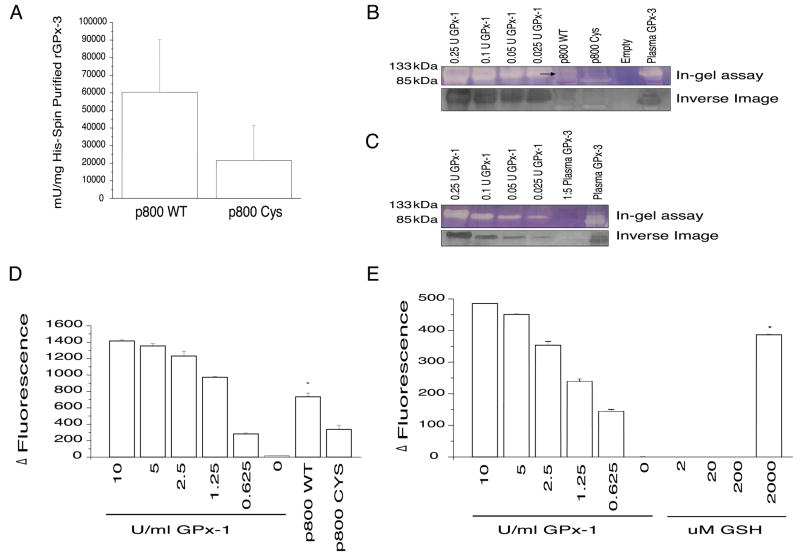
In order to compare enzymatic activity of the p800 WT and p800 Cys mutant, we performed a spectrophotometric activity assay of large-scale His-Gravi Trap purified rGPx-3 with GSH and t-BuOOH. The relationship between partially purified GPx3, rGPx-3 WT, and rGPx-3 Cys from Western blot band intensity and protein determinations were established to quantify rGPx-3 specific activity. The p800 WT protein showed approximately 3-fold greater specific activity than the p800 Cys mutant protein (Fig. 6a). Partially purified plasma GPx-3 had similar specific activity as the rGPx-3 WT (data not shown).
Fig. 6. Plasma GPx-3 Forms and Effect on GPx-3 Activity.
a) rGPx-3 Activity of His-Gravi Trap Purified Cos7 Cell Media. p800 WT rGPx-3 was His-Gravi Trap purified from transiently transfected Cos7 cell media and activity was determined spectrophotometrically and compared to the activity of rGPx-3 Cys. The p800 WT bar represents the average of N=17 experiments and the p800 Cys represents the average of N=8 (p<0.05), with values are given as mean +/− SD. b) In-gel Activity of rGPx-3. rGPx -3 activity was compared to GPx-1 from bovine erythrocytes using GSH and t-BuOOH as substrates. Partially purified GPx-3 from plasma was used as a control. The first panel represents the in-gel assay of the tetrameric of the standard GPx-1 compared to rGPx-3 WT, rGPx-3 Cys, and empty vector. The second panel represents the inverse image of the in-gel assay. The in-gel assay represents three independent experiments. c) In-gel Assay of Partially Purified GPx-3. A 5-fold dilution of the sample was compared to demonstrate a limit of detection with the in-gel assay. The first panel represents the in-gel assay of standard GPx-1 and GPx-3. The second panel represents the inverse image of the in-gel assay. d) rGPx-3 Activity by the Amplex Red Assay. His-Gravi Trap purified and concentrated rGPx-3 WT and rGPx-3 Cys activity using GSH and t-BuOOH (p<0.005). Each bar represents the average of three experiments, with values are given as mean +/− SD. e)Partially purified plasma GPx-3 activity using 2 μM, 20 μM, 200 μM and 2000 μM GSH (p<0.02). Each bar represents the average of three experiments, with values are given as mean +/− SD.
In order to examine the tetrameric form of rGPx-3 WT and rGPx-3, we employed the in-gel assay where the activity of each form can be visualized for GPx activity. Both GSH and t-BuOOH were used as substrates to detect non-denatured reduced GPx-1 and GPx-3. GPx-1 from bovine erythrocytes showed activity of its tetrameric form at approximately 100 kDa. rGPx-3 WT appeared to show modest activity at approximately the same size, but at a slightly greater size than that of partially purified plasma GPx-3 (Fig. 6b). The same size band did not appear in the p800 Cys and empty vector control lanes. A dilution of the partially purified form of GPx-3 from plasma (to concentrations equivalent to that of the most concentrated GPx-3 achieved) also showed modest and similar activity in the tetramer (Fig. 6c).
In order to compare the effect of the role of reducing cofactors on the enzymatic activity of GPx-3, wedeveloped a sensitive Amplex Red/GPx fluor ometric endpoint assay to detect rGPx-3 activity from cell media and partially purified GPx-3 from human plasma. GPx-3 has a 10-fold lower Km for GSH than GPx-1 [37]. GSH at 2, 20, 200, and 2000 μM, as well as proposed alternative reducing cofactors, such as thioredoxin reductase (TR), thioredoxin (Trx), and glutaredoxin (Grx), were tested at concentrations based on earlier work from Bjornstedt and colleagues [38] (0.05 μM, 2.5 μM, and 2.5 μM, respectively). An examination of rGPx-3 activity demonstrates that the rGPx-3 WT form has significantly more activity than the rGPx-3 Cys mutant, confirming the less sensitive assays; the difference is approximately 2-fold, which is similar to what was observed spectrophotometrically (Fig. 6d). Partially purified plasma GPx-3 showed activity only with GSH at 2 mM GSH (Fig. 6e), having none with TR, Trx, or Grx (data not shown).
Discussion
Selenoprotein expression, including GPx-3 regulation at the translational level, may be influenced by several factors, including a) mRNA stability, which may be affected by mRNA sequence and RNA binding proteins [39]; b) expression hierarchy of the selenoproteins [40]; c) the ability of the SECIS element to reinterpret the stop codon for Sec [41], allowing for the completion of polypeptide translation; and d) the cell or tissue type, and the cell-dependent availability of translational cofactors or selenium uptake mechanisms.
To study the contributors to GPx-3 expression, we relied on transient and/or stable transfection systems. This allowed us to analyze the contributions of the Sec codon, the 3′-UTR, as well as other factors, such as selenium and known translational cofactors, on GPx-3 expression. We found that stable cell lines demonstrated lower expression of rGPx-3 in the media compared to transiently transfected cells, possibly influenced by the number of GPx-3 cDNA that may have integrated into the cellular genome. Owing to this finding, transient cell lines were used for some of our studies.
Human liver (HepG2 [16], Hep3B [16, 42], human intestinal epithelial (Caco-2 [16, 42] and human kidney (Caki-2 [16], HEK293 [23] cell lines have previously been shown to secrete endogenous GPx-3 [16], and GPx-1 mRNA has been detected in HepG2 cells [42]. The human intestinal epithelialn Caco-2 cell line was also shown to secrete endogenous GPx-3 [17]. Our study is the first to use Cos7 cells for production of the selenoprotein GPx-3. We have found that Cos7, Caki-2, and HepG2 cells all can support the expression of mature, secreted GPx-3; however, owing to the very low levels of expression in HepG2 and Caki-2 cells, we primarily used Cos7 in our studies. As shown in other studies, conversion of the Sec codon to a Cys codon resulted in increased (i.e., more efficient translation of) protein synthesis [43–48].
Following transfection, we measured both rGPx-3 mRNA and protein. In this way, we could determine whether rGPx-3 expression was affected by facors influencing transcript levels or by translational mechanisms. The lack of difference in mRNA levels and mRNA stability between the p800 WT and p800 Cys constructs suggests that GPx-3 expression is primarily regulated at the translational level in this system. The differences in protein expression between the p800 WT and p800 Cys mutant could be explained by the differences in the selenol and thiol forms of the protein. Selenoprotein synthesis may be limited by translational cofactors in the p800 WT; synthesis of the Sec73Cys mutant, however, does not require any special translational mechanism.
Adequate selenium is important for the formation of Sec on the tRNAsec. In selenium starved cells, supplementation with inorganic selenium as well as organified forms of selenium, seleno-L-methionine and Se-methylselenocysteine, appears to enhance rGPx-3 protein expression. Seleno-L-methionine is also known to be incorporated in proteins in place of methionine [49] and must be metabolized to provide selenium exclusively for Sec incorporation into selenoproteins. Se-methylselenocysteine, in contrast, is not directly incorporated into proteins and, theoretically, is available exclusively for Sec incorporation [50] or can be rapidly excreted lowering the risk of oxidative toxicity associated with inorganic selenium (selenite) use.
The various responses to selenium treatment, particularly at higher concentrations of the selenocompounds, suggest that Cos7 cells may respond differently to different concentrations and different forms. High molar concentrations of sodium selenite may be cytotoxic, as selenite is a potent oxidant at these concentrations. Selenium deficiency is also cytotoxic in culture [51]. Zhong and colleagues demonstrated that excess sodium selenite induced apoptosis and cell cycle arrest after chronic exposure in human prostate cancer cells [52]. The same group demonstrated apoptosis by selenomethionine in the same cell lines [53]. In our cell system, the concentrations of selenium ranged from antioxidant levels to those found to be toxic or pro-oxidant. Each of the selenocompounds used had a unique effect on rGPx-3 production in this system. These findings may relate to differences in cellular uptake or metabolism of each compound.
The effects of selenium on mRNA and protein expression may be different in cultured cells than in tissues. Several studies have shown GPx-1 activity and transcript levels are sensitive to selenium deprivation [54, 55]; however, the decrease in GPx-1 activity did not always coincide with alterations in transcript levels [16]. Another group found that selenium deficiency reduces serum GPx activity in rats, while kidney GPx-3 mRNA levels remain constant [56].
In contrast, Reszka and colleagues showed that changes in GPx-3 mRNA levels in mouse fibrosarcoma cells were affected by culturing cells in low selenite conditions (levels as low as 1–5 ng Se/ml medium) [57]. These data suggest a low selenium range for optimal selenium expression for these particular cells. Based on our data, it is possible that Cos7 cells regulate selenium availability over a narrow range (levels such as 10–50 ng Se/ml) that affects translation but not mRNA stability. In the studies presented here, we also observed a dose-dependent increase of rGPx-3 expression in Cos7 cells after seleno-L-methionine and Se-methylselenocysteine treatment, suggesting that these compounds are also bioavailable for uptake into Cos7 cells. We are currently exploring the hierarchy of effects of selenium supplementation on GPx-3 and other selenoproteins in cell culture and in vivo systems.
Evidence on the regulation of selenoprotein synthesis is preliminary, and little is known about the efficiency of selenocysteine incorporation [25]. Additional translational cofactors have recently been shown to be important regulators of selenoprotein synthesis in eukaryotes, such as ribosomal protein L30 and nuclease sensitive binding protein 1 [58, 59]. Previous experiments in our laboratory showed that GPx-3 expression is affected by SBP2, EFsec, and tRNAsec [23]. Data presented here with the Sec translational cofactors demonstrate that SBP2 is important for optimal rGPx-3 expression in Cos7 cells. In contrast, Copeland and colleagues also demonstrated the importance of SBP2 in transfected rat hepatoma cells in vitro [27], but no further enhancement was seen in rat hepatoma cells using a luciferase reporter system [60]. In other studies, tRNAsec was shown to affect Sec incorporation greatly [61]. The importance of SBP2 in GPx-3 expression was also shown in studies of human subjects who carry a mutation in SBP2. These individuals had decreased serum GPx-3 activity (13% of normal) and decreased serum selenoprotein P levels [62], suggesting that SBP2 plays a direct role on GPx-3 and other selenoprotein expression in vivo. Interestingly, a recent study by Squires and colleagues provided further support for a hierarchy of selenoprotein synthesis. They demonstrated that SBP2 showed preferential binding to certain selenoprotein mRNAs, and may play a key role in preventing nonsense-mediated decay [63]. The levels of GPx-3 mRNA were too low to be reliably quantified; however, GPx-3 was predicted to be susceptible to nonsense-mediated decay owing to the position of an intron near the UGA (Sec) codon [63].
Our laboratory has also previously shown a trend toward increased expression of GPx-3 using SelD and tRNAsec [23] in human Caki-2 cells; however, these cofactors did not augment rGPx-3 protein levels when compared to SBP2 co-transfection alone in Cos7 cells. Several factors may influence these differences, including endogenous levels of expression of these factors or other differential features of these model systems.
Regulation of Sec incorporation at a UGA also requires the presence of the SECIS element in the 3′UTR of the transcript. Several studies have demonstrated that a 3′UTR from a selenocysteine gene is sufficient to promote selenocysteine incorporation in a heterologous protein containing a UGA in the open reading frame [64, 65]. These and other studies have defined sequence and structural characteristics of the stem-loop structure that are required for a functional SECIS element [66, 67]. For the rat type I deoidinase [67], rat thioredoxin reductase [68], and phospholipid hydroperoxide glutathione peroxidase [69], minimal functional SECIS elements of 45, 59, and 82 nt have been defined. We found, however, that a 100 nt piece overlapping the entire stem-loop-SECIS element of the human GPx-3 was insufficient for UGA-read-through in our cell system.
The formation of the tetrameric and monomeric rGPx-3 forms under non-reduced conditions suggests a potential limitation with the synthesis, processing, and formation of the tetramer that may affect its net enzymatic activity. GPx-3 is thought to exist primarily as a tetramer in plasma. It is possible that dimeric and monomeric forms may contribute to GPx-3 activity; however, the tetramer is considered the active form of the enzyme [36]. In purifying rGPx-3 from media, we found that a greater proportion was monomeric rather than tetrameric compared with the purified plasma form. It is not clear whether this difference is due to the expression system (V5/His-tag, the low levels of GPx-3 produced) or the purification process. Alternatively, the cell type may not have provided all the necessary factors for optimal translational conditions and tetramer assembly. Another possibility is that in vivo plasma kinetics may differ for the monomeric and tetrameric forms such that the monomeric form is preferentially removed from the circulation. While a barely detectable monomeric form still existed under non-reducing conditions in partially purified GPx-3 from plasma, rGPx-3 in our system was principally monomeric. A putative GPx-3 crystal structure suggest that two asymmetric units form a tetramer [36]; GPx-4 is the only known GPx family member that functions as monomer. It would be interesting to understand further if and how quaternary structure influences activity; data presented here, however, suggest that specific activity is not influenced significantly by quaternary structure, as specific activity of the rGPx-3 was similar to that of partially purified human plasma GPx-3.
Spectrophotometric and Amplex Red rGPx-3 activity measurements show differences in catalytic activities between a Sec-containing and Cys-containing GPx-3 active site likely as a consequence of the Sec representing the more catalytically active form based on differences in its pKa 5.2 for a selenol and compared with the pKa of a thiol (8.5) [70]. Based on the Amplex Red assay, partially purified plasma GPx-3 reduced t-BuOOH with only 2 mM GSH and not at physiological extracellular GSH concentrations. One study has shown purified GPx-3 may utilize other substrates such as TR, Trx, and Grx [38] in the physiological range. In our hands, neither TR or TRx or Grx had any effect as reducing substrates.
We determined the kcat/Km of a purified GPx-3 to be approximately 1.8 × 108 M−1 S−1 using GSH as a substrate, suggesting a catalytically efficient enzyme. Previous kinetic assays suggest plasma GPx-3 has a 10-fold weaker (i.e., lower) affinity for the substrate (Km of 5.3) [37] than for GSH than cellular GPx-1 due to two less GSH binding sites [36], with a specific activity of approximately 20–26 U/mg of protein [32], suggesting extracellular GSH concentrations cannot provide the necessary reducing equivalents for plasma GPx-3 activity. Theoretically, GPx-3’s secretion into plasma and other extracellular fluids suggests it may function as an antioxidant in the extracellular environment. Thus far, GPx-3 has only been demonstrated to act as a peroxidase in lung lining fluid where the concentration of GSH is significantly higher than in plasma [71]; however, the concentration of GPx-3 is higher in plasma than in epithelial lining fluid [72].
It has been suggested that GPx-3 may not function as a glutathione peroxidase, but as a TRx peroxidase since it reacted with a higher affinity using reducing equivalents from the thioredoxin system [38] in in vitro assays. The crystal structure of GPx-3 shows a thioredoxin fold [36] similar to other enzymes like Trx and Grx [73] and, yet, GPx-3 may be less reactive toward H2O2 than GPx-1 owing to the fact that GPx-3 has two less GSH binding sites than GPx-1 [36]. Trx has been suggested to be a more effective electron donor for SelP [74]. Since TR, Trx, and Grx are found in the extracellular compartment and protein disulfide isomerase, TR, and Trx are also found on the cell surface [75–78], it may be possible that the major extracellular selenoproteins, GPx-3 and SelP, work with these or other unknown reducing equivalents found on or near the cell membrane that may have effluxed from the intracellular compartment [79] to produce effective peroxidase activity in that microenvironment. Future studies are needed to address this issue.
Acknowledgments
This work is supported by National Institutes of Health grants HL61795, HL58976, HL28178, and HL81587. The authors would like to acknowledge and thank Barbara Voestch and Ying-Yi Zhang for their helpful advice and suggestions, and Stephanie Tribuna for excellent secretarial assistance.
Disclaimer: The project described was supported by Award numbers HL 61795, HL 58976, N01 HV 28178, and P01 HL 81587 from the National Heart, Lung, and Blood Institute. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Heart, Lung, and Blood Institute or the National Institutes of Health.
Abbreviations
- GPx-3
Plasma glutathione peroxidase
- UTR
3′untranslated region
- SECIS
selenocysteine insertion sequence
- rGPx-3
recombinant GPx-3
- GPx
glutathione peroxidase
- GSH
glutathione
- Sec
selenocysteine
- eEFSec
Sec-elongation factor
- SBP2
SECIS binding protein 2
- qRT-PCR
qualitative real-time polymerase chain reaction
- SelD
human selenophosphate synthetase D
- G3PDH
glyceraldehyde-3-phosphate dehydrogenase
- rLacZ
recombinant Lac Z
- TRx
thioredoxin
References
- 1.Gromer S, Eubel JK, Lee BL, Jacob J. Human selenoproteins at a glance. Cell Mol Life Sci. 2005;62:2414–2437. doi: 10.1007/s00018-005-5143-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.www.expasy.org.
- 3.Behne D, Kyriakopoulos A. Mammalian selenium-containing proteins. Annu Rev Nutr. 2001;21:453–473. doi: 10.1146/annurev.nutr.21.1.453. [DOI] [PubMed] [Google Scholar]
- 4.Ghyselinck NB, Dufaure I, Lareyre JJ, Rigaudiere N, Mattei MG, Dufaure JP. Structural organization and regulation of the gene for the androgen-dependent glutathione peroxidase-like protein specific to the mouse epididymis. Mol Endocrinol. 1993;7:258–272. doi: 10.1210/mend.7.2.8469239. [DOI] [PubMed] [Google Scholar]
- 5.Yoshimura S, Suemizu H, Taniguchi Y, Arimori K, Kawabe N, Moriuchi T. The human plasma glutathione peroxidase-encoding gene: organization, sequence and localization to chromosome 5q32. Gene. 1994;145:293–297. doi: 10.1016/0378-1119(94)90023-x. [DOI] [PubMed] [Google Scholar]
- 6.Avissar N, Eisenmann C, Breen JG, Horowitz S, Miller RK, Cohen HJ. Human placenta makes extracellular glutathione peroxidase and secretes it into maternal circulation. Am J Physiol. 1994;267:E68–76. doi: 10.1152/ajpendo.1994.267.1.E68. [DOI] [PubMed] [Google Scholar]
- 7.Avissar N, Slemmon JR, Palmer IS, Cohen HJ. Partial sequence of human plasma glutathione peroxidase and immunologic identification of milk glutathione peroxidase as the plasma enzyme. J Nutr. 1991;121:1243–1249. doi: 10.1093/jn/121.8.1243. [DOI] [PubMed] [Google Scholar]
- 8.Chu FF, Esworthy RS, Doroshow JH, Doan K, Liu XF. Expression of plasma glutathione peroxidase in human liver in addition to kidney, heart, lung, and breast in humans and rodents. Blood. 1992;79:3233–3238. [PubMed] [Google Scholar]
- 9.Tham DM, Whitin JC, Kim KK, Zhu SX, Cohen HJ. Expression of extracellular glutathione peroxidase in human and mouse gastrointestinal tract. Am J Physiol. 1998;275:G1463–1471. doi: 10.1152/ajpgi.1998.275.6.G1463. [DOI] [PubMed] [Google Scholar]
- 10.Comhair SA, Bhathena PR, Farver C, Thunnissen FB, Erzurum SC. Extracellular glutathione peroxidase induction in asthmatic lungs: evidence for redox regulation of expression in human airway epithelial cells. FASEB J. 2001;15:70–78. doi: 10.1096/fj.00-0085com. [DOI] [PubMed] [Google Scholar]
- 11.Maeda K, Okubo K, Shimomura I, Mizuno K, Matsuzawa Y, Matsubara K. Analysis of an expression profile of genes in the human adipose tissue. Gene. 1997;190:227–235. doi: 10.1016/s0378-1119(96)00730-5. [DOI] [PubMed] [Google Scholar]
- 12.Avissar N, Finkelstein JN, Horowitz S, Willey JC, Coy E, Frampton MW, Watkins RH, Khullar P, Xu YL, Cohen HJ. Extracellular glutathione peroxidase in human lung epithelial lining fluid and in lung cells. Am J Physiol. 1996;270:L173–182. doi: 10.1152/ajplung.1996.270.2.L173. [DOI] [PubMed] [Google Scholar]
- 13.Haung W, Koralewska-Makar A, Bauer B, Akesson B. Extracellular glutathione peroxidase and ascorbic acid in aqueous humor and serum of patients operated on for cataract. Clin Chim Acta. 1997;261:117–130. doi: 10.1016/s0009-8981(97)06520-0. [DOI] [PubMed] [Google Scholar]
- 14.Oshima G, Kunimoto M, Nakagawa Y. Appearance of extracellular glutathione peroxidase (eGPx) in the ascite fluid of casein-elicited rats. Biol Pharm Bull. 2000;23:532–536. doi: 10.1248/bpb.23.532. [DOI] [PubMed] [Google Scholar]
- 15.Whitin JC, Bhamre S, Tham DM, Cohen HJ. Extracellular glutathione peroxidase is secreted basolaterally by human renal proximal tubule cells. Am J Physiol Renal Physiol. 2002;283:F20–28. doi: 10.1152/ajprenal.00014.2001. [DOI] [PubMed] [Google Scholar]
- 16.Avissar N, Kerl EA, Baker SS, Cohen HJ. Extracellular glutathione peroxidase mRNA and protein in human cell lines. Arch Biochem Biophys. 1994;309:239–246. doi: 10.1006/abbi.1994.1108. [DOI] [PubMed] [Google Scholar]
- 17.Whitin JC, Tham DM, Bhamre S, Ornt DB, Scandling JD, Tune BM, Salvatierra O, Avissar N, Cohen HJ. Plasma glutathione peroxidase and its relationship to renal proximal tubule function. Mol Genet Metab. 1998;65:238–245. doi: 10.1006/mgme.1998.2760. [DOI] [PubMed] [Google Scholar]
- 18.Freedman JE, Frei B, Welch GN, Loscalzo J. Glutathione peroxidase potentiates the inhibition of platelet function by S-nitrosothiols. J Clin Invest. 1995;96:394–400. doi: 10.1172/JCI118047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Freedman JE, Loscalzo J, Benoit SE, Valeri CR, Barnard MR, Michelson AD. Decreased platelet inhibition by nitric oxide in two brothers with a history of arterial thrombosis. J Clin Invest. 1996;97:979–987. doi: 10.1172/JCI118522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kenet G, Freedman J, Shenkman B, Regina E, Brok-Simoni F, Holzman F, Vavva F, Brand N, Michelson A, Trolliet M, Loscalzo J, Inbal A. Plasma glutathione peroxidase deficiency and platelet insensitivity to nitric oxide in children with familial stroke. Arterioscler Thromb Vasc Biol. 1999;19:2017–2023. doi: 10.1161/01.atv.19.8.2017. [DOI] [PubMed] [Google Scholar]
- 21.Voetsch B, Jin RC, Bierl C, Benke KS, Kenet G, Simioni P, Ottaviano F, Damasceno BP, Annichino-Bizacchi JM, Handy DE, Loscalzo J. Promoter polymorphisms in the plasma glutathione peroxidase (GPx-3) gene: a novel risk factor for arterial ischemic stroke among young adults and children. Stroke. 2007;38:41–49. doi: 10.1161/01.STR.0000252027.53766.2b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brigelius-Flohe R, Banning A, Schnurr K. Selenium-dependent enzymes in endothelial cell function. Antioxid Redox Signal. 2003;5:205–215. doi: 10.1089/152308603764816569. [DOI] [PubMed] [Google Scholar]
- 23.Bierl C, Voetsch B, Jin RC, Handy DE, Loscalzo J. Determinants of human plasma glutathione peroxidase (GPx-3) expression. J Biol Chem. 2004;279:26839–26845. doi: 10.1074/jbc.M401907200. [DOI] [PubMed] [Google Scholar]
- 24.Comhair SA, Erzurum SC. The regulation and role of extracellular glutathione peroxidase. Antioxid Redox Signal. 2005;7:72–79. doi: 10.1089/ars.2005.7.72. [DOI] [PubMed] [Google Scholar]
- 25.Driscoll DM, Copeland PR. Mechanism and regulation of selenoprotein synthesis. Annu Rev Nutr. 2003;23:17–40. doi: 10.1146/annurev.nutr.23.011702.073318. [DOI] [PubMed] [Google Scholar]
- 26.Low SC, Grundner-Culemann E, Harney JW, Berry MJ. SECIS-SBP2 interactions dictate selenocysteine incorporation efficiency and selenoprotein hierarchy. Embo J. 2000;19:6882–6890. doi: 10.1093/emboj/19.24.6882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Copeland PR, Stepanik VA, Driscoll DM. Insight into mammalian selenocysteine insertion: domain structure and ribosome binding properties of Sec insertion sequence binding protein 2. Mol Cell Biol. 2001;21:1491–1498. doi: 10.1128/MCB.21.5.1491-1498.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fagegaltier D, Hubert N, Yamada K, Mizutani T, Carbon P, Krol A. Characterization of mSelB, a novel mammalian elongation factor for selenoprotein translation. EMBO J. 2000;19:4796–4805. doi: 10.1093/emboj/19.17.4796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Low SC, Harney JW, Berry MJ. Cloning and functional characterization of human selenophosphate synthetase, an essential component of selenoprotein synthesis. J Biol Chem. 1995;270:21659–21664. doi: 10.1074/jbc.270.37.21659. [DOI] [PubMed] [Google Scholar]
- 30.Lee BJ, Rajagopalan M, Kim YS, You KH, Jacobson KB, Hatfield D. Selenocysteine tRNA[Ser]Sec gene is ubiquitous within the animal kingdom. Mol Cell Biol. 1990;10:1940–1949. doi: 10.1128/mcb.10.5.1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Copeland PR, Fletcher JE, Carlson BA, Hatfield D, Driscoll DM. A novel RNA binding protein, SBP2, is required for the translation of mammalian selenoprotein mRNAs. EMBO J. 2000;19:304–314. doi: 10.1093/emboj/19.2.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maddipati KR, Marnett LJ. Characterization of the major hydroperoxide-reducing activity of human plasma. Purification and properties of a selenium-dependent glutathione peroxidase. J Biol Chem. 1987;262:17398–17403. [PubMed] [Google Scholar]
- 33.Lin CL, Chen HJ, Hou WC. Activity staining of glutathione peroxidase after electrophoresis on native and sodium dodecyl sulfate polyacrylamide gels. Electrophoresis. 2002;23:513–516. doi: 10.1002/1522-2683(200202)23:4<513::AID-ELPS513>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 34.Handy DE, Zhang Y, Loscalzo J. Homocysteine down-regulates cellular glutathione peroxidase (GPx1) by decreasing translation. J Biol Chem. 2005;280:15518–15525. doi: 10.1074/jbc.M501452200. [DOI] [PubMed] [Google Scholar]
- 35.Weiss Sachdev S, Sunde RA. Selenium regulation of transcript abundance and translational efficiency of glutathione peroxidase-1 and -4 in rat liver. Biochem J. 2001;357:851–858. doi: 10.1042/0264-6021:3570851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ren B, Huang W, Akesson B, Ladenstein R. The crystal structure of seleno-glutathione peroxidase from human plasma at 2.9 A resolution. J Mol Biol. 1997;268:869–885. doi: 10.1006/jmbi.1997.1005. [DOI] [PubMed] [Google Scholar]
- 37.Takahashi K, Avissar N, Whitin J, Cohen H. Purification and characterization of human plasma glutathione peroxidase: a selenoglycoprotein distinct from the known cellular enzyme. Arch Biochem Biophys. 1987;256:677–686. doi: 10.1016/0003-9861(87)90624-2. [DOI] [PubMed] [Google Scholar]
- 38.Bjornstedt M, Xue J, Huang W, Akesson B, Holmgren A. The thioredoxin and glutaredoxin systems are efficient electron donors to human plasma glutathione peroxidase. J Biol Chem. 1994;269:29382–29384. [PubMed] [Google Scholar]
- 39.Muller C, Wingler K, Brigelius-Flohe R. 3′UTRs of glutathione peroxidases differentially affect selenium-dependent mRNA stability and selenocysteine incorporation efficiency. Biol Chem. 2003;384:11–18. doi: 10.1515/BC.2003.002. [DOI] [PubMed] [Google Scholar]
- 40.Wingler K, Bocher M, Flohe L, Kollmus H, Brigelius-Flohe R. mRNA stability and selenocysteine insertion sequence efficiency rank gastrointestinal glutathione peroxidase high in the hierarchy of selenoproteins. Eur J Biochem. 1999;259:149–157. doi: 10.1046/j.1432-1327.1999.00012.x. [DOI] [PubMed] [Google Scholar]
- 41.Bermano G, Arthur JR, Hesketh JE. Role of the 3′ untranslated region in the regulation of cytosolic glutathione peroxidase and phospholipid-hydroperoxide glutathione peroxidase gene expression by selenium supply. Biochem J. 1996;320:891–895. doi: 10.1042/bj3200891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Avissar N, Whitin JC, Allen PZ, Wagner DD, Liegey P, Cohen HJ. Plasma selenium-dependent glutathione peroxidase. Cell of origin and secretion. J Biol Chem. 1989;264:15850–15855. [PubMed] [Google Scholar]
- 43.Axley MJ, Bock A, Stadtman TC. Catalytic properties of an Escherichia coli formate dehydrogenase mutant in which sulfur replaces selenium. Proc Natl Acad Sci USA. 1991;88:8450–8454. doi: 10.1073/pnas.88.19.8450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Berry MJ, Kieffer JD, Harney JW, Larsen PR. Selenocysteine confers the biochemical properties characteristic of the type I iodothyronine deiodinase. J Biol Chem. 1991;266:14155–14158. [PubMed] [Google Scholar]
- 45.Berry MJ, Kieffer JD, Larsen PR. Evidence that cysteine, not selenocysteine, is in the catalytic site of type II iodothyronine deiodinase. Endocrinology. 1991;129:550–552. doi: 10.1210/endo-129-1-550. [DOI] [PubMed] [Google Scholar]
- 46.Berry MJ, Maia AL, Kieffer JD, Harney JW, Larsen PR. Substitution of cysteine for selenocysteine in type I iodothyronine deiodinase reduces the catalytic efficiency of the protein but enhances its translation. Endocrinology. 1992;131:1848–1852. doi: 10.1210/endo.131.4.1396330. [DOI] [PubMed] [Google Scholar]
- 47.Berry MJ, Banu L, Chen YY, Mandel SJ, Kieffer JD, Harney JW, Larsen PR. Recognition of UGA as a selenocysteine codon in type I deiodinase requires sequences in the 3′ untranslated region. Nature. 1991;353:273–276. doi: 10.1038/353273a0. [DOI] [PubMed] [Google Scholar]
- 48.Kim IY, Guimaraes MJ, Zlotnik A, Bazan JF, Stadtman TC. Fetal mouse selenophosphate synthetase 2 (SPS2): characterization of the cysteine mutant form overproduced in a baculovirus-insect cell system. Proc Natl Acad Sci USA. 1997;94:418–421. doi: 10.1073/pnas.94.2.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Patterson BH, Levander OA. Naturally occurring selenium compounds in cancer chemoprevention trials: a workshop summary. Cancer Epidemiol Biomarkers Prev. 1997;6:63–69. [PubMed] [Google Scholar]
- 50.Ip C. Lessons from basic research in selenium and cancer prevention. J Nutr. 1998;128:1845–1854. doi: 10.1093/jn/128.11.1845. [DOI] [PubMed] [Google Scholar]
- 51.Saito Y, Yoshida Y, Akazawa T, Takahashi K, Niki E. Cell death caused by selenium deficiency and protective effect of antioxidants. J Biol Chem. 2003;278:39428–39434. doi: 10.1074/jbc.M305542200. [DOI] [PubMed] [Google Scholar]
- 52.Zhong W, Oberley TD. Redox-mediated effects of selenium on apoptosis and cell cycle in the LNCaP human prostate cancer cell line. Cancer Res. 2001;61:7071–7078. [PubMed] [Google Scholar]
- 53.Zhao R, Domann FE, Zhong W. Apoptosis induced by selenomethionine and methioninase is superoxide mediated and p53 dependent in human prostate cancer cells. Mol Cancer Ther. 2006;5:3275–3284. doi: 10.1158/1535-7163.MCT-06-0400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bermano G, Nicol F, Dyer JA, Sunde RA, Beckett GJ, Arthur JR, Hesketh JE. Tissue-specific regulation of selenoenzyme gene expression during selenium deficiency in rats. Biochem J. 1995;311:425–430. doi: 10.1042/bj3110425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Christensen MJ, Burgener KW. Dietary selenium stabilizes glutathione peroxidase mRNA in rat liver. J Nutr. 1992;122:1620–1626. doi: 10.1093/jn/122.8.1620. [DOI] [PubMed] [Google Scholar]
- 56.Suzuki KT, Ishiwata K, Ogra Y. Incorporation of selenium into selenoprotein P and extracellular glutathione peroxidase: HPLC-ICPMS data with enriched selenite. Analyst. 1999;124:1749–1753. doi: 10.1039/a906521k. [DOI] [PubMed] [Google Scholar]
- 57.Reszka E, Gromadzinska J, Stanczyk M, Wasowicz W. Effect of selenium on expression of selenoproteins in mouse fibrosarcoma cells. Biol Trace Elem Res. 2005;104:165–172. doi: 10.1385/BTER:104:2:165. [DOI] [PubMed] [Google Scholar]
- 58.Chavatte L, Brown BA, Driscoll DM. Ribosomal protein L30 is a component of the UGA-selenocysteine recoding machinery in eukaryotes. Nat Struct Mol Biol. 2005;12:408–416. doi: 10.1038/nsmb922. [DOI] [PubMed] [Google Scholar]
- 59.Shen Q, Fan L, Newburger PE. Nuclease sensitive element binding protein 1 associates with the selenocysteine insertion sequence and functions in mammalian selenoprotein translation. J Cell Physiol. 2006;207:775–783. doi: 10.1002/jcp.20619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mehta A, Rebsch CM, Kinzy SA, Fletcher JE, Copeland PR. Efficiency of mammalian selenocysteine incorporation. J Biol Chem. 2004;279:37852–37859. doi: 10.1074/jbc.M404639200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Berry MJ, Harney JW, Ohama T, Hatfield DL. Selenocysteine insertion or termination: factors affecting UGA codon fate and complementary anticodon:codon mutations. Nucleic Acids Res. 1994;22:3753–3759. doi: 10.1093/nar/22.18.3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dumitrescu AM, Liao XH, Abdullah MS, Lado-Abeal J, Majed FA, Moeller LC, Boran G, Schomburg L, Weiss RE, Refetoff S. Mutations in SECISBP2 result in abnormal thyroid hormone metabolism. Nat Genet. 2005;37:1247–1252. doi: 10.1038/ng1654. [DOI] [PubMed] [Google Scholar]
- 63.Squires JE, Stoytchev I, Forry EP, Berry MJ. SBP2 binding affinity is a major determinant in differential selenoprotein mRNA translation and sensitivity to nonsense-mediated decay. Mol Cell Biol. 2007;27:7848–7855. doi: 10.1128/MCB.00793-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shen Q, Chu FF, Newburger PE. Sequences of the 3′-untranslated region of the human cellular glutathone peroxidase gene are necessary and sufficient for selenocysteine incorporation at the UGA codon. J Biol Chem. 1993;268:11463–11469. [PubMed] [Google Scholar]
- 65.Lesoon A, Mehta A, Singh R, Chisolm GM, Driscoll DM. An RNA-binding protein recognizes a mammalian selenocysteine insertion sequence element required for cotranslational incorporation of selenocysteine. Mol Cell Biol. 1997;17:1977–1985. doi: 10.1128/mcb.17.4.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Grundner-Culemann E, Martin GWr, Harney JW, Berry MJ. Two distinct SECIS structures capable of directing selenocysteine incorporation in eukaryotes. RNA. 1999;5:625–635. doi: 10.1017/s1355838299981542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Martin GWr, Harney JW, Berry MJ. Selenocysteine incorporation in eukaryotes: insights into mechanism and efficiency from sequence, structure, and spacing proximity studies of the type1 deiodinase SECIS element. RNA. 1996;2:171–182. [PMC free article] [PubMed] [Google Scholar]
- 68.Fujiwara N, Fujii T, Fujii J, Taniguchi N. Functional expression of rat thioredoxin reductase: selenocysteine insertion sequence element for the active enzyme. Biochem J. 1999;340:439–444. [PMC free article] [PubMed] [Google Scholar]
- 69.Copeland PR, Driscoll DM. Purification, redox sensitivity, and RNA binding properties of SECIS-binding protein 2, a protein involved in selenoprotein biosynthesis. J Biol Chem. 1999;274:25447–25454. doi: 10.1074/jbc.274.36.25447. [DOI] [PubMed] [Google Scholar]
- 70.Ambrogelly A, Palioura S, Soll D. Natural expansion of the genetic code. Nat Chem Biol. 2007;3:29–35. doi: 10.1038/nchembio847. [DOI] [PubMed] [Google Scholar]
- 71.Cantin AM, North SL, Hubbard RC, Crystal RG. Normal alveolar epithelial lining fluid contains high levels of glutathione. J Appl Physiol. 1987;63:152–157. doi: 10.1152/jappl.1987.63.1.152. [DOI] [PubMed] [Google Scholar]
- 72.Avissar N, Whitin JC, Allen PZ, Palmer IS, Cohen HJ. Antihuman plasma glutathione peroxidase antibodies: immunologic investigations to determine plasma glutathione peroxidase protein and selenium content in plasma. Blood. 1989;73:318–323. [PubMed] [Google Scholar]
- 73.Martin JL. Thioredoxin--a fold for all reasons. Structure. 1995;3:245–250. doi: 10.1016/s0969-2126(01)00154-x. [DOI] [PubMed] [Google Scholar]
- 74.Takebe G, Yarimizu J, Saito Y, Hayashi T, Nakamura H, Yodoi J, Nagasawa S, Takahashi K. A comparative study on the hydroperoxide and thiol specificity of the glutathione peroxidase family and selenoprotein P. J Biol Chem. 2002;277:41254–41258. doi: 10.1074/jbc.M202773200. [DOI] [PubMed] [Google Scholar]
- 75.Chen K, Detwiler TC, Essex DW. Characterization of protein disulphide isomerase released from activated platelets. Br J Haematol. 1995;90:425–431. doi: 10.1111/j.1365-2141.1995.tb05169.x. [DOI] [PubMed] [Google Scholar]
- 76.Kroning H, Kahne T, Ittenson A, Franke A, Ansorge S. Thiol-proteindisulfide-oxidoreductase (proteindisulfide isomerase): a new plasma membrane constituent of mature human B lymphocytes. Scand J Immunol. 1994;39:346–350. doi: 10.1111/j.1365-3083.1994.tb03384.x. [DOI] [PubMed] [Google Scholar]
- 77.Martin H, Dean M. Identification of a thioredoxin-related protein associated with plasma membranes. Biochem Biophys Res Commun. 1991;175:123–128. doi: 10.1016/s0006-291x(05)81209-4. [DOI] [PubMed] [Google Scholar]
- 78.Schallreuter KU, Wood JM. The activity and purification of membrane-associated thioredoxin reductase from human metastatic melanotic melanoma. Biochim Biophys Acta. 1988;967:103–109. doi: 10.1016/0304-4165(88)90194-8. [DOI] [PubMed] [Google Scholar]
- 79.Ottaviano FG, Handy DE, Loscalzo J. Redox regulation in the extracellular environment. Circ J. 2008;72:1–16. doi: 10.1253/circj.72.1. [DOI] [PubMed] [Google Scholar]