Abstract
The LARK RNA-binding protein (RBP) has well documented roles in the circadian systems of Drosophila and mammals. Recent studies have demonstrated that the Drosophila LARK RBP is associated with many mRNA targets, in vivo, including those that regulate either neurophysiology or development of the nervous system. In the present study, we have employed conditional expression techniques to distinguish developmental and physiological functions of LARK for a defined class of neurons: the Pigment Dispersing Factor (PDF)-containing LNv clock neurons. We found that increased LARK expression during development dramatically alters the small LNv class of neurons with no obvious effects on the large LNv cells. Conversely, conditional expression of LARK at the adult stage results in altered clock protein rhythms and circadian locomotor activity, even though neural morphology is normal in such animals. Electrophysiological analyses at the larval neuromuscular junction indicate a role for LARK in regulating neuronal excitability. Altogether, our results demonstrate that LARK activity is critical for neuronal development and physiology.
Keywords: Drosophila, circadian, post-transcriptional, development, neuronal excitability, RBM4
Introduction
Post-transcriptional regulatory events are essential for the function of the molecular oscillator governing circadian behavior. These include post-translational modifications by kinases, phosphatases and other regulatory factors that modulate clock protein intracellular localization, stability and activities (Young and Kay, 2001; Lee et al., 2003b; Harms et al., 2004; Liu, 2005; Vanselow and Kramer, 2007; Zheng and Sehgal, 2008). In addition, it has become apparent that circadianly regulated RNA-binding proteins (RBPs) have important post-transcriptional circadian functions in several different species (Heintzen et al., 1997; McNeil et al., 1998; Dockendorff et al., 2002; Morales et al., 2002; Liu, 2005; Iliev et al., 2006; Garbarino-Pico and Green, 2007). The Drosophila LARK RBP and its mammalian homolog mLARK (a.k.a RBM4) display circadian changes in abundance and mediate post-transcriptional events that are critical for circadian timing in flies and mammals, respectively (Newby and Jackson, 1993, 1996; McNeil et al., 1998; Zhang et al., 2000; Schroeder et al., 2003; Kojima et al., 2007). Our recent studies demonstrate that fly LARK is associated with a large number of mRNA target molecules, in vivo, and suggest that the RBP regulates certain targets by interacting with the microRNA (miRNA) pathway (Huang et al., 2007); mouse LARK has also been implicated in miRNA-mediated gene regulation (Hock et al., 2007). Surprisingly, one recent study demonstrates in vivo interactions of LARK with the Fragile X Mental Retardation Protein (FMRP) – another RBP implicated in microRNA pathway function – suggesting that the two RBPs might cooperate to regulate certain target RNAs (Sofola et al., 2008). LARK target mRNAs encode proteins important for many developmental and physiological processes including neuronal survival, neurite growth and pathfinding, neuronal excitability, synaptic function, circadian biology and others. Although the circadian role of LARK is well established (Newby and Jackson, 1993, 1996; McNeil et al., 1998; Zhang et al., 2000; McNeil et al., 2001; Schroeder et al., 2003), a mechanistic understanding of this function remains a goal of current work. Furthermore, despite the identification of potential LARK targets that function in neuronal growth and differentiation (Huang et al., 2007), the role of LARK in neural development has not been studied.
In this report, we have chosen a specific group of neurons in the Drosophila brain known as the Pigment-Dispersing Factor (PDF)-expressing ventral lateral neurons (PDF-positive LNvs, hereafter referred to as “PDF neurons”) as a model to study the diverse functions of LARK. The PDF neurons were chosen for several reasons. They represent a small group of neurons with well characterized anatomy (Helfrich-Forster, 1997, 2003; Helfrich-Forster et al., 2007). In addition, the LNvs are known to be critical components of the circadian circuitry regulating locomotor activity. LARK overexpression in the LNvs was previously shown to cause arrhythmic locomotor activity, but it was not determined whether this phenotype was a consequence of altered clock neuron physiology or development (Schroeder et al., 2003). Both developmental and physiological alterations of the LNvs have dramatic effects on circadian function. Previous studies have shown that genetic ablation of the PDF neurons causes defects in circadian locomotor activity (Renn et al., 1999). Furthermore, a number of manipulations that alter the physiology of the LNvs, including the elimination of PDF neuropeptide (Renn et al., 1999), overexpression of the PER clock protein (Blanchardon et al., 2001), blockade of synaptic transmission by expression of tetanus toxin (Blanchardon et al., 2001), and modification of neuronal membrane excitability (Nitabach et al., 2002; Nitabach et al., 2006; Wu et al., 2008), all dramatically affect the circadian control of locomotor activity. Here we show that LARK overexpression affects development of the PDF neurons. Surprisingly, however, increased LARK expression exclusively at the adult stage abolishes rhythmic locomotor activity without affecting neuronal morphology, suggesting a dual role for LARK in development and physiology of the LNvs. In addition, we show that the effect of LARK overexpression on two key components of the molecular clock, PERIOD (PER) and PAR Domain Protein1ε (PDP1ε), resembles that caused by electrical silencing of the PDF neurons. To determine if LARK might regulate neuronal activity, we recorded from the larval neuromuscular junctions (NMJs) of lark null and overexpression mutants; those studies indicated that changes in the abundance of this RBP can regulate excitability. Taken together with previous findings that LARK is associated with mRNAs encoding potassium channels, an intriguing model is that an increased amount of the RBP promotes expression of these channels, resulting in decreased PDF cell membrane excitability and abolition of the circadian locomotor activity rhythm.
Results
Dosage-dependent effects of LARK expression on development of the PDF-expressing small lateral neurons (s-LNvs)
Overexpression of LARK in the PDF neurons was previously found to cause behavioral arrhythmicity without altering survival of the neurons or the gross morphology of neuronal projections (Schroeder et al., 2003). Those studies, however, utilized flies carrying only single copies of the pdf-gal4 driver and uas-lark transgenes. In our recent studies, we found that increasing the dosage of either the pdf-gal4 driver or uas-lark responder transgene caused obvious developmental defects. For example, male flies carrying a pdf-gal4 insertion on the X chromosome (which is dosage-compensated and therefore equivalent to two copies in females) and a single copy of uas-lark on the third chromosome displayed obvious defects in the elaboration of the s-LNv dorsal projections. In control flies, the four s-LNvs on each side of the brain send tightly fasciculated projections towards dorsal regions of the brain, branching and forming varicosities in the dorsal brain (Fig. 1A). In contrast, the projections of pdf-gal4; uas-lark/+ males appeared de-fasciculated and in one brain (out of 16 brains examined) they failed to reach dorsal brain areas (Fig. 1B and data not shown). Furthermore, those reaching the dorsal brain failed to branch and form varicosities (Fig. 1B). We note that the cell bodies of the s-LNvs are present and appear grossly normal in these LARK-overexpressing animals (hereafter referred to as LARK OE flies); i.e., the effect of LARK overexpression at this dosage seems to be restricted to effects on projections of the s-LNvs. Similarly, the cell bodies and processes of the l-LNvs were found to be grossly normal in males of this genotype (data not shown).
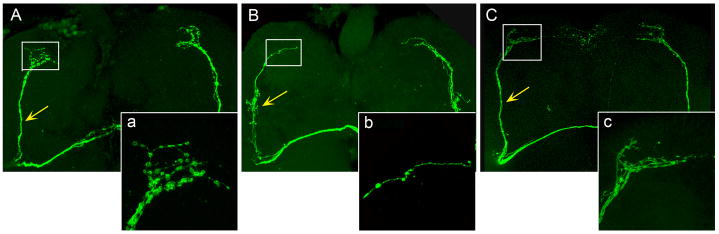
Figure 1.
Morphology of the PDF projections in adult brains of control and lark overexpression animals. The PDF projections were labeled with pdf>mCD8GFP. Representative images show PDF projections of A. control animal (pdf-gal4; uas-mCD8GFP/+), B. LARK overexpression animal (pdf-gal4; uas-mCD8GFP/+; uas-lark/+), and C, an animal that overexpresses a mutant form of LARK protein with defective RRM domains (pdf-gal4; uas-mCD8GFP uas-larkΔRRM/+). The images in a, b and c show the nerve terminals of the projections in greater detail. Arrows point to the axon bundles of the dorsal projections. At least 8 brains were examined for each genotype and similar phenotypes were observed for all brains within each genotype group.
When the dosage of LARK was further increased, i.e. when pdf-gal4 and uas-lark transgenes were present in two copies each, all 4 s-LNv cells and their projections were missing in most flies (Fig. 2A); in 19.4 % (7 out of 36) of the brain hemispheres of LARK OE animals one or two remaining s-LNvs could still be visualized. Thus, the average number of s-LNv cells per brain hemisphere was dramatically reduced in LARK high-dose OE flies (Fig. 2C). Interestingly, this effect on cell number was completely restricted to the s-LNvs – the l-LNvs and their projections were normal in such flies (Fig. 2A, C). Although we cannot exclude the possibility that the missing s-LNv phenotype is merely a result of dramatic down-regulation of all three markers employed in our studies [(GFP, PDF, and PERIOD (PER)], rendering the neurons undetectable, other observations suggested that the cells were actually eliminated during development. In the brains of third instar larvae of the same genotype, we did not detect PDF or PER in the s-LNvs, but the GFP signal was weakly detected. In addition, the larval cells were smaller in size and irregular in shape relative to the larger, round cells of control brains (Fig. 2B, D). As the cells were eliminated in LARK OE flies by the pupal stage (data not shown), these observations suggest that the s-LNvs are beginning to undergo cell death in third instar larvae. In future experiments, it will be interesting to test whether co-overexpression of the baculovirus P35 protein, a potent inhibitor of apoptotic cell death (Hay et al., 1994), can suppress the cell death phenotype.
Figure 2.
Overexpression of LARK causes death of the small PDF neurons. PDF neurons were labeled with pdf>mCD8GFP. Brains were also stained with antibodies against PDF and PER (to assist accurate counting of cell numbers). A. Adult brain. B. Third instar larval brain. C. Average number of large and small PDF cells per brain hemisphere for various genotypes. Control: pdf-gal4 uas-mCD8GFP, n = 23. LARK OE: pdf-gal4 uas-mCD8GFP; uas-lark, n = 36. Mutant OE: pdf-gal4 uas-mCD8GFP; uas-larkΔRRM, n = 38. D. Average cell diameter of the small PDF cells in the third instar larval brains of control and LARK overexpression animals. Control: n = 28, LARK OE: n = 26. *: P < 0.000001, Student’s t-test. Error bars represent SEM.
Intact RNA Recognition Motifs within LARK, and presumably its RNA-binding activities, are required for normal function of the protein (McNeil et al., 2001). High-level overexpression of a doubly mutant LARK protein, with single mutations in each RRM that are predicted to decrease or eliminate RNA-binding (McNeil et al., 2001), had no effect on either the morphology of projections (Fig. 1C ) or survival (Fig. 2C) of the s-LNv neurons. This result strongly indicates that the RNA-binding activity of the LARK RBP is required for production of the LNv mutant phenotypes we have described.
Induction of LARK expression exclusively at the adult stage disrupts free-running circadian locomotor activity rhythms
Although LARK overexpression using one copy each of the pdf-gal4 and uas-lark transgenes does not cause obvious LNv cell developmental defects (Schroeder et al., 2003) and data not shown), we wished to completely exclude the possibility that subtle developmental anomalies caused the behavioral phenotype observed in such flies. In order to dissociate the developmental and physiological functions of LARK, we utilized the TARGET method – with a temperature-sensitive GAL80 (GAL80ts) to inhibit GAL4 activity (McGuire et al., 2004) – to overexpress LARK exclusively at the adult stage. To perform these experiments, a tub-gal80ts transgene (GAL80ts under the control of a tubulin promoter) was introduced into the pdf-gal4; uas-lark background and flies were reared under a permissive temperature (23°C) throughout development prior to being transferred to a non-permissive temperature (30°C) at the adult stage. Immunohistochemistry using anti-LARK antibody showed that GAL80ts effectively blocked LARK overexpression at 23°C and that high temperature (30°C) relieved the repression by GAL80ts, inducing LARK overexpression (Fig. 3B). As expected, we found that pdf-gal4; uas-lark, tub-gal80ts flies, when reared at 23°C, developed normal LNvs with wild-type-like dorsal projections, in marked contrast to pdf-gal4; uas-lark OE flies, which were missing the s-LNvs and their projections (Fig. 3A). When tested at 23°C, these flies also had robust circadian locomotor activity rhythms that were indistinguishable from those of the control flies (Fig. 3C, lower left).
Figure 3.
Overexpression of LARK specifically at the adult stage impairs free-running circadian locomotor activity. A. GAL80ts effectively blocked developmental defects caused by overexpression from the pdf-gal4; uas-lark constructs when flies were raised at 23°C during development. Note the cell bodies and projections of the s-LNvs are missing in the LARK OE brain and restored in the LARK OE + G80ts brain. B. Induction of LARK overexpression specifically in adults causes behavioral arrhythmicity. Brains were stained with anti-PDF (green) and anti-LARK antibodies (red) to visualize the small PDF neurons. Representative actograms are shown for each genotype. Note that at 30°C the control animal shows a shorter active phase, which is also observed in flies of similar genetic background, w1118 (not shown). C. Quantification of the average Rhythmicity Index (RI) for various genotypes. WT (w1118), n = 43 for 23°C, n = 53 for 30°C. Control: w1118; pdf-gal4; Tub-gal80ts, n = 29 for 23°C, n = 48 for 30°C. LARK OE: w1118; pdf-gal4; uas-lark, n = 25 for 23°C, n = 36 for 30°C. LARK OE + G80ts: w1118; pdf-gal4;uas-lark Tub-gal80ts, n = 55 for 23°C, n = 53 for 30°C.
Despite proper development of the LNvs and their projections, induction of LARK overexpression at the adult stage caused arrhythmic locomotor activity in free-running conditions. In those experiments, control and LARK OE flies were transferred to 30°C and then entrained to a light-dark (LD) cycle consisting of 12 hours light and 12 hours dark for at least 4 days before being released into constant darkness (DD). Remarkably, we found that 100% (n=53) of the flies acutely overexpressing LARK became arrhythmic, compared to 6.5% (n= 48) of the control flies (Fig. 3B). Comparison of the average Rhythmicity Indices (RIs), a measure of robustness, demonstrated a dramatic difference between LARK OE and control flies (Fig. 3C). Similar to our past studies (Schroeder et al, 2003), expression of the LARK RRM-mutant protein did not affect rhythmicity under the same conditions (data not shown).
We excluded the possibility that the observed arrhythmicity results from cell death of the LNvs during adulthood by examining the brains of LARK OE flies – the LNvs had normal morphology and projections 8 days after the acute induction of LARK expression (Supplemental Figure 1). These results strongly suggest that LARK, independent of its developmental role, also regulates the expression of molecules that are required for the physiology of the adult LNv neurons.
LARK expression in adults alters the free-running molecular clock
There are two possible mechanisms by which LARK overexpression in the LNvs might cause arrhythmic locomotor activity. First, LARK might act cell autonomously to disrupt the molecular clock in the LNvs. Since the LNvs are a major neuronal group driving rhythmic locomotor activity, disruption of the molecular clock in these neurons is expected to cause arrhythmicity. Alternatively, LARK overexpression in the LNvs might leave the molecular clock intact while disrupting only the output signals from these neurons, which are known to be required for sustaining robust rhythms in constant conditions [for recent reviews, see (Taghert and Shafer, 2006; Nitabach and Taghert, 2008)]. In order to distinguish between these possibilities, we first tested whether the molecular clock in the LNvs is disrupted by examining the PERIOD (PER) clock protein in these neurons. In wild-type flies, PER protein has a nuclear localization in most clock neurons during the late night coincident with feedback repression. Following nuclear entry, PER level gradually decreases, becoming nearly undetectable by the end of the day.
As shown in Figure 4, we detected a failure of PER nuclear entry specifically in the large PDF neurons (l-LNvs) of the LARK OE flies under free-running conditions. Figure 4 depicts the intracellular localization of PER at different times of the circadian cycle in the l-LNv (A) and s-LNv (B) neurons during the first day of constant darkness (DD). We found that in the l-LNvs, PER begins to enter the nucleus at ~CT21 in the control brains. PER is observed to have a nuclear localization in control brains at CT1, CT4 and CT9, whereas PER staining is decreased and cytoplasmic at CT13. By contrast, in flies overexpressing LARK during adulthood, PER was predominantly cytoplasmic within the l-LNvs at all times of the circadian cycle (note the characteristic “donut” pattern of staining in these cells; Figure 4A, right panel). This phenotype – cytoplasmic retention of PER at all times of the cycle – was observed in 2 independent experiments in which multiple brains were examined at 6 different times of day. At CT 9, we occasionally detected nuclear PER staining in one single large PDF cell in some LARK OE brains. However, this was not observed at other circadian times. The defective nuclear entry observed for LARK OE flies is expected to lead to a disruption of the PER rhythm as the repression phase of the cycle is short circuited. Although we cannot exclude the possibility that LARK OE flies have a long-period PER rhythm, i.e. PER nuclear entry might have occurred after the latest time examined in our study (CT21), the arrhythmic behavior of LARK OE adults is more consistent with a lack of PER cycling. Whereas PER intracellular localization appeared to be altered in the l-LNvs, the protein had a normal cycle of nuclear entry and abundance in the s-LNvs of LARK OE flies (Figure 4B).
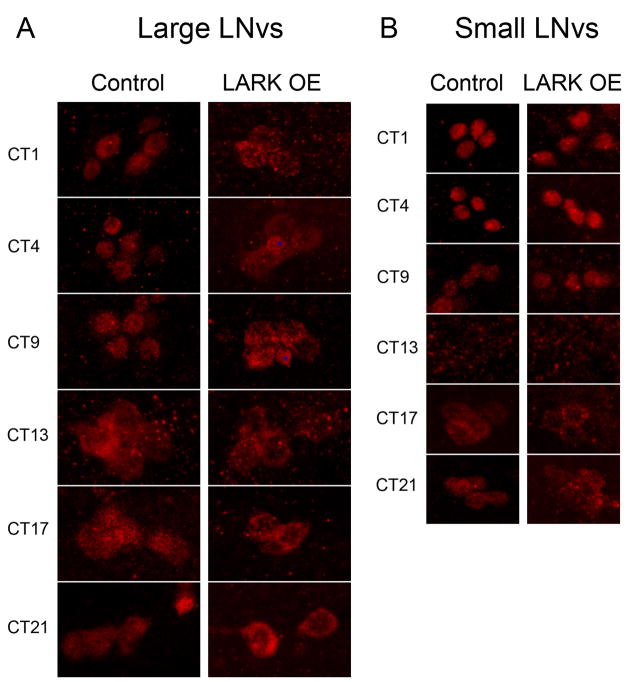
Figure 4.
Overexpression of LARK in adult flies alters PER cycling in l-LNv but not s-LNv neurons under free-running conditions. Control: pdf-gal4; Tub-gal80ts, LARK OE: pdf-gal4; uas-lark Tub-gal80ts. Anti-PER antibody was used to localize PER. Flies were raised at 23°C and entrained at 30°C for at least 4 days before release into constant darkness (DD). Images were acquired on the first day of DD. * indicates the fifth s-LNv that is known to be PER positive and PDF negative, which occasionally is seen located adjacent to l-LNvs.
In independent experiments, we examined PDP1ε, a rhythmically expressed transcription factor thought to be required for the clock mechanism or output (Glossop et al., 1999; Cyran et al., 2003; Benito et al., 2007; Lim et al., 2007). In wild-type files, PDP1ε is known to cycle in abundance within the s-LNvs under free running condition (Benito et al., 2007) and we observed similar results in our control flies; i.e., PDP1ε displayed differences in abundance between CT17 and CT5 (Figure 5). In contrast, PDP1ε cycling was not observed in the s-LNvs of LARK OE flies in the same conditions (Figure 5).
Figure 5.
LARK overexpression affects cycling of PDP1ε. Panels show representative images of PDP1ε immunoreactivity in the control and LARK OE animals at CT 17 and CT 5. Brains were also stained with anti-PDF antibody to identify the LNvs. Flies were raised at 23°C, entrained to LD 12:12 at 30°C for at least 4 days before release into constant darkness (DD). Images were acquired on the first day of DD. Control: pdf-gal4; Tub-gal80ts, LARK OE: pdf-gal4; uas-lark Tub-gal80ts. Arrows point to large PDF neurons. Arrowhead points to small PDF neurons. Note that PDP1ε ceases to cycle in abundance in the l-LNvs on the first day of DD even in control flies.
We note that these PER and PDP1 molecular phenotypes are quite similar to those reported in previous experiments in which clock neuronal membrane excitability was perturbed (Nitabach et al., 2002; Wu et al., 2008). Those studies showed that electrical silencing of the LNvs – by ectopic overexpression of a “leak” potassium channel – stops the free-running molecular clock and severely impairs circadian locomotor activity rhythms (Nitabach et al., 2002; Wu et al., 2008). In the previous studies, clock protein abundance was assayed at only two times of day – the beginning of subjective day and near lights-off – and a comparison of our results from CT1 or 4 and CT13 are quite similar, suggesting that LARK overexpression might perturb clock neuronal membrane excitability (see later section). This is an intriguing possibility given the observation that certain RNA targets of LARK are predicted to encode ion channels.
LARK expression does not alter locomotor activity rhythms or the molecular oscillator in light-dark conditions
Nitabach et al. (2002) reported that electrical silencing of the LNvs stops the molecular clock in free-running conditions but has no apparent effect when flies are entrained to a light-dark (LD) cycle (Nitabach et al., 2002). In order to test whether LARK overexpression has a similar impact on oscillator function, we examined behavioral rhythms and cycling of clock proteins in the LNvs of flies entrained to LD. Similar to the results of Nitabach et al. (2002), flies with increased LARK amounts displayed apparently wild-type locomotor activity rhythms in LD (data not shown), with normal anticipation of the lights-on and lights-off signals. In addition, rhythms in PER abundance and nuclear entry were observed within both the s-LNv and l-LNv neurons of LARK OE flies maintained in LD, although the amplitude of the molecular rhythm was reduced in the l-LNvs due to a decrease in PER amount at ZT1 (Figure 6A).
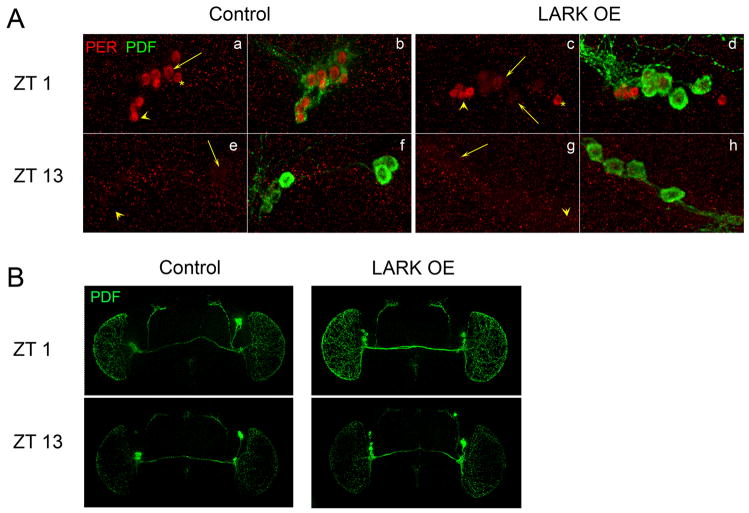
Figure 6.
LARK overexpression has no effect on the circadian oscillation of PER abundance or rhythmic release of PDF under light-dark conditions. A. Representative images showing levels of PER immunoreactivity in the l- and s-LNvs at ZT1 and ZT13 for LARK overexpression and control flies. Brains were also stained with anti-PDF antibody to identify the LNvs. B. Representative images showing level of the PDF neuropeptide in the control and LARK OE flies at ZT1 and ZT13. Control: pdf-gal4; Tub-gal80ts. LARK OE: pdf-gal4; uas-lark Tub-gal80ts. Flies were raised at 23°C, entrained to LD 12:12 at 30°C for at least 4 days before the images were acquired. Arrows point to large PDF neurons. Arrowhead points to small PDF neurons. * indicates the fifth LNv that is known to be PER positive and PDF negative.
To assay output, we examined the effect of LARK overexpression on the rhythmic abundance of PDF neuropeptide in light-dark conditions. In wild-type flies, the PDF immunoreactive signal in the dorsal projections of the s-LNvs is known to be under clock control; immunoreactivity displays a daily rhythm with a peak during the early subjective day and a trough in the early night (Park et al., 2000). We assessed PDF levels at ZT1 and ZT13 in LARK OE and control flies using anti-PDF antibodies. In contrast to previous reports that PDF cycling can be observed only in the dorsal projections of the s-LNvs, we found that PDF level in the projections of both the s-LNvs and l-LNvs, which cover the optic lobes, displays rhythmic daily changes (Figure 6B). The difference between our studies and the past report may be a consequence of different genetic backgrounds or antibody staining conditions. Nonetheless, LARK overexpression had no effect on the cycling of PDF in the projections of either the l-LNvs or the s-LNvs (Figure 6B).
These results suggest that LARK overexpression has a minimal effect on LNv function in the entrained state despite the dramatic phenotypes observed in free-running conditions. Similar observations were made by Nitabach et al. (2002) for flies with electrically silenced LNvs, further supporting the idea that LARK overexpression decreases membrane excitability of the LNvs. In addition, these results also provide additional evidence that LARK overexpression during adulthood does not alter neuronal morphology, as the cells appear to be functioning normally in light-dark conditions.
LARK regulates neuronal excitability
The results described above suggest that LARK may regulate neuronal excitability. Consistent with this idea, our past work indicates that many of the putative LARK target mRNAs encode ion channels or other proteins which regulate neuronal excitability (Huang et al., 2007). To test whether LARK can regulate neuronal excitability, we examined the effects of LARK overexpression and loss-of-function on the electrophysiological properties of the larval neuromuscular junction (NMJ). Larvae carrying the lark1null allele survive until pupation, facilitating studies of the null phenotype. In addition, the D42-gal4 transgene (Yeh et al., 1995) was used to drive LARK overexpression specifically in motor neurons.
We found that LARK loss-of-function and overexpression conferred opposite effects on excitability of the larval motor neurons. At all calcium concentrations tested (0.1 mM – 1 mM), the loss-of-function larvae displayed increased excitatory junctional potential (EJP) amplitude and an increased success rate for EJPs. The mutant larvae also show faster onset of long-term facilitation (LTF), a phenomenon that occurs after repetitive stimulation of the motor neurons at frequencies of 5–10 Hz (Jan and Jan, 1978) (Figure 7). It is noteworthy that at a calcium concentration of 0.1 mM, a concentration known to result in either EJP failure or fusion of a single vesicle in the wild type, lark1 mutants displayed a dramatically reduced rate of failures. In addition, the amplitude of the successful EJPs was higher than in the wild type, indicating release of multiple vesicles. These electrophysiological phenotypes closely resemble those of Drosophila mutants with increased neuronal excitability such as Shaker (Sh) (Jan et al., 1977; Mallart et al., 1991), Hyperkinetic (Hk) (Stern and Ganetzky, 1989; Chouinard et al., 1995), frequenin (frq) (Poulain et al., 1994), pumilio (pum) (Schweers et al., 2002), duplication of the paralytic gene (Dp para+) (Stern et al., 1990), and inebriated (ine) (Stern and Ganetzky, 1992; Huang and Stern, 2002), suggesting that LARK loss-of-function is associated with increased neuronal excitability.
Figure 7.
The lark1null mutation and LARK OE have oppos ite effects on the excitability of larval motor neurons. A. Mean EJP amplitudes (Y ax is) at the indicated [Ca2+] (X axis), from the indicated genotypes. Larval nerves were stimulated at a frequency of 1 Hz, and 30 responses were measured from each of six larvae. Means ± SEMs are shown. B. Number of stimulations required to induce LTF (Y axis) at the indicated stimulus frequencies (X axis) in the indicated genotypes. Geometric means ± SEMs are shown. n = 6 for each genotype. For LTF measurements, the bath solution contained 0.15 mM [Ca2+] and 100 μM quinidine. C. Effects of the lark1 mutationand LARK OE on the probability of transmitter release. Mean transmitter release success rates ± SEMs (Y axis) at the indicated Ca2+ concentration (X axis) for the indicated genotypes. Larval nerves were stimulated at 1 Hz. Thirty responses were collected per nerve from each of 6 larvae for the given genotype and at the given Ca2+ concentration.
In contrast to loss-of-function, LARK OE caused reduced EJP amplitude, reduced EJP success rate, and a slower onset of LTF, phenotypes which are typical of flies with reduced neuronal excitability such as paralytic (para) (Stern et al., 1990) or inebriated overexpression flies (Overine+) (Huang and Stern, 2002). We note that the miniature EJPs (mEJPs), which result from spontaneous release of single vesicles – and thus reflect the sensitivity of the post-synaptic membrane to neurotransmitter – were not altered by loss-of-function or overexpression (data not shown), indicating a presynaptic origin of the mutant phenotypes. Taken together, these results suggest that LARK can regulate neuronal excitability, possibly by controlling the expression of mRNAs encoding ion channels.
Discussion
Diverse roles of LARK in neural development and physiology
Post-transcriptional regulation of gene expression, via RNA-binding proteins (RBPs), is a hall-mark of nervous system development and function (Robinow and White, 1991; Antic et al., 1999; Keene and Lager, 2005; Keene, 2007; Thyagarajan et al., 2007; Zearfoss et al., 2008). For example, the HuD RBP regulates neuronal differentiation, maintenance and plasticity (reviewed in (Deschenes-Furry et al., 2006)). Another RBP, the Fragile X mental retardation protein, FMRP, is critical for synapse differentiation and circadian functions (Nimchinsky et al., 2001; Zhang et al., 2001; Dockendorff et al., 2002; Morales et al., 2002; Lee et al., 2003a; Pan et al., 2004) and a recent study indicates that LARK interacts with FMRP (Sofola et al., 2008). Our previous work shows that the Drosophila LARK RBP is associated with a large number of distinct RNA targets, in vivo (Huang et al., 2007), including many that are involved in cell proliferation, fate determination, axon guidance, synapse formation and cell death. LARK overexpression causes defasciculation of the s-LNv dorsal projections and failure of the projections to reach their targets or form synaptic boutons, phenotypes that might reflect misregulation of a set of LARK targets required for axonal development (e.g., NetB and Fas2). High level LARK expression results in the elimination of the s-LNvs, which may come about through an abnormal regulation of targets mediating apoptotic or autophagic cell death (e.g., Aac11, Mmp1, Eip74EF, bun and l(2)01424).
In addition to targets governing neural development, LARK is also associated with RNAs encoding proteins that are critical for neural physiology including ion channels such as Shaker (Sh), Hyperkinetic (Hk),, small conductance calcium-activated potassium channel (SK) and GluClα, signaling molecules such as Calmodulin (Cam) and no receptor potential A (norpA), and molecules critical for synaptic function such as Synapsin (Syn), Syntaxin 1A (Syx1A) and shibire (shi) (Huang et al., 2007). The diversity of LARK targets suggests that the RBP has many distinct roles in the nervous system, consistent with results reported here that implicate the protein in both developmental and physiological processes.
How does LARK expression affect the circadian regulation of locomotor activity?
By using an inducible GAL80ts transgene, we demonstrate that conditional overexpression of LARK in all PDF-positive LNv neurons of the adult results in arrhythmic locomotor activity during free-running conditions. This result indicates a physiological function for the RBP that is independent of its roles in neural development. Given the similarity of our results to those obtained with perturbations of clock neuronal membrane excitability (Nitabach et al., 2002; Wu et al., 2008), it is possible that increased LARK amounts alters expression of one or more ion channels in the LNvs, leading to decreased neuronal excitability and arrhythmic behavior. It is known that deficits for specific ion channels within the LNvs or other clock neurons lead to altered rhythmic behavior (Nitabach et al., 2002; Hodge and Stanewsky, 2008; Wu et al., 2008). Moreover, normal LNv excitability is known to be critical for maintaining molecular oscillator function and rhythmic locomotor activity, and several recent studies have demonstrated that electrical silencing and other perturbations of clock neuronal excitability severely disrupt locomotor activity rhythms in constant conditions (Nitabach et al., 2002; Nitabach et al., 2006; Wu et al., 2008). Interestingly, Nitabach et al (2002) also reported that normal LNv membrane excitability is required for sustaining the molecular clock under free-running conditions but not when the clock is entrained to light-dark cues (Nitabach et al., 2002). They proposed that electrical activity might act through voltage-gated calcium channels to regulate calcium entry, and hence calcium signaling pathways which are known to have important roles in the clock system (Belvin et al., 1999; Harrisingh et al., 2007) as a reinforcing feedback mechanism in the absence of environmental cues. Whereas the s-LNv population is known to be crucial for free-running circadian behavior (Helfrich-Forster et al., 2007), the functions of the l-LNv cells are not understood in the same detail. However, recent studies show that the l-LNv cells exhibit circadian changes in electrical activity and they are directly responsive to light stimulation in a Crytochrome (Cry)-dependent manner (Sheeba et al., 2008). Other results indicate that the l-LNv cells mediate light-dependent arousal and perhaps contribute to light resetting of the circadian clock (Shang et al., 2008).
The hypothesis that LARK overexpression alters LNv membrane excitability is supported by several lines of evidence: (1) several LARK targets encode ion channels including two well-known voltage-gated potassium channels, Shaker (Sh) and Hyperkinetic (Hk), as well as a calcium-activated potassium channel called SK. In addition, another target of LARK, pumilio (pum), is known to negatively regulate the paralytic (para) voltage-gated sodium channel (Mee et al., 2004; Muraro et al., 2008). Thus, the effects of LARK overexpression may be indirectly mediated through pum. Regulation by the RBP may also involve microRNA-dependent mechanisms (Hock et al., 2007; Huang et al., 2007) (2) Electrophysiological experiments performed using the larval NMJ clearly demonstrate that a lark null mutation is associated with increased neuronal excitability whereas overexpression of LARK causes decreased excitability. (3) LARK overexpression phenocopies the electrical silencing of the LNvs at both behavioral and molecular levels. In additional to behavioral arrhythmicity, LARK overexpression also abolishes PER cycling in the l-LNvs under free-running conditions, consistent with a previous report showing that electrical silencing of the LNvs stops PER and TIM cycling in these neurons (Nitabach et al., 2002), although that report did not specify whether clock protein cycling is abolished in the l-LNvs, the s-LNvs or both. In addition, that publication showed that electrical silencing of the LNvs has no apparent effect on rhythmicity when the clock is entrained to a light-dark cycle, similar to the results with LARK overexpression. Further, a recent report shows that electrical silencing of the LNvs abolishes PDP1ε cycling in the s-LNvs (Wu et al., 2008) which is also phenocopied by LARK overexpression (Figure 5).
While LARK may regulate neuronal activity directly by modulating ion channel targets, it is also possible that the RBP modulates targets encoding signaling molecules (e.g., kinases or phosphatases) which in turn control ion channels and excitability. Alternatively the RBP might regulate targets that control PDP1ε, given our results that this factor is nearly undetectable in the s-LNvs of LARK OE flies at CT17, a time when its level peaks in the wild type (Figure 5). Benito et al (2007) have suggested that PDP1ε is a clock output factor rather than a core clock component and loss of the molecule is known to cause behavioral arrhythmicity (Glossop et al., 1999; Cyran et al., 2003; Benito et al., 2007; Lim et al., 2007).
Differential effects of LARK expression on the l-LNv and s-LNv neurons
Finally, our results reveal a differential affect of LARK overexpression on the s-LNv and l-LNv neurons. Increased LARK expression within the PDF-containing LNvs causes developmental defects and cell death exclusively in the s-LNv population with no apparent effects on development of the l-LNvs. In contrast, the conditional overexpression of LARK in adult LNv neurons perturbs PER cycling specifically in the l-LNvs but does not impair cycling of this clock protein in the s-LNvs (Figure 4). Interestingly, however, PDP1 cycling is altered in the s-LNvs with conditional overexpression, indicative of effects on output (Benito et al., 2007) (Figure 5). These differential effects suggest that distinct mRNA targets are regulated in the small versus the large LNvs during development and in adulthood. For example, it is possible that LARK-regulated targets relevant for neuronal differentiation are expressed in the s-LNvs but not in the l-LNvs during development. Similarly, the adult l-LNv and s-LNv populations may differentially express target mRNAs which regulate particular circadian functions. Given the observed effects of LARK expression on PER cycling in the l-LNv neurons and PDP1 in the s-LNv cells, it is possible that physiological changes in both populations contribute to altered behavior. Future studies that manipulate LARK amounts selectively in only the s-LNvs or l-LNvs may determine the population in which function of the RBP is most relevant for behavior.
Experimental Methods
Fly strains
The pdf-gal4 lines were provided by Jae Park (University of Tennessee, Knoxville, TN). w1118, UAS-mCD8GFP, and ts-tubulin-gal80 were obtained from the Bloomington Stock Center. UAS-lark (23A) was described previously (Schroeder et al., 2003). All GAL4, UAS, and GAL80 transgenes were crossed onto the w1118 genetic background. Flies were reared on a modified cornmeal/agar medium (medium1 of (Newby and Jackson, 1991)) in an 12:12 light-dark cycle at 23°C with constant humidity (~60%) unless otherwise indicated.
Behavioral Analysis
Locomotor activity was assayed using 2- to 3-day-old males and the Drosophila Activity Monitoring (DAM) system (Trikinetics, Waltham, MA). After loading flies into monitors, they were entrained to LD 12:12 for 4–5 days and then transferred to constant darkness (DD) for an additional 7–10 days. Visualization of actograms and the analysis of rhythmicity were performed using a signal processing toolbox (Levine et al., 2002) within the MATLAB software package (MathWorks).
Immunohistochemistry
Drosophila adults or larva were were fixed in 4% paraformaldehyde solution and then washed in PBS and PBS-T (0.05% Triton X-100). For immunostaining procedures, we employed the primary antibodies at the following dilutions: Rabbit anti-PER (1:10000, R. Stanewsky), mouse anti-PDF (1:10, DSHB), guinea pig anti-PDP1 (1:30000, Hardin, P), rabbit anti-PDH (1:1000, K.R. Rao). Rabbit anti-LARK (1:1000, (McNeil et al., 1998). Secondary antibodies – goat anti-mouse IgG (Alexa-488 conjugated, Molecular Probes), goat anti-rabbit (Cy3 conjugated or Alexa-488 conjugated, Molecular Probes), goat-anti-guinea pig (Cy3-conjugated, Molecular Probes) – were all used at a dilution of 1:300 and an incubation time of at least 5 hours. Confocal images were acquired from brain whole mounts using a Leica TCS SP2 AOBS microscope within the Tufts Center for Neuroscience Research (CNR) Imaging Core. Measurement of cell diameter was conducted using the LCS Leica confocal software. As most cells are not perfectly circular, the longest distance across a cell was used as the diameter.
Electrophysiology
Larvae were grown to the wandering third-instar stage in uncrowded bottles at room temperature and dissected as described (Jan and Jan, 1978; Stern and Ganetzky, 1989) in standard saline solution (128 mM NaCl, 2.0 mM KCl, 4.0 mM MgCl2, 340 mM sucrose, 5.0 mM HEPES, pH 7.1, and CaCl2 as specified in the text). Peripheral nerves were cut posterior to the ventral ganglion and were stimulated using a suction electrode. Muscle recordings were taken from muscle 6 in abdominal sections 3–5. Recording electrodes were pulled using a Flaming/Brown micropipette puller to a tip resistance of 10–40 MΩ and filled with 3M KCl.
Supplementary Material
Supplemental Figure 1. Cell bodies and projections of the PDF neurons are normal 8 days after induction of LARK overexpression in adult flies. Flies were raised at 23°C and transferred into 30°C after eclosion to induce LARK overexpression. They were entrained to a LD 12:12 schedule at 30°C. Brains were dissected on the 8th day of 30°C incubation and stained with anti-LARK (red) and anti-PDF (green) antibodies. Control: pdf-gal4; Tub-gal80ts. LARK OE: pdf-gal4; uas-lark Tub-gal80ts. A low concentration of LARK antibody was used such that the control flies showed minimal LARK immunoreactivity. In contrast, the LARK OE flies showed dramatically increased LARK staining. Note that despite induction of LARK overexpression, the cell bodies and projections of the PDF neurons were intact.
Acknowledgments
We thank Jae Park for providing the pdf-gal4 lines, Dr. Lai Ding and Dr. Alenka Lovy-Wheeler (Tufts Imaging Facility) for assistance with confocal imaging, and Ralf Stanewsky, Paul Hardin, K. Ranga Rao, and Gerry McNeil for providing the anti-PER, anti-PDP1ε, anti-PDH and anti-LARK antibodies, respectively. This research was supported by NIH R01 HL59873 (FRJ), Department of Defense W81XWH-04-1-0272 (MS), and center grant NIH P30 NS047243 to the Tufts Center for Neuroscience Research (PI, FRJ).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Antic D, Lu N, Keene JD. ELAV tumor antigen, Hel-N1, increases translation of neurofilament M mRNA and induces formation of neurites in human teratocarcinoma cells. Genes Dev. 1999;13:449–461. doi: 10.1101/gad.13.4.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belvin MP, Zhou H, Yin JC. The Drosophila dCREB2 gene affects the circadian clock. Neuron. 1999;22:777–787. doi: 10.1016/s0896-6273(00)80736-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benito J, Zheng H, Hardin PE. PDP1epsilon functions downstream of the circadian oscillator to mediate behavioral rhythms. J Neurosci. 2007;27:2539–2547. doi: 10.1523/JNEUROSCI.4870-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchardon E, Grima B, Klarsfeld A, Chelot E, Hardin PE, Preat T, Rouyer F. Defining the role of Drosophila lateral neurons in the control of circadian rhythms in motor activity and eclosion by targeted genetic ablation and PERIOD protein overexpression. Eur J Neurosci. 2001;13:871–888. doi: 10.1046/j.0953-816x.2000.01450.x. [DOI] [PubMed] [Google Scholar]
- Chouinard SW, Wilson GF, Schlimgen AK, Ganetzky B. A potassium channel beta subunit related to the aldo-keto reductase superfamily is encoded by the Drosophila hyperkinetic locus. Proc Natl Acad Sci U S A. 1995;92:6763–6767. doi: 10.1073/pnas.92.15.6763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyran SA, Buchsbaum AM, Reddy KL, Lin MC, Glossop NR, Hardin PE, Young MW, Storti RV, Blau J. vrille, Pdp1, and dClock form a second feedback loop in the Drosophila circadian clock. Cell. 2003;112:329–341. doi: 10.1016/s0092-8674(03)00074-6. [DOI] [PubMed] [Google Scholar]
- Deschenes-Furry J, Perrone-Bizzozero N, Jasmin BJ. The RNA-binding protein HuD: a regulator of neuronal differentiation, maintenance and plasticity. Bioessays. 2006;28:822–833. doi: 10.1002/bies.20449. [DOI] [PubMed] [Google Scholar]
- Dockendorff TC, Su HS, McBride SM, Yang Z, Choi CH, Siwicki KK, Sehgal A, Jongens TA. Drosophila lacking dfmr1 activity show defects in circadian output and fail to maintain courtship interest. Neuron. 2002;34:973–984. doi: 10.1016/s0896-6273(02)00724-9. [DOI] [PubMed] [Google Scholar]
- Garbarino-Pico E, Green CB. Posttranscriptional regulation of mammalian circadian clock output. Cold Spring Harb Symp Quant Biol. 2007;72:145–156. doi: 10.1101/sqb.2007.72.022. [DOI] [PubMed] [Google Scholar]
- Glossop NR, Lyons LC, Hardin PE. Interlocked feedback loops within the Drosophila circadian oscillator. Science. 1999;286:766–768. doi: 10.1126/science.286.5440.766. [DOI] [PubMed] [Google Scholar]
- Harms E, Kivimae S, Young MW, Saez L. Posttranscriptional and posttranslational regulation of clock genes. J Biol Rhythms. 2004;19:361–373. doi: 10.1177/0748730404268111. [DOI] [PubMed] [Google Scholar]
- Harrisingh MC, Wu Y, Lnenicka GA, Nitabach MN. Intracellular Ca2+ regulates free-running circadian clock oscillation in vivo. J Neurosci. 2007;27:12489–12499. doi: 10.1523/JNEUROSCI.3680-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay BA, Wolff T, Rubin GM. Expression of baculovirus P35 prevents cell death in Drosophila. Development. 1994;120:2121–2129. doi: 10.1242/dev.120.8.2121. [DOI] [PubMed] [Google Scholar]
- Heintzen C, Nater M, Apel K, Staiger D. AtGRP7, a nuclear RNA-binding protein as a component of a circadian-regulated negative feedback loop in Arabidopsis thaliana. Proc Natl Acad Sci U S A. 1997;94:8515–8520. doi: 10.1073/pnas.94.16.8515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfrich-Forster C. Development of pigment-dispersing hormone-immunoreactive neurons in the nervous system of Drosophila melanogaster. J Comp Neurol. 1997;380:335–354. doi: 10.1002/(sici)1096-9861(19970414)380:3<335::aid-cne4>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- Helfrich-Forster C. The neuroarchitecture of the circadian clock in the brain of Drosophila melanogaster. Microsc Res Tech. 2003;62:94–102. doi: 10.1002/jemt.10357. [DOI] [PubMed] [Google Scholar]
- Helfrich-Forster C, Shafer OT, Wulbeck C, Grieshaber E, Rieger D, Taghert P. Development and morphology of the clock-gene-expressing lateral neurons of Drosophila melanogaster. J Comp Neurol. 2007;500:47–70. doi: 10.1002/cne.21146. [DOI] [PubMed] [Google Scholar]
- Hock J, Weinmann L, Ender C, Rudel S, Kremmer E, Raabe M, Urlaub H, Meister G. Proteomic and functional analysis of Argonaute-containing mRNA-protein complexes in human cells. EMBO Rep. 2007;8:1052–1060. doi: 10.1038/sj.embor.7401088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodge JJ, Stanewsky R. Function of the Shaw potassium channel within the Drosophila circadian clock. PLoS ONE. 2008;3:e2274. doi: 10.1371/journal.pone.0002274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Stern M. In vivo properties of the Drosophila inebriated-encoded neurotransmitter transporter. J Neurosci. 2002;22:1698–1708. doi: 10.1523/JNEUROSCI.22-05-01698.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Genova G, Roberts M, Jackson FR. The LARK RNA-binding protein selectively regulates the circadian eclosion rhythm by controlling E74 protein expression. PLoS ONE. 2007;2:e1107. doi: 10.1371/journal.pone.0001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iliev D, Voytsekh O, Schmidt EM, Fiedler M, Nykytenko A, Mittag M. A heteromeric RNA-binding protein is involved in maintaining acrophase and period of the circadian clock. Plant Physiol. 2006;142:797–806. doi: 10.1104/pp.106.085944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan YN, Jan LY. Genetic dissection of short-term and long-term facilitation at the Drosophila neuromuscular junction. Proc Natl Acad Sci U S A. 1978;75:515–519. doi: 10.1073/pnas.75.1.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan YN, Jan LY, Dennis MJ. Two mutations of synaptic transmission in Drosophila. Proc R Soc Lond B Biol Sci. 1977;198:87–108. doi: 10.1098/rspb.1977.0087. [DOI] [PubMed] [Google Scholar]
- Keene JD. RNA regulons: coordination of post-transcriptional events. Nat Rev Genet. 2007;8:533–543. doi: 10.1038/nrg2111. [DOI] [PubMed] [Google Scholar]
- Keene JD, Lager PJ. Post-transcriptional operons and regulons co-ordinating gene expression. Chromosome Res. 2005;13:327–337. doi: 10.1007/s10577-005-0848-1. [DOI] [PubMed] [Google Scholar]
- Kojima S, Matsumoto K, Hirose M, Shimada M, Nagano M, Shigeyoshi Y, Hoshino S, Ui-Tei K, Saigo K, Green CB, Sakaki Y, Tei H. LARK activates posttranscriptional expression of an essential mammalian clock protein, PERIOD1. Proc Natl Acad Sci U S A. 2007;104:1859–1864. doi: 10.1073/pnas.0607567104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A, Li W, Xu K, Bogert BA, Su K, Gao FB. Control of dendritic development by the Drosophila fragile X-related gene involves the small GTPase Rac1. Development. 2003a;130:5543–5552. doi: 10.1242/dev.00792. [DOI] [PubMed] [Google Scholar]
- Lee K, Dunlap JC, Loros JJ. Roles for WHITE COLLAR-1 in circadian and general photoperception in Neurospora crassa. Genetics. 2003b;163:103–114. doi: 10.1093/genetics/163.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim C, Lee J, Koo E, Choe J. Targeted inhibition of Pdp1epsilon abolishes the circadian behavior of Drosophila melanogaster. Biochem Biophys Res Commun. 2007;364:294–300. doi: 10.1016/j.bbrc.2007.10.009. [DOI] [PubMed] [Google Scholar]
- Liu Y. Analysis of posttranslational regulations in the Neurospora circadian clock. Methods Enzymol. 2005;393:379–393. doi: 10.1016/S0076-6879(05)93017-6. [DOI] [PubMed] [Google Scholar]
- Mallart A, Angaut-Petit D, Bourret-Poulain C, Ferrus A. Nerve terminal excitability and neuromuscular transmission in T(X;Y)V7 and Shaker mutants of Drosophila melanogaster. J Neurogenet. 1991;7:75–84. doi: 10.3109/01677069109066212. [DOI] [PubMed] [Google Scholar]
- McGuire SE, Mao Z, Davis RL. Spatiotemporal gene expression targeting with the TARGET and gene-switch systems in Drosophila. Sci STKE. 2004;2004:pl6. doi: 10.1126/stke.2202004pl6. [DOI] [PubMed] [Google Scholar]
- McNeil GP, Zhang X, Genova G, Jackson FR. A molecular rhythm mediating circadian clock output in Drosophila. Neuron. 1998;20:297–303. doi: 10.1016/s0896-6273(00)80457-2. [DOI] [PubMed] [Google Scholar]
- McNeil GP, Schroeder AJ, Roberts MA, Jackson FR. Genetic analysis of functional domains within the Drosophila LARK RNA-binding protein. Genetics. 2001;159:229–240. doi: 10.1093/genetics/159.1.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mee CJ, Pym EC, Moffat KG, Baines RA. Regulation of neuronal excitability through pumilio-dependent control of a sodium channel gene. J Neurosci. 2004;24:8695–8703. doi: 10.1523/JNEUROSCI.2282-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales J, Hiesinger PR, Schroeder AJ, Kume K, Verstreken P, Jackson FR, Nelson DL, Hassan BA. Drosophila fragile X protein, DFXR, regulates neuronal morphology and function in the brain. Neuron. 2002;34:961–972. doi: 10.1016/s0896-6273(02)00731-6. [DOI] [PubMed] [Google Scholar]
- Muraro NI, Weston AJ, Gerber AP, Luschnig S, Moffat KG, Baines RA. Pumilio binds para mRNA and requires Nanos and Brat to regulate sodium current in Drosophila motoneurons. J Neurosci. 2008;28:2099–2109. doi: 10.1523/JNEUROSCI.5092-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newby LM, Jackson FR. Drosophila ebony mutants have altered circadian activity rhythms but normal eclosion rhythms. J Neurogenet. 1991;7:85–101. doi: 10.3109/01677069109066213. [DOI] [PubMed] [Google Scholar]
- Newby LM, Jackson FR. A new biological rhythm mutant of Drosophila melanogaster that identifies a gene with an essential embryonic function. Genetics. 1993;135:1077–1090. doi: 10.1093/genetics/135.4.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newby LM, Jackson FR. Regulation of a specific circadian clock output pathway by lark, a putative RNA-binding protein with repressor activity. J Neurobiol. 1996;31:117–128. doi: 10.1002/(SICI)1097-4695(199609)31:1<117::AID-NEU10>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Nimchinsky EA, Oberlander AM, Svoboda K. Abnormal development of dendritic spines in FMR1 knock-out mice. J Neurosci. 2001;21:5139–5146. doi: 10.1523/JNEUROSCI.21-14-05139.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitabach MN, Taghert PH. Organization of the Drosophila circadian control circuit. Curr Biol. 2008;18:R84–93. doi: 10.1016/j.cub.2007.11.061. [DOI] [PubMed] [Google Scholar]
- Nitabach MN, Blau J, Holmes TC. Electrical silencing of Drosophila pacemaker neurons stops the free-running circadian clock. Cell. 2002;109:485–495. doi: 10.1016/s0092-8674(02)00737-7. [DOI] [PubMed] [Google Scholar]
- Nitabach MN, Wu Y, Sheeba V, Lemon WC, Strumbos J, Zelensky PK, White BH, Holmes TC. Electrical hyperexcitation of lateral ventral pacemaker neurons desynchronizes downstream circadian oscillators in the fly circadian circuit and induces multiple behavioral periods. J Neurosci. 2006;26:479–489. doi: 10.1523/JNEUROSCI.3915-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan L, Zhang YQ, Woodruff E, Broadie K. The Drosophila fragile X gene negatively regulates neuronal elaboration and synaptic differentiation. Curr Biol. 2004;14:1863–1870. doi: 10.1016/j.cub.2004.09.085. [DOI] [PubMed] [Google Scholar]
- Park JH, Helfrich-Forster C, Lee G, Liu L, Rosbash M, Hall JC. Differential regulation of circadian pacemaker output by separate clock genes in Drosophila. Proc Natl Acad Sci U S A. 2000;97:3608–3613. doi: 10.1073/pnas.070036197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulain C, Ferrus A, Mallart A. Modulation of type A K+ current in Drosophila larval muscle by internal Ca2+; effects of the overexpression of frequenin. Pflugers Arch. 1994;427:71–79. doi: 10.1007/BF00585944. [DOI] [PubMed] [Google Scholar]
- Renn SC, Park JH, Rosbash M, Hall JC, Taghert PH. A pdf neuropeptide gene mutation and ablation of PDF neurons each cause severe abnormalities of behavioral circadian rhythms in Drosophila. Cell. 1999;99:791–802. doi: 10.1016/s0092-8674(00)81676-1. [DOI] [PubMed] [Google Scholar]
- Robinow S, White K. Characterization and spatial distribution of the ELAV protein during Drosophila melanogaster development. J Neurobiol. 1991;22:443–461. doi: 10.1002/neu.480220503. [DOI] [PubMed] [Google Scholar]
- Schroeder AJ, Genova GK, Roberts MA, Kleyner Y, Suh J, Jackson FR. Cell-specific expression of the lark RNA-binding protein in Drosophila results in morphological and circadian behavioral phenotypes. J Neurogenet. 2003;17:139–169. [PubMed] [Google Scholar]
- Schweers BA, Walters KJ, Stern M. The Drosophila melanogaster translational repressor pumilio regulates neuronal excitability. Genetics. 2002;161:1177–1185. doi: 10.1093/genetics/161.3.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Y, Griffith LC, Rosbash M. Light-arousal and circadian photoreception circuits intersect at the large PDF cells of the Drosophila brain. Proc Natl Acad Sci U S A. 2008;105:19587–19594. doi: 10.1073/pnas.0809577105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheeba V, Fogle KJ, Kaneko M, Rashid S, Chou YT, Sharma VK, Holmes TC. Large ventral lateral neurons modulate arousal and sleep in Drosophila. Curr Biol. 2008;18:1537–1545. doi: 10.1016/j.cub.2008.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sofola O, Sundram V, Ng F, Kleyner Y, Morales J, Botas J, Jackson FR, Nelson DL. The Drosophila FMRP and LARK RNA-binding proteins function together to regulate eye development and circadian behavior. J Neurosci. 2008;28:10200–10205. doi: 10.1523/JNEUROSCI.2786-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern M, Ganetzky B. Altered synaptic transmission in Drosophila hyperkinetic mutants. J Neurogenet. 1989;5:215–228. doi: 10.3109/01677068909066209. [DOI] [PubMed] [Google Scholar]
- Stern M, Ganetzky B. Identification and characterization of inebriated, a gene affecting neuronal excitability in Drosophila. J Neurogenet. 1992;8:157–172. doi: 10.3109/01677069209083445. [DOI] [PubMed] [Google Scholar]
- Stern M, Kreber R, Ganetzky B. Dosage effects of a Drosophila sodium channel gene on behavior and axonal excitability. Genetics. 1990;124:133–143. doi: 10.1093/genetics/124.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taghert PH, Shafer OT. Mechanisms of clock output in the Drosophila circadian pacemaker system. J Biol Rhythms. 2006;21:445–457. doi: 10.1177/0748730406293910. [DOI] [PubMed] [Google Scholar]
- Thyagarajan A, Strong MJ, Szaro BG. Post-transcriptional control of neurofilaments in development and disease. Exp Cell Res. 2007;313:2088–2097. doi: 10.1016/j.yexcr.2007.02.014. [DOI] [PubMed] [Google Scholar]
- Vanselow K, Kramer A. Role of phosphorylation in the mammalian circadian clock. Cold Spring Harb Symp Quant Biol. 2007;72:167–176. doi: 10.1101/sqb.2007.72.036. [DOI] [PubMed] [Google Scholar]
- Wu Y, Cao G, Nitabach MN. Electrical silencing of PDF neurons advances the phase of non-PDF clock neurons in Drosophila. J Biol Rhythms. 2008;23:117–128. doi: 10.1177/0748730407312984. [DOI] [PubMed] [Google Scholar]
- Yeh E, Gustafson K, Boulianne GL. Green fluorescent protein as a vital marker and reporter of gene expression in Drosophila. Proc Natl Acad Sci U S A. 1995;92:7036–7040. doi: 10.1073/pnas.92.15.7036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young MW, Kay SA. Time zones: a comparative genetics of circadian clocks. Nat Rev Genet. 2001;2:702–715. doi: 10.1038/35088576. [DOI] [PubMed] [Google Scholar]
- Zearfoss NR, Farley BM, Ryder SP. Post-transcriptional regulation of myelin formation. Biochim Biophys Acta. 2008;1779:486–494. doi: 10.1016/j.bbagrm.2008.06.003. [DOI] [PubMed] [Google Scholar]
- Zhang X, McNeil GP, Hilderbrand-Chae MJ, Franklin TM, Schroeder AJ, Jackson FR. Circadian regulation of the lark RNA-binding protein within identifiable neurosecretory cells. J Neurobiol. 2000;45:14–29. doi: 10.1002/1097-4695(200010)45:1<14::aid-neu2>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- Zhang YQ, Bailey AM, Matthies HJ, Renden RB, Smith MA, Speese SD, Rubin GM, Broadie K. Drosophila fragile X-related gene regulates the MAP1B homolog Futsch to control synaptic structure and function. Cell. 2001;107:591–603. doi: 10.1016/s0092-8674(01)00589-x. [DOI] [PubMed] [Google Scholar]
- Zheng X, Sehgal A. Probing the relative importance of molecular oscillations in the circadian clock. Genetics. 2008;178:1147–1155. doi: 10.1534/genetics.107.088658. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Cell bodies and projections of the PDF neurons are normal 8 days after induction of LARK overexpression in adult flies. Flies were raised at 23°C and transferred into 30°C after eclosion to induce LARK overexpression. They were entrained to a LD 12:12 schedule at 30°C. Brains were dissected on the 8th day of 30°C incubation and stained with anti-LARK (red) and anti-PDF (green) antibodies. Control: pdf-gal4; Tub-gal80ts. LARK OE: pdf-gal4; uas-lark Tub-gal80ts. A low concentration of LARK antibody was used such that the control flies showed minimal LARK immunoreactivity. In contrast, the LARK OE flies showed dramatically increased LARK staining. Note that despite induction of LARK overexpression, the cell bodies and projections of the PDF neurons were intact.