Abstract
Background
Head and neck squamous cell carcinoma (HNSCC) is a heterogenous disease evolving through multi-step carcinogenesis, one of the steps being genetic alterations. Non-invasive identification of HNSCC-specific genetic alterations using saliva would have immense potential in early diagnosis and screening, particularly among high-risk patients.
Design
In this exploratory study, a prospective cohort of 27HNSCC and 10 healthy controls was examined to determine whether genetic alterations (losses and gains) in saliva DNA differentiated HNSCC patients from normal controls. Saliva DNA was interrogated by a candidate gene panel comprising 82 genes using the multiplex ligation-dependent probe amplification (MLPA) assay.
Results
Eleven genes showed some predictive ability in identifying HNSCC cases from normal controls: PMAIP1, PTPN1 ERBB2, ABCC4, UTY, DNMT1, CDKN2B, CDKN2D, NFKB1, TP53 and DCC. Statistical analysis using the Classification and Regression Tree (CART) identified 2 genes, PMAIP1 and PTPN1, which correctly discriminated all 27 HNSCC patients (100%) from normal controls. Results were validated using the leave-one-out validation approach.
Conclusions
Noninvasive high-throughput MLPA identified discrete gene signatures that differentiated HNSCC patients from normal controls providing poof-of-concept for noninvasive HNSCC detection.
Keywords: Non-invasive, Head and neck squamous cell carcinoma, genetic alterations, Multiplex Ligation-dependent Probe Amplification Assay (MLPA), Classification and Regression Tree (CART)
INTRODUCTION
Head and neck squamous cell carcinoma (HNSCC) is a significant health concern worldwide with a prevalence of more than 1.6 million (1). In the United States, its estimated incidence of HNSCC for 2006 was 30, 990, and caused over 7,400 deaths (2). Despite rapid advances in the treatment of HNSCC, the 5-year survival has only marginally improved from 54.4% to 59.4% over the past two decades (1, 3). Survivors suffer serious and devastating morbidities including speech and swallowing problems, disfigurement and exorbitant healthcare costs (4).
The poor outcome for HNSCC has been explained predominantly on the basis of late detection of cases (5). Early detection through screening seems an obvious solution (6). Currently used screening by clinical examination is complex and presents with several challenges even in a primary care setting (7, 8, 9). Hence, there is a need to develop screening tests which are effective in routine clinical practice. Molecular approaches appear promising in this regard.
Molecular alterations occur early in HNSCC (10). Acquisition of a fully malignant phenotype is a stepwise progression defined by several genetic alterations at 3p, 4q, 5q, 6p, 9p, 11q, 13q, 17p, 18q and 20q (11–14). Genetic alterations confer upon the malignant cells characteristics which determine tumor behavior (14). However, no single molecular event is sufficient to accurately predict the pathobiology of HNSCC. Recently developed high-throughput assays permit detection of alterations in a large number of gene targets. Identification of HNSCC-specific genetic alterations can potentially serve as clonal molecular signatures to differentiate tumor cells from their normal counterparts.
Once established, these molecular HNSCC signatures may have clinical utility as diagnostic, prognostic and therapeutic biomarkers. This approach can produce validated marker panels for screening purposes, comprising of candidate gene probes specific for HNSCC (15). Since genetic alterations occur before phenotypic expression of cancer (5), they have the potential to serve as biomarkers for early detection (10).
Head and neck cancers bathe in readily accessible saliva secretions. Saliva being a non-invasive patient sample has immense potential for use in screening programs. Since there is no patient discomfort, repeat samples can be collected which are important for screening and tumor surveillance. In HNSCC patients, saliva contains exfoliated cancer cells harboring genetic alterations (15). Thus it offers a potential non-invasive source to examine genetic alterations in HNSCC patients.
Multiplex ligation-dependent probe amplification assay (MLPA) is a recent high throughput genetic technique allowing simultaneous interrogation of 41 genes using scant amounts (20 ng) of DNA (14,16). Validated using real-time PCR, it has been used in a wide variety of samples including cell lines, tissue specimens and even saliva (14, 16–21). It can be used for detection of genetic alterations (losses and gains) (14,16, 21) as well as to evaluate epigenetic alterations (17, 18). It is distinctly advantageous over currently available genetic technologies like comparative genomic hybridization or DNA chip hybridization methods (14, 19, 20). Requiring minimal sample preparation, it is easy to perform, cost-effective, timesaving and reproducible one-tube assay (in comparison to array-based technology). As each component of the test is fluid, quality control is simpler in comparison to DNA microarrays.
In this exploratory analysis, we analyzed a non-invasive saliva approach to detect HNSCC-specific genetic alterations using MLPA.
MATERIALS AND METHODS
Study subjects
Thirty-seven subjects, 27 HNSCC patients and 10 normal controls were studied. Of the 27 HNSCC patients, 11 were early stage (I/II) and 16 were late stage (III/IV) tumors located in the upper aero-digestive tract, including 7 in the oral cavity, 2 in the orophaynx, 8 in the larynx, 5 on the tonsil, 1 in the pyriform sinus; and 4 cases had unknown primary squamous cell carcinoma metastatic to the neck. Only untreated patients were included in the study. Normal controls were healthy volunteers with no benign/malignant lesions. All study subjects were over 18 years of age. Informed consent was obtained according to the institutional review board–approved protocols.
Sample collection
Two ml saliva was collected from each study subject, in Oragene kits (DNA Genotek Inc, Ontario) and saliva DNA was extracted according to the manufacturer’s instructions.
Genetic analysis
Saliva DNA was interrogated for gene copy number alterations (losses and gains) using MLPA assay. Two gene probe panels, p005 and p006 (www.mlpa.com), comprising of 82 genes were examined. The panel detects primarily oncogenes and tumor suppressor genes that are located at chromosomal segments that have been implicated in cancer and distributed throughout the genome (21,22)
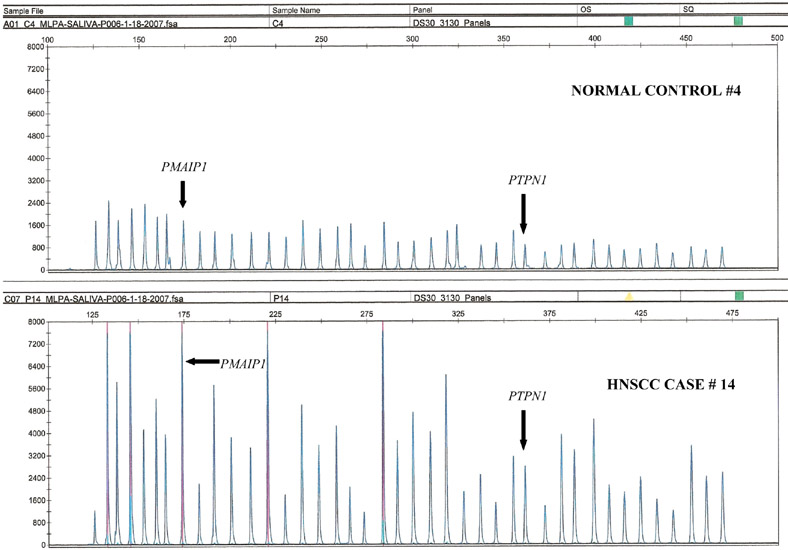
MLPA analysis was done as outlined previously (14, 16). Briefly, probes added to the samples were amplified and quantified instead of target nucleic acids. Amplification of probes by PCR depends on the presence of probe target sequences in the sample. Each probe consists of two oligonucleotides, one synthetic and one M13-derived, each hybridizing to adjacent sites of the target sequence. Such hybridized probe oligonucleotides, when ligated, permit subsequent amplification. Ligated probes have identical end sequences, permitting simultaneous PCR amplification using only one primer pair. Each probe gives rise to an amplification product of unique size between 130 and 480 bp. Probe target sequences are small (50–70 nucleotides). The prerequisite of a ligation reaction provides the opportunity to discriminate single nucleotide differences. The amplified fragments are separated on a DNA sequencer (Figure 1). Quantification of loss or gain of gene loci is determined through a process of normalization (14, 16). The peak area for each probe is expressed as a percent of the total surface area of all peaks of a sample in an assay run (Figure 1). Relative copy number for each probe is obtained as a ratio of the normalized value for each locus (peak) of the sample to that of the normal control, and in general the copy numbers in a range of 0.75 to 1.3 is regarded as normal, <0.75 as loss and >1.3 as gain (14, 21).
Figure 1.
Saliva MLPA: gain of PMAIP1 and PTPN1 genes seen in HNSCC case 14 compared to normal control.
Data Collection and Statistical analysis
To obtain saliva MLPA norms, we estimated 99.99% confidence intervals (CI’s) on normal controls. Given the result, we decided to use the numerical copy numbers for analysis to obtain more information from the data collected.
To discriminate HNSCC patients from normal controls, genetic alterations identified in saliva DNA, were analyzed using the Classification and Regression Tree (CART®) statistical tool to generate gene-based algorithms. CART methodology, known as binary recursive partitioning, uses non-parametric approaches (16,23,24).
Given the large number of genes (82 variables) and a small data set with 37 subjects, to avoid the over fitting, we first performed CART on all genes and ranked them based on their individual importance from high (e.g., 100%) to low (0%), which is similar to conducting univariate analyses prior to multivariable modeling using logistic regression. Variables with a ranking of 20% or higher were included in the CART model for discrimination analysis.
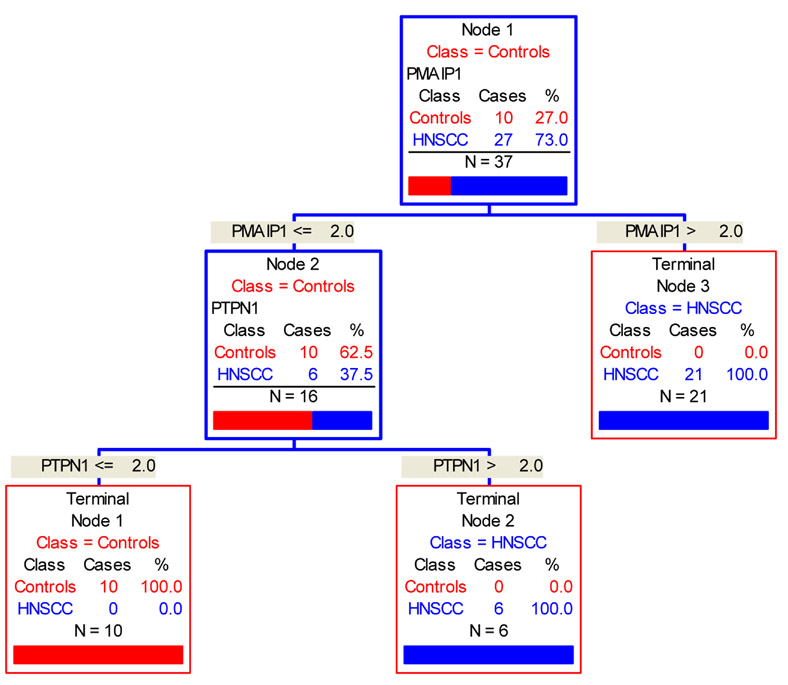
In the multivariable modeling process, CART identifies the first gene probe variable with the greatest predictive power and divides the subjects into two groups of HNSCC and normal (Figure 2). It will then identify a second gene probe with the next highest predictive power to further partition subjects. The process continues until further partitioning is exhausted.
Figure 2.
CART statistical analysis: Starting with 37 study subjects (Node1; blue box), gain of PMAIP1 gene partitioned 21 HNSCC patients (Node 3; red box); gain of PTPN1 gene separated the remaining 6 HNSCC patients ( terminal Node 2; red box) from all 10 normal controls (terminal Node 1; red box).
After CART created a discrimination model based on all subjects, referred as “learning data”, this model was validated using the leave-one-out validation approach, called the “testing” data. Leave-one-out cross-validation is the statistical practice of partitioning a sample of data into subsets such that the analysis is initially performed on a single subset, while the other subsets are retained for subsequent use in confirming and validating the initial analysis. A single observation from the original sample is used as the validation data, and the remaining observations as the training data. This is repeated such that each observation in the sample is used once as the validation data.
Sensitivity and specificity measures and their 95% confidence interval (CI) were calculated based on the “testing” dataset using the Exact analytical approach.
RESULTS
Of the 37 subjects analyzed, 27 were HNSCC patients, 85 % males; age 22–81 years (mean 57; median 56); 11(41%) early stage (I/II) tumors. Ten were healthy normal controls, 20% males, age 20–50 years (mean 37.2; median 35). The 99.99 confidence intervals (CI’s) of the saliva gene copy numbers are presented in Table 1 based on 10 normal controls. The 99.9% confidence interval for normality of each gene probe in the normal control group remained within the expected range for normal copy number (21), but were much tighter norms supporting the use of the numerical raw data rather than the categorical data (based on arbitrary cutoffs of 1.33 {gain} and 0.75 {loss}) (21).
Table 1.
99.99% confidence intervals (CI’s) of the normal saliva gene data
| GENE | Chr pos | 99.99% CI | GENE | Chr pos | 99.99%CI |
|---|---|---|---|---|---|
| NRAS | 01p13.2 | (0.85, 1.04) | CCND1_1 | 11q13 | (0.90, 1.10) |
| BCAR3 | 01p22.1 | (0.97, 1.15) | EHF | 11p13 | (0.87, 1.10) |
| F3 | 01p22-p21 | (1.01, 1.15) | HIPK3 | 11p13 | (0.83, 1.05) |
| IL10 | 01q31 | (0.85, 1.18) | LMO2 | 11p13 | (1.00, 1.20) |
| IL1A | 02q14 | (0.86, 1.05) | HRAS | 11p15.5 | (1.00, 1.13) |
| TANK | 02q24 | (0.78, 0.99) | ATM | 11q22-q23 | (1.00, 1.15) |
| ERBB4 | 02q33 | (0.93, 1.18) | LRMP | 12p12.1 | (0.86, 0.95) |
| CTNNB1 | 03p21 | (0.87, 1.24) | TNFRSF7 | 12p13 | (0.91, 1.01) |
| MLH1 | 03p21.3 | (0.90, 1.05) | CCND2 | 12p13 | (0.93, 1.14) |
| IL12A | 03q25.33 | (0.90, 1.30) | BCL7A | 12q24.13 | (0.75, 1.20) |
| PIK3CA | 03q26.3 | (0.74, 1.10) | BRCA2 | 13q12.3 | (0.77, 1.01) |
| ABCG2 | 04q22 | (0.92, 1.18) | RB1 -1 | 13q14.2 | (0.76, 1.23) |
| NFKB1 | 04q24 | (0.92, 1.10) | RB1 -2 | 13q14.3 | (0.89, 1.03) |
| CASP6 | 04q25 | (0.90, 1.30) | ABCC4 | 13q32 | (0.97, 1.06) |
| RAD17 | 05q13 | (0.87, 1.07) | MESDC1 | 15q13 | (0.91, 1.24) |
| IL13 | 05q31 | (0.91, 1.14) | CDH1 | 16q22.1 | (0.87, 1.04) |
| IL4 | 05q31.1 | (0.82, 0.98) | TP53 | 17p13.1 | (0.83, 1.20) |
| VEGF | 06p12 | (0.76, 1.13) | CRK | 17p13.3 | (0.93, 1.52) |
| CDKN1A | 06p21.2 | (1.01, 1.10) | BRCA1- 1 | 17q21 | (0.65, 1.08) |
| IER3 | 06p21.3 | (0.82, 1.21) | BRCA1- 2 | 17q21 | (0.85, 1.06) |
| TNF | 06p21.3 | (0.84, 1.18) | ERBB2- 1 | 17q21.1 | (0.97, 1.16) |
| BAK1 | 06p21.3 | (0.87, 1.09) | ERBB2- 2 | 17q21.1 | (0.91, 1.09) |
| KIAA0170 | 06p21.31 | (0.78, 1.18) | AXIN2 | 17q23-q24 | (0.73, 1.14) |
| MYB | 06q22 | (0.70, 1.00) | TIMP2 | 17q25 | (0.88, 1.21) |
| ABCB1 | 07q21.1 | (0.88, 1.11) | CDH2 | 18q11.2 | (0.62, 0.88) |
| MET | 07q31 | (0.88, 1.06) | BCL2 | 18q21.3 | (0.98, 1.12) |
| FGFR1 | 08p11.2- | (1.04, 1.33) | PMAIP1 | 18q21.31 | (0.91, 1.00) |
| MYC | 08q24.12 | (1.00, 1.19) | BCL2 | 18q21.3 | (0.88, 1.09) |
| PTP4A3 | 08q24.3 | (0.87, 1.18) | DCC | 18q21.3 | (1.01, 1.26) |
| RECQL4 | 08q24.3 | (1.02, 1.35) | CDKN2D | 19p13 | (0.87, 1.15) |
| CDKN2A | 09p21 | (0.93, 1.17) | DNMT1 | 19p13.2 | (0.89, 1.08) |
| CDKN2B | 09p21 | (0.97, 1.06) | KLK3 | 19q13.41 | (0.87, 1.07) |
| CREM | 10p11.21 | (0.90, 1.06) | BAX | 19q13.3 | (0.89, 1.05) |
| RENT2 | 10p14 | (0.70, 1.09) | STK15_STK | 20q13 | (0.83, 0.95) |
| CCND1 | 11q13 | (0.82, 1.24) | PTPN1 | 20q13.13 | (0.87, 1.01) |
| EMS1 | 11q13 | (1.08, 1.19) | STCH | 21q11 | (0.60, 0.90) |
| RELA | 11q13 | (1.04, 1.18) | MIF | 22q11.23 | (0.89, 1.57) |
| FGF3 | 11q13 | (0.85, 1.04) | SIM2 | 21q22.13 | (0.83, 1.08) |
Eleven genes (ERBB2, ABCC4, UTY, DNMT1, PMAIP1, CDKN2B, CDKN2D, NFKB1, TP53, PTPN1, DCC), with the highest ranking (20% or higher, Table 2), were included in the multivariable classification modeling.
Table 2.
Gene variable ranking (percent importance) for individual predictive ability (univariate approach for gene probe selection)
| Variable | Score |
|---|---|
| NFKB1 | 100.00 |
| PMAIP1 | 78.18 |
| CDKN2D | 47.25 |
| TP53_1 | 47.25 |
| PTPN1 | 44.68 |
| DCC | 43.00 |
| DNMT1 | 35.90 |
| CDKN2B | 32.92 |
| ERBB2 | 27.73 |
| ABCC4 | 27.73 |
| UTY | 27.73 |
CART identified two gene probes (PMAIP1 and PTPN1) which classified the study subjects into either the HNSCC group or the normal control group with 0 error rate on the learning samples (Table 3 ). Assuming a value of 2 as the cut-off-point for gain, based on “learning” samples, subjects with gain of PMAIP1 solely, or with concomitant gain of PTPNI, were 100% correctly classified into the HNSCC group (Figure 2). Subjects, on the other hand, who had loss or normal copy of PTPNI and PMAIP1 were 100% correctly classified into the normal control group (Figure 3). The sensitivity and specificity were 100% for both with 95% lower bound interval as 87.2% and 69.2% respectively.
Table 3.
Misclassification for Learned and Testing Data
| Class | # of study subjects | # Mis-classified | % Error |
|---|---|---|---|
| Learning Data | |||
| Healthy controls | 10 | 0 | 0.00 |
| HNSCC | 27 | 0 | 0.00 |
| Testing Data | |||
| Healthy controls | 10 | 2 | 20.00 |
| HNSCC | 27 | 1 | 3.70 |
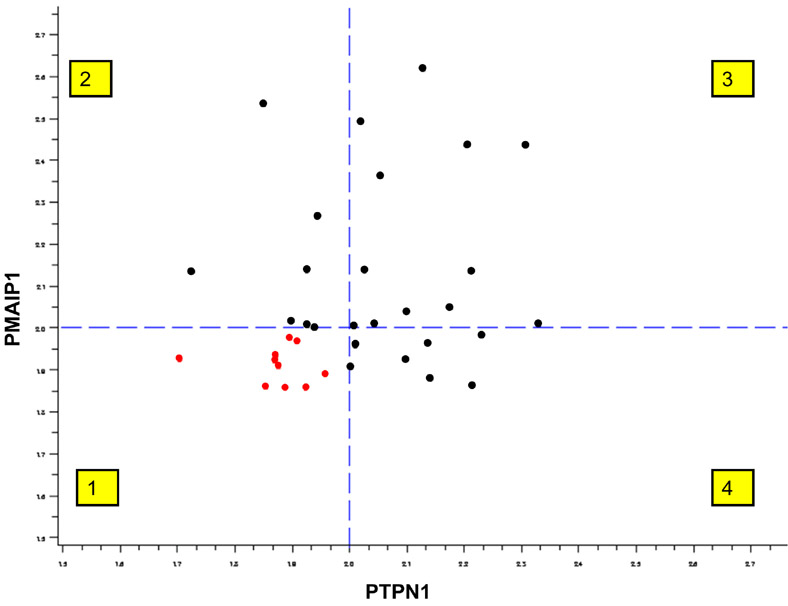
Figure 3.
PMAIP1 and PTPN1 gene status: all HNSCC patients (black dots) showed gain (>2) of one/both genes, quadrants 2, 3 and 4; 27 observations; in contrast to <2 seen in normal controls (red dots) in quadrant 1; 10 observations.
The leave-one-out validation miss-classified three subjects; two normal controls were miss-classified as HNSCC and one HNSCC was miss-classified as a normal control. Based on these results, the sensitivity and specificity were 96.3% with 95% CI of .81.0% to 99.5%, and 80% and 95% CI of 44.4% to 97.5%, respectively.
DISCUSSION
HNSCC continues to pose a clinical challenge despite rapid advances in therapeutic options. Late detection is the single most important factor in the poor prognosis of HNSCC. Early diagnosis will significantly help in reducing the mortality and morbidity associated with HNSCC.
Several consistent genetic alterations have been described in HNSCC at 3p, 4q, 5q, 6p, 9p, 11q, 13q, 17p, 18q and 20q (11–14). However no single gene alteration is exclusive for HNSCC. There is a need to identify HNSCC-specific genetic alterations which can potentially be used as biomarkers for early diagnosis, screening, prognosis and treatment of HNSCC. They can also assess risk for HNSCC, assist in tumor surveillance and monitor disease progression.
Saliva, a non-invasive patient sample promises to revolutionize diagnostic medicine as it can reflect the entire spectrum of health and disease states (25). Its advantages over serum samples, include ease of collection, storing, shipping and handling, as it does not clot lessening the manipulations required. It dramatically reduces patient anxiety and discomfort allowing repeated samples for monitoring over time. The initial notion that informative analytes are generally present in lower amounts in the saliva than in serum (26), have now been allayed with new and highly sensitive analytical techniques (25).
Newly developed high throughput analytical methods in the fields of molecular biology and genetic medicine have enabled detailed molecular characterization of cancer. The present study demonstrates the efficacy of MLPA for simultaneous interrogation of several genes in scant amounts of saliva DNA. In our study, the 99.9% confidence interval for normality of each probe in the normal control group remained in large part, within the expected range for normal copy number, and is consistent with published results (21). Our confidence limits were narrower with less variation in the confidence intervals when compared to other studies (21), stressing reliability of the data.
Classification and Regression Tree (CART) analysis is a statistical tool used to extract pertinent information from extremely complex datasets. In the current analysis, its ability to separate data sets on the basis of numeric or categorical variables and delineating the useful genes from the insignificant ones was emphasized, as its utility has been demonstrated in previous studies (16). It produces decision trees, based on simple yes/no questions, revealing relationships that are sometimes hidden. CART is more likely to be practical in a clinical setting as it is much simpler to interpret than multivariate logistic regression models. The advantages of CART, compared to the usual logistic regression are: (i) there is no assumption requirement for the covariate (e.g. the linear of log odd required for a logistic model); (ii) there is no pre-specified cut off point for each covariate (CART will explore all the possible cut offs for each gene and generate the best cut off point based on predictive ability); (iii) it allows the assessment of gene by gene variable interaction if any; and (iv) it generates discriminatory models, preferable to association studies. In the leave-one-out validation, one of CART’s validation methods, all of the data are used for fitting (but not at the same time) providing prediction capabilities usually requiring larger data sets, leading to smaller prediction errors, and a superior outcome for small data sets, compared to split-sample validation (27).
The present study demonstrates proof of concept that saliva gene-based algorithms can differentiate HNSCC patients from normal controls using high throughput MLPA assays.. In our ongoing study, we are currently analyzing genetic alterations in HNSCC patients and comparing them with high risk patient controls who are smokers and alcohol consumers.
In the present analysis, CART discriminated HNSCC from normal controls based on two gene variables (Figure 2). Gain of PMAIP1 (18q21.31) solely, and in conjunction with gain of PTPN1 (20q13.13), with 100% accuracy based on the learning dataset, and 96% sensitivity (81%, 99.5%) and 80% specificity (44.4%, 97.5%) based on the “testing” datasets. The wide range of specificity reflects small size of normal controls. Univariate analysis for the variable selection prior to multivariable modeling given the sample dataset avoids model over-fitting. The 20% of variable importance in CART was an arbitrary cut-off-point, and it can be varied based on sample size. Cut-offs of 40% or more, yield only 6 gene variables for inclusion in the multivariable model with a resulting sensitivity and specificity of 100% (87.2% to 100%) and 80% (44%, 97.5) based on the “testing” data set. When all 82 gene variables, as an extreme scenario, are included, the same two gene classification model is identified with 100% accuracy based on the “learning” dataset, but sensitivity and specificity are reduced to 77.8% (57.7% to 91.4%) and 80% (44.4%, 97.5). In the present study, the sample size, especially the sample size for controls is small and the analysis results should be regarded as exploratory.
The present study included 41% early stage lesions indicating the ability of this non-invasive approach to identify both early and late stage HNSCC. Although as yet not tested, there is great potential that these methods may provide the ability to identify early cancers prior to their gross clinical manifestations. This may be particularly important in high-risk patients. The study cases included 74% non-oral cavity HNSCC; providing proof of concept that saliva based genetic test has the potential to detect non-oral cavity cancers.
In summary, the combination of two genes, gain of PMAIP1 (18q21.31) solely or in conjunction with gain of PTPN1 (20q13.13), in saliva DNA can differentiate HNSCC cases from normal controls with high sensitivity and specificity. Our study demonstrates that saliva genomics may have clinical utility for noninvasive HNSCC detection and screening. Molecular targets contained within CART gene algorithms would emphasize these genes as relevant biomarkers for screening, early detection, prognosis and treatment in HNSCC. Additionally, these findings would enhance our understanding of the pathogenesis of HNSCC.
Acknowledgments
SUPPORT
HFHS A10236; R01 NIH DE 15990
References
- 1.Ries LAGHD, Krapcho M, Mariotto A, et al. SEER Cancer Statistics Review, 1975–2003. Bethesda, MD: National Cancer Institute; 2006. [Google Scholar]
- 2.Oral Cancer Facts and Figures. Atlanta, GA: American Cancer Society; 2006. [Google Scholar]
- 3.Forastiere A, Koch W, Trotti A, et al. Head and neck cancer. N Engl J Med. 2001;345(26):1890–900. doi: 10.1056/NEJMra001375. [DOI] [PubMed] [Google Scholar]
- 4.Franzmann EJ, Reategui EP, Carraway KL, et al. Salivary soluble CD44: a potential molecular marker for head and neck cancer. Cancer Epidemiol Biomarkers Prev. 2005;14(3):735–9. doi: 10.1158/1055-9965.EPI-04-0546. [DOI] [PubMed] [Google Scholar]
- 5.Li Y, St John MA, Zhou X, et al. Salivary transcriptome diagnostics for oral cancer detection. Clin Cancer Res. 2004;15(10):24–8442. 50. doi: 10.1158/1078-0432.CCR-04-1167. [DOI] [PubMed] [Google Scholar]
- 6.O’Hara J, Bradley P. Head and neck cancer: a screening strategy. Clin Otolaryngol. 2002;27:133 – 4. doi: 10.1046/j.1365-2273.2002.00547.x. [DOI] [PubMed] [Google Scholar]
- 7.Neville BW, Day TA. Oral cancer and precancerous lesions. CA Cancer J Clin. 2002;52:195 – 215. doi: 10.3322/canjclin.52.4.195. [DOI] [PubMed] [Google Scholar]
- 8.Dolan RW, Vaughan CW, Fuleihan N. Symptoms in early head and neck cancer: an inadequate indicator. Otolaryngol Head Neck Surg. 1998;119:463 – 7. doi: 10.1016/S0194-5998(98)70102-0. [DOI] [PubMed] [Google Scholar]
- 9.National Cancer Institute. Oral cancers: research report. NIH publication no. 92–2876. U.S. Department of Health and Human Services, Public Health Service, NIH; 1991. [Google Scholar]
- 10.Sidransky D. Emerging molecular markers of cancer. Nat Rev Cancer. 2002;2:210–9. doi: 10.1038/nrc755. [DOI] [PubMed] [Google Scholar]
- 11.Choi P, Chen C. Genetic expression profiles and biologic pathway alterations in head and neck squamous cell carcinoma. Cancer. 2005;104(6):1113–28. doi: 10.1002/cncr.21293. [DOI] [PubMed] [Google Scholar]
- 12.Akervall J. Gene profiling in squamous cell carcinoma of the head and neck. Cancer Metastasis Rev. 2005;24(1):87–94. doi: 10.1007/s10555-005-5049-z. [DOI] [PubMed] [Google Scholar]
- 13.Jeon GA, Lee JS, Patel V, Gutkind JS, et al. Global gene expression profiles of human head and neck squamous carcinoma cell lines. Int J Cancer. 2004;112 (2):249–58. doi: 10.1002/ijc.20399. [DOI] [PubMed] [Google Scholar]
- 14.Worsham MJ, Chen KM, Tiwari N, et al. Fine-mapping loss of gene architecture at the CDKN2B (p15INK4b), CDKN2A (p14ARF, p16INK4a), and MTAP genes in head and neck squamous cell carcinoma. Arch Otolaryngol Head Neck Surg. 2006;132(4):409–15. doi: 10.1001/archotol.132.4.409. [DOI] [PubMed] [Google Scholar]
- 15.Boyle JO, Mao L, Brennan JA, et al. Gene mutations in saliva as molecular markers for head and neck squamous cell carcinomas. Am J Surg. 1994;168(5):429–32. doi: 10.1016/s0002-9610(05)80092-3. [DOI] [PubMed] [Google Scholar]
- 16.Raju U, Lu M, Sethi S, et al. Molecular Classification of Breast Carcinoma in Situ. Curr Genomics. 2006;7(8):523–532. doi: 10.2174/138920206779315719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen KM, SR, Khan M, Benninger MS, et al. Methylation of multiple genes as diagnostic and therapeutic markers in primary HNSCC. Arch Otolaryngol Head Neck Surg. 2007;133(11):1131–1138. doi: 10.1001/archotol.133.11.1131. [DOI] [PubMed] [Google Scholar]
- 18.Stephen JKVL, Chen KM, Sethi S, et al. Epigenetic events underlie the pathogenesis of sinonasal papillomas. Mod Pathol. 2007;20(10):1–9. doi: 10.1038/modpathol.3800944. [DOI] [PubMed] [Google Scholar]
- 19.Fernandez L, Lapunzina P, Arjona D, et al. Comparative study of three diagnostic approaches (FISH, STRs and MLPA) in 30 patients with 22q11.2 deletion syndrome. Clin Genet. 2005;68(4):373–8. doi: 10.1111/j.1399-0004.2005.00493.x. [DOI] [PubMed] [Google Scholar]
- 20.Douglas J, Tatton-Brown K, Coleman K, et al. Partial NSD1 deletions cause 5% of Sotos syndrome and are readily identifiable by multiplex ligation dependent probe amplification. J Med Genet. 2005;42(9):56. doi: 10.1136/jmg.2005.031930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bremmer JF, Braakhuis BJ, Ruijter-Schippers HJ, et al. A noninvasive genetic screening test to detect oral preneoplastic lesions. Lab Invest. 2005;85(12):1481–8. doi: 10.1038/labinvest.3700342. [DOI] [PubMed] [Google Scholar]
- 22.Schouten JP, McElgunn CJ, Waaijer R, Zwijnenburg D, Diepvens F, Pals G. Relative quantification of 40 nucleic acid sequences by multiplex ligation dependent probe amplification. Nucleic Acids Res. 2002;30:e57. doi: 10.1093/nar/gnf056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Breiman L, Friedman J, Stone CJ. Classification and Regression Trees. 1. New York: Chapman and Hall; 1984. [Google Scholar]
- 24.Frankel MR, Morgenstern LB, Kwiatkowski T, et al. Predicting prognosis after stroke: a placebo group analysis from the National Institute of Neurological Disorders and Stroke rt-PA Stroke Trial. Neurology. 2000;55:952–959. doi: 10.1212/wnl.55.7.952. [DOI] [PubMed] [Google Scholar]
- 25.Wong DT. Salivary diagnostics powered by nanotechnologies, proteomics and genomics. J Am Dent Assoc. 2006;137(3):313–21. doi: 10.14219/jada.archive.2006.0180. [DOI] [PubMed] [Google Scholar]
- 26.Miller SM. Saliva testing--a nontraditional diagnostic tool. Clin Lab Sci. 1994;7 (1):39–44. [PubMed] [Google Scholar]
- 27.Goutte C. Note on free lunches and cross-validation. Neural Computation. 1997;9:1211–1215. [Google Scholar]