Abstract
Characterizing cotinine pharmacokinetics is a useful way to study nicotine metabolism because the same liver enzyme is primarily responsible for the metabolism of both, and the clearances of nicotine and cotinine are highly correlated. We conducted a whole-genome linkage analysis to search for candidate regions influencing quantitative variation in cotinine pharmacokinetics in a large-scale pharmacokinetic study with 61 families containing 224 healthy adult participants. The strongest linkage signal was identified at 135 cM of chromosome 9 with LOD=2.81 and P=0.0002; two other suggestive linkage peaks appear at 31.4 and 73.5 cM of chromosome 11 with LOD=1.96 (P=0.0013) and 1.94 (P=0.0014). The confidence level of the linkage between the three genome regions and cotinine pharmacokinetics is statistically significant with a genome-wide empirical probability of P=0.029.
Keywords: pharmacokinetics, nicotine, dependence, linkage analysis
INTRODUCTION
Nicotine addiction, most commonly in the form of chronic cigarette smoking, is the largest modifiable risk factor for morbidity and mortality in developed countries [Bergen and Caporaso, 1999]. Smoking increases the risk of cardiovascular and pulmonary disease, lung and other cancers, and a number of infectious diseases. The attributable risk of lung cancer and chronic obstructive pulmonary disease due to smoking ranges between 80% and 90% [Thun et al., 2002].
Nicotine metabolism is a complex trait, influenced by sex, environmental influences such as diet and medications, as well as hepatic enzyme variation [Johnstone et al., 2006]. The rate of nicotine metabolism influences smoking intensity [Hukkanen et al., 2005]. The influence of genetics and genetic variation on the rate of nicotine metabolism has been estimated as being close to 60% [Swan et al., 2005]. The influence of variation attributable to common variants of the gene for the liver enzyme CYP2A6, considered the principal enzyme involved in the metabolism of nicotine, has been estimated to account for a small fraction (10-14%) of the variation in total nicotine clearance [Nakajima et al., 1996; Swan et al., 2005]. Thus, the identification of genes influencing nicotine clearance promises to improve our understanding of the determinants of individual differences in nicotine pharmacokinetics.
Cotinine is the major proximate metabolite of nicotine, where ∼75% of nicotine is eliminated after it is transformed to cotinine and cotinine-derived metabolites [Hukkanen et al., 2005]. Within individuals the clearance of nicotine and cotinine are highly correlated [Zevin et al., 2000]. The concentration of plasma cotinine over time (the Area Under the concentration-time Curve (AUC), from time zero to infinity) in relationship to the dose of cotinine administered provides a measure of the total clearance of cotinine [Zevin et al., 2000]. We used cotinine clearance as a surrogate for nicotine clearance in the linkage study because the longer half-life of cotinine makes it is easier to study the pharmacokinetics of cotinine than nicotine noninvasively.
MATERIALS AND METHODS
Sample set
For the investigation of cotinine pharmacokinetics, we utilized a family sample (Smoking in Families study, known as SMOFAM (DA03706, Hyman Hops, PI, Oregon Research Institute). The SMOFAM study is a comprehensive, repeated measures cohort study of environmental and psychosocial risk factors for adolescent and young adult substance use, including tobacco. The original SMOFAM study, initiated in 1984, recruited 763 families, with at least one adolescent age 11 or older through advertisements in the newspaper, on television and radio, and flyers distributed at middle and high schools in four mid-sized and small urban and rural Pacific Northwest cities with populations ranging from 30,000 to 175,000. Families with smoking parents and/or adolescents were of special interest since the adolescents were at risk for tobacco and other substance use. Within each family an adolescent was designated as the “target” if s/he had previously tried a substance. Each target had to have at least one parent agree to participate. An attempt was made to encourage both parents and all sibs over the age of 11 to participate.
The only other requirement was that all participants needed to be able to read basic level English. Seventy-five percent of the original number of families included at least one smoking parent and 41.8% included an ever-smoking teen. Contrast families with nonsmoking parents and adolescents were also recruited. At the beginning of the study, target adolescents were ages 11-15 (mean = 13.2 years), 49% were males, 92% were Caucasian, 2% Hispanic, 3% African American, and 2% Native American, and 52% were from two-parent families.
Over a 17-year period, 15 annual assessments have been completed and as of the last assessment, there were 465 target participants ranging from 27 to 31 years of age remaining in the study. The repeated assessment of target participants facilitated characterization of longitudinal phenotypes for tobacco use, including the acquisition and maintenance of smoking, as well as many potential psychosocial and environmental predictors of substance [Hops et al., 2000].
Probands and family members for the present study were recruited from those SMOFAM families where the proband had completed at least seven of the first ten assessments on tobacco use and elected to provide a blood sample for DNA analysis [Swan et al., 2003]. Families selected for analysis of nicotine and cotinine metabolism and other tobacco phenotypes consisted of the proband, and at least two living, ever-smoking (>100 cigarettes in the lifetime), first-degree relatives. Individuals who had: 1) impaired renal or hepatic function, or 2) were taking anticonvulsants, phenobarbital, or other agents that were suspected to alter the metabolism of nicotine or cotinine were not eligible for participation in the pharmacokinetic assessment. A total of 61 families containing 224 healthy adults participated in the cotinine kinetics study. The participants included 106 males and 118 females with an average age of 39.2 (range 18 to 66) years and an average body weight of 85.5 (standard deviation 20.9) kilograms. The majority of participants (212 individuals, 94.6%) identified themselves as Caucasian with the remainder reporting non-Caucasian or mixed origin. The Institutional Review Boards of SRI International, the University of California at San Francisco, and at the Oregon Research Institute reviewed and approved the study in accordance with the code of ethics of the World Medical Association (Declaration of Helsinki).
Data collection
Because grapefruit juice affects the metabolism of cotinine, participants were asked to refrain from grapefruit and grapefruit juice consumption for at least 48 hours prior to and for the duration of the pharmacokinetic study [Hukkanen et al., 2006]. On the day of the study participants remained on-site with the research nurse for nine hours. Breakfast, lunch, snacks, and drinks were provided to the participants. All participants were given 1000 mg of ammonium chloride to reduce variability in the renal clearance of nicotine and cotinine with water one hour before dosing. After one hour, 10 mg of deuterium-labeled cotinine (COT-2, 4, 5, 6-d4) was administered to smokers, as these individuals already had natural cotinine in their bodies as a metabolite of nicotine derived from either active tobacco use or passive exposure to tobacco smoke. Unlabeled cotinine was supplied to non-smokers in the same dosage. Two hours following dosing, the participant ate breakfast. Three hours after initial dosing, a second dose of 1000 mg of ammonium chloride was administered in water.
Concentrations of cotinine in plasma and saliva are highly correlated, and Zevin et al. showed that measurement of cotinine in saliva provides an accurate estimate of systemic clearance of cotinine [Zevin et al., 2000]. We collected saliva samples (3-5 ml each) in labeled containers from participants before dosing and at 6, 12, 24, 36, 48, and 60 hours following dosing. Cotinine concentration was measured by gas chromatography-mass spectrometry [Jacob P3rd et al., 1991]. AUC cotinine (ng/ml*hr) was calculated using the trapezoidal rule over the interval of time (6, 12, 24, 36, 48, and 60 hours) when saliva samples were measured, and using the terminal half-life to extrapolate the AUC after the end of sampling [Zevin et al., 2000].
Genotyping and Linkage analysis
Genomic DNA was extracted from venous blood samples (Puregene, Gentra Systems, Inc., Minneapolis, MN) and the concentration measured via optical density. Genotypes were determined for 739 dinucleotide microsatellite markers [Swan et al. 2006] on 211 individuals from 61 families (an average of 3.5 genotyped individuals per family, with 485 family members in total). There were N=93 parents with AUC data and N=92 parents with genotypes. A sex-averaged genetic map developed by Applied Biosystems (Foster City, CA), using 763 autosomal map positions generated from CEPH genotype data (http://www3.appliedbiosystems.com/cms/groups/mcb_support/documents/generaldocuments/cms_041230.pdf) was used in the linkage analysis.
All available genotypes were analyzed for each family using PREST to validate structure of pedigree [Sun et al., 2002]. Pedcheck was used to detect non-Mendelian inheritance patterns [O'Connell et al., 1998]. The probability that each genotype was correct was assessed in the context of all other available genotypes using the error-checking algorithm implemented in Merlin [Abecasis et al., 2002]. Less than 0.5% of all genotypes were excluded after these quality controls were applied.
Autosomal multipoint non-parametric linkage analysis (NPL) was performed on the final genotype data in Merlin with the Kong and Cox exponential model with AUC cotinine as the phenotype [Abecasis et al., 2002; Kong and Cox, 1997]. For evaluating impact on the linkage by other factors, such as age, body weight, gender, and cigarette smoking (both cigarette smoking status and cigarettes smoked per day), four covariates commonly considered in pharmacokinetic research of nicotine, a linear regression was performed for AUC cotinine along with covariates in R (The R Project for Statistical Computing, release 2.4.1, http://www.r-project.org/index.html). Akaike's information criterion (AIC) and multistep optimization were used to build a best fitting model while considering possible interaction of the covariates [Insightful Corporation, 2002]. Based on the optimized regression model, residuals of the regression resulted in AUC cotinine values adjusted for the effect of the covariates: age (P=0.09), gender (P=0.08) and body weight (P<0.001). Neither cigarette smoking status nor cigarettes per day were included in the final regression model (P>0.10). We repeated the original linkage analysis using the AUC cotinine values adjusted for the effect of the covariates.
RESULTS
In all participants, the average AUC cotinine = 2341 ng/ml*hr, standard derivation = 977, skew = 1.17, and kurtosis = 2.52. Normality of AUC cotinine could not be improved with commonly used transformation functions (data not shown). However, non-normality of AUC cotinine would be expected to have only a small impact on a linkage analysis utilizing the Kong and Cox exponential model [Kong and Cox, 1997], because the score statistic used in the exponential model is minimally affected by non-normality of data [Feingold, 2002].
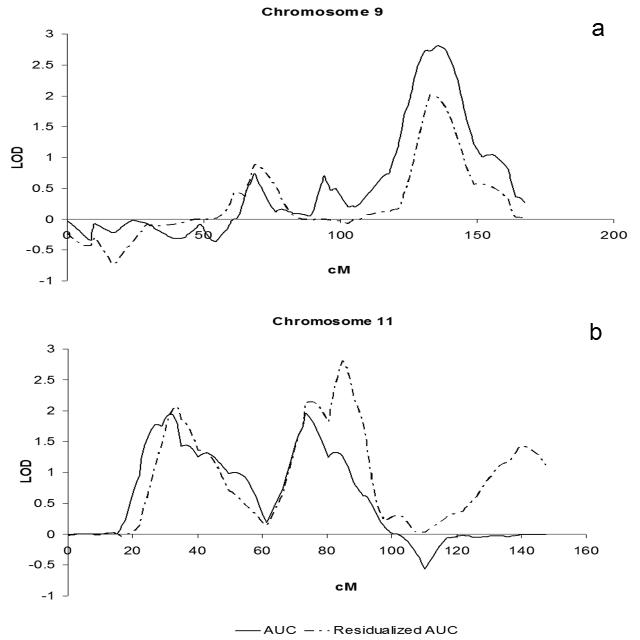
Three suggestive linkage peaks with LOD ≥ 1.86 were identified in Merlin for AUC cotinine [Abecasis et al., 2002; Lander and Kruglyak, 1995]. The strongest linkage signal appeared at 135 cM of chromosome 9 with LOD=2.81 (P=0.0002) (Fig. 1 a), between markers D9S1682 and D9S290. The support interval with LOD ≥ 2 extends 17 cM from 126 to 143 cM, between markers D9S289 and D9S164. The two other suggestive linkage peaks for AUC cotinine occur on chromosome 11 (Fig. 1 b). The first one is located at 31.4 cM with LOD=1.94 and P=0.0014 between markers D11S4190 and D11S915. The second one is located at 73.5 cM with LOD=1.96 and P=0.0013 at marker D11S1314.
Fig. 1.
Linkage peaks on Chromosomes 9 and 11. Multipoint LOD traces for AUC cotinine and residualized AUC cotinine of a) chromosome 9 and b) chromosome 11.
We performed a simulation in Merlin to estimate the empirical P-value of the three suggestive linkage peaks [Abecasis et al., 2002; Sawcer et al., 1997]. Under the null hypothesis with no linkage, the number of simulations with at least one linkage peak with LOD ≥ 1.94, ≥ 1.96 or ≥ 2.81 were 467, 455, or 75, respectively, in 1000 whole-genome linkage analysis simulations. Overall statistical significance of our linkage findings for AUC cotinine (all three linkage peaks) is the probability of at least three linkage peaks with LOD ≥ 1.94, 1.96 and 2.81, appearing by chance in one whole genome linkage analysis. We estimated the empirical probability of suggestive linkage between AUC cotinine and regions of chromosomes 9 and 11 as P=0.029 since only 29 such cases were observed in 1000 simulations.
False discovery rate (FDR) was defined as the expected proportion of incorrectly rejected null hypotheses [Benjamini and Hochberg, 1995]. In the present case, FDR indicates the proportion of expected false linkage peaks among our three linkage peaks that are supported using both conventional criteria for suggestive linkage and through simulation analysis. We observed a total of 620 linkage peaks with LOD ≥ 1.94 in our 1000 simulations. The number is larger than 467 (mentioned above) because more than one peak could be observed in a single simulation. Thus, we expect one or less false linkage peak among our three identified linkage peaks. The FDR of the three linkage peaks was 0.620/3 ≈ 0.206 if the observed linkage peaks are considered independently.
Analysis of regression residuals with the Kong and Cox exponential model in Merlin identified linkage of AUC cotinine after adjustment for the covariates. Using the same model and statistic enabled the comparison of the linkage results with or without adjustment for covariates. Furthermore, this approach does not appear to be affected by possible type I error inflation due to a non-Gaussian distribution [Feingold, 2002; Kong and Cox, 1997; Sham et al., 2002]. The linkage signal of the residuals was compared to the three unadjusted linkage peaks of AUC cotinine to evaluate the impact of covariates and to identify stable linkage signatures. The analysis of residuals of AUC cotinine supported the extended region of linkage on chromosome 9, with a peak located at 132.8 cM with LOD=2.01 (P=0.0012) (Fig. 1 a), at marker D9S1682. The LOD score of the regression residuals on chromosome 9 is generally lower than that of the unadjusted AUC cotinine for the same region. There was a minor shift in location of the linkage peak for adjusted AUC cotinine, which was only 2 cM away from the linkage peak of unadjusted AUC cotinine. The 2 LOD supporting interval of AUC cotinine completely overlaps the 1 LOD interval identified in the linkage analysis of the residuals.
Both suggestive linkage regions on chromosome 11 exhibited increased LOD scores with the residuals of AUC cotinine (Fig. 1b). The location and magnitude of the first peak of the residual linkage analysis, was located at 33.9 cM with LOD=2.07 and P=0.0011 at marker D11S915, just distal to that found in the analysis of unadjusted AUC cotinine. In the second extended linkage region, both linkage analyses exhibited two peaks, i.e., at 74 cM between markers D11S1314 and D11S4207 and 85 cM, at D11S901. The LOD score of the proximal peak in this region changed minimally from the unadjusted to adjusted AUC cotinine. However, the distal peak in this region increased by nearly one LOD unit between the unadjusted and adjusted AUC cotinine linkage analyses, with a peak LOD of 2.81 (p=0.0002) at 84.8 cM, with a LOD ≥ 2.0 support interval from 81 cM to 89 cM, between markers D11S937 and D11S4147. Our initial AUC cotinine linkage findings are fully supported by the linkage analysis of residualized AUC cotinine because the LOD scores of all three linkage peaks are larger than the criterion for suggestive linkage (LOD ≥ 1.86) [Lander and Kruglyak, 1995].
We further note the presence of linkage peaks for AUC cotinine (unadjusted and adjusted) with LOD scores ≥ 1 distributed on eleven chromosomes (Table I and Figure S1), and note that the largest extended regions of linkage appeared on chromosomes 9 and 11.
TABLE I.
Chromosome Regions with LOD ≥ 1.
| Chr and region* | AUC cotinine (cM) | Residualized AUC cotinine (cM) |
|---|---|---|
| Chr 4 | 179.30-187.00 | --- |
| Chr 7 I | 17.45-31.58 | 15.23-27.30 |
| Chr 7 II | 70.58-74.73 | --- |
| Chr 9 | 120.00-156.45 | 127.73-143.33 |
| Chr 10 | --- | 149.70-151.66 |
| Chr 11 I | 22.30-49.23 | 26.87-46.97 |
| Chr 11 II | 68.36-87 | 68.37-93.25 |
| Chr 11 III | --- | 131.88-141.70 |
| Chr 14 | --- | 123.83-131.60 |
| Chr 15 | 13.83-14.10 | 0.00-19.43 |
| Chr 18 | 52.00-55.75 | 52-55.76 |
| Chr 19 | 0.00-2.43 | 0.00-9.73 |
| Chr 21 I | 0.00-7.4 | 0.00-12.27 |
| Chr 21 II | 20.80-21.60 | 15.80-28.6 |
| Chr 22 | 8.33-16.05 | --- |
The Roman numerals denote different regions of linkage on the same chromosome.
DISCUSSION
Due to the labor-intensive nature of the protocol, sample size is limited in most pharmacokinetic studies and, therefore it may be difficult to apply whole genome-wide association methods with adequate statistical power. Linkage analysis is more powerful than whole genome association study in many genetic scenarios and has successfully identified risk loci for several complex traits in the past twenty years [Blangero, 2004; Tu and Whittemore, 1999]. The three linkage regions identified in this analysis overlapped with chromosome regions that were identified previously as “significant” or “suggestive” linkages in at least two different well-performed linkage studies of nicotine dependence [Li, 2008]. Specifically, the chr9q region identified in this study overlaps suggestive linkage peaks identified in linkage analyses of nicotine dependence measures in both European and African-American families ascertained on alcohol and nicotine dependence criteria, respectively, and the chr11 regions identified in this study are flanked by two chr11 regions that have been identified in linkage analyses of nicotine dependence in six family samples from the United States, Finland and Australia, ascertained using a variety of clinical and/or population criteria [Bergen et al., 1999; Li et al., 2006; Li, 2008].
With preliminary evidence from a linkage study such as the present one, a candidate gene-based approach may represent a better choice for further genetic exploration. The loci for hepatic enzymes considered to be involved in nicotine and cotinine metabolism include the chr19q13.2 p450 cytochrome loci, the flavin containing monooxygenase loci, found on chr1q21.1 and 1q24.3, and the uridine diphosphate-glycosyltransferase loci, found on chr2q37 and chr4q13, respectively [Davis et al., 1986; McCombie et al., 1996; Monaghan et al., 1992; Van Es et al., 1993]. However, no LOD score ≥ 1 was found at these loci in our study. A limited effect size of functional variations affecting cotinine clearance at these loci in this particular sample, or a sample size limiting statistical power, may explain this result [Swan et al., 2005].
There are a total of 561 genes under the 9q and chr11 linkage peaks (Human Genome Build 36.2, CHR9:115455388-135245933, CHR11:17445082-31319866, CHR11:69627739-94298127), but none currently with annotation indicating a role in hepatic metabolism, suggesting that additional molecular genetic analyses will be necessary to develop evidence for candidate genes involved in cotinine and nicotine clearance in these regions [Maglott et al., 2007]. We have performed multiple comparison-adjusted analyses of gene functional annotation [Mi et al., 2007] and note that olfactory receptors and ubiquitin-protein ligases are significantly overrepresented in these three regions. The ubiquitin-proteosome system has been shown to be down-regulated by nicotine [Kane et al., 2004; Ficklin et al., 2005; Rezvani et al., 2007], a potential mechanism affecting nicotine metabolism that could be evaluated using candidate gene approaches.
In summary, we have conducted a linkage study of cotinine pharmacokinetics using unadjusted and residualized AUC cotinine and have identified three chromosomal regions that may contain candidate loci influencing the pharmacokinetics of nicotine and cotinine with genome-wide statistical significance (P=0.029, FDR=0.206).
ACKNOWLEDGMENTS
We thank Kirk C. Wilhelmsen for the SMOFAM whole genome scan genotypes. Research supported by grants U01 DA020830-03, DA03706 and DA12393 from the National Institutes of Health, and 7PT2000-2004 from the University of California Tobacco-Related Disease Research Program. We especially thank the individuals from the SMOFAM families for their participation in the cotinine pharmacokinetic study.
Supplementary Material
REFERENCES
- Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin--rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Statist Soc B. 1995;57:89–300. [Google Scholar]
- Bergen AW, Caporaso N. Cigarette smoking. J Natl Cancer Inst. 1999;91:1365–1375. doi: 10.1093/jnci/91.16.1365. [DOI] [PubMed] [Google Scholar]
- Bergen AW, Korczak JF, Weissbecker KA, Goldstein AM. Genome-wide search for loci contributing to smoking and alcoholism. Genet Epidemiol. 1999;17(Suppl 1):S55–60. doi: 10.1002/gepi.1370170710. [DOI] [PubMed] [Google Scholar]
- Blangero J. Localization and identification of human quantitative trait loci: king harvest has surely come. Curr Opin Genet Dev. 2004;14:233–240. doi: 10.1016/j.gde.2004.04.009. [DOI] [PubMed] [Google Scholar]
- Davis MB, West LF, Shephard EA, Phillips IR. Regional localization of a human cytochrome P-450 (CYP1) to chromosome 19q13.1-13.3. Ann Hum Genet. 1986;50:237–240. doi: 10.1111/j.1469-1809.1986.tb01044.x. [DOI] [PubMed] [Google Scholar]
- Feingold E. Regression-based quantitative-trait-locus mapping in the 21st century. Am J Hum Genet. 2002;71:217–222. doi: 10.1086/341964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ficklin MB, Zhao S, Feng G. Ubiquilin-1 regulates nicotine-induced up-regulation of neuronal nicotinic acetylcholine receptors. J Biol Chem. 2005;280:34088–34095. doi: 10.1074/jbc.M506781200. [DOI] [PubMed] [Google Scholar]
- Hops H, Andrews JA, Duncan SC, Duncan TE, Tildesley E. Adolescent drug use development: A social interactional and contextual perspective. In: Sameroff AJ, Lewis M, Miller SM, editors. Handbook of developmental psychopathology. 2nd ed. Kluwer Academic/Plenum; New York: 2000. pp. 589–605. [Google Scholar]
- Hukkanen J, Jacob P, 3rd, Benowitz NL. Effect of grapefruit juice on cytochrome P450 2A6 and nicotine renal clearance. Clin Pharmacol Ther. 2006;80:522–530. doi: 10.1016/j.clpt.2006.08.006. [DOI] [PubMed] [Google Scholar]
- Hukkanen J, Jacob P, 3rd, Benowitz NL. Metabolism and disposition kinetics of nicotine. Pharmacol Rev. 2005;57:79–115. doi: 10.1124/pr.57.1.3. [DOI] [PubMed] [Google Scholar]
- Insightful Corporation . Regression and Smoothing for Continuous Response Data. In: Insightful Corporation, editor. S-plus 6 Gide to Statistics for Windows. Insightful Corporation; 2002. pp. 235–330. [Google Scholar]
- Jacob P, 3rd, Yu L, Wilson M, Benowitz NL. Selected ion monitoring method for determination of nicotine, cotinine and deuterium-labeled analogs: absence of an isotope effect in the clearance of (S)-nicotine-3′,3′-d2 in humans. Biol Mass Spectrom. 1991;20:247–252. doi: 10.1002/bms.1200200503. [DOI] [PubMed] [Google Scholar]
- Johnstone E, Benowitz N, Cargill A, Jacob R, Hinks L, Day I, Murphy M, Walton R. Determinants of the rate of nicotine metabolism and effects on smoking behavior. Clin Pharmacol Ther. 2006;80:319–330. doi: 10.1016/j.clpt.2006.06.011. [DOI] [PubMed] [Google Scholar]
- Kane JK, Konu O, Ma JZ, Li MD. Nicotine coregulates multiple pathways involved in protein modification/degradation in rat brain. Brain Res Mol Brain Res. 2004;132:181–191. doi: 10.1016/j.molbrainres.2004.09.010. [DOI] [PubMed] [Google Scholar]
- Kong A, Cox NJ. Allele-sharing models: LOD scores and accurate linkage tests. Am J Hum Genet. 1997;61:1179–1188. doi: 10.1086/301592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander E, Kruglyak L. Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet. 1995;11:241–247. doi: 10.1038/ng1195-241. [DOI] [PubMed] [Google Scholar]
- Li MD, Payne TJ, Ma JZ, Lou XY, Zhang D, Dupont RT, Crews KM, Somes G, Williams NJ, Elston RC. A genomewide search finds major susceptibility loci for nicotine dependence on chromosome 10 in African Americans. Am J Hum Genet. 2006;79:745–751. doi: 10.1086/508208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MD. Identifying susceptibility loci for nicotine dependence: 2008 update based on recent genome-wide linkage analyses. Hum Genet. 2008;123:119–131. doi: 10.1007/s00439-008-0473-0. [DOI] [PubMed] [Google Scholar]
- Maglott D, Ostell J, Pruitt KD, Tatusova T. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 2007;35:D26–31. doi: 10.1093/nar/gkl993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCombie RR, Dolphin CT, Povey S, Phillips IR, Shephard EA. Localization of human flavin-containing monooxygenase genes FMO2 and FMO5 to chromosome 1q. Genomics. 1996;34:426–429. doi: 10.1006/geno.1996.0308. [DOI] [PubMed] [Google Scholar]
- Mi H, Guo N, Kejariwal A, Thomas PD. PANTHER version 6: protein sequence and function evolution data with expanded representation of biological pathways. Nucleic Acids Res. 2007;35:D247–52. doi: 10.1093/nar/gkl869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaghan G, Povey S, Burchell B, Boxer M. Localization of a bile acid UDP-glucuronosyltransferase gene (UGT2B) to chromosome 4 using the polymerase chain reaction. Genomics. 1992;13:908–909. doi: 10.1016/0888-7543(92)90188-x. [DOI] [PubMed] [Google Scholar]
- Nakajima M, Yamamoto T, Nunoya K, Yokoi T, Nagashima K, Inoue K, Funae Y, Shimada N, Kamataki T, Kuroiwa Y. Role of human cytochrome P4502A6 in C-oxidation of nicotine. Drug Metab Dispos. 1996;24:1212–1217. [PubMed] [Google Scholar]
- O'Connell JR, Weeks DE. PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet. 1998;63:259–266. doi: 10.1086/301904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezvani K, Teng Y, Shim D, De Biasi M. Nicotine regulates multiple synaptic proteins by inhibiting proteasomal activity. J Neurosci. 2007;27:10508–10519. doi: 10.1523/JNEUROSCI.3353-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawcer S, Jones HB, Judge D, Visser F, Compston A, Goodfellow PN, Clayton D. Empirical genomewide significance levels established by whole genome simulations. Genet Epidemiol. 1997;14:223–229. doi: 10.1002/(SICI)1098-2272(1997)14:3<223::AID-GEPI1>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Sham PC, Purcell S, Cherny SS, Abecasis GR. Powerful regression-based quantitative-trait linkage analysis of general pedigrees. Am J Hum Genet. 2002;71:238–253. doi: 10.1086/341560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Wilder K, McPeek MS. Enhanced pedigree error detection. Hum Hered. 2002;54:99–110. doi: 10.1159/000067666. [DOI] [PubMed] [Google Scholar]
- Swan GE, Benowitz NL, Lessov CN, Jacob P, 3rd, Tyndale RF, Wilhelmsen K. Nicotine metabolism: the impact of CYP2A6 on estimates of additive genetic influence. Pharmacogenet Genomics. 2005;15:115–125. doi: 10.1097/01213011-200502000-00007. [DOI] [PubMed] [Google Scholar]
- Swan GE, Hops H, Wilhelmsen KC, Lessov-Schlaggar CN, Cheng LS, Hudmon KS, Amos CI, Feiler HS, Ring HZ, Andrews JA, Tildesley E, Benowitz N. A genome-wide screen for nicotine dependence susceptibility loci. Am J Med Genet B (Neuropsychiatr Genet) 2006;141:354–360. doi: 10.1002/ajmg.b.30315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swan GE, Hudmon KS, Jack LM, Hemberger K, Carmelli D, Khroyan TV, Ring HZ, Hops H, Andrews JA, Tildesley E, McBride D, Benowitz N, Webster C, Wilhelmsen KC, Feiler HS, Koenig B, Caron L, Illes J, Cheng LS. Environmental and genetic determinants of tobacco use: methodology for a multidisciplinary, longitudinal family-based investigation. Cancer Epidemiol Biomarkers Prev. 2003;12:994–1005. [PMC free article] [PubMed] [Google Scholar]
- Thun MJ, Henley SJ, Calle EE. Tobacco use and cancer: an epidemiologic perspective for geneticists. Oncogene. 2002;21:7307–7325. doi: 10.1038/sj.onc.1205807. [DOI] [PubMed] [Google Scholar]
- Tu IP, Whittemore AS. Power of association and linkage tests when the disease alleles are unobserved. Am J Hum Genet. 1999;64:641–649. doi: 10.1086/302253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Es HH, Bout A, Liu J, Anderson L, Duncan AM, Bosma P, Oude Elferink R, Jansen PL, Chowdhury JR, Schurr E. Assignment of the human UDP glucuronosyltransferase gene (UGT1A1) to chromosome region 2q37.1. Cytogenet Cell Genet. 1993;63:114–116. doi: 10.1159/000133513. [DOI] [PubMed] [Google Scholar]
- Zevin S, Jacob P, Geppetti P, Benowitz NL. Clinical pharmacology of oral cotinine. Drug Alcohol Depend. 2000;60:13–18. doi: 10.1016/s0376-8716(99)00135-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.