Abstract
Tobacco smoking continues to be a leading cause of preventable death. Recent research has underscored the important role of specific cholinergic nicotinic receptor subunit (CHRN) genes in risk for nicotine dependence and smoking. To detect and characterize the influence of genetic variation on vulnerability to nicotine dependence, we analyzed 226 SNPs covering the complete family of 16 CHRN genes, which encode the nicotinic acetylcholine receptor (nAChR) subunits, in a sample of 1050 nicotine-dependent cases and 879 non-dependent controls of European descent. This expanded SNP coverage has extended and refined the findings of our previous large scale genome-wide association and candidate gene study. After correcting for the multiple tests across this gene family, we found significant association for two distinct loci in the CHRNA5-CHRNA3-CHRNB4 gene cluster, one locus in the CHRNB3-CHRNA6 gene cluster, and a fourth, novel locus in the CHRND-CHRNG gene cluster. The two distinct loci in CHRNA5-CHRNA3-CHRNB4 are represented by the non-synonymous SNP rs16969968 in CHRNA5 and by rs578776 in CHRNA3, respectively, and joint analyses show that the associations at these two SNPs are statistically independent. Nominally significant single-SNP association was detected in CHRNA4 and CHRNB1. In summary, this is the most comprehensive study of the CHRN genes for involvement with nicotine dependence to date. Our analysis reveals significant evidence for at least four distinct loci in the nicotinic receptor subunit genes that each influence the transition from smoking to nicotine dependence and may inform the development of improved smoking cessation treatments and prevention initiatives.
Keywords: cholinergic nicotinic receptors, nicotinic acetylcholine receptors, smoking, genetic association
INTRODUCTION
Tobacco use continues to be a leading cause of preventable death. Smoking causes an estimated 21% of yearly cancer deaths worldwide (Danaei et al., 2005), currently kills approximately 5 million people a year, and is projected to be responsible for 10% of all deaths globally by the year 2015 (Mathers and Loncar, 2006). Despite heightened public awareness of the negative health consequences, smoking remains prevalent in the United States, and rates are increasing in some developing countries (Mackay et al., 2006). In 2005, 20.9% of US adults were current cigarette smokers, the same percentage as in 2004 (Centers for Disease Control and Prevention, 2006). This lack of decline underscores the limitations of current treatments and policy approaches aimed at curbing tobacco use.
Nicotine dependence is a leading predictor of smoking continuation (Breslau et al., 2001; Johnson et al., 2002). Thus an improved understanding of the biological underpinnings of nicotine dependence may be key to the success of further smoking cessation efforts. Twin studies find significant genetic influences on nicotine dependence (Lessov et al., 2004; Maes et al., 2004; Pergadia et al., 2006; Prescott and Kendler, 1995), motivating the search for genetic variants that influence risk for nicotine dependence.
Nicotine is a naturally occurring alkaloid in tobacco. Nicotine is highly addictive (Davis, 1988), and the nicotinic acetylcholine receptors, in part, mediate the effects of nicotine in the body (Lindstrom, 2003; Tapper et al., 2004). These receptors are pentameric molecular assemblies of nicotinic acetylcholine receptor (nAChR) subunits, which are coded by a family of distinct cholinergic nicotinic receptor (CHRN) genes. These subunits, and the receptors assembled from them, fall into two classes, neuronal and neuromuscular. In humans the neuronal subunits consist of α2 through α7, α9, α10, and β2 through β4; the muscle nAChRs are α1, β1, δ, ε and γ. The α8 subunit has been detected in avian tissue but not in mammals.
The muscle-type nicotinic receptor is composed of two alpha subunits, one beta, one delta and either one gamma or one epsilon subunit (Karlin, 2002). Neuronal-type receptors can be homopentamers or heteropentamers. The predominant heteropentameric nAChRs are α4β2* in brain and α3β4* in the peripheral nervous system (Gotti et al., 2006; Mao et al., 2006). In some instances, other subunits (as indicated by the asterisk), principally α5, also contribute to these two major heteromeric receptor subtypes. The α7 subunit is the only nAChR subunit confirmed to form homomeric nAChRs in mammalian brain (Chen and Patrick, 1997). The dopamine reward pathway, which is thought to play a critical role in addiction, is modulated by multiple nAChR subtypes, including α4β2, α4β2α5, α6β2β3, α4α6β2β3, and α7 (Klink et al., 2001; Pidoplichko et al., 2004; Salminen et al., 2004; Wooltorton et al., 2003; Wu et al., 2004).
Early genetic association studies of nicotine dependence and smoking have looked at some of the nicotinic subunit genes (Greenbaum et al., 2006; Lou et al., 2006), with CHRNA4 and CHRNB2 (coding α4 and β2 respectively) receiving the most attention (Ehringer et al., 2007; Feng et al., 2004; Li et al., 2005; Lou et al., 2007; Lueders et al., 2002; Silverman et al., 2000) until our reports of strong association within the CHRNA5-CHRNA3-CHRNB4 cluster and the CHRNB3-CHRNA6 cluster (Bierut et al., 2007; Saccone et al., 2007a). More recently, our association with rs16969968, a nonsynonymous coding SNP in CHRNA5, has been replicated in several independent datasets using the same SNP (Bierut et al., In press) (Stevens et al., unpublished data) or proxy SNPs having very high r2 with it (Berrettini et al., 2008; Thorgeirsson et al., 2008). Furthermore, this SNP or its r2 proxies have now been associated with lung cancer (Amos et al., 2008; Hung et al., 2008; Thorgeirsson et al., 2008). These findings underscore the nicotinic receptors as important targets for further indepth study.
Here we present a comprehensive association study of nicotine dependence with SNP-based variation in the complete family of 16 nicotinic receptor subunits. This targeted study of the CHRN genes expands upon the coverage we previously attained in NICSNP, a large-scale case-control nicotine dependence association study consisting of the first high-density genome-wide association screen (GWAS) for nicotine dependence (Bierut et al., 2007) paired with an extensive candidate gene (CG) survey of 348 genes (Saccone et al., 2007a).
MATERIALS AND METHODS
Study design and sample
All individuals in this case/control sample have reported smoking at least 100 cigarettes lifetime, the threshold classically used to define a smoker (Centers for Disease Control and Prevention, 2006). Cases have lifetime FTND scores of 4 or greater, while controls have never exhibited symptoms of dependence (lifetime FTND = 0). This sample therefore is designed to examine the transition from nicotine use to dependence. All subjects (1050 cases and 879 controls) were recruited by two ongoing studies: the Collaborative Genetic Study of Nicotine Dependence (COGEND), a US-based sample (N = 797 cases, N= 813 controls), and the Nicotine Addiction Genetics (NAG) study (Saccone et al., 2007b), an Australian-based sample (N= 253 cases, N= 66 controls). All subjects were self-identified as being of European ancestry. Table I gives demographic details including the distribution of cases and controls, by gender, and mean age and FTND, in the US and Australian samples. The table shows that the mean FTND in cases is very similar across the two recruiting sites (US and Australia), although the proportion of cases and controls recruited differs. Also, the proportion of women differs in cases and controls. Therefore the variables site and sex are included as covariates in the genetic association analyses. Analyses using STRUCTURE (Pritchard et al., 2000) demonstrating no evidence of confounding population substructure are published elsewhere (Bierut et al., 2007). The study was carried out in compliance with the Code of Ethics of the World Medical Association and obtained informed consent from all participants and approval from the appropriate institutional review boards.
Table I.
Distribution of cases (by sex), age and mean FTND, by recruitment site
| United States | Australia | Total sample | |
|---|---|---|---|
| Cases (% Male) | 797 (44.0%) | 253 (45.1%) | 1050 (44.3%) |
| Controls (% Male) | 813 (30.9%) | 66 (25.8%) | 879 (30.5%) |
| Mean age | 36.3 ± 5.42 | 41.8 ± 11.7 | 37.2 ± 7.2 |
| Mean FTND in cases | 6.4 ± 1.7 | 6.1 ± 1.6 | 6.3 ± 1.7 |
Genotyping and quality control
226 SNPs covering the 16 CHRN genes were analyzed in this study. Initial genotyping was carried out by Perlegen Sciences using custom arrays as previously detailed (Bierut et al., 2007; Saccone et al., 2007a), leading to 119 CHRN SNPs passing quality control (QC) measures including reliability of genotype calls and Hardy-Weinberg equilibrium (HWE) > 0.01. Additional genotyping was then performed by the Center for Inherited Disease Research (CIDR) using Illumina Golden Gate technology (http://www.illumina.com/downloads/GOLDENGATEASSAY.pdf). These additional SNPs were selected for the following purposes. First, SNPs with poor or marginal genotype call rates (<98%) in our initial data were selected for regenotyping; these included some of the 119 SNPs and also SNPs that had already been attempted but did not meet the prior QC requirements. Second, SNPs were selected using our custom tag SNP programs which use r2 bin tagging (Carlson et al., 2004) to enhance coverage of the CHRN genes. Third, we chose additional SNPs to fine-map the strong association signals already seen in the CHRNA5-CHRNA3-CHRNB4 and CHRNB3-CHRNA6 clusters. Prior to analysis, all SNPs were required to have call rates ≥ 98%. SNPs having r2 ≥ 0.8 with CHRN SNPs (e.g. in IREB2, LOC123688, and PSMA4 which flank the CHRNA5-CHRNA3-CHRNB4 cluster) were also included in the final analysis set of 226 SNPs, of which 118 are newly genotyped by CIDR. Map positions and genomic annotations were obtained from the National Center for Biotechnology Information (NCBI) Human Reference Build 36.2 and dbSNP build 127.
Association analyses
Linkage disequilibrium
Linkage disequilibrium (LD) between SNPs was calculated for cases and controls using the verbose option of ldmax (Abecasis and Cookson, 2000). Signed D’ coefficients were calculated to retain LD phase information (Saccone et al., 2006). LD plots were generated using a custom program.
Single SNP analyses
Our primary single SNP association analyses of case-control status use logistic regression models to correct for significant covariates of gender and sample source site (US or Australia) (Table 1). The non-genetic base model is: ln, where P is the probability of being a case, g is gender (0=male, 1=female) and s is site (0 for US, 1 for Australia). Genotype status at each marker, coded as an ordinal variable, is then added to the model and tested for significance by the standard likelihood chi-square statistic with one degree of freedom (df). The ordinal coding corresponds to a log-additive (multiplicative) model for the number of copies of the risk allele, defined as the allele more common in cases than controls. This primary test differs from the test used in (Bierut et al., 2007; Saccone et al., 2007a), which included a genotype by gender interaction term to allow detection of loci having differential effect in males versus females. In our focused nAChR study here, we first tested for SNP effect only; then, for SNPs significant in that primary test, we investigated possible gender effects and alternative modes of inheritance.
To determine a “best model” for nominally significant SNPs, we evaluated whether an added gender-by-genotype interaction term is itself significant and also whether modeling a log-linear trend effect of the genotype is sufficient. If the gender-by-genotype interaction term was significant at the p < 0.05 level, the logistic regression was repeated with that term, and the “best model” p-value reported for that two df test. To determine whether modeling a non-multiplicative genotype effect gives a better fit than the primary model, we added a single dichotomous variable coding for heterozygote status, allowing deviation from an ordinal model. When this variable was significant, we evaluated other modes of inheritance: recessive (1 df), dominant (1 df), or genotype coded with 2 degrees of freedom to allow an arbitrary model, and report the “best model” p-value for the appropriate alternative model.
To interpret results, we ordered SNPs by primary test p-value and then formed r2 bins across the 226 SNPs, by chromosome, as follows. The correlation coefficients were computed separately in cases (r2case) and controls (r2control). The most significant SNP and all SNPs having min(r2case , r2control) ≥ 0.8 with it defined the first bin; these were removed from the binning process, which continued with the next most significant SNP that was not already binned. By ordering the SNPs by significance to define the bins, we enable the bins to better reflect the correlations with the most significant SNPs. Using min(r2case , r2control) is slightly more conservative (potentially leading to more bins) than using r2 from the combined sample, but in practice makes little difference. Alternative algorithms may be efficient for selecting tag SNPs, for example the greedy algorithm of (Carlson et al., 2004). However, such an algorithm might then relegate two SNPs into separate bins even though they are both correlated with a very significant SNP, giving the illusion that they are two distinct signals.
Multiple test correction
A Bonferroni correction for our 226 SNPs is overly conservative due to LD among SNPs and would require an individual p-value of 0.000221 for an experiment-wide significance level of 0.05. A more precise Sidak correction would require a p-value of α’ = 0.000227, so that 0.05 = 1 - (1-α’)226.
However, we may take LD into account by selecting an r2 tagging subset with an r2 threshold of 0.8 as described above. This approach results in 111 proxy SNPs, corresponding to no more than 111 distinct tests and a Sidak corrected p-value of 0.000462.
Multilocus tests in the CHRNA5-CHRNA3-CHRNB4 cluster
We performed selected multilocus analyses of case-control status. The purpose of these analyses is to help clarify and distinguish among the effects of multiple significant SNPs within an associated region but not in strong LD. The non-synonymous SNP rs16969968 in CHRNA5 is a biologically promising finding in our analysis, and other studies have shown evidence for replication via our analysis of rs16969968 in independent samples of families (Bierut et al., In press) and heavy smokers and controls (Stevens et al., unpublished data in review), and via analyses of strongly correlated SNPs by independent groups studying cigarettes-per-day (Berrettini et al., 2008) and cigarettes-per-day and nicotine dependence (Thorgeirsson et al., 2008). Furthermore, a new functional study has demonstrated that the risk allele at rs16969968 decreases response to a nicotine agonist (Bierut et al., In press). Therefore we targeted rs16969968 for these joint analyses; results for SNPs having high r2 with it will necessarily be similar.
Extensive LD across the CHRNA5-CHRNA3-CHRNB4 cluster led to multiple significant SNPs in our data. After accounting for the LD structure in this region, we identified two distinct, significant associations tagged by rs16969968 and rs578776. Therefore, genotypes at these SNPs were entered together into a multivariate logistic regression model to evaluate the significance of the effect of one locus when controlling for genotype at the other locus. Additionally, the combined influence of the two-locus genotype was examined in a 3×3 table.
The full logistic model has the form ln, where G1 codes genotype status at rs16969968 with a recessive model (determined from the best model analysis) and G2 codes the second SNP with the primary, log-additive model. We test for the effect of each SNP by removing it from the model and calculating the likelihood ratio chi-square with one df.
RESULTS
Characterization of the sample
The covariates recorded included sex, age, and site (US or Australia). Forward selection using logistic regression indicated that sex and site were significant predictors of case/control status, as expected from Table I. Thus the covariate-corrected genetic analyses include these variables in the baseline model. With these main effects in the model, additional interaction terms such as age*sex were not significant. Figure 1 gives the distribution of FTND scores in male and female cases.
Figure 1.
Distribution of FTND scores in cases. Displayed by percentages in men (N=464) and women (N=586) separately.
Genetic association analyses
Twenty-one SNPs were significant after multiple test correction (p < 0.000462), using the primary 1 df logistic regression test. These 21 SNPs form five distinct r2 bins in the following genes or gene clusters: CHRNA5-CHRNA3-CHRNB4 (3 bins), CHRNB3-CHRNA6 (1 bin), and CHRND, (1 bin). Of the three bins in the CHRNA5-CHRNA3-CHRNB4 cluster, two are still correlated with each other as detailed below, leading to two distinct signals in the cluster.
Eight of the significant SNPs are on chromosome 8 in the β3-α6 gene cluster, twelve are on chromosome 15 in or near the α5—α3—β4 cluster, and one is on chromosome 2 in CHRND. These significant results are in bold in Table II, which presents association results for all SNPs genes or gene clusters that contained at least one SNP with primary p-value ≤ 0.05. Association results for all 226 SNPs sorted by chromosome and position are in supplementary table S1, so that the coverage for every nAChR gene may be seen. To aid interpretation, the tables also include the LD bin membership of the SNPs, as described in the methods. Of the 226 SNPs, 84 are nominally significant. For the nominally significant SNPs, supplementary table S1 gives p-values for the “best model” determined as described in the methods.
Table II.
Primary (log-additive) model results for CHRN genes containing at least one nominally significant SNP
| rs number |
Chr | Position (bp) |
Gene | Lab1 | SNP function | Risk allele (case allele frequency, control allele frequency) |
Primary p-value |
Odds Ratio (95% CI) | Rank by primary p-value |
LD bin2 |
|---|---|---|---|---|---|---|---|---|---|---|
| CHRND-CHRNG | ||||||||||
|
| ||||||||||
| 3791729 | 2 | 233103541 | CHRND | PSC | INTRON | A (0.365 , 0.318) | 1.299E-03 | 1.25 (1.09 , 1.44) | 30 | 8 |
| 3762528 | 2 | 233104619 | CHRND | PSC | INTRON | G (0.069 , 0.065) | 9.953E-01 | 1 (0.77 , 1.31) | 225 | 111 |
| 12466358 | 2 | 233105769 | CHRND | CIDR | INTRON | C (0.277 , 0.233) | 2.725E-04 | 1.32 (1.14 , 1.54) | 17 | 5 |
| 2767 | 2 | 233108318 | CHRND | PSC | mRNA-UTR | G (0.387 , 0.341) | 1.246E-03 | 1.25 (1.09 , 1.44) | 27 | 8 |
| 2289080 | 2 | 233114422 | CHRNG | PSC | NONSYNON | G (0.981 , 0.978) | 4.838E-01 | 1.18 (0.74 , 1.9) | 166 | 81 |
| 1881492 | 2 | 233115242 | CHRNG | CIDR | INTRON | C (0.82 , 0.777) | 6.945E-04 | 1.33 (1.13 , 1.57) | 23 | 6 |
| 2853462 | 2 | 233115364 | CHRNG | PSC | INTRON | A (0.252 , 0.231) | 1.506E-01 | 1.12 (0.96 , 1.3) | 100 | 39 |
| 6761667 | 2 | 233115655 | CHRNG | CIDR | INTRON | A (0.481 , 0.454) | 6.850E-02 | 1.13 (0.99 , 1.29) | 87 | 29 |
| 2099489 | 2 | 233118538 | CHRNG | PSC | SYNON | G (0.803 , 0.783) | 6.061E-02 | 1.17 (0.99 , 1.38) | 86 | 28 |
| 11690038 | 2 | 233118747 | CHRNG | CIDR | mRNA-UTR | A (0.981 , 0.978) | 5.551E-01 | 1.15 (0.72 , 1.85) | 172 | 81 |
| 6749955 | 2 | 233146161 | PSC | A (0.769 , 0.733) | 1.244E-02 | 1.21 (1.04 , 1.41) | 65 | 19 | ||
| 2276560 | 2 | 233159163 | PSC | A (0.773 , 0.736) | 8.839E-03 | 1.23 (1.05 , 1.43) | 61 | 19 | ||
|
| ||||||||||
| CHRNB3-CHRNA6 | ||||||||||
|
| ||||||||||
| 5005909 | 8 | 42647824 | PSC | INTRON | A (0.838 , 0.801) | 1.976E-03 | 1.3 (1.1 , 1.54) | 37 | 10 | |
| 10958726 | 8 | 42655066 | PSC | INTRON | A (0.809 , 0.761) | 9.636E-05 | 1.38 (1.17 , 1.62) | 5 | 1 | |
| 1530847 | 8 | 42667396 | PSC | INTRON | A (0.865 , 0.83) | 1.257E-03 | 1.36 (1.13 , 1.64) | 28 | 9 | |
| 1955186 | 8 | 42668648 | CIDR | INTRON | C (0.808 , 0.76) | 8.252E-05 | 1.38 (1.18 , 1.63) | 4 | 1 | |
| 1955185 | 8 | 42668804 | PSC | INTRON | A (0.809 , 0.76) | 1.010E-04 | 1.38 (1.17 , 1.62) | 7 | 1 | |
| 13277254 | 8 | 42669139 | PSC | INTRON | A (0.809 , 0.758) | 4.022E-05 | 1.4 (1.19 , 1.65) | 1 | 1 | |
| 13280301 | 8 | 42669174 | PSC | INTRON | G (0.866 , 0.831) | 1.468E-03 | 1.35 (1.12 , 1.63) | 31 | 9 | |
| 13277524 | 8 | 42669214 | PSC | INTRON | A (0.809 , 0.759) | 6.043E-05 | 1.39 (1.18 , 1.64) | 2 | 1 | |
| 6474412 | 8 | 42669655 | PSC | INTRON | A (0.808 , 0.76) | 1.126E-04 | 1.38 (1.17 , 1.62) | 9 | 1 | |
| 6474413 | 8 | 42670221 | PSC | INTRON | A (0.809 , 0.759) | 6.260E-05 | 1.39 (1.18 , 1.64) | 3 | 1 | |
| 4950 | 8 | 42671790 | CHRNB3 | CIDR | mRNA-UTR | A (0.808 , 0.76) | 1.001E-04 | 1.38 (1.17 , 1.62) | 6 | 1 |
| 7838246 | 8 | 42704454 | CHRNB3 | CIDR | INTRON | A (0.967 , 0.952) | 7.582E-03 | 1.59 (1.13 , 2.23) | 60 | 14 |
| 4952 | 8 | 42706222 | CHRNB3 | PSC | SYNON | G (0.968 , 0.952) | 4.128E-03 | 1.64 (1.17 , 2.31) | 48 | 14 |
| 4953 | 8 | 42706816 | CHRNB3 | PSC | SYNON | C (0.968 , 0.952) | 4.055E-03 | 1.64 (1.17 , 2.31) | 47 | 14 |
| 17621256 | 8 | 42709065 | CHRNB3 | CIDR | INTRON | C (0.972 , 0.966) | 3.080E-01 | 1.22 (0.83 , 1.78) | 123 | 59 |
| 7012713 | 8 | 42711460 | CHRNB3 | CIDR | INTRON | A (0.037 , 0.031) | 3.475E-01 | 1.19 (0.83 , 1.72) | 133 | 66 |
| 6987323 | 8 | 42716389 | CIDR | INTRON | G (0.961 , 0.95) | 3.270E-02 | 1.42 (1.03 , 1.96) | 74 | 24 | |
| 9298628 | 8 | 42725148 | PSC | INTRON | G (0.813 , 0.782) | 2.229E-02 | 1.21 (1.03 , 1.43) | 71 | 20 | |
| 9298629 | 8 | 42725343 | PSC | INTRON | C (0.815 , 0.783) | 1.899E-02 | 1.22 (1.03 , 1.44) | 69 | 20 | |
| 2304297 | 8 | 42727356 | CHRNA6 | PSC | mRNA-UTR | G (0.79 , 0.751) | 4.777E-03 | 1.26 (1.07 , 1.47) | 53 | 15 |
| 892413 | 8 | 42733535 | CHRNA6 | PSC | INTRON | C (0.818 , 0.791) | 3.254E-02 | 1.2 (1.02 , 1.42) | 73 | 15 |
| 10087172 | 8 | 42736025 | CHRNA6 | PSC | INTRON | A (0.817 , 0.79) | 3.702E-02 | 1.2 (1.01 , 1.41) | 76 | 15 |
| 2196128 | 8 | 42737443 | CHRNA6 | CIDR | INTRON | A (0.785 , 0.763) | 8.455E-02 | 1.15 (0.98 , 1.35) | 91 | 33 |
| 1072003 | 8 | 42739158 | CHRNA6 | PSC | INTRON | C (0.836 , 0.804) | 8.959E-03 | 1.26 (1.06 , 1.5) | 62 | 20 |
| 7828365 | 8 | 42748471 | PSC | INTRON | G (0.885 , 0.859) | 2.368E-02 | 1.26 (1.03 , 1.54) | 72 | 23 | |
| 10107450 | 8 | 42749052 | CIDR | INTRON | G (0.796 , 0.758) | 4.409E-03 | 1.26 (1.08 , 1.48) | 50 | 15 | |
|
| ||||||||||
| CHRNA5-CHRNA3-CHRNB4 | ||||||||||
|
| ||||||||||
| 17483548 | 15 | 76517368 | IREB2 | PSC | LOCUS | A (0.369 , 0.308) | 1.623E-04 | 1.3 (1.14 , 1.5) | 12 | 3 |
| 17405217 | 15 | 76518204 | IREB2 | CIDR | INTRON | A (0.37 , 0.308) | 1.821E-04 | 1.3 (1.13 , 1.5) | 14 | 3 |
| 924840 | 15 | 76518863 | IREB2 | CIDR | INTRON | A (0.806 , 0.772) | 3.471E-02 | 1.19 (1.01 , 1.4) | 75 | 25 |
| 17483686 | 15 | 76520445 | IREB2 | PSC | INTRON | T (0.373 , 0.314) | 3.205E-04 | 1.29 (1.12 , 1.48) | 19 | 3 |
| 2568488 | 15 | 76523648 | IREB2 | CIDR | INTRON | A (0.806 , 0.773) | 4.372E-02 | 1.18 (1 , 1.39) | 84 | 25 |
| 2656052 | 15 | 76527987 | IREB2 | CIDR | INTRON | C (0.375 , 0.32) | 1.141E-03 | 1.26 (1.09 , 1.44) | 26 | 3 |
| 2656073 | 15 | 76529331 | IREB2 | CIDR | INTRON | C (0.806 , 0.772) | 3.889E-02 | 1.19 (1.01 , 1.39) | 81 | 25 |
| 2958719 | 15 | 76530084 | IREB2 | CIDR | INTRON | A (0.806 , 0.772) | 3.889E-02 | 1.19 (1.01 , 1.39) | 82 | 25 |
| 2656071 | 15 | 76532398 | IREB2 | CIDR | INTRON | T (0.806 , 0.772) | 3.889E-02 | 1.19 (1.01 , 1.39) | 80 | 25 |
| 2656069 | 15 | 76532762 | IREB2 | CIDR | INTRON | A (0.806 , 0.772) | 3.889E-02 | 1.19 (1.01 , 1.39) | 79 | 25 |
| 2568485 | 15 | 76539169 | IREB2 | CIDR | INTRON | A (0.805 , 0.772) | 4.030E-02 | 1.18 (1.01 , 1.39) | 83 | 25 |
| 2568483 | 15 | 76539398 | IREB2 | CIDR | INTRON | A (0.805 , 0.772) | 3.776E-02 | 1.19 (1.01 , 1.4) | 77 | 25 |
| 17484235 | 15 | 76548469 | IREB2 | PSC | INTRON | C (0.37 , 0.308) | 1.244E-04 | 1.31 (1.14 , 1.51) | 10 | 3 |
| 1504549 | 15 | 76553684 | IREB2 | PSC | INTRON | A (0.651 , 0.604) | 6.163E-03 | 1.21 (1.06 , 1.38) | 56 | 17 |
| 7164594 | 15 | 76590112 | LOC123688 | CIDR | INTRON | G (0.807 , 0.763) | 2.848E-03 | 1.28 (1.09 , 1.5) | 43 | 12 |
| 8034191 | 15 | 76593078 | LOC123688 | PSC | INTRON | G (0.381 , 0.323) | 5.836E-04 | 1.27 (1.11 , 1.46) | 22 | 4 |
| 10519203 | 15 | 76601101 | LOC123688 | CIDR | INTRON | G (0.386 , 0.329) | 9.378E-04 | 1.26 (1.1 , 1.44) | 24 | 4 |
| 4461039 | 15 | 76604502 | LOC123688 | CIDR | INTRON | T (0.809 , 0.767) | 4.236E-03 | 1.27 (1.08 , 1.49) | 49 | 7 |
| 2036534 | 15 | 76614003 | CIDR | A (0.81 , 0.768) | 4.555E-03 | 1.27 (1.08 , 1.49) | 51 | 7 | ||
| 12916483 | 15 | 76619452 | PSMA4 | CIDR | LOCUS | G (0.582 , 0.571) | 5.108E-01 | 1.05 (0.91 , 1.2) | 168 | 68 |
| 3813570 | 15 | 76619887 | PSMA4 | CIDR | mRNA-UTR | A (0.81 , 0.767) | 3.589E-03 | 1.27 (1.08 , 1.5) | 44 | 7 |
| 11858230 | 15 | 76622607 | PSMA4 | CIDR | INTRON | G (0.582 , 0.569) | 4.291E-01 | 1.06 (0.92 , 1.21) | 156 | 68 |
| 880395 | 15 | 76631411 | PSC | G (0.581 , 0.568) | 4.274E-01 | 1.06 (0.92 , 1.21) | 155 | 68 | ||
| 7164030 | 15 | 76631716 | PSC | A (0.58 , 0.565) | 3.713E-01 | 1.06 (0.93 , 1.22) | 141 | 68 | ||
| 905739 | 15 | 76632165 | PSC | A (0.812 , 0.766) | 2.131E-03 | 1.29 (1.1 , 1.52) | 39 | 7 | ||
| 2036527 | 15 | 76638670 | CIDR | A (0.387 , 0.324) | 2.233E-04 | 1.29 (1.13 , 1.48) | 16 | 4 | ||
| 503464 | 15 | 76644951 | CHRNA5 | CIDR | LOCUS | A (0.802 , 0.757) | 5.047E-03 | 1.26 (1.07 , 1.47) | 54 | 7 |
| 684513 | 15 | 76645455 | CHRNA5 | CIDR | INTRON | G (0.821 , 0.779) | 5.315E-03 | 1.26 (1.07 , 1.49) | 55 | 7 |
| 667282 | 15 | 76650527 | CHRNA5 | CIDR | INTRON | A (0.807 , 0.759) | 1.258E-03 | 1.3 (1.11 , 1.53) | 29 | 7 |
| 588765 | 15 | 76652480 | CHRNA5 | CIDR | INTRON | G (0.582 , 0.567) | 3.653E-01 | 1.06 (0.93 , 1.21) | 139 | 51 |
| 6495306 | 15 | 76652948 | CHRNA5 | PSC | INTRON | A (0.581 , 0.567) | 4.024E-01 | 1.06 (0.93 , 1.21) | 148 | 51 |
| 17486278 | 15 | 76654537 | CHRNA5 | CIDR | INTRON | C (0.387 , 0.322) | 1.690E-04 | 1.3 (1.13 , 1.49) | 13 | 4 |
| 601079 | 15 | 76656634 | CHRNA5 | PSC | INTRON | T (0.578 , 0.564) | 3.813E-01 | 1.06 (0.93 , 1.21) | 143 | 51 |
| 680244 | 15 | 76658343 | CHRNA5 | CIDR | INTRON | G (0.579 , 0.563) | 3.229E-01 | 1.07 (0.94 , 1.22) | 127 | 51 |
| 621849 | 15 | 76659916 | CHRNA5 | CIDR | INTRON | A (0.58 , 0.564) | 3.632E-01 | 1.06 (0.93 , 1.21) | 138 | 51 |
| 637137 | 15 | 76661031 | CHRNA5 | CIDR | INTRON | A (0.808 , 0.759) | 9.515E-04 | 1.31 (1.12 , 1.54) | 25 | 7 |
| 692780 | 15 | 76663560 | CHRNA5 | CIDR | INTRON | C (0.63 , 0.629) | 8.931E-01 | 1.01 (0.88 , 1.16) | 209 | 96 |
| 11637635 | 15 | 76664205 | CHRNA5 | CIDR | INTRON | G (0.63 , 0.628) | 8.686E-01 | 1.01 (0.88 , 1.16) | 202 | 96 |
| 951266 | 15 | 76665596 | CHRNA5 | CIDR | INTRON | A (0.384 , 0.321) | 2.730E-04 | 1.29 (1.12 , 1.48) | 18 | 4 |
| 555018 | 15 | 76666297 | CHRNA5 | CIDR | INTRON | A (0.582 , 0.567) | 3.593E-01 | 1.06 (0.93 , 1.22) | 136 | 51 |
| 647041 | 15 | 76667536 | CHRNA5 | CIDR | INTRON | G (0.584 , 0.569) | 3.614E-01 | 1.06 (0.93 , 1.22) | 137 | 51 |
| 2229961 | 15 | 76667807 | PSC | A (0.023 , 0.014) | 5.315E-02 | 1.64 (0.99 , 2.77) | 85 | 27 | ||
| 16969968 | 15 | 76669980 | CHRNA5 | PSC | NONSYNON | A (0.383 , 0.318) | 1.298E-04 | 1.31 (1.14 , 1.5) | 11 | 4 |
| 514743 | 15 | 76671282 | CHRNA5 | PSC | INTRON | A (0.629 , 0.629) | 8.950E-01 | 1.01 (0.88 , 1.16) | 211 | 96 |
| 660652 | 15 | 76674887 | CHRNA3 | PSC | mRNA-UTR | G (0.63 , 0.628) | 8.220E-01 | 1.02 (0.89 , 1.17) | 195 | 96 |
| 472054 | 15 | 76675049 | CHRNA3 | PSC | mRNA-UTR | G (0.629 , 0.628) | 8.165E-01 | 1.02 (0.89 , 1.17) | 193 | 96 |
| 578776 | 15 | 76675455 | CHRNA3 | CIDR | mRNA-UTR | G (0.759 , 0.697) | 1.064E-04 | 1.34 (1.16 , 1.56) | 8 | 2 |
| 6495307 | 15 | 76677376 | CHRNA3 | PSC | INTRON | G (0.585 , 0.573) | 4.638E-01 | 1.05 (0.92 , 1.2) | 162 | 51 |
| 1051730 | 15 | 76681394 | CHRNA3 | PSC | SYNON | A (0.382 , 0.318) | 2.007E-04 | 1.3 (1.13 , 1.49) | 15 | 4 |
| 3743078 | 15 | 76681814 | CHRNA3 | CIDR | INTRON | C (0.8 , 0.754) | 2.727E-03 | 1.27 (1.09 , 1.49) | 42 | 7 |
| 3743077 | 15 | 76681951 | CHRNA3 | PSC | INTRON | G (0.584 , 0.569) | 3.292E-01 | 1.07 (0.94 , 1.22) | 130 | 51 |
| 1317286 | 15 | 76683184 | CHRNA3 | CIDR | INTRON | G (0.388 , 0.326) | 3.396E-04 | 1.28 (1.12 , 1.47) | 21 | 4 |
| 938682 | 15 | 76683602 | CHRNA3 | CIDR | INTRON | A (0.806 , 0.759) | 1.839E-03 | 1.29 (1.1 , 1.52) | 32 | 7 |
| 11637630 | 15 | 76686774 | CHRNA3 | CIDR | INTRON | A (0.806 , 0.759) | 1.839E-03 | 1.29 (1.1 , 1.52) | 33 | 7 |
| 7177514 | 15 | 76694461 | CHRNA3 | CIDR | INTRON | C (0.804 , 0.757) | 2.027E-03 | 1.29 (1.1 , 1.51) | 38 | 7 |
| 6495308 | 15 | 76694711 | CHRNA3 | CIDR | INTRON | A (0.804 , 0.757) | 1.858E-03 | 1.29 (1.1 , 1.51) | 34 | 7 |
| 8042059 | 15 | 76694914 | CHRNA3 | CIDR | INTRON | A (0.804 , 0.757) | 1.858E-03 | 1.29 (1.1 , 1.51) | 35 | 7 |
| 8042374 | 15 | 76695087 | CHRNA3 | CIDR | INTRON | A (0.804 , 0.757) | 1.901E-03 | 1.29 (1.1 , 1.51) | 36 | 7 |
| 4887069 | 15 | 76696125 | CHRNA3 | CIDR | INTRON | A (0.801 , 0.753) | 2.144E-03 | 1.28 (1.09 , 1.5) | 40 | 7 |
| 8192479 | 15 | 76696453 | CHRNA3 | PSC | SYNON | A (0.024 , 0.019) | 3.954E-01 | 1.22 (0.77 , 1.95) | 145 | 71 |
| 3743075 | 15 | 76696507 | CHRNA3 | PSC | SYNON | A (0.373 , 0.372) | 9.576E-01 | 1 (0.87 , 1.14) | 221 | 96 |
| 3743074 | 15 | 76696535 | CHRNA3 | PSC | INTRON | G (0.374 , 0.373) | 8.984E-01 | 0.99 (0.86 , 1.14) | 212 | 96 |
| 3743073 | 15 | 76696594 | CHRNA3 | PSC | INTRON | A (0.628 , 0.627) | 8.343E-01 | 1.01 (0.88 , 1.16) | 196 | 96 |
| 8192475 | 15 | 76698285 | CHRNA3 | CIDR | NONSYNON | G (0.954 , 0.941) | 1.197E-01 | 1.26 (0.94 , 1.69) | 94 | 31 |
| 1878399 | 15 | 76699058 | CHRNA3 | CIDR | INTRON | G (0.58 , 0.561) | 2.305E-01 | 1.08 (0.95 , 1.24) | 114 | 51 |
| 6495309 | 15 | 76702300 | CHRNB4 | CIDR | LOCUS | G (0.817 , 0.776) | 6.526E-03 | 1.25 (1.07 , 1.48) | 57 | 7 |
| 1948 | 15 | 76704454 | CHRNB4 | CIDR | mRNA-UTR | G (0.669 , 0.663) | 5.329E-01 | 1.05 (0.91 , 1.2) | 169 | 82 |
| 12914008 | 15 | 76710560 | CHRNB4 | PSC | NONSYNON | G (0.959 , 0.945) | 7.811E-02 | 1.31 (0.97 , 1.78) | 89 | 31 |
| 17487223 | 15 | 76711042 | CHRNB4 | PSC | INTRON | A (0.406 , 0.344) | 3.274E-04 | 1.28 (1.12 , 1.47) | 20 | 4 |
| 950776 | 15 | 76713073 | CHRNB4 | CIDR | INTRON | A (0.661 , 0.654) | 6.469E-01 | 1.03 (0.9 , 1.19) | 183 | 82 |
| 17408858 | 15 | 76713073 | PSC | A (0.66 , 0.655) | 7.019E-01 | 1.03 (0.89 , 1.18) | 186 | 82 | ||
| 12440014 | 15 | 76713781 | CHRNB4 | CIDR | INTRON | C (0.794 , 0.753) | 1.247E-02 | 1.22 (1.04 , 1.43) | 66 | 22 |
| 11636605 | 15 | 76715933 | CHRNB4 | CIDR | INTRON | G (0.83 , 0.791) | 6.562E-03 | 1.27 (1.07 , 1.5) | 58 | 13 |
| 12441998 | 15 | 76716427 | CHRNB4 | CIDR | INTRON | A (0.829 , 0.787) | 3.807E-03 | 1.28 (1.08 , 1.52) | 45 | 13 |
| 1316971 | 15 | 76717565 | CHRNB4 | CIDR | INTRON | G (0.831 , 0.791) | 3.910E-03 | 1.29 (1.08 , 1.53) | 46 | 13 |
| 3813567 | 15 | 76721606 | PSC | A (0.828 , 0.793) | 1.411E-02 | 1.24 (1.04 , 1.47) | 68 | 13 | ||
| 3971872 | 15 | 76729090 | PSC | A (0.09 , 0.08) | 1.976E-01 | 1.17 (0.92 , 1.48) | 109 | 47 | ||
|
| ||||||||||
| CHRNB1 | ||||||||||
|
| ||||||||||
| 4796305 | 17 | 7276779 | PSC | A (0.914 , 0.913) | 8.708E-01 | 1.02 (0.8 , 1.29) | 204 | 101 | ||
| 4151125 | 17 | 7285221 | FGF11 | CIDR | INTRON | G (0.858 , 0.854) | 7.688E-01 | 1.03 (0.85 , 1.24) | 189 | 84 |
| 3829603 | 17 | 7287766 | FGF11 | CIDR | mRNA-UTR | A (0.273 , 0.268) | 2.096E-01 | 1.1 (0.95 , 1.27) | 111 | 48 |
| 17856697 | 17 | 7289349 | CHRNB1 | CIDR | NONSYNON | G (0.388 , 0.364) | 3.152E-01 | 1.07 (0.93 , 1.23) | 125 | 61 |
| 7215056 | 17 | 7290774 | CHRNB1 | PSC | INTRON | C (0.808 , 0.781) | 2.013E-02 | 1.21 (1.03 , 1.43) | 70 | 21 |
| 2302765 | 17 | 7291699 | CHRNB1 | CIDR | INTRON | A (0.861 , 0.854) | 5.525E-01 | 1.06 (0.88 , 1.28) | 171 | 84 |
| 7210231 | 17 | 7298588 | CHRNB1 | PSC | INTRON | C (0.806 , 0.777) | 1.372E-02 | 1.23 (1.04 , 1.45) | 67 | 21 |
| 4796418 | 17 | 7298724 | CHRNB1 | PSC | INTRON | C (0.861 , 0.854) | 5.858E-01 | 1.05 (0.87 , 1.27) | 175 | 84 |
| 2302761 | 17 | 7299244 | CHRNB1 | CIDR | INTRON | G (0.805 , 0.775) | 9.904E-03 | 1.24 (1.05 , 1.46) | 64 | 21 |
| 2302762 | 17 | 7299585 | CHRNB1 | CIDR | INTRON | A (0.278 , 0.271) | 1.985E-01 | 1.1 (0.95 , 1.27) | 110 | 48 |
| 2302764 | 17 | 7300834 | CHRNB1 | PSC | mRNA-UTR | G (0.158 , 0.155) | 3.727E-01 | 1.09 (0.91 , 1.3) | 142 | 69 |
| 3855924 | 17 | 7301047 | CHRNB1 | PSC | mRNA-UTR | A (0.86 , 0.854) | 6.129E-01 | 1.05 (0.87 , 1.27) | 176 | 84 |
| 17732878 | 17 | 7303083 | ZBTB4 | PSC | LOCUS | A (0.804 , 0.775) | 9.540E-03 | 1.24 (1.05 , 1.46) | 63 | 21 |
| 9217 | 17 | 7303812 | ZBTB4 | CIDR | mRNA-UTR | A (0.664 , 0.628) | 7.035E-03 | 1.22 (1.05 , 1.4) | 59 | 18 |
| 9901643 | 17 | 7360548 | CIDR | G (0.865 , 0.859) | 6.283E-01 | 1.05 (0.87 , 1.27) | 181 | 84 | ||
|
| ||||||||||
| CHRNA4 | ||||||||||
|
| ||||||||||
| 4603829 | 20 | 61439336 | PSC | A (0.648 , 0.625) | 1.768E-01 | 1.1 (0.96 , 1.26) | 105 | 43 | ||
| 2236196 | 20 | 61448000 | CHRNA4 | CIDR | mRNA-UTR | G (0.272 , 0.236) | 4.767E-03 | 1.25 (1.07 , 1.45) | 52 | 16 |
| 3787137 | 20 | 61449544 | CHRNA4 | CIDR | INTRON | A (0.57 , 0.566) | 8.940E-01 | 0.99 (0.87 , 1.13) | 210 | 92 |
| 3787138 | 20 | 61449668 | CHRNA4 | PSC | INTRON | G (0.133 , 0.11) | 3.875E-02 | 1.24 (1.01 , 1.52) | 78 | 26 |
| 1044396 | 20 | 61451578 | CHRNA4 | PSC | SYNON | A (0.54 , 0.538) | 7.211E-01 | 0.98 (0.86 , 1.11) | 187 | 92 |
| 2229959 | 20 | 61451998 | CHRNA4 | CIDR | SYNON | C (0.124 , 0.106) | 1.098E-01 | 1.19 (0.96 , 1.46) | 93 | 26 |
| 2273500 | 20 | 61457393 | CHRNA4 | PSC | INTRON | G (0.085 , 0.071) | 8.109E-02 | 1.23 (0.98 , 1.55) | 90 | 32 |
| 2273504 | 20 | 61458505 | CHRNA4 | CIDR | INTRON | G (0.843 , 0.803) | 2.266E-03 | 1.31 (1.1 , 1.55) | 41 | 11 |
| 3787140 | 20 | 61459555 | CHRNA4 | PSC | INTRON | G (0.072 , 0.061) | 9.522E-02 | 1.25 (0.96 , 1.63) | 92 | 32 |
| 755203 | 20 | 61464708 | PSC | A (0.557 , 0.543) | 9.411E-01 | 1.01 (0.88 , 1.15) | 219 | 106 | ||
| 1107953 | 20 | 61471990 | PSC | G (0.524 , 0.513) | 4.023E-01 | 1.06 (0.93 , 1.21) | 147 | 73 | ||
SNPs significant after multiple test correction (p ≤ 0.000462) are indicated with boldface.
Genotyping lab: PSC = Perlegen Sciences, CIDR = Center for Inherited Disease Research
Bins defined using r2 ≥ 8 after sorting SNPs by significance, as described in the Methods. Bins are numbered by the significance (primary p-value) of the tag.
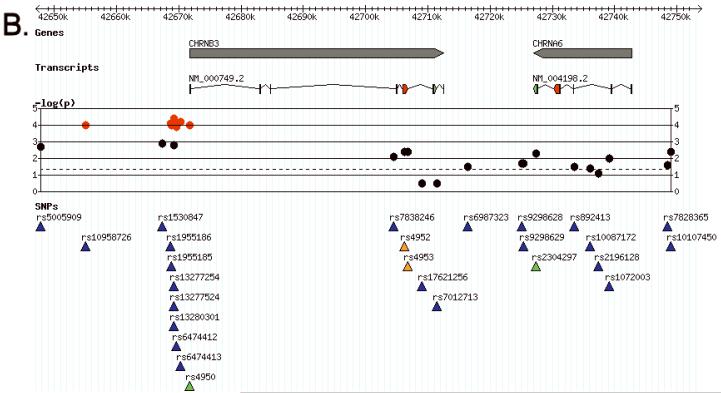
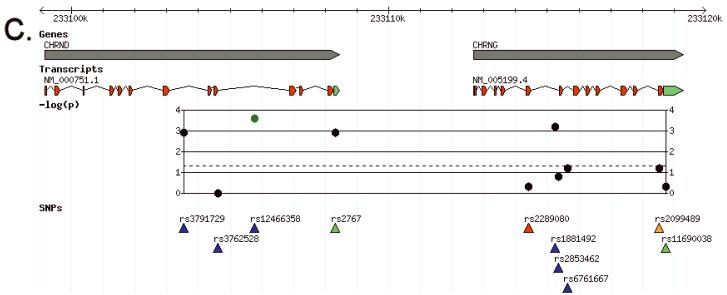
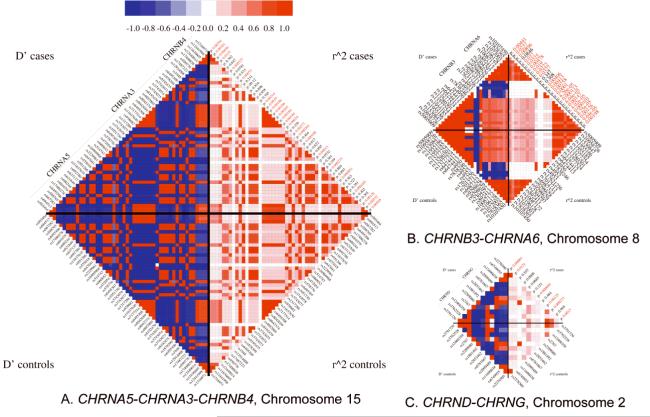
Results were entered into our custom installation of the Generic Genome Browser (Stein et al., 2002). Panels A, B and C of Figure 2 display plots of the negative logarithm of the primary (log-additive model) p-values for the three gene clusters that contained SNPs significant after multiple test correction (α5-α3-β4 and β3-α6, δ-γ). Figure 3A, 3B and 3C display the pairwise LD patterns (signed D’ and r2) for these gene clusters in cases and controls, labeled by rs number and primary p-value. Supplementary figure S2 extends the CHRNA5-CHRNA3-CHRNB4 LD plot to include the neighboring genes PSMA4, LOC123688 and IREB2.
Figure 2.
Association results aligned with genes and transcripts from NCBI. The −log(p) track plots the negative logarithm of the primary p-value, color coded by LD bin: red = bin 1, yellow = bin 2, cyan = bin 3, blue = bin 4, green = bin 5. Panel A: the CHRNA5-CHRNA3-CHRNB4 gene cluster on chromosome 15. Panel B: the CHRNB3-CHRNA6 cluster on chromosome 8. Panel C: CHRND-CHRNG on chromosome 2. Panel D: Key for transcript and SNP function color codings.
Figure 3.
Linkage disequilibrium plots. Pairwise signed D’ values indicate the phase of LD, so that a positive value denotes coupling (the minor alleles tend to co-occur) and a negative value denotes repulsion (the minor allele at one locus tends to co-occur with the common allele at the other locus). Primary p-values are also displayed. Panel A: CHRNA5-CHRNA3-CHRNB4. Panel B: CHRNB3-CHRNA6. Panel C: CHRND-CHRNG.
The two most significant SNPs by best model p-value are in the CHRNA5-CHRNA3-CHRNB4 cluster: rs17487223 (best model p = 0.0000266) in CHRNB4 and rs16969968 (best model p = 0.0000284) in CHRNA5. The latter is a non-synonymous SNP in CHRNA5, and we consider it the most biologically promising finding. Both SNPs are in the same r2 bin that includes 7 additional SNPs (5 surpassing the multiple test criterion of 0.000462) extending both upstream into the gene LOC123688 and downstream through CHRNA3 and CHRNB4 (Table II and Figure 2A). These seven SNPs therefore represent a single signal. The additional 4 significant SNPs in the neighboring gene IREB2 form a separate r2 ≥ 0.8 bin but have r2 ≥ 0.68 with rs16969968 (0.78 for rs1504549, 0.694 for rs17405217, 0.690 for rs17484235, 0.697 for rs17483548 0.697, and 0.679 for rs17483686); thus these are still highly correlated and likely constitute the same signal. In this bin, an intronic SNP in IREB2, rs17484235, has a primary p-value of 0.0001244 compared to 0.0001298 for the non-synonymous SNP rs16969968.
In contrast, the remaining significant SNP in the CHRNA5-CHRNA3-CHRNB4 cluster, rs578776, is not correlated with rs16969968 or its bin. The LD between rs578776 and rs16969968 is r2 = 0.20 overall (0.196 in cases, 0.205 in controls) with D’ = −1.0, due to the LD being in repulsion phase. We thus observed two distinct loci in the CHRNA5-CHRNA3-CHRNB4 gene cluster whose significant association with nicotine dependence is not simply explained by LD between these loci. Table II, column 7 clarifies that for rs16969968 the “risk” allele (meaning the allele more common in cases than in controls) is the minor allele, while for rs578776 the risk allele is the major allele. Thus, recoding the genotypes with the major allele as the reference allele would give an odds ratio of 1.31 for rs16969968 versus a “protective” odds ratio of 0.75 for rs578776.
The CHRNB3-CHRNA6 gene cluster on chromosome 8 contains eight SNPs surpassing the multiple test criterion. These constitute a single r2 bin and a single association signal after LD is taken into account: rs13277254 is in nearly perfect LD with the other SNPs (Figure 3B), with each pairwise r2 exceeding 0.99. Even after the increased CIDR SNP coverage of this gene cluster, all significant SNPs are in the same LD bin as the previous associated SNPs we reported in (Saccone et al., 2007a), and no new distinct signals emerged.
The CHRND gene contains one SNP significant after multiple test correction. This SNP, rs12466358 (p = 0.00027), is a singleton bin in our data, although other SNPs have r2 ≥ 0.6 (but less than 0.8) with it (Figure 3C). It is noteworthy that the next most significant SNP in the CHRND-CHRNG cluster, rs1881492 (p = 0.00069) is uncorrelated with rs12466358 (r2 = 0.086), suggesting that there is a second distinct risk allele in this gene cluster, although this second signal does not meet our multiple test correction criterion.
For the SNPs significant after multiple test correction, the odds ratios (ORs) for the primary genetic model are modest, ranging up to 1.4 (for SNPs in CHRNB3) for the effect of a single copy of the risk allele. Among the SNPs covering the CHRNA5-CHRNA3-CHRNB4 region, the strongest alternative odds ratio is found for rs16969968 when modeled recessively, with an OR of 1.83 for the effect of having 2 copies of the risk allele (Table S1). This OR is similar to but larger than the OR for two risk alleles under the primary model: (1.31)2 = 1.7. For rs16969968, both the recessive-model and primary model ORs are higher than the corresponding ORs for the eight additional correlated SNPs in bin 4, further suggesting the importance of this non-synonymous variant relative to these alternative loci.
In a few additional genes, we observed nominally significant SNPs that did not meet our multiple test significance criterion. These were in CHRNA4, and CHRNB1. There is borderline nominal significance at a nonsynonymous SNP, rs12914008, in CHRNB4 (p = 0.078); this suggestive signal is distinct from the experiment-wide significant findings at rs16969968 and rs578776. Another SNP in CHRNA5, rs2229961, is a nonsynonymous change according to NCBI build 35.1 and has a borderline p-value of 0.053 but is rarer than the other nonsynonymous SNPs in the cluster, rs16969968 and rs12914008.
Multilocus analysis in CHRNA5-CHRNA3-CHRNB4
The CHRNA5-CHRNA3-CHRNB4 gene cluster is of particular interest after our report of association with nicotine dependence (Saccone et al., 2007a) and subsequent evidence of replication in independent samples (Berrettini et al., 2008; Bierut et al., In press; Thorgeirsson et al., 2008). We carried out joint analyses of the two distinct, uncorrelated findings in the CHRNA5-CHRNA3-CHRNB4 cluster, represented by rs16969968 coded recessively and rs578776 coded log-additively to match their best models.
In the joint logistic regression analysis, CHRNA5 SNP rs16969968 remains significant (p = 0.0000645) with a similar order of magnitude as its single SNP best model result of p = 0.000028. When controlling for rs16969968 genotype, rs578776 is reduced by an order of magnitude compared to its single SNP result, but it still remains significant in the joint model (p=0.00543). This gives further evidence that there are two distinct susceptibility loci in this gene cluster.
Table III presents the joint genotype analysis. From the allele frequencies in Table II (column 7), we see that rs16969968 represents a locus whose minor allele (A) confers risk for addiction, while rs578776 represents a locus whose minor allele (A) is protective. Thus, as expected, the GG/AA genotype combination for rs16969968/rs578776 confers the greatest protection from nicotine dependence, while the AA/GG is associated with the highest risk (Table III). The odds of nicotine dependence relative to non-dependence among those with the highest risk joint genotype is 2.5 higher than in those with the lowest risk. It is interesting to note the zero cells for the AG/AA, AA/AG and AA/AA combinations, which clarifies that the high risk genotype AA at rs16969968 occurs only in combination with the homozygous “risk” GG genotype at rs578776. We thus cannot observe the effect of the AA genotype at rs16969968 on other background genotypes at rs578776, and similarly the A-A haplotype is not observed (double heterozygotes being extremely likely to carry A-G and G-A). However, the effect on the observed background is clear from the cells of the first row and first column of Table III, where GG/GG is set as the reference genotype. We see that on the background of the low risk (protective) genotype at rs16969968 (GG), there is significantly increased protection for one and two copies of the A allele at rs578776 along row 1 (OR = 0.68 and 0.60 respectively). On the background of the risk genotype at rs578776 (GG), there is significantly increased risk for two copies of the A allele at rs16969968 along column 1 (OR = 1.49).
Table III.
Joint analysis of rs16969968 and rs578776.
| rs578776 | |||
|---|---|---|---|
| rs16969968 | GG | AG | AA |
| GG | 151/110 1.00 (ref) |
201/214 0.68 (0.50-0.93) |
60/72 0.60 (0.40-0.93) |
| AG | 286/221 0.94 (0.70-1.28) |
182/171 0.77 (0.56-1.07) |
0/0 -- |
| AA | 168/82 1.49 (1.04-2.14) |
0/0 -- |
0/0 -- |
The first line of the cell indicates the number of cases and controls with the specific genotype combination; the second line indicates the OR and 95% CI when GG/GG for rs16969968/rs578776 is the reference genotype.
DISCUSSION AND CONCLUSIONS
This is the most comprehensive survey of the nicotinic receptor subunit genes for involvement in nicotine dependence to date. We have further explored the relationship between nicotine dependence and this gene family using 226 SNPs genotyped in the NICSNP sample of nicotine dependent cases and non-dependent smoking controls. Four distinct findings, two in the CHRNA5-CHRNA3-CHRNB4 cluster, one in CHRNB3-CHRNA6, and one in CHRND-CHRNG are significant after multiple test correction across the CHRN gene family. Additional genes CHRNA4 and CHRNB1 harbor nominally significant SNPs. Those CHRN genes that were not nominally associated in our initial reports (Bierut et al., 2007; Saccone et al., 2007a) still lack significant association with nicotine dependence with the denser SNP coverage.
This study improved our coverage of an important gene family using a sample previously genotyped for a joint GWAS and large-scale candidate gene study (Bierut et al., 2007; Saccone et al., 2007a). Given the current popularity and importance of the GWAS design, the question of how best to interpret new analyses of GWAS datasets or samples in the context of the original large-scale GWAS is a challenge facing not only this study but the field as a whole. In the present case, although the NICSNP sample was used for a large-scale GWAS and candidate gene study, the CHRN gene family was in fact given the highest priority in our original design and would have been targeted even if our resources had been limited to only a few hundred SNPs. Therefore we feel it is appropriate and useful to report significant results based on multiple test correction for the 226 SNPs (111 r2 bins) tested here. However, these findings are not significant after formal multiple test correction for all the genotyping performed on this sample to date. In general, clearly citing any previous, overlapping GWAS or large-scale candidate gene studies is important to allow fully informed interpretation of results.
Our results continue to support an important role for the nonsynonymous CHRNA5 SNP rs16969968 in determining vulnerability to nicotine dependence, as first reported in (Saccone et al., 2007a), and separate in vitro evidence indicates that rs16969968 may indeed be the functional variant explaining the association across the other correlated members of this LD bin (Bierut et al., In press). Due to the extensive LD encompassing not only the CHRNA5-CHRNA3-CHRNB4 cluster but also the neighboring genes LOC123688, PSMA4 and IREB2 (Figures 2A, 3A and S2), further work to definitively identify a causal variant from among all SNPs correlated with rs16969968 will be important.
A second variant in the CHRNA5-CHRNA3-CHRNB4 cluster, rs578776, constitutes a distinct, significantly associated locus that also warrants further study. This SNP was previously noted to have a false discovery rate of 0.09 (Saccone et al., 2007a); with our denser coverage it remains the most significant representative of this second locus. Joint analysis of the uncorrelated SNPs rs578776 and rs16969968 indicates that these two variants each exert independent influence on nicotine dependence vulnerability. Though |D’| between these SNPs is 1, the low r2 between them means that the disease association at one does not statistically explain the disease association at the other. These two variants demonstrate an interesting evolutionary history, with the risk allele for rs16969968 occurring on the background of the higher risk variant for rs578776 (Table III). Although not all genotype combinations occur because of this history, the three genotypes at one locus on a fixed background for the other demonstrate a clear pattern of altered risk (Table III, first row and first column). It is unclear from our genetic data whether the functional variants underlying this susceptibility reside within the same CHRN gene or reflect variation in two different genes.
Our third significant single-SNP finding is at the 5′ end of CHRNB3-CHRNA6, tagged by rs13277254, and here again the functional source of this signal is unclear. Interestingly, a study in non-small cell cancer tumors found higher expression of CHRNA6 and CHRNB3 in non-smokers compared to smokers (Lam et al., 2007), suggesting potential mechanisms of action related to exposure to smoking and nicotine dependence.
CHRND on chromosome 2 harbors a fourth significant locus, with an additional interesting, though only nominally significant, uncorrelated locus in the neighboring gene CHRNG. Efforts to replicate these findings in independent samples would be of great interest. The CHRND-CHRNG cluster lies at the end of chromosome 2q in a region of linkage that has been persistently reported in the literature for nicotine dependence (Straub et al., 1999) and other addiction phenotypes (Agrawal et al., 2008; Gelernter et al., 2005; Gelernter et al., 2006). The γ subunit is known only to be fetally expressed and replaced by ε in late fetal development (Mishina et al., 1986), suggesting an unexpected mechanism for influencing addiction risk if indeed CHRNG is involved.
Our CIDR genotyping included the CHRNA3 SNPs rs1317286 and rs6495308 recently reported by Berrettini et al. to be associated with cigarettes-per-day with p-values of 0.0000026 and 0.000069 respectively (Berrettini et al., 2008). With our comprehensive coverage we are thus able to make a direct comparison and confirm that their results are in line with our two distinct findings represented by rs16969968 and rs578776. In our sample, rs1317286 (primary p = 0.00034, OR = 1.28) is in the same LD bin and highly correlated (r2 = 0.975) with the slightly more significant rs16969968 (p = 0.00013, OR = 1.30), confirming that these findings coincide. We believe that rs16969968 is the best functional candidate among the correlated SNPs representing this locus, and further work to test this is underway. The second SNP rs6495308 is correlated (r2 = 0.76) with our separate significant finding at rs578776. This suggests that the signal at rs6495308 (p = 0.0019, OR = 1.29) may be an attenuation of the stronger association at rs578776 (p = 0.00011, OR = 1.34). The correlation between rs16969968 and rs6495308 is low (r2 = 0.15), again indicating that these represent two different signals.
The CHRNA5-CHRNA3-CHRNB4 cluster is of further interest because of recent reports of significant association with lung cancer (Amos et al., 2008; Hung et al., 2008; Thorgeirsson et al., 2008), a disease for which cigarette smoking is known to be the major risk factor. The associations with lung cancer were either at the non-synonymous CHRNA5 SNP rs16969968 (p-value 1 × 10−20 (Hung et al., 2008)), or at SNPs highly correlated with it (p = 1.5 × 10−8 at rs1051730 (Thorgeirsson et al., 2008); p = 7× 10−18 at rs1051730 and p = 3× 10−18 at rs8034191 (Amos et al., 2008)). The lung cancer risk allele matches the risk allele for nicotine dependence.
Those reports differed in their interpretation of this association with lung cancer — that is, whether it is evidence of a direct effect on lung cancer vulnerability, or whether it can be explained entirely through the indirect effect of increased risk for smoking. What is clear is that this locus is a risk factor for nicotine dependence and smoking quantity. However, it is interesting to note that for lung cancer, the odds ratios for the effect of one copy of the risk allele were 1.30 (95% confidence interval 1.23-1.38) for rs16969968 (Hung et al., 2008), 1.32 (1.24-1.41) for rs8034191 and 1.32 (1.23-1.39) for rs1051730 (Amos et al., 2008), and 1.31 (1.19-1.44) for rs1051730 (Thorgeirsson et al., 2008). These therefore match the odds ratio we see for nicotine dependence: 1.31 (1.14-1.5) for rs16969968. The lung cancer studies highlighted this single risk locus in this region, and appear to indicate weaker association between lung cancer and rs578776, which tags our second nicotine dependence locus in the CHRNA5-CHRNA3-CHRNB4 cluster. In our nicotine dependence study, rs16969968 (p = 1.3×10−4, OR 1.31 (1.14-1.5)) and rs578776 (p = 1.1×10−4, OR 1.34 (1.16-1.56)) have comparable evidence and effect size for association. In contrast, (Hung et al., 2008) typed both SNPs in the discovery subset of their sample and in that subsample found lung cancer association p-values of 5×10−9 (OR 1.32 (1.2-1.44)) for rs16969968 versus 2.5×10−4 (OR 1.2 (1.08-1.33)) for rs578776. Future work to clarify the direct versus indirect effects of these variants on lung cancer will be of great interest.
An important, unique feature of our sample is the definition of controls: smokers (> 100 cigarettes lifetime) who have never exhibited symptoms of dependence. Genetic associations from this study thus provide insight into the transition from smoking to nicotine dependence. This design also circumvents noise that may occur if the control group includes non-smoking (unexposed) but genetically vulnerable individuals. We expect our results to overlap with those from studies of alternative phenotypes such as cigarettes-per-day, but differences in genetic findings may reflect important differences in the phenotypes.
Recently, (Zeiger et al., 2008) demonstrated that the CHRNB3-CHRNA6 cluster is associated with early subjective responses to tobacco in adolescents. One of their most significant results is tagged by rs4950 in CHRNB3; this SNP is significantly associated with nicotine dependence in our sample (p = 0.00010) and is in the LD bin tagged by rs13277254 (r2 = 0.995). This concordance of genetic findings for these two phenotypes suggests that subjective effects may mediate the association between this locus and addiction, and raises the possibility that early interventions may be effective in disrupting this particular pathway leading to increased addiction risk.
Other groups have reported significant associations between measures of cigarette use or nicotine dependence and some CHRN genes. Nicotine dependence was associated with two CHRNA4 SNPs (rs1044396 and rs1044397) in a family-based study of Chinese male smokers (Feng et al., 2004). We tested both of these SNPs and did not replicate the univariate findings, perhaps because of ethnic differences across our samples.
A study of six CHRNA4 SNPs in European Americans (EA) and African Americans (AA) reported various nominally significant findings for specific phenotypes or samples, with rs2236196 highlighted in AA women after correction for multiple testing (Li et al., 2005). Our current study provides some support for rs2236196 (p = 0.0048), although we see a stronger association with nicotine dependence at rs2273504 (p = 0.0023). Importantly, these two SNPs are uncorrelated (r2 = 0.07), indicating that we have modest evidence for potentially two distinct loci affecting nicotine dependence risk, one which is novel and the other which replicates the earlier finding in (Li et al., 2005).
A follow-up to the above cited study in the same sample reported association between rs2302763 in CHRNB1 and smoking quantity in EA (Lou et al., 2006). We did not test this specific SNP, but genotyped other highly correlated SNPs according to HapMap Centre d’Etudie du Polymorphisme Humain (CEPH) from Utah (CEU) data (The International HapMap Consortium, 2005) (e.g. rs2302765, rs4796418, rs3855924). None were associated with nicotine dependence in our sample. We do observe a separate, nominally significant signal in CHRNB1 tagged by rs17732878; this LD bin is not correlated with rs2302763 (HapMap CEU r2 < 0.06).
A study of 39 SNPs in 11 CHRN genes in Israeli women reported main effects of rs2072660 (CHRNB2) with history of daily smoking, and of rs1909884 (CHRNA7), rs4861065 (CHRNA9), and rs9298629 (CHRNB3) with nicotine dependence (Greenbaum et al., 2006). Our sample provides weak support for rs9298629 (p = 0.019). Another study reported associations between subjective reactions to cigarettes and rs2072660 and rs2072658 in CHRNB2, in Caucasian and Hispanic young adults (Ehringer et al., 2007). We do not see association between these SNPs and nicotine dependence in our sample.
In summary, we have strong evidence that at least four distinct variants in CHRNA5-CHRNA3-CHRNB4, CHRNB3-CHRNA6 and CHRND-CHRNG influence nicotine dependence risk. These four significant nicotinic receptor findings, though important, together account for less than 10% of the phenotypic variance in our sample, indicating that additional risk factors are yet to be discovered and illustrating the challenge of complex disease genetics. Our ongoing work will continue the search for other genes and epistatic effects; we will also extend our analyses to diverse population samples having contrasting LD structure, as such studies can help narrow down among correlated SNPs and localize the most likely functional source of an association signal. Ultimately, determining the mechanism of action for these variants via functional studies can help improve prevention and cessation therapies.
ACKNOWLEDGMENTS
We thank Dennis Ballinger for advice on genotyping and Louis Fox for database and analytic support. The NICSNP project is a collaborative research group and part of the NIDA Genetics Consortium. Subject collection was supported by NIH grants CA89392 (COGEND, PI - L Bierut) from the National Cancer Institute and DA012854 (NAG, PI - P Madden) from the National Institute on Drug Abuse. Genotyping work at Perlegen Sciences was performed under NIDA Contract HHSN271200477471C. Additional genotyping services were provided by the Center for Inherited Disease Research (CIDR). CIDR is fully funded through a federal contract from the National Institutes of Health to The Johns Hopkins University, contract number HHSN268200782096C. Phenotypic and genotypic data are stored in the NIDA Center for Genetic Studies (NCGS) at http://zork.wustl.edu/. We are grateful to Gary Chase for his many contributions to this project and remember with great appreciation his important and diverse scientific accomplishments. This work is in his memory, and in memory of Theodore Reich, founding Principal Investigator of COGEND.
Grant Support: P01 CA89392 (LJB) from the National Cancer Institute, R01 DA012854 (PAFM), R01 DA014369 (JAS), K08 DA019951 (MLP), K01 DA015129 (NLS) from the National Institute on Drug Abuse, K01 AA015572 (ALH) from the National Institute on Alcohol Abuse and Alcoholism, and IRG-58-010-50 from the American Cancer Society (SFS).
REFERENCES
- Abecasis GR, Cookson WO. GOLD--graphical overview of linkage disequilibrium. Bioinformatics. 2000;16(2):182–3. doi: 10.1093/bioinformatics/16.2.182. [DOI] [PubMed] [Google Scholar]
- Agrawal A, Hinrichs AL, Dunn G, Bertelsen S, Dick DM, Saccone SF, Saccone NL, Grucza RA, Wang JC, Cloninger CR, et al. Linkage scan for quantitative traits identifies new regions of interest for substance dependence in the Collaborative Study on the Genetics of Alcoholism (COGA) sample. Drug Alcohol Depend. 2008;93(1-2):12–20. doi: 10.1016/j.drugalcdep.2007.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amos CI, Wu X, Broderick P, Gorlov IP, Gu J, Eisen T, Dong Q, Zhang Q, Gu X, Vijayakrishnan J, et al. Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nat Genet. 2008;40(5):616–22. doi: 10.1038/ng.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berrettini W, Yuan X, Tozzi F, Song K, Francks C, Chilcoat H, Waterworth D, Muglia P, Mooser V. alpha-5/alpha-3 nicotinic receptor subunit alleles increase risk for heavy smoking. Mol Psychiatry. 2008;(13):368–373. doi: 10.1038/sj.mp.4002154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierut LJ, Madden PA, Breslau N, Johnson EO, Hatsukami D, Pomerleau OF, Swan GE, Rutter J, Bertelsen S, Fox L, et al. Novel genes identified in a high-density genome wide association study for nicotine dependence. Hum Mol Genet. 2007;16(1):24–35. doi: 10.1093/hmg/ddl441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierut LJ, Stitzel JA, Wang JC, Hinrichs AL, Bertelsen S, Fox L, Grucsa RA, Horton WJ, Kauwe JS, Morgan SF, et al. In press. Variants in the nicotinic receptors alter the risk for nicotine dependence American Journal of Psychiatry [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breslau N, Johnson EO, Hiripi E, Kessler R. Nicotine dependence in the United States: prevalence, trends, and smoking persistence. Arch Gen Psychiatry. 2001;58(9):810–6. doi: 10.1001/archpsyc.58.9.810. [DOI] [PubMed] [Google Scholar]
- Carlson CS, Eberle MA, Rieder MJ, Yi Q, Kruglyak L, Nickerson DA. Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am J Hum Genet. 2004;74(1):106–20. doi: 10.1086/381000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention Tobacco Use Among Adults - United States, 2005. Morbidity and Mortality Weekly Report. 2006;55:1145–1148. [PubMed] [Google Scholar]
- Chen D, Patrick JW. The alpha-bungarotoxin-binding nicotinic acetylcholine receptor from rat brain contains only the alpha7 subunit. J Biol Chem. 1997;272(38):24024–9. doi: 10.1074/jbc.272.38.24024. [DOI] [PubMed] [Google Scholar]
- Danaei G, Vander Hoorn S, Lopez AD, Murray CJ, Ezzati M. Causes of cancer in the world: comparative risk assessment of nine behavioural and environmental risk factors. Lancet. 2005;366(9499):1784–93. doi: 10.1016/S0140-6736(05)67725-2. [DOI] [PubMed] [Google Scholar]
- Davis RM, editor. A report of the Surgeon General. U.S. Department of Health and Human Services; 1988. The Health Consequences of Smoking: Nicotine Addiction. [Google Scholar]
- Ehringer MA, Clegg HV, Collins AC, Corley RP, Crowley T, Hewitt JK, Hopfer CJ, Krauter K, Lessem J, Rhee SH, et al. Association of the neuronal nicotinic receptor beta2 subunit gene (CHRNB2) with subjective responses to alcohol and nicotine. Am J Med Genet B Neuropsychiatr Genet. 2007;144(5):596–604. doi: 10.1002/ajmg.b.30464. [DOI] [PubMed] [Google Scholar]
- Feng Y, Niu T, Xing H, Xu X, Chen C, Peng S, Wang L, Laird N, Xu X. A common haplotype of the nicotine acetylcholine receptor alpha 4 subunit gene is associated with vulnerability to nicotine addiction in men. Am J Hum Genet. 2004;75(1):112–21. doi: 10.1086/422194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelernter J, Panhuysen C, Weiss R, Brady K, Hesselbrock V, Rounsaville B, Poling J, Wilcox M, Farrer L, Kranzler HR. Genomewide linkage scan for cocaine dependence and related traits: significant linkages for a cocaine-related trait and cocaine-induced paranoia. Am J Med Genet B Neuropsychiatr Genet. 2005;136(1):45–52. doi: 10.1002/ajmg.b.30189. [DOI] [PubMed] [Google Scholar]
- Gelernter J, Panhuysen C, Wilcox M, Hesselbrock V, Rounsaville B, Poling J, Weiss R, Sonne S, Zhao H, Farrer L, et al. Genomewide linkage scan for opioid dependence and related traits. Am J Hum Genet. 2006;78(5):759–69. doi: 10.1086/503631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotti C, Zoli M, Clementi F. Brain nicotinic acetylcholine receptors: native subtypes and their relevance. Trends Pharmacol Sci. 2006;27(9):482–91. doi: 10.1016/j.tips.2006.07.004. [DOI] [PubMed] [Google Scholar]
- Greenbaum L, Kanyas K, Karni O, Merbl Y, Olender T, Horowitz A, Yakir A, Lancet D, Ben-Asher E, Lerer B.Why do young women smoke? I. Direct and interactive effects of environment, psychological characteristics and nicotinic cholinergic receptor genes Mol Psychiatry 2006113312–22., 223. [DOI] [PubMed] [Google Scholar]
- Hung RJ, McKay JD, Gaborieau V, Boffetta P, Hashibe M, Zaridze D, Mukeria A, Szeszenia-Dabrowska N, Lissowska J, Rudnai P, et al. A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature. 2008;452(7187):633–7. doi: 10.1038/nature06885. [DOI] [PubMed] [Google Scholar]
- Johnson EO, Chase GA, Breslau N. Persistence of cigarette smoking: familial liability and the role of nicotine dependence. Addiction. 2002;97(8):1063–70. doi: 10.1046/j.1360-0443.2002.00211.x. [DOI] [PubMed] [Google Scholar]
- Karlin A. Emerging structure of the nicotinic acetylcholine receptors. Nat Rev Neurosci. 2002;3(2):102–14. doi: 10.1038/nrn731. [DOI] [PubMed] [Google Scholar]
- Klink R, de Kerchove d’Exaerde A, Zoli M, Changeux JP. Molecular and physiological diversity of nicotinic acetylcholine receptors in the midbrain dopaminergic nuclei. J Neurosci. 2001;21(5):1452–63. doi: 10.1523/JNEUROSCI.21-05-01452.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam DC, Girard L, Ramirez R, Chau WS, Suen WS, Sheridan S, Tin VP, Chung LP, Wong MP, Shay JW, et al. Expression of nicotinic acetylcholine receptor subunit genes in non-small-cell lung cancer reveals differences between smokers and nonsmokers. Cancer Res. 2007;67(10):4638–47. doi: 10.1158/0008-5472.CAN-06-4628. [DOI] [PubMed] [Google Scholar]
- Lessov CN, Martin NG, Statham DJ, Todorov AA, Slutske WS, Bucholz KK, Heath AC, Madden PA. Defining nicotine dependence for genetic research: evidence from Australian twins. Psychol Med. 2004;34(5):865–79. doi: 10.1017/s0033291703001582. [DOI] [PubMed] [Google Scholar]
- Li MD, Beuten J, Ma JZ, Payne TJ, Lou XY, Garcia V, Duenes AS, Crews KM, Elston RC. Ethnic- and gender-specific association of the nicotinic acetylcholine receptor alpha4 subunit gene (CHRNA4) with nicotine dependence. Hum Mol Genet. 2005;14(9):1211–9. doi: 10.1093/hmg/ddi132. [DOI] [PubMed] [Google Scholar]
- Lindstrom JM. Nicotinic acetylcholine receptors of muscles and nerves: comparison of their structures, functional roles, and vulnerability to pathology. Ann N Y Acad Sci. 2003;998:41–52. doi: 10.1196/annals.1254.007. [DOI] [PubMed] [Google Scholar]
- Lou XY, Chen GB, Yan L, Ma JZ, Zhu J, Elston RC, Li MD. A generalized combinatorial approach for detecting gene-by-gene and gene-by-environment interactions with application to nicotine dependence. Am J Hum Genet. 2007;80(6):1125–37. doi: 10.1086/518312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou XY, Ma JZ, Payne TJ, Beuten J, Crew KM, Li MD. Gene-based analysis suggests association of the nicotinic acetylcholine receptor beta1 subunit (CHRNB1) and M1 muscarinic acetylcholine receptor (CHRM1) with vulnerability for nicotine dependence. Hum Genet. 2006;120(3):381–9. doi: 10.1007/s00439-006-0229-7. [DOI] [PubMed] [Google Scholar]
- Lueders KK, Hu S, McHugh L, Myakishev MV, Sirota LA, Hamer DH. Genetic and functional analysis of single nucleotide polymorphisms in the beta2-neuronal nicotinic acetylcholine receptor gene (CHRNB2) Nicotine Tob Res. 2002;4(1):115–25. doi: 10.1080/14622200110098419. [DOI] [PubMed] [Google Scholar]
- Mackay J, Eriksen M, Shafey O, editors. The Tobacco Atlas. 2 American Cancer Society; Atlanta, GA: 2006. [Google Scholar]
- Maes HH, Sullivan PF, Bulik CM, Neale MC, Prescott CA, Eaves LJ, Kendler KS. A twin study of genetic and environmental influences on tobacco initiation, regular tobacco use and nicotine dependence. Psychol Med. 2004;34(7):1251–61. doi: 10.1017/s0033291704002405. [DOI] [PubMed] [Google Scholar]
- Mao D, Yasuda RP, Fan H, Wolfe BB, Kellar KJ. Heterogeneity of nicotinic cholinergic receptors in rat superior cervical and nodose Ganglia. Mol Pharmacol. 2006;70(5):1693–9. doi: 10.1124/mol.106.027458. [DOI] [PubMed] [Google Scholar]
- Mathers CD, Loncar D. Projections of global mortality and burden of disease from 2002 to 2030. PLoS Med. 2006;3(11):e442. doi: 10.1371/journal.pmed.0030442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishina M, Takai T, Imoto K, Noda M, Takahashi T, Numa S, Methfessel C, Sakmann B. Molecular distinction between fetal and adult forms of muscle acetylcholine receptor. Nature. 1986;321(6068):406–11. doi: 10.1038/321406a0. [DOI] [PubMed] [Google Scholar]
- Pergadia ML, Heath AC, Agrawal A, Bucholz KK, Martin NG, Madden PA. The implications of simultaneous smoking initiation for inferences about the genetics of smoking behavior from twin data. Behav Genet. 2006;36(4):567–76. doi: 10.1007/s10519-005-9042-7. [DOI] [PubMed] [Google Scholar]
- Pidoplichko VI, Noguchi J, Areola OO, Liang Y, Peterson J, Zhang T, Dani JA. Nicotinic cholinergic synaptic mechanisms in the ventral tegmental area contribute to nicotine addiction. Learn Mem. 2004;11(1):60–9. doi: 10.1101/lm.70004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott CA, Kendler KS. In: Genetic and environmental influences on alcohol and tobacco dependence among women. Fertig JB, Allen JP, editors. Bethesda, MD: 1995. pp. 59–87. [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155(2):945–59. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccone SF, Hinrichs AL, Saccone NL, Chase GA, Konvicka K, Madden PA, Breslau N, Johnson EO, Hatsukami D, Pomerleau O, et al. Cholinergic nicotinic receptor genes implicated in a nicotine dependence association study targeting 348 candidate genes with 3713 SNPs. Hum Mol Genet. 2007a;16(1):36–49. doi: 10.1093/hmg/ddl438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccone SF, Pergadia ML, Loukola A, Broms U, Montgomery GW, Wang JC, Agrawal A, Dick DM, Heath AC, Todorov AA, et al. Genetic linkage to chromosome 22q12 for a heavy-smoking quantitative trait in two independent samples. Am J Hum Genet. 2007b;80(5):856–66. doi: 10.1086/513703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccone SF, Rice JP, Saccone NL. Power-based, phase-informed selection of single nucleotide polymorphisms for disease association screens. Genet Epidemiol. 2006;30(6):459–70. doi: 10.1002/gepi.20159. [DOI] [PubMed] [Google Scholar]
- Salminen O, Murphy KL, McIntosh JM, Drago J, Marks MJ, Collins AC, Grady SR. Subunit composition and pharmacology of two classes of striatal presynaptic nicotinic acetylcholine receptors mediating dopamine release in mice. Mol Pharmacol. 2004;65(6):1526–35. doi: 10.1124/mol.65.6.1526. [DOI] [PubMed] [Google Scholar]
- Silverman MA, Neale MC, Sullivan PF, Harris-Kerr C, Wormley B, Sadek H, Ma Y, Kendler KS, Straub RE. Haplotypes of four novel single nucleotide polymorphisms in the nicotinic acetylcholine receptor beta2-subunit (CHRNB2) gene show no association with smoking initiation or nicotine dependence. Am J Med Genet. 2000;96(5):646–53. [PubMed] [Google Scholar]
- Stein LD, Mungall C, Shu S, Caudy M, Mangone M, Day A, Nickerson E, Stajich JE, Harris TW, Arva A, et al. The generic genome browser: a building block for a model organism system database. Genome Res. 2002;12(10):1599–610. doi: 10.1101/gr.403602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub RE, Sullivan PF, Ma Y, Myakishev MV, Harris-Kerr C, Wormley B, Kadambi B, Sadek H, Silverman MA, Webb BT, et al. Susceptibility genes for nicotine dependence: a genome scan and followup in an independent sample suggest that regions on chromosomes 2, 4, 10, 16, 17 and 18 merit further study. Mol Psychiatry. 1999;4(2):129–44. doi: 10.1038/sj.mp.4000518. [DOI] [PubMed] [Google Scholar]
- Tapper AR, McKinney SL, Nashmi R, Schwarz J, Deshpande P, Labarca C, Whiteaker P, Marks MJ, Collins AC, Lester HA. Nicotine activation of alpha4* receptors: sufficient for reward, tolerance, and sensitization. Science. 2004;306(5698):1029–32. doi: 10.1126/science.1099420. [DOI] [PubMed] [Google Scholar]
- The International HapMap Consortium A haplotype map of the human genome. Nature. 2005;437(7063):1299–320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorgeirsson TE, Geller F, Sulem P, Rafnar T, Wiste A, Magnusson KP, Manolescu A, Thorleifsson G, Stefansson H, Ingason A, et al. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature. 2008;452(7187):638–42. doi: 10.1038/nature06846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wooltorton JR, Pidoplichko VI, Broide RS, Dani JA. Differential desensitization and distribution of nicotinic acetylcholine receptor subtypes in midbrain dopamine areas. J Neurosci. 2003;23(8):3176–85. doi: 10.1523/JNEUROSCI.23-08-03176.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, George AA, Schroeder KM, Xu L, Marxer-Miller S, Lucero L, Lukas RJ. Electrophysiological, pharmacological, and molecular evidence for alpha7-nicotinic acetylcholine receptors in rat midbrain dopamine neurons. J Pharmacol Exp Ther. 2004;311(1):80–91. doi: 10.1124/jpet.104.070417. [DOI] [PubMed] [Google Scholar]
- Zeiger JS, Haberstick BC, Schlaepfer I, Collins AC, Corley RP, Crowley TJ, Hewitt JK, Hopfer CJ, Lessem J, McQueen MB, et al. The neuronal nicotinic receptor subunit genes (CHRNA6 and CHRNB3) are associated with subjective responses to tobacco. Hum Mol Genet. 2008;17(5):724–34. doi: 10.1093/hmg/ddm344. [DOI] [PubMed] [Google Scholar]