Abstract
The SIL gene expression is increased in multiple cancers and correlates with the expression of mitotic spindle checkpoint genes and with increased metastatic potential. SIL regulates mitotic entry, organization of the mitotic spindle and cell survival. The E2F transcription factors regulate cell cycle progression by controlling the expression of genes mediating the G1/S transition. More recently E2F has been shown to regulate mitotic spindle checkpoint genes as well. As SIL expression correlates with mitotic checkpoint genes we hypothesized that SIL is regulated by E2F. We mined raw data of published experiments and performed new experiments by modification of E2F expression in cell lines, reporter assays and chromatin immunoprecipitation. Ectopic expression or endogenous activation of E2F induced the expression of SIL, while knockdown of E2F by shRNA, downregulated SIL expression. E2F activated SIL promoter by reporter assay and bound to SIL promoter in-vivo. Taken together these data demonstrate that SIL is regulated by E2F. As SIL is essential for mitotic entry, E2F may regulate G2/M transition through the induction of SIL. Furthermore, as silencing of SIL cause apoptosis in cancer cells, these finding may have therapeutic relevance in tumors with constitutive activation of E2F.
Keywords: SIL, E2F, Mitosis
The SIL gene, (SCL Interrupting Locus), located on chromosome 1, encodes a cytosolic protein that have a critical role in cell growth and proliferation 1 2 3. SIL is tightly regulated during the cell cycle. Its mRNA expression is higher in rapidly proliferating cells 4 and tissues, and it decreases rapidly during terminal differentiation 5. The SIL protein a is phopshorylated upon transition into mitosis and is degraded upon exit from mitosis4, 6 SIL is necessary for mitotic entry and proper organization of the mitotic spindle 3 7.
SIL critical role in cell growth and proliferation lead us to examine its role in tumorgenesis. Increased expression of SIL is observed in multiple types of cancer and is associated with metastatic spread8. SIL is essential for cancer cells survival as knockdown of SIL resulted in marked delay in mitotic entry coupled with apoptosis, in-vitro and in-vivo 3. SIL expression in tumors is correlates significantly with the expression pattern of the mitotic spindle checkpoint genes and with, high histopathological mitotic index. We were interested in elucidating the transcriptional basis for the co-regulation of SIL with the expression of other mitotic spindle checkpoint genes.
Numerous studies have shown the E2F family of proteins to be aberrantly activated in multiple cancers. The E2F transcription factors have been demonstrated to control gene expression necessary for cell cycle progression, particularly for the G1/S transition. Recently a role for E2F in regulating mitosis has emerged. Specifically, E2F1 was shown to induce the transcription of genes coding mitotic checkpoint regulator proteins, such as MAD2. As SIL expression is highly correlated with the expression of MAD2, 3 8, we hypothesized that SIL may be a target of E2F.
Materials and Methods
SIL regulation by E2F induction
WI38 human embryonic lung fibroblasts stably expressing E2F1 fused to the ligand binding domain of estrogen receptor were induced for E2F1 activation with 4-hydroxytamoxifen (OHT) (300 nM) as published 9.
PCR
In each 50 µL PCR reaction; 50–100 ng DNA sample, 15 pmol of the 5' and 3' oligonucleotide primers, and 1.5 U Super-Therm DNA polymerase were used. Conditions: initial denaturation for 5 minutes at 95°C, followed by 35 cycles of 60 seconds at 95°C, 60 seconds at 58°C, and 90 seconds at 72°C using a MJ Research PTC-100 thermal cycler. After the last cycle, an additional extension step of 10 minutes at 72°C was performed. Appropriate positive and negative controls were included in all experiments
The oligonucleotides used for amplification:
Hu-SIL
F- 5'–GACTACTTCAGGCACAGATTC-3' R- 5' –ATGCATGCCAACACACTG-3'
Cell culture
Drosophilla melangogaster SL2 cells (obtained from A.Black) were cultured at room temperature in Schneider’s Drosophila medium (Life Technologies) supplemented with 10% fetal calf serum (Rehatuin Intergen). 293T cell line was grown in DMEM supplemented with 10% heatinactivated fetal bovine serum, penicillin–streptomycin (100U and 100 mcg/ml, respectively) and 2mM glutamine (Life Technologies). WI38 human embryonic lung fibroblasts as published 9.
Infection of the shRNA for E2F1
293T cell line was transfected with shRNA for E2F1 (5’ GACGTGTCAGGACCTTCGT) 9. in pSuper reterovirus plasmid together with the gag pole of the Maloni virus, The viral supernated was harvested 48 hours later, filtered through 0.45 micron mixed with u 8 mcg/ml polybrene and added to a T-Cell leukemic cell- line, plated a day before on a 6 well plate at a concentration of 2× 10 5. The cells were centrifuged 45 minutes, 1800 RPM at room temperature. The procedure was repeated 24 hours later. The infected cells were selected with Puromycin 10 mcg/ml for 10 days before the extraction of RNA.
Reporter Assay
Plasmids
a 2.3 kb fragment encompassing the SIL transcription initiation site was subcloned into the Xba I site of the chloramphenicol acyltransferase (CAT) reporter plasmid, p CAT Basic (Promega) to generate pCATSIL. PMINCATSIL was prepared by exonuclease III digestion.
Transfection of SL2 Cells
SL2 cells were transected by calcium- phosphate coprecipitation with five mcg of DNA from each plasmid (with the exception of E2F1- O.2 mcg). A truncated adenovirus 10 was cotransfected as control for transfection efficiency. The total amount of transfected DNA was adjusted to 20 mcg with pBLUESCRIPT II. Cells were harvested after 2 days, pelleted at 1000 × g and resuspended in Reporter Lysis Buffer (Promega). The lysates were analyzed according to the protocol. All CAT data was normalized to luciferase activity to control for transfection efficiency. The fold activation was calculated based on pCAT Basic expression.
Chromatin immunoprecipitation
was performed as described 9 The PCR was preformed using these oligos:
SIL promoter
F- 5'- CCGCAGTTCTCCAAGAAGAC- 3' R-5'GAACTGAGGCGGCAAAC- 3'
Exon 12 of SIL
F- 5’GAGACACTGCAAAGTAAGACAG3’ R-5’GTGGAGGGTCTTATAGGATACTC3’
Results
Bioinformatic analysis of published data suggests that SIL is an E2F target gene
The finding that MAD2, a well characterized mitotic check point gene that is co-expressed with SIL in cancer cells, is a target of E2F1 11 lead us to hypothesize that SIL might be an E2F1 target gene. We interrogated the raw microarray data of several studies done on both human and mice cell lines, designed to find E2F1 direct target genes 12 13 14. We found that either ectopic overexpression of E2F1 or silencing the Rb gene in several cell lines, result in a rise of both mouse and human SIL, together with other known E2F1 target genes. Table 1 summarizes the results from Semizarov paper focusing on E2F induced mitotic genes in human cell lines together with SIL results retrieved by us from the supplementary data (table 1) 13. Thus, mining of publicly available data reveals that SIL is induced in a similar pattern of response with known E2F1 induced genes necessary to insure a proper G2/M transition.
Table 1. Selected Genes induced upon E2F Activation.
Selected genes induced upon Rb knockdown: The table is based on the analysis of microarray data from the Semizarov paper 2004, concerning genes induced upon Rb knockdown and focusing specifically on SIL and known mitotic genes. The table shows the increase in gene expression levels represented by the fold change (FC), and the standard deviation, (SD) post suppression of Rb. The last column indicates the presence of an E2F binding site in the promoter as analyzed by Semizarov [9].
| Name | FC | SD | E2F site |
|---|---|---|---|
| SIL | 2.93 | 0.36 | + |
| ZWINT | 3.64 | 0.24 | + |
| TTK | 3.6 | 0.44 | + |
| MAD2 | 4.16 | 0.95 | + |
| Cyclin B | 3.66 | 0.22 | − |
| Cyclin E | 5.27 | 0.81 | + |
SIL is induced by E2F in vitro
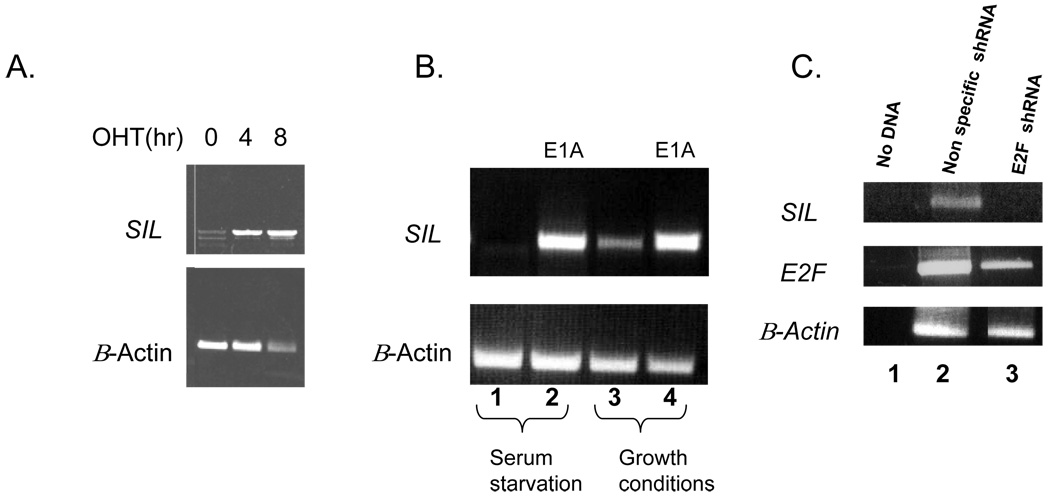
To examine if SIL is induced by activation of E2F we have utilized our previously published human WI-38, embryonic lung fibroblasts engineered to conditionally express E2F1 after treatment 4-hydroxytamoxifen (OHT) 9. Activation of E2F by OHT was associated with increased mRNA levels of SIL (Figure 1A). As an alternative to ectopic expression of E2F we wished to examine the effect of activation of the endogenous E2F1 on SIL. E1A is a viral oncoprotein that, when expressed, binds to and inhibits Rb and consequently activates the endogenous E2Fs. As we have previously shown, SIL is not expressed in serum starved growth arrested fibroblasts (Figure 1B, lane 1). Transfection of E1A resulted in marked expression of SIL mRNA, despite the serum starvation (Figure 1B, lane 2). Similarly the expression of SIL markedly increased in proliferating fibroblasts after activation of the endogenous E2F1 by E1A (Figure 1B, lane 4 in comparison to lane 3). To further substantiate the relationship between E2F1 and SIL we infected T- cell leukemic cell line with either shRNA for E2F1 or with a nonspecific shRNA. As can be seen in Figure 1C knockdown of E2F was associated with reduction in SIL expression. Taken together, these results suggest that expression of SIL is regulated by E2F1.
Figure 1. SIL is regulated by E2F1 in vitro.
A. SIL levels rise post induction with ectopic E2F1; PCR results WII38, embryonic lung fibroblasts, expressing ER-E2F1 were kept in serum starvation in 0.1% FCS containing medium for 24 hours after which E2f1 E2F1 was activated for the indicated times-0, 4hr, 8hr.
B. SIL levels rise post induction of endogenous E2F by E1A; In the first two lanes, WI38 cells were grown under serum starvation conditions; in 0.1% FCS containing medium, while in lanes 3 and 4, cells were grown under growth conditions, (MEM plus 7.5% FCS, 2 mM L-glutamine, and 1 mM sodium pyruvate). Lane 2 and 4 show SIL levels in WI-38 post induction with E1A.
C. shRNA for E2F1 downregulates SIL 1. No DNA 2.Control- unspecific shRNA 3. shRNA for E2F1.
E2F binding sites in SIL promoter are functional in vitro and in vivo
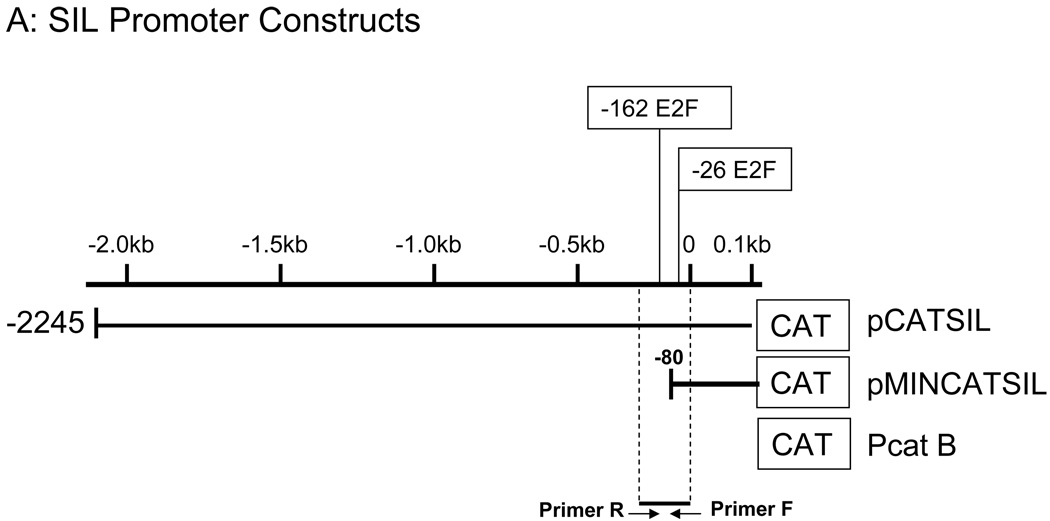
We next wanted to examine if E2F induce SIL expression directly through activation and binding to its promoter. SIL promoter 15 has two potential E2F1 binding sites in a region conserved between human and mouse at −162 and −26 from the transcription initiation site. The upstream site is a 6/8 match to the murine sequence while the downstream site is not well conserved. However, this binding site is a 7/8 match with E2F binding site in N-MYC. 16.
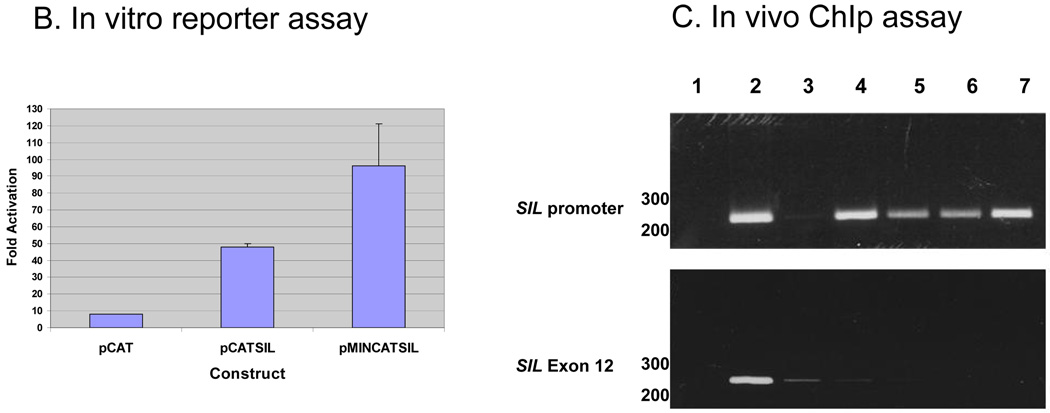
To determine if these binding sites are functionally active on SIL promoter, we first performed a reporter assay using either SIL whole promoter, pCATSIL, or a deleted construct possessing only −80 nucleotide before the start transcription initiation site, pMINCATSIL (Figure 2A). Only one E2F site is positioned within the SIL minimal promoter vector. To determine whether E2F could modulate SIL promoter activity we transfected either pCATSIL or PMINCATSIL into D. melangaster SL2 cells either alone or with an E2F1 coexpression vector. The SIL vectors alone had very low activity in SL2 cells in the absence of E2F. After correcting for the fold activation by the promotorless construct, pCATBasic, the coexpression of E2F1 with pCATSIL resulted in a 6-fold increased activation while coexpression of E2F1 with pMINCATSIL lead to 12-fold increased in CAT activity (Figure 2B).
Figure 2. SIL Promoter is regulated by E2F1 In vitro and In vivo.
A. SIL promoter constructs and designed primers
B. Reporter Assay- E2F1 overexpression activates SIL promoter. 5 mcg of either pCATSIL or pMINCATSIL with the minimal promoter were transfected in Drosophila melanogaster SL2 cells alone or with 0.2 mcg of an E2F1 coexpression vector. CAT activity was measured, background activity from mock transfection controls was substracted and the adjusted activity was normalized to luciferase activity. Each bar represents the mean of at least three transfections.
C. Chromatin immunoprecipitation - E2F transcription factors bind SIL promoter in vivo; PCR for SIL promoter and exon 12 as control 1. no DNA 2. Input before IP 3. anti- HA 4. anti- E2F1 5. anti- E2F2 6. anti- E2F3 7. anti- E2F4.
To examine if E2F transcription factors are bound to the SIL promoter in-vivo we performed a chromatin immunoprecipitation assay (ChIp). Chromatin from Jurkat cells (human T lymphocyte cell line) crosslinked and immunoprecipitated with antibodies against E2F1, E2F2, E2F3, E2F4, and HA was used as a template for PCR amplification of SIL. Negative (no DNA) and positive (input DNA representing 0.2% of total input chromatin) control amplifications are shown (Figure 2C). SIL primers were constructed from the promoter containing an E2F1 binding site, and also from exon 12, not containing an E2F1 binding site (negative control). All E2F factors, in particular E2F1 and E2F4 were found to be bound to the SIL promoter. The results show that E2F binding sites in SIL promoter are functional in vitro and in-vivo, especially for E2F1 and E2F4.
Discussion
The E2F transcription factors are aberrantly activated in cancer. Recent studies have revealed an expanded role of the E2F in cell cycle regulation beyond its function in regulating G1 to S transition and suggest it plays a role also in regulating G2/M transition 17.18. Specifically it has been shown to regulate the MAD2 spindle checkpoint gene. MAD2 is an essential component of the spindle checkpoint that blocks premature sister chromatid separation during metaphase. E2F1 activation of MAD2 expression has been shown to be critically important for the cancer phenotype. We have recently shown that SIL is overexpressed in a large variety of cancers and that its expression is essential for mitotic entry and cell survival 3 8. SIL expression is highly correlated with the expression of other mitotic checkpoint genes, such as MAD2. Here we confirmed the hypothesis that E2F regulates the expression of SIL.
Analysis of raw databases of published micro-array experiments revealed that SIL could be a target gene of E2F1 together with other genes upregulated during the G2/M phase of the cell cycle 12 13 14 8. We then experimentally demonstrated that SIL is regulated by E2F1 and that E2F1 and E2F4 bind to SIL promoter.
Although SIL general role in growth and development has been observed in mouse and zebra fish mutants 2, 19, its function has been unknown. We have recently shown that SIL is an important mitotic regulator 3 6. It is phosphorylated during mitosis and degraded upon transition to G1. Silencing of SIL demonstrated that it is needed for mitotic entry and activation of the CDK1/Cyclin B complex. A recent study by Pfaff et al,7 demonstrated that SIL localizes to the mitotic spindle poles during metaphases and is essential for proper organization of the mitotic spindle. Together these studies establish SIL as a major mitotic regulator.
SIL is the second mitotic regulator, after MAD2, which has been unequivocally shown to be regulated by E2F1. Co-regulation of S phase genes and G2/M genes by the same transcription factor ensures proper coordination of DNA replication with preparation of mitosis. Accordingly, deregulation of this activity in cancer leads to enhanced DNA replication and defects in the mitotic checkpoint. SIL regulation by E2F1 could have implications in many types of cancers, since most cancers display an aberrant expression of the E2F pathway. Since SIL is required for survival of cancer cells 3, drugs targeting E2F1 may mediate their anticancer activity through silencing of SIL. More specifically, in T-Cell-ALL where SIL promoter causes aberrant expression of the oncogenic SCL, blockage of E2F1 may reverse the leukemia by counteracting the activity of the SIL promoter.
Supplementary Material
Acknowledgments
Source of Support- This study was supported by research grants from the Israel Science Foundation, the Israel Cancer Research Foundation, the US-Israel Binational Science Foundation and the Intramural Research Program of the NIH, NCI
This work was preformed in partial fulfillment of the requirements for the PhD degree of Ayelet Erez, Sackler Faculty of Medicine, Tel-Aviv University
Bibliography
- 1.Aplan PDLD, Ginsberg AM, Cossman J, Bertness VL, Kirsch IR. Disruption of the human SCL locus by "illegitimate" V-(D)-J recombinase activity. SCIENCE. 1990;250(4986):1426–1429. doi: 10.1126/science.2255914. [DOI] [PubMed] [Google Scholar]
- 2.Izraeli S, Lowe LA, Bertness VL, Good DJ, Dorward DW, Kirsch IR, et al. The SIL gene is required for mouse embryonic axial development and left- right specification. Nature. 1999;399(6737):691–694. doi: 10.1038/21429. [DOI] [PubMed] [Google Scholar]
- 3.Erez A, Castiel A, Trakhtenbrot L, Perelman M, Rosenthal E, Goldstein I, et al. The SIL gene is essential for mitotic entry and survival of cancer cells. Cancer Res. 2007;67(9):4022–4027. doi: 10.1158/0008-5472.CAN-07-0064. [DOI] [PubMed] [Google Scholar]
- 4.Izraeli S, Colaizzo-Anas T, Bertness VL, Mani K, Aplan PD, Kirsch IR. Expression of the SIL gene is correlated with growth induction and cellular proliferation. Cell Growth Differ. 1997;8(11):1171–1179. [PubMed] [Google Scholar]
- 5.Collazo-Garcia NSP, Aplan PD. Cloning and characterization of a murine SIL gene. Genomics. 1995;30(3):506–513. doi: 10.1006/geno.1995.1271. [DOI] [PubMed] [Google Scholar]
- 6.Campaner SKP, Izraeli S, Kirsch IR. Sil phosphorylation in a Pin1 binding domain affects the duration of the spindle checkpoint. Mol Cell Biol. 2005;25(15):6660–6672. doi: 10.1128/MCB.25.15.6660-6672.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pfaff KL, Straub CT, Chiang K, Bear DM, Zhou Y, Zon LI. The Zebra fish cassiopeia Mutant Reveals that SIL Is Required for Mitotic Spindle Organization. Mol Cell Biol. 2007;27(16):5887–5897. doi: 10.1128/MCB.00175-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Erez APM, Hewitt SM, Cojacaru G, Goldberg I, Shahar I, Yaron P, Muler I, Campaner S, Amariglio N, Rechavi G, Kirsch IR, Krupsky M, Kaminski N, Izraeli S. Sil overexpression in lung cancer characterizes tumors with increased mitotic activity. Oncogene. 2004;23(31):5371–5377. doi: 10.1038/sj.onc.1207685. [DOI] [PubMed] [Google Scholar]
- 9.Chaussepied M, Ginsberg D. Transcriptional regulation of AKT activation by E2F. Mol Cell. 2004;16(5):831–837. doi: 10.1016/j.molcel.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 10.Shiaw-Yih Lin ARB, Kostic Dusan, Pajovic Sanja, Hoover Carol N, Azizkhan JC. Cell Cycle-Regulated Association of E2F1 and Sp1 Is Related to Their Functional Interaction. Molecular and Cellular Biology. 1996:1668–1675. doi: 10.1128/mcb.16.4.1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hernando ENZ, Juan G, Diaz-Rodriguez E, Alaminos M, Hemann M, Michel L, Mittal V, Gerald W, Benezra R, Lowe SW, Cordon-Cardo C. Rb inactivation promotes genomic instability by uncoupling cell cycle progression from mitotic control. nature. 2004;12(430):797–802. doi: 10.1038/nature02820. [DOI] [PubMed] [Google Scholar]
- 12.Muller HBA, Vernell R, Moroni MC, Christians F, Grassilli E, Prosperini E, Vigo E, Oliner JD, Helin K. E2Fs regulate the expression of genes involved in differentiation, development, proliferation, and apoptosis. Genes Dev. 2001;15(3):267–285. doi: 10.1101/gad.864201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Semizarov DKP, Fesik S. siRNA-mediated gene silencing: a global genome view. Nucleic Acids Res. 2004;32(13):3836–3845. doi: 10.1093/nar/gkh714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vernell RHK, Muller H. Identification of target genes of the p16INK4A-pRB-E2F pathway. Biol Chem. 2003;278(46):46124–46137. doi: 10.1074/jbc.M304930200. [DOI] [PubMed] [Google Scholar]
- 15.Colaizzo-Anas TAP. Cloning and characterization of the SIL promoter. Biochim Biophys Acta. 2003;1625(2):207–213. doi: 10.1016/s0167-4781(02)00597-3. [DOI] [PubMed] [Google Scholar]
- 16.JR N. E2F: a link between the Rb tumor suppressor protein and viral oncoproteins. Science. 1992;258(5081):424–429. doi: 10.1126/science.1411535. [DOI] [PubMed] [Google Scholar]
- 17.Ishida SHE, Zuzan H, Spang R, West M, Nevins JR. Role for E2F in control of both DNA replication and mitotic functions as revealed from DNA microarray analysis. Mol Cell Biol. 2001;21(14):4684–4699. doi: 10.1128/MCB.21.14.4684-4699.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Polager SKY, Berkovich E, Ginsberg D. E2F up-regulate expression of genes involved in DNA replication, DNA repair and mitosis. Oncogene. 2002;21(3):437–446. doi: 10.1038/sj.onc.1205102. [DOI] [PubMed] [Google Scholar]
- 19.Golling GAA, Sun Z, Antonelli M, Maldonado E, Chen W, Burgess S, Haldi M, Artzt K, Farrington S, Lin SY, Nissen RM, Hopkins N. Insertional mutagenesis in zebrafish rapidly identifies genes essential for early vertebrate development. Nat Genet. 2002;31(2):135–140. doi: 10.1038/ng896. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.