Abstract
The cellular metabolism of doxorubicin generates reactive oxygen species with significant potential to damage DNA. Such DNA damage can result in mutations if not adequately repaired by cellular DNA repair pathways. Secondary malignancies have been reported in patients who have received doxorubicin-containing chemotherapeutic regimens; however, the underlying molecular mechanism(s) to explain the development of these tumors remains under active investigation. We have previously demonstrated the presence of DNA bases modified by oxidation in the peripheral blood mononuclear cells of patients with breast cancer following treatment with doxorubicin. In those studies, doxorubicin was administered by continuous infusion over 96 hours to minimize the risk of cardiac toxicity. To evaluate potential mechanisms underlying doxorubicin-induced DNA base oxidation in non-malignant tissues, MCF-10A breast epithelial cells were cultured for 96 hours with the same doxorubicin concentration achieved in vivo (0.1 μM). During doxorubicin exposure, MCF-10A cells underwent growth arrest and apoptosis, developed elevated levels of reactive oxygen species, and demonstrated a time-dependent and significant increase in the levels of 11 oxidized DNA bases, as determined by gas chromatography/mass spectroscopy. Diminished expression of DNA repair enzymes was also observed over the same time course. Thus, clinically achievable concentrations of doxorubicin induce a level of oxidative stress in MCF-10A cells that is capable of oxidizing DNA bases and significantly altering cellular proliferation.
Keywords: doxorubicin, DNA damage, reactive oxygen species, apoptosis, chemotherapy, secondary malignancy
1. Introduction
The anthracycline antibiotic doxorubicin (DOX) is widely used in the treatment of adult and pediatric solid tumors and hematologic malignancies, especially breast cancer and lymphoma. Unfortunately, the cytotoxic effects of DOX, although desirable in malignant cells, may also cause delayed damage to normal tissues, such as the bone marrow. Secondary malignancies, primarily acute myelogenous leukemia (AML), have been reported 1 to 3 years following treatment of primary tumors with DOX in combination with other chemotherapeutic agents or radiotherapy [1] [2] [3].
DOX has long been recognized as a mutagen and carcinogen [4]. Possible mechanisms of DOX cytotoxicity for tumor cells that may also be related to the mutagenic potential of the drug include the formation of DNA-DOX-topoisomerase II cleavable complexes, and DOX-related inhibition of DNA helicases [4]. DOX is also subject to cycles of reduction and oxidation by flavin dehydrogenases that, in the presence of molecular oxygen and transition metals (particularly iron), lead to the formation of reactive oxygen species (ROS) [5] [6] [7]. ROS (including the superoxide anion, O2−•; hydrogen peroxide, H2O2; and hydroxyl radical, •OH) are also generated endogenously as a by-product of normal cellular metabolism [8]; however, DOX exposure can dramatically enhance intracellular reactive oxygen formation [4].
The more pathological aspects of ROS production are related to their ability to cause oxidative damage to nuclear and mitochondrial DNA [9]. Hydroxyl radicals can either abstract a hydrogen atom from DNA bases or sugars or undergo addition to double bonds of DNA bases. Unstable intermediates formed in this way produce a number of stable, modified DNA bases after consecutive molecular transformations. DNA base radicals formed by the attack of the hydroxyl radical may also react with amino acids of neighboring proteins to form covalent DNA–protein crosslinks [10]. Efficient and accurate repair of such DNA damage is critical to the prevention of mutations that may initiate carcinogenesis. Non-malignant tissues utilize an extensive system of antioxidant molecules, ROS catalytic enzymes, and DNA repair enzymes to limit the DNA damage caused by ROS [11].
In prior experiments, we used gas chromatography/mass spectrometry (GC/MS) to demonstrate that redox cycling of DOX catalyzed by NADH dehydrogenase resulted in significantly elevated levels of 12 promutagenic oxidized DNA bases in isolated human chromatin [12]. Each of these 12 oxidized bases had been identified previously as products formed by the interaction of ionizing radiation-generated free radicals with DNA bases [13]. More recently, we observed elevated levels of nine oxidized DNA bases in the peripheral blood mononuclear cells (PBMCs) of patients with breast cancer following a 96-hour intravenous infusion of DOX as the first agent of a multi-agent chemotherapy regimen [14]. The steady-state plasma level of DOX achieved in these patients, who received a total DOX dose of 165 mg/m2, was 0.1 μM. DOX was administered over this extended period to minimize the risk of cardiac toxicity, a potentially life-threatening toxicity associated with DOX treatment [15].
Based on these results and the association of DOX therapy with the development of secondary malignancies, we sought to determine in the current study whether DOX exposure produced DNA base oxidation in non-malignant cells, and, if products of DNA base oxidation were observed, to gain insights into the mechanism(s) of base damage production. We found that immortalized, non-transformed MCF-10A breast epithelial cells cultured for 96 hours in clinically achievable concentrations of DOX did not proliferate and contained both elevated amounts of ROS and of 11modified DNA bases. Furthermore, the expression of several DNA base excision repair (BER) enzymes was diminished in DOX-exposed cells at the same time that oxidized DNA bases were measurable. These results demonstrate that treatment with pharmacologically relevant concentrations of DOX leads to the formation of ROS in normal breast epithelial cells, as well as DNA damage typical of ROS exposure, suggesting an important potential mechanism of DOX-mediated mutagenesis and cell death.
2. Materials and Methods
2.1. Reagents
Pure samples of 5-hydroxy-5-methylhydantoin (5-OH-5-MeHyd), 5-hydroxyuracil (5-OH-Ura), 5-hydroxycytosine (5-OH-Cyt), thymine glycol (ThyGly), 5,6-dihydroxyuracil (5,6diOH-Ura), 4,6-diamino-5-formamidopyrimidine (FapyAde), 8-hydroxyadenine (8-OH-Ade), Xanthine (Xan), and 2,6-diamino-4-hydroxy-5-formamidopyrimidine (FapyGua) were kindly provided by Dr. Miral Dizdaroglu from the National Institute of Standards and Technology, Gaithersburg, MD. 2-amino-6, 8-dihydroxyguanine (8-OH-Gua) was purchased from Sigma Chemical Co. (St. Louis, MO). 2-Hydroxy-6-aminopurine (2-OH-Ade) was purchased from ACROS Organic (Fairlawn, NJ). N, O-bis (trimethylsilyl) trifluoroacetamide (BSTFA) and acetonitrile were purchased from Pierce (Rockford, IL). Thymine-alfa1, alfa2, alfa3, 6-d4 was obtained from MSD Isotopes, Merck Chemical Division (St. Louis, MO) and alfa-32P-uridine 5′-triphosphate (10 mCi/mL) was obtained from PerkinElmer Life Sciences (Boston, MA). Non-itemized chemicals were all of the highest possible quality. Only de-ionized, glass-distilled water was used in this study. Unless specified, antibodies were obtained from Novus Biologicals Inc., (Littleton, CO). Anti-actin polyclonal antibody and HRP-conjugated anti-goat IgG were obtained from Santa Cruz Biotechnology, Inc., (Santa Cruz, CA).
2.2. Cell Culture for DNA Preparation and Flow Cytometry
MCF-10A cells were obtained from ATCC (Manassas, VA). The cells form an adherent monolayer when cultured in DMEM/F-12 medium enriched with 5% equine serum and supplemented with antibiotics (penicillin, streptomycin, and amphotericin), epidermal growth factor, cholera toxin, insulin, and hydrocortisone. The doubling time of the cells is approximately 48 hours. Cell synchronization in the G0/G1 phase was achieved by culturing cells to 100% confluence for 1 to 3 days in the same medium and was confirmed by flow cytometric analysis. Synchronized cells were subsequently trypsinized and split into T-75 flasks. All cells remained sub-confluent during the subsequent culture period. Split cells were cultured for 2 to 3 hours until attached, trypsinized again, collected by centrifugation at 1000 × g, rinsed once with phosphate buffered saline (PBS), and centrifuged again. Nuclear lysis buffer (NLB) was added (NLB: 10 mM Tris HCl, pH 8.0, 2 mM EDTA, 0.4 M NaCl; 0.6 mL of NLB for each 3 × 106 cells), and the samples were kept at −80 °C until further processing. Culture medium containing unattached cells was divided into normal medium or normal medium supplemented with 0.1μM DOX. At the indicated time intervals, control and DOX-treated cells were collected as described and stored at −80 °C until use. For flow cytometric analysis, at each time point 1 × 106 trypsinized cells were collected by centrifugation for 6 minutes at 200 × g, resuspended in 5 mL PBS, and centrifuged again. Cells were resuspended in 0.5 mL PBS and stored in 70% ethanol at 4 °C until use.
2.3. Flow Cytometric Analysis
All flow cytometric analyses were performed using a MoFlo MLS flow cytometer with Summit software (DakoCytomation, Fort Collins, CO). Data were acquired using dual laser excitation. Light scatter signals were acquired after excitation from a HeNe laser (Melles Griot, Carlsbad, CA). Filters were purchased from Omega Optical (Brattleboro, VT). Cell pellets were suspended in 1 mL propidium iodide (PI) staining solution (10 mL of 0.1% [v/v] Triton X-100 in PBS with 2 mg DNase-free RNase A, 200 μL of 1 mg/mL PI). Fluorescent excitation was performed at 488nm with an Innova-90 Argon laser (Coherent, Santa Clara, CA) at 500mW. The PI signal was collected with a 640LP filter.
2.4. Detection of DOX-Induced Intracellular ROS in MCF-10A Cells
Oxidative products represented by free 2′, 7′-dichlorofluorescein (DCF) fluorescence were measured by flow cytometry [16]. G0/G1 phase-synchronized control cells and DOX-treated cells, each cultured for 24, 48, 72, and 96 hours, were harvested and washed once with Ringers’ solution (137 mM NaCl, 2.68 mM KCl, 1.84 mM CaCl, 1.03 mM MgCl2, 5.55 mM dextrose, 11.91 mM NaHCO3, 0.44 mM NaH2PO4). Cell pellets were resuspended in 2 mL of Ringers’ solution and then divided into two 1-mL aliquots. 10 μL of 1 μM 5-(and –6)-chloromethyl- 2′, 7′-dichlorodihydrofluorescin diacetate and mixed isomers (DCF-DA; Molecular Probes, Eugene, OR) in DMSO was added to one aliquot. All samples were kept at room temperature for 60 minutes; fluorescence of the samples without DCF-DA probe was also measured. DCF emission was measured through a 530DF30 filter.
2.5. DNA Preparation
The procedure for DNA preparation without exposure to phenol was adapted from Cheng et al. [17]. Briefly, control and DOX-treated cells stored in NLB were thawed and mixed with 40 μL of 10% SDS and 100 μL of 2 mg/mL proteinase K, freshly prepared in 1% SDS and 2 mM EDTA (amounts for 3 × 106 cells), and then digested overnight at 37 °C with gentle shaking. Solubilized nuclei were incubated with boiled RNase (final concentration 0.27 mg/mL) at 50 °C for 2 hours. Proteins were precipitated by adding 6 M NaCl (saturated solution) to a final concentration of 1.5 M. Samples were shaken vigorously for 30 seconds and then centrifuged at 10,000 × g for 10 minutes at room temperature. DNA was precipitated in ethanol, dried, and resuspended in water at room temperature.
2.6. Measurement of Oxidative DNA Base Modifications by GC/MS
DNA was prepared for modified base analysis according to a well-described GC/MS protocol [18]. Each set of cell samples contained the same amount of DNA by OD260 estimation. To establish the amount of DNA in each sample, a known amount of thymine-d4 (T-d4) was added before hydrolysis. The relative molecular response factor (RMRF) for thymine using T-d4 as internal standard was determined, and then the amount of thymine (nanomoles) in each hydrolyzed sample was calculated as described [14]. A known amount of azaT was also added to each sample before hydrolysis as an internal standard for quantitative determination of the modified DNA bases 5-OH-Hyd, 5-OH-5-MeHyd, 5-OH-Ura, 5-OH-Cyt, Thy Gly, 5,6diOH-Ura, FapyAde, 8-OH-Ade, Xan, FapyGua, 2-OH-Ade, and 8-OH-Gua. In separate experiments, RMRFs for all damaged bases were obtained using azaT and pure samples of modified bases as described [14]. For GC/MS measurements, all samples in a series were lyophilized, hydrolyzed with 60% formic acid at 140 °C for 45 minutes, lyophilized again, and derivatized with a mixture of BSTFA/acetonitrile (4:1) at 130 °C for 45 minutes. A Shimadzu GC/MS-QP5000 with GC-17A gas chromatograph equipped with auto injector/auto sampler AOC-20 and CLASS-5000 software was used for these studies. The injector port was kept at 250 °C, interface at 280 °C, and oven temperature at 150 °C for 2 minutes followed with 8 °C/minute gradient up to 260 °C; the oven was then kept at 260 °C for an additional 2 minutes. A Hewlett-Packard Ultra 2 (cross-linked methyl-siloxane) column (12 meters, 0.20 mm ID, 0.33 μm film) was used with helium as carrier gas. Column head pressure was 50 kPa with a 10:1 split. The 12 oxidative products of DNA bases, azaT, thymine, and T-d4 were detected simultaneously in a single chromatogram.
2.7. RNA Preparation and Ribonuclease Protection Assay (RPA)
G0/G1-phase synchronized cells cultured with or without DOX were collected after 2, 8, 16, 24, 48, 72, and 96 hours and immediately suspended in 3 mL of RNAzol B (Tel-Test, Inc., Friendswood, TX). RNA was then extracted, dissolved in autoclaved water, and stored at −80 °C. The Multi-Probe RNase Protection Assay System and Human Base Excision Repair Multi-Probe Template Set (BD PharMingen, San Diego, CA) were used to measure changes in mRNA expression levels of DNA repair enzymes G/T mismatch-specific thymine-DNA glycosylase (TDG), uracil-DNA glycosylase (UDG), apurinic/apyrimidinic endonuclease (APE), O(6)-methylguanine-DNA methyltransferase (MGMT), methylpurine-DNA glycosylase (MPG), 8-oxoguanine-DNA glycosylase (hOGG1), and thymine glycol-DNA glycosylase/AP lyase (ENTG), and two cellular controls, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and ribosomal L32. Protected mRNAs were separated by electrophoresis on 5% acrylamide gels. Gels adsorbed to filter paper were exposed to a Phosphor Screen (Molecular Dynamics, Sunnyvale, CA) overnight at room temperature. To quantify mRNA expression, the phosphoimage of an exposed gel was scanned with a Storm 860 scanner (GE Healthcare, Piscataway, NJ), and the density of each protected RNA band was analyzed with ImageQuant software (Molecular Dynamics). Relative abundance of each RNA band was quantified as the ratio to GAPDH or L32 within each sample.
2.8. Preparation of Cell Extracts for Western Analysis
Harvested cells were resuspended in PBS and immediately stored at −80 °C. Once cells from all the time points were collected, lysate buffer (PBSTDS; 1 mL per 20 × 106 cells) with protease inhibitors was added to each sample at room temperature. The composition of PBSTDS buffer with protease inhibitors included the following: 25 mL of 10X PBS, 2.5 mL Triton X-100, 1.25 g sodium deoxycholate, 0.25 g SDS, 12.5 μL leupeptin hemisulfate (10 mg/mL), 10 μL pepstatin A (25 mg/mL), 0.5 mL of 0.5M EDTA, 0.5 mL of 100 mM phenylmethylsulfonyl fluoride (PMSF). PBSTDS buffer was made up to 250 mL with water. Cells were incubated on ice for 60 minutes and then rocked at 4 °C for 1 hour. DNA was sheared by passing the cell suspension through a 21-gauge needle. Samples were centrifuged at 16,000 × g for 10 minutes at 4 °C, and the protein concentration of the supernatant was determined using Advanced Protein Assay Reagent (Cytoskeleton Inc., Denver, CO). Samples were stored at −80 °C until use.
2.9. Western Blot Analysis
The amounts of APE, NTH-1, hOGG1, mtOGG1, MGMT, DNA polymerase ε, and DNA ligase I in the cell extracts were quantified using β-actin as an internal standard. Secondary antibodies used included HRP-conjugated anti-rabbit, anti-mouse, and anti-goat IgG, as appropriate. HeLa whole cell extract (Novus Biologicals) was used as a positive control. Cell extract proteins were separated by electrophoresis on 10% bis-Tris gels with MOPS running buffer. Equal amounts (between 5–10 μg) of whole cell extract protein were loaded for each time point. Proteins were electroblotted onto PVDF transfer membranes and probed using the ECL-Plus Western blotting detection system and Hyperfilm ECL (GE Healthcare). Bands corresponding to DNA repair proteins and actin were quantified using an Eagle Eye instrument and EagleSight Software from Stratagene (La Jolla, CA).
2.10. Monitoring Apoptosis in MCF-10A Cells Using Terminal Transferase dUTP-Biotin Nick End Labeling (TUNEL) Assay Relative to Cell Cycle Progression
For this experiment, subconfluent, unsynchronized cells were employed. Control and DOX-treated cells cultured for 96 hours were collected and washed with 0.1% BSA in Hanks balanced salt solution (HBSS), and then incubated with biotinylated dUTP and terminal transferase in a cobalt-based buffer for 30 minutes at 37°C. Supernatant was removed, and cells were washed with 0.1% BSA-HBSS before incubation with Avidin-FITC reagent (2–4 mole fluorescein isothiocyanate per mole avidin) at room temperature for 30 minutes. After washing with 0.1% Triton X-100 in HBSS, the cells were resuspended in PI solution in HBSS and left at room temperature for 30 minutes before analysis. FITC emission (representing DNA strand breaks related to apoptosis) was measured through a 530DF30 filter. Cell cycle status was determined using PI emission measured through a 640EFLP filter. The two fluorescent signals were separated with 580DRLP and 630DRLP dichroic filters.
2.11. Statistical Methods
Samples from all time points for each experiment were analyzed on the same day to minimize inter-assay variability. Levels of change were normalized to pretreatment values. A two-tailed, paired t-test was used to compare the fold change of treated samples versus control samples. P<0.05 was considered significant.
3. Results
3.1. ROS in MCF-10A Cells During Exposure to 0.1 μM DOX
Levels of ROS were elevated in MCF-10A cells exposed to DOX. Arbitrary fluorescence unit values for DOX-treated and control cells after 24 hours were 3290 and 3097, respectively. After 48 hours, the relative fluorescence unit values for DOX-treated vs. control cells were 2033 vs. 1164. By 72 hours this difference in fluorescence unit values for DOX-treated vs. control cells was 3529 vs. 2619, and by 96 hours the relative ROS levels were 2571 vs. 2545. Cells cultured with and without DOX in the absence of DCF-DA also exhibited fluorescence (autofluorescence in the case of control cells and DOX fluorescence for drug-treated cells); relative fluorescence values for control and DOX-treated MCF-10A cells at 24, 48, 72, and 96 hours were 3.19 vs. 2.52, 2.74 vs. 1.87, 6.29 vs. 1.79, and 6.26 vs. 1.67, respectively.
3.2. GC/MS Identification and Quantitation of Modified DNA Bases in DOX-Treated Cells
The modified DNA base content before DOX treatment is presented with RMRF measurements in Table 1. Average fold increases (±SD) in modified DNA base levels normalized to control values after 24, 48, 72, and 96 hours of DOX treatment from three separate experiments are presented in Table 2. Statistically significant (P<0.05) increases in 2-OH-Ade and FapyGua were observed after 24 hours of DOX treatment and in Xan, 2-OH-Ade, 5-OH-Cyt, and 5-OH-Ura after 48 hours of treatment. The levels of many bases increased with time of exposure; by 96 hours, statistically significant increases were noted at one or more time points for all of the modified bases with the exception of FapyAde.
Table 1.
Experimentally derived relative molar response factors and modified DNA base content in MCF-10A cells prior to doxorubicin exposure
| DNA base | Ion used | RMRF | Modified DNA base level (nM/mg DNA)* | Modified DNA base level (modified bases/106 bases)† |
|---|---|---|---|---|
| 5-OH-5-MeHyd | 331 | 0.51±0.05 | 0.887±0.971 | 284.±311 |
| 5-OH-Hyd | 317 | - | 0.527±0.492 | 169±157 |
| 5-OH-Ura | 329 | 0.61±0.05 | 0.253±0.243 | 81.0±77.8 |
| 5-OH-Cyt | 328 | 0.65±0.16 | 0.407±0.255 | 130±81.6 |
| Thy Gly | 259 | 0.36±0.05 | 0.608±0.493 | 194±158 |
| 5, 6diOH-Ura | 417 | 4.58±0.44 | 1.16±1.14 | 371±365 |
| FapyAde | 354 | 2.02±0.06 | 1.28±0.539 | 410±172 |
| 8-OH-Ade | 352 | 0.88±0.21 | 2.13±0.1.49 | 682±477 |
| Xan | 353 | 0.70±0.27 | 16.8±5.55 | 5376±1776 |
| 2-OH-Ade | 352 | 0.29±0.08 | 0.509±0.379 | 163±121 |
| FapyGua | 442 | 8.09±1.82 | 7.62±4.88 | 2438±1562 |
| 8-OH-Gua | 440 | 15.84±4.86 | 38.5±18.0 | 12320±5760 |
Mean ± SD.
1 nmole modified base/mg DNA is 320 molecules of modified base/106DNA bases.
Table 2.
Modified DNA base content in MCF-10A cells cultured for up to 96 hours with 0.1 μM doxorubicin
| DNA base | 0 h | 24 h | 48 h | 72 h | 96 h |
|---|---|---|---|---|---|
| 5OH-5-MeHyd | 1 | 1.72±0.44 | 2.24±0.88 | 2.24±0.49* | 2.45±0.10* |
| 5-OH-Hyd | 1 | 1.58±0.45 | 1.94±0.72 | 2.14±0.59 | 2.36±0.50* |
| 5-OH-Ura | 1 | 1.36±0.29 | 1.82±0.27* | 2.21±0.30* | 2.21±0.20* |
| 5-OH-Cyt | 1 | 1.29±0.3 | 1.34±0.08* | 1.88±0.25* | 1.56±0.24 |
| Thy Gly | 1 | 1.48±0.31 | 1.14±0.19 | 2.00±0.09* | 3.08±1.39 |
| 5,6diOH-Ura | 1 | 1.63±0.36 | 1.65±0.26 | 2.22±0.48* | 1.73±0.06* |
| FapyAde | 1 | 1.28±0.17 | 1.44±0.43 | 1.65±0.45 | 1.46±0.31 |
| 8-OH-Ade | 1 | 1.55±0.41 | 1.69±0.26* | 2.10±0.43* | 1.87±0.40 |
| Xan | 1 | 1.71±0.43 | 2.05±0.10* | 2.48±0.80 | 2.59±0.74 |
| 2-OH-Ade | 1 | 1.49±0.09* | 1.34±0.36 | 1.84±0.55 | 2.15±0.37* |
| FapyGua | 1 | 1.33±0.12* | 1.23±0.22 | 1.68±0.20* | 2.27±0.62* |
| 8-OH-Gua | 1 | 1.10±0.29 | 1.11±0.22 | 1.80±0.16* | 1.96±0.45 |
P < 0.05 versus control DNA.
3.3. BER Gene Expression and Cell Cycle Progression in MCF-10A Cells During 96 Hours of DOX Treatment
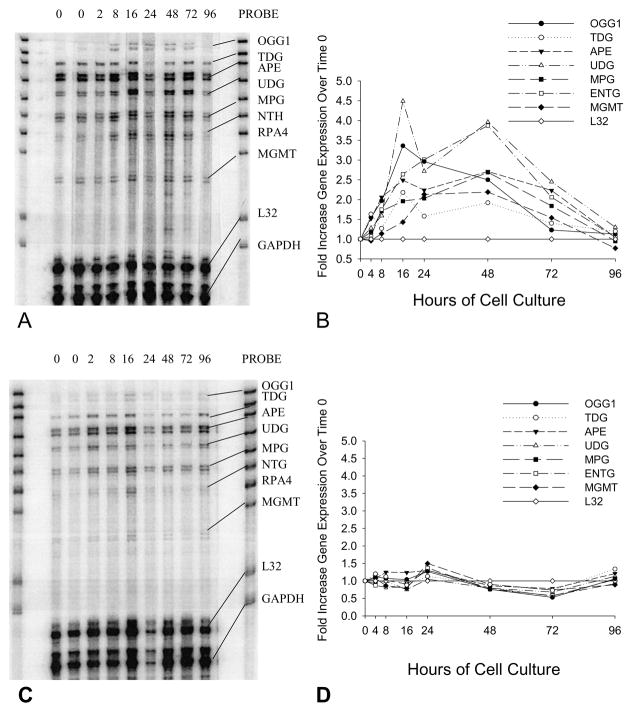
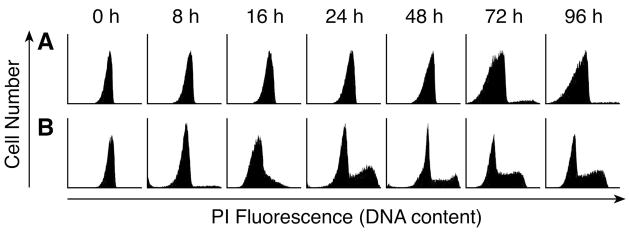
RPA analysis demonstrated that, following release from cell cycle arrest by addition of fresh serum, the expression of seven DNA repair genes increased up to 4.5-fold over the subsequent 96 hours, peaking at 12 to 48 hours following the end of cell cycle synchronization (Fig. 1A and 1B). On the other hand, the expression of the same DNA repair genes in MCF-10A cells cultured in 0.1 μM DOX did not increase following exposure to fresh serum and medium. In fact, their expression appeared to decrease by the 72 hour time point (Fig. 1C and 1D); however, this decreased expression was not statistically significant. Flow cytometric analysis demonstrated that cells synchronized in the G0/G1 phase and then cultured in media without DOX rapidly re-entered the cell cycle and exhibited a cell cycle phase distribution characteristic of exponentially growing cells within 24 hours; this cell cycle phase distribution was maintained for the subsequent 72 hours of culture (Fig. 2B). Cells grown in DOX were viable as determined by trypan blue exclusion and adherence to plastic; however, they remained in the G0/G1 phase during the entire 96-hour treatment period (Fig. 2A). Replacing the media after 96 hours with DOX-free media was not sufficient to allow these cells to recover.
Fig. 1.
RPA results showing changes in the expression of seven base excision repair genes in MCF-10A cells treated without (A) or with (C) 0.1μM DOX for up to 96 hours. Results are representative of multiple experiments. Both cultures were started with cells synchronized in G0/G1 that did not achieve confluence. The PROBE lane shows the bands for each of the probes used in the assay. The probe lengths are greater than the “protected” fragment lengths because probes containing flanking sequences derived from the plasmid do not hybridize with target mRNAs. RP4 (subunit of RPA holoenzyme) expression was not detected in MCF-10A cells under the described experimental conditions. All gene expression values were normalized to the expression of L32 at the 0-hour point and fold increases/decreases over the expression of each gene at time zero were calculated for 2, 8, 16, 24, 48, 72, and 96 hours. (B) Quantification of selected DNA repair gene expression in MCF-10A cells cultured for 2, 8, 16, 24, 48, 72, and 96 hours following release from synchronization by addition of fresh medium. In these control cells, the expression of certain genes (mean ± SD; n=3) increased significantly following release from G1 block, including: OGG1 (2.5 ± 0.1 fold at 48 hours, P<0.01); TDG (2.2 ± 0.4 fold at 16 hours, P<0.05); APE (2.5 ± 0.3 fold at 16 hours, P<0.01); and UDG (4.0 ± 0.7 fold at 48 hours, P<0.02). (D) Quantification of the expression of selected DNA repair genes in MCF-10A cells cultured in the presence of 0.1μM DOX in fresh medium for 2, 8, 16, 24, 48, 72, and 96 hours.
Fig. 2.
Flow cytometric analysis of MCF-10A cells synchronized in the G1/G0 phase and then cultured for up to 96 hours in fresh medium. Increasing PI fluorescence reflects increasing DNA content and is indicative of cell cycle progression. Times for each peak represent the duration since medium was changed. (A) Cell cycle distribution with 0.1μM DOX. (B) Cell cycle distribution with normal medium.
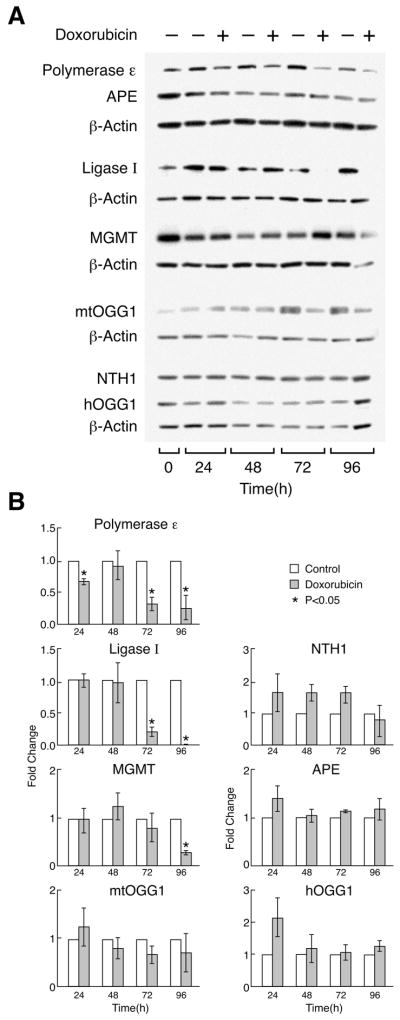
3.4. Levels of DNA Repair Enzyme Protein Expression in MCF-10A Cells During 96 Hours of DOX Treatment
Levels of three of the seven DNA repair enzymes measured by Western analysis (DNA polymerase ε, DNA ligase I, and MGMT), were significantly lower in cells treated with DOX than in control cells by the end of 96 hours of drug exposure (Fig. 3A and 3B). Expression of the other enzymes measured was not significantly different in DOX-treated cells compared to control cells. The largest differences in DNA repair enzyme expression levels were found for DNA polymerase ε and DNA ligase I. After 72 hours, expression of DNA polymerase ε in DOX-treated cells decreased to approximately 33% of the expression level measured in the control cells, and, after 96 hours, it decreased to approximately 25% of that observed with the control cells. For DNA ligase I, decreased expression was more remarkable; after 72 hours, expression in DOX-treated cells was approximately 20% of that measured in control cells, and, after 96 hours, the expression level was approximately 10% of that of control cells.
Fig. 3.
(A) Representative Western blot analysis of seven DNA repair enzyme expression levels in MCF-10A cells treated with or without 0.1 μM DOX. Enzymes measured included APE, NTH1, hOGG1, mtOGG1, MGMT, DNA polymerase ε, DNA ligase I, and anti-actin control. Point 0 indicates the expression of proteins in cells synchronized in G0/G1 before the addition of DOX. (B) Average quantitative representation of Western blot results from three independent experiments. For quantitative analysis, protein bands for the particular enzyme at each time point were normalized to the same protein concentration using actin content as an internal standard. Fold changes in protein expression for each enzyme over the expression levels in the control cells were calculated for each time point. Error bars represent standard deviation.
3.5. Effect of 0.1 μM DOX Treatment for 96 Hours on Apoptosis and Cell Cycle Progression in MCF-10A Cells
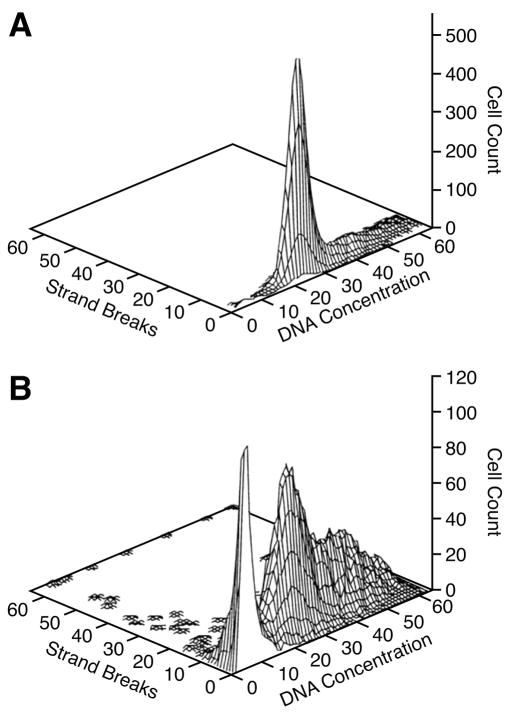
Apoptosis, as determined by TUNEL assay, was observed in MCF10-A cells treated with DOX for 96 hours when compared to untreated cells (Fig. 4); apoptotic cells were found throughout the cell cycle. Concurrent staining with PI also demonstrated a large sub-G0 peak in DOX-treated cells indicative of apoptosis, and a G2 block in cell cycle progression consistent with exposure to DOX [4].
Fig. 4.
Death of unsynchronized MCF-10A cells treated for 96 hours with 0.1 μM DOX relative to the cell cycle. Apoptosis, measured by TUNEL assay, is represented as strand breaks. (A) MCF-10A cells cultured for 96 hours in standard DMEM/F-12 medium. Unsynchronized cells did not reach confluence, and no DNA strand breaks were detected. (B) Apoptotic cells were detected in DOX-treated cells from all phases of the cell cycle; a large sub-G0 peak of apoptotic cells is also visible.
4. Discussion
We previously demonstrated that PBMCs from patients with breast cancer who were treated with a 96-hour infusion of DOX had significantly increased levels of nine oxidized DNA base lesions [14]. The steady-state plasma level of DOX achieved in those patients was 0.1 μM. During DOX treatment of MCF-10A cells in the current experiments, we have attempted to mirror the systemic exposures achieved in our clinical trial. It is recognized, however, that during a continuous DOX infusion in patients with breast cancer that the intravascular serum concentration is much higher than that employed in vitro. On the other hand, the DOX concentration achieved in solid tumors or in bone marrow cells in vivo is altered by DOX binding to elements of the extracellular matrix, a condition which occurs to much lesser extent under cell culture conditions. However, both under cell culture conditions, and in vivo, DOX is concentrated in mammalian cells through avid binding to a wide variety of intracellular constituents [4]. With these considerations in mind, the present study demonstrates that this clinically relevant concentration of DOX caused similar oxidative damage to DNA in non-transformed MCF-10A breast epithelial cells.
Increased amounts of ROS were measured in MCF-10A cells relative to control cells following DOX exposure, with the largest relative increase occurring after 48 hours. Based on earlier studies of the redox cycling of DOX, it can be inferred that the ROS measured in these experiments were produced following an initial one-electron reduction of the DOX quinone by flavin dehydrogenases at multiple intracellular sites, including the nucleus [4]. The nature of the DCF flow cytometric assay for ROS only allows the conclusion that, at the time points measured, and particularly after 48 to 72 hours of DOX exposure, the net production of ROS exceeded detoxification, increasing the overall level of oxidant species in DOX-treated MCF-10A cells.
DNA from normal, untreated MCF-10A cells contained an intrinsic level of modified DNA bases consistent with base lesions observed in human breast cells [19] [20] [21]. The levels of 11 modified DNA bases (of the 12 measured) were significantly increased in MCF-10A cells cultured for 96 hours with 0.1 μM DOX. Thus, an increase in DNA base oxidation in MCF-10A cells was present intracellularly under the same conditions of DOX exposure that yielded an increase in ROS.
All of the modified DNA bases identified in this study can be generated via the interaction of hydroxyl radicals with DNA, and all have been well documented in the literature [10] [13]. During the generation of ROS by routine cellular respiration, a pleiotropic array of antioxidant and DNA repair systems, as well as the cellular apoptosome, minimize the opportunity for sustained DNA injury [22]. However, previous studies from our group have demonstrated that the levels of oxidized DNA base damage observed in these experiments increase mutation rates in mammalian cell reporter systems [23]. Thus, if the modified DNA bases we observed are not repaired before the next cycle of DNA replication, mutations could be induced that, if tolerated by the cell, might result in malignant transformation.
MCF-10A cells synchronized in G0/G1 phase and cultured in the presence of DOX remained in G0/G1 during the entire 96 hours of DOX exposure, while cells cultured without DOX emerged from G1 phase after 16 hours, exhibiting the typical cell cycle progression observed in proliferating cells by 24 hours. Cells exposed to oxidants or other DNA damaging agents often experience growth arrest to allow for DNA repair; however, cells unable to undergo successful DNA repair may sustain malignant transformation. It is of interest that a malignant phenotype has been observed in MCF-10A cells exposed to cigarette smoke condensate for 72 hours [24]. Recent work has implicated impaired BER as a contributor to this transformation [25].
Treatment of unsynchronized MCF-10A cells with 0.1 μM DOX for 96 hours was sufficient to prevent cell proliferation, induce apoptosis, and produce the G2/M arrest typical of the anthracycline antibiotics, such as DOX [4]. This result is consistent with earlier reports showing that H2O2 induces the formation of DNA strand breaks and oxidized DNA bases in MCF-10A cells resulting in growth inhibition [20]. Thus, in addition to mutagenic potential, the oxidized DNA base lesions that we observed following DOX exposure might also contribute to the antiproliferative effects of the anthracyclines.
Differences in the mRNA expression levels of seven BER enzymes were observed in MCF-10A cells cultured with or without 0.1 μM DOX. Expression of OGG1, MGMT, TDG, UDG, APE, MPG, and ENTG mRNA in untreated cells increased by up to 4.5-fold during the 96-hour culture period, as expected during peak rates of DNA synthesis in S-phase [26]. In contrast, expression of the seven BER genes was either unchanged or decreased over the same time frame in DOX-treated cells. From this observation it might be argued that DOX-enhanced reactive-oxygen formation was only capable of producing the observed 2.5- to 3-fold increases in oxidized DNA base content when the DNA repair capacity of the MCF-10A cells was exceeded. It is also possible that the damage inflicted on our MCF-10A cells was so extensive that it not only stopped DNA replication, but it also halted or significantly retarded BER gene transcription. The respective genes for these enzymes are located on five different chromosomes [27]. The fact that the expression of each of the BER genes studied was inhibited to a similar degree despite the fact that the genes reside on different chromosomes suggests that damage to genomic DNA caused by 0.1μM DOX was extensive.
Protein levels of selected human DNA repair enzymes were also significantly lower in cells treated with DOX, compared with control cells, following 72 to 96 hours of drug exposure. It is possible that the continuous exposure to DOX over 96 hours resulted in certain aspects of cellular repair capacity being overwhelmed by ROS, leading to the degradation of specific repair enzymes. In this regard, although the mechanism(s) through which polymerase ε, DNA ligase I, and MGMT protein levels were decreased following DOX treatment are unknown, the activities of other DNA repair proteins, including Fpg (formamidopyrimidine-DNA-glycosylase) and hOGG1, have been shown to be inhibited in mammalian cells by reactive oxygen and nitrogen species [28] [29]. It is also possible that activation of proteasomal pathways by DOX-enhanced oxygen radical production could have contributed to the decrease in polymerase ε, DNA ligase I, and MGMT protein levels we observed.
In conclusion, a 96-hour exposure to a pharmacologically relevant concentration of DOX produced a spectrum of oxidative, and potentially mutagenic, DNA damage in non-malignant MCF-10A breast epithelial cells consistent with that measured in PBMCs during single-agent, DOX-containing, breast cancer chemotherapy. The DNA base damage that occurs both in vitro and in patients may contribute to both the antiproliferative and mutagenic potential of the anthracycline antibiotics. Unfortunately, little is currently known regarding the mechanism(s) by which secondary malignancies occur in patients following DOX chemotherapy. If oxidative DNA damage is related to the etiology of these tumors, monitoring of patients’ blood or urine for sensitivity or resistance to the accumulation of DNA base modifications might be advantageous during and after DOX therapy [30]. It may also be of interest to determine whether there is a therapeutic dosing regimen of DOX which can achieve plasma levels necessary for tumor cell killing while sparing the DNA repair pathways of healthy cells. Evaluation of different dosing regimens and DNA damage in healthy cells may also be relevant for the preclinical and clinical development of newer DOX analogs with more potent cytotoxic effects [31] [32].
Acknowledgments
We wish to thank Dr. Mel Simpson, SAIC-Frederick, Inc., for editorial assistance in the preparation of this manuscript. This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract N01-CO-12400, and grant CA33572. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government. These results were reported, in part, in the Proceedings of the American Association for Cancer Research. 2001;42: 201.
Abbreviations
- DOX
doxorubicin
- AML
acute myelogenous leukemia
- ROS
reactive oxygen species
- GC/MS
gas chromatography/mass spectroscopy
- PBMCs
peripheral blood mononuclear cells
- BER
base excision repair
- NLB
nuclear lysis buffer
- RMRF
Relative Molecular Response Factor
- RPA
ribonuclease protection assay
- RPA4
subunit of the RPA haloenzyme
- L32
ribosomal protein L32
- GAPDH
glyceraldehydes-3-phosphate dehydrogenase
- 5-OH-Hyd
5-hydroxyhydantoin
- 5-OH-5-MeHyd
5-hydroxy-5-methylhydantoin
- 5-OH-Ura
5-hydroxyuracil
- 5-OH-Cyt
5-hydroxycytosine
- ThyGly
thymine glycol
- 5,6diOH-Ura
5,6-dihydroxyuracil
- FapyAde
4,6-diamino-5-formamidopyrimidine
- 8-OH-Ade
8-hydroxyadenine
- Xan
Xanthine
- FapyGua
2,6-diamino-4-hydroxy-5-formamidopyrimidine
- 2-OH-Ade
2-Hydroxy-6-aminopurine
- 8-OH-Gua
2-amino-6, 8-dihydroxyguanine
- APE
apurinic/apyrimidinic endonuclease
- hOGG1
8-oxoguanine DNA-glycosylase
- mtOGG1
mitochondrial-8-oxoguanine DNA-glycosylase
- MGMT
O6-methylguanine-DNA methyltransferase
- TDG
G/T mismatch-specific thymine-DNA glycosylase
- UDG
uracil-DNA glycosylase
- MPG
methylpurine-DNA glycosylase
- ENTG
thymine glycol-DNA glycosylase/AP lyase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Le Deley MC, Leblanc T, Shamsaldin A, Raquin MA, Lacour B, Sommelet D, et al. Risk of secondary leukemia after a solid tumor in childhood according to the dose of epipodophyllotoxins and anthracyclines: a case-control study by the Societe Francaise d’Oncologie Pediatrique. J Clin Oncol. 2003;21:1074–1081. doi: 10.1200/JCO.2003.04.100. [DOI] [PubMed] [Google Scholar]
- 2.Smith RE, Bryant J, DeCillis A, Anderson S. Acute myeloid leukemia and myelodysplastic syndrome after doxorubicin-cyclophosphamide adjuvant therapy for operable breast cancer: the National Surgical Adjuvant Breast and Bowel Project Experience. J Clin Oncol. 2003;21:1195–1204. doi: 10.1200/JCO.2003.03.114. [DOI] [PubMed] [Google Scholar]
- 3.Pedersen-Bjergaard J, Sigsgaard TC, Nielsen D, Gjedde SB, Philip P, Hansen M, et al. Acute monocytic or myelomonocytic leukemia with balanced chromosome translocations to band 11q23 after therapy with 4-epi-doxorubicin and cisplatin or cyclophosphamide for breast cancer. J Clin Oncol. 1992;10:1444–1451. doi: 10.1200/JCO.1992.10.9.1444. [DOI] [PubMed] [Google Scholar]
- 4.Doroshow JH. Anthracyclines and anthracenediones. In: Chabner BA, Longo DL, editors. Cancer Chemotherapy and Biotherapy: Principles and Practice. Williams and Wilkins, Pubs.; Philadelphia: 2001. pp. 500–537. [Google Scholar]
- 5.Doroshow JH, Davies KJ. Redox cycling of anthracyclines by cardiac mitochondria. II Formation of superoxide anion, hydrogen peroxide, and hydroxyl radical. J Biol Chem. 1986;261:3068–3074. [PubMed] [Google Scholar]
- 6.Malisza KL, Hasinoff BB. Production of hydroxyl radical by iron (III)-anthraquinone complexes through self-reduction and through reductive activation by the xanthine oxidase/hypoxanthine system. Arch Biochem Biophys. 1995;321:51–60. doi: 10.1006/abbi.1995.1367. [DOI] [PubMed] [Google Scholar]
- 7.Muller I, Niethammer D, Bruchelt G. Anthracycline-derived chemotherapeutics in apoptosis and free radical cytotoxicity (Review) Int J Mol Med. 1998;1:491–494. doi: 10.3892/ijmm.1.2.491. [DOI] [PubMed] [Google Scholar]
- 8.Rhee SG. Cell signaling. H2O2, a necessary evil for cell signaling. Science. 2006;312:1882–1883. doi: 10.1126/science.1130481. [DOI] [PubMed] [Google Scholar]
- 9.Valko M, Rhodes CJ, Moncol J, Izakovic M, Mazur M. Free radicals, metals and antioxidants in oxidative stress-induced cancer. Chem-Biol Interact. 2006;160:1–40. doi: 10.1016/j.cbi.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 10.Evans MD, Dizdaroglu M, Cooke MS. Oxidative DNA damage and disease: induction, repair and significance. Mutation Res/Reviews in Mutat Res. 2004;567:1–61. doi: 10.1016/j.mrrev.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 11.Storz P. Reactive oxygen species in tumor progression. Front Biosci. 2005;10:1881–1896. doi: 10.2741/1667. [DOI] [PubMed] [Google Scholar]
- 12.Akman SA, Doroshow JH, Burke TG, Dizdaroglu M. DNA base modifications induced in isolated human chromatin by NADH dehydrogenase-catalyzed reduction of doxorubicin. Biochemistry. 1992;31:3500–3506. doi: 10.1021/bi00128a026. [DOI] [PubMed] [Google Scholar]
- 13.Gajewski E, Rao G, Nackerdien Z, Dizdaroglu M. Modification of DNA bases in mammalian chromatin by radiation-generated free radicals. Biochemistry. 1990;29:7876–7882. doi: 10.1021/bi00486a014. [DOI] [PubMed] [Google Scholar]
- 14.Doroshow JH, Synold TW, Somlo G, Akman SA, Gajewski E. Oxidative DNA base modifications in peripheral blood mononuclear cells of patients treated with high-dose infusional doxorubicin. Blood. 2001;97:2839–2845. doi: 10.1182/blood.v97.9.2839. [DOI] [PubMed] [Google Scholar]
- 15.Legha SS, Benjamin RS, Mackay B, Ewer M, Wallace S, Valdivieso M, et al. Reduction of doxorubicin cardiotoxicity by prolonged continuous intravenous infusion. Ann Intern Med. 1982;96:133–139. doi: 10.7326/0003-4819-96-2-133. [DOI] [PubMed] [Google Scholar]
- 16.Ubezio P, Civoli F. Flow cytometric detection of hydrogen peroxide production induced by doxorubicin in cancer cells. Free Radic Biol Med. 1994;16:509–516. doi: 10.1016/0891-5849(94)90129-5. [DOI] [PubMed] [Google Scholar]
- 17.Cheng S, Chen Y, Monforte JA, Higuchi R, Van HB. Template integrity is essential for PCR amplification of 20- to 30-kb sequences from genomic DNA. PCR Methods Appl. 1995;4:294–298. doi: 10.1101/gr.4.5.294. [DOI] [PubMed] [Google Scholar]
- 18.Dizdaroglu M. Chemical determination of oxidative DNA damage by gas chromatography-mass spectrometry. Meth Enzymol. 1994;234:3–16. doi: 10.1016/0076-6879(94)34072-2. [DOI] [PubMed] [Google Scholar]
- 19.Musarrat J, Rezina-Wilson J, Wani AA. Prognostic and aetiological relevance of 8-hydroxyguanosine in human breast carcinogenesis. Eur J Cancer. 1996;32A:1209–1214. doi: 10.1016/0959-8049(96)00031-7. [DOI] [PubMed] [Google Scholar]
- 20.Djuric Z, Everett CK, Luongo DA. Toxicity, single-strand breaks, and 5-hydroxymethyl-2′-deoxyuridine formation in human breast epithelial cells treated with hydrogen peroxide. Free Radic Biol Med. 1993;14:541–547. doi: 10.1016/0891-5849(93)90111-7. [DOI] [PubMed] [Google Scholar]
- 21.Malins DC, Holmes EH, Polissar NL, Gunselman SJ. The etiology of breast cancer. Characteristic alteration in hydroxyl radical-induced DNA base lesions during oncogenesis with potential for evaluating incidence risk. Cancer. 1993;71:3036–3043. doi: 10.1002/1097-0142(19930515)71:10<3036::aid-cncr2820711025>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 22.Roos WP, Kaina B. DNA damage-induced cell death by apoptosis. Trends Molec Med. 2006;12:440–450. doi: 10.1016/j.molmed.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 23.Akman SA, Forrest GP, Doroshow JH, Dizdaroglu M. Mutation of potassium permanganate- and hydrogen peroxide-treated plasmid pZ189 replicating in CV-1 monkey kidney cells. Mutat Res. 1991;261:123–130. doi: 10.1016/0165-1218(91)90058-t. [DOI] [PubMed] [Google Scholar]
- 24.Narayan S, Jaiswal AS, Kang D, Srivastava P, Das GM, Gairola CG. Cigarette smoke condensate-induced transformation of normal human breast epithelial cells in vitro. Oncogene. 2004;23:5880–5889. doi: 10.1038/sj.onc.1207792. [DOI] [PubMed] [Google Scholar]
- 25.Kundu CN, Balusu R, Jaiswal AS, Gairola CG, Narayan S. Cigarette smoke condensate-induced level of adenomatous polyposis coli blocks long-patch base excision repair in breast epithelial cells. Oncogene. doi: 10.1038/sj.onc.1209925. advance online publication, 21 August 2006. [DOI] [PubMed] [Google Scholar]
- 26.van der Meijden CMJ, Lapointe DS, Luong MX, Peric-Hupkes D, Cho B, Stein JL, et al. Gene Profiling of Cell Cycle Progression through S-Phase Reveals Sequential Expression of Genes Required for DNA Replication and Nucleosome Assembly. Cancer Res. 2002;62:3233–3243. [PubMed] [Google Scholar]
- 27.Wood RD, Mitchell M, Lindahl T. Human DNA repair genes, 2005. Mutat Res. 2005;577:275–283. doi: 10.1016/j.mrfmmm.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 28.Wink DA, Laval J. Fpg protein, a DNA repair enzyme, is inhibited by the biomediator nitric oxide in vitro and in vivo. Carcinogenesis. 1994;15:2125–2129. doi: 10.1093/carcin/15.10.2125. [DOI] [PubMed] [Google Scholar]
- 29.Mistry P, Herbert KE. Modulation of hOGG1 DNA repair enzyme in human cultured cells in response to pro-oxidant and antioxidant challenge. Free Radic Biol Med. 2003;35:397–405. doi: 10.1016/s0891-5849(03)00319-8. [DOI] [PubMed] [Google Scholar]
- 30.Cooke MS, Olinski R, Evans MD. Does measurement of oxidative damage to DNA have clinical significance? Clinica Chimica Acta. 2006;365:30–49. doi: 10.1016/j.cca.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 31.Ciesielska E, Studzian K, Wasowska M, Oszczapowicz I, Szmigiero L. Cytotoxicity, cellular uptake and DNA damage by daunorubicin and its new analogues with modified daunosamine moiety. Cell Biol Toxicol. 2005;21:139–147. doi: 10.1007/s10565-005-0142-1. [DOI] [PubMed] [Google Scholar]
- 32.Kaushal V, Kaushal GP, Mehta P. Differential toxicity of anthracyclines on cultured endothelial cells. Endothelium. 2004;11:253–258. doi: 10.1080/10623320490904124. [DOI] [PubMed] [Google Scholar]