Abstract
Critical molecular and cellular biological factors impacting design of licensable DNA vaccine vectors that combine high yield and integrity during bacterial production with increased expression in mammalian cells are reviewed. Food and Drug Administration (FDA), World Health Organization (WHO) and European Medical Agencies (EMEA) regulatory guidance’s are discussed, as they relate to vector design and plasmid fermentation. While all new vectors will require extensive preclinical testing to validate safety and performance prior to clinical use, regulatory testing burden for follow-on products can be reduced by combining carefully designed synthetic genes with existing validated vector backbones. A flowchart for creation of new synthetic genes, combining rationale design with bioinformatics, is presented. The biology of plasmid replication is reviewed, and process engineering strategies that reduce metabolic burden discussed. Utilizing recently developed low metabolic burden seed stock and fermentation strategies, optimized vectors can now be manufactured in high yields exceeding 2 g/L, with specific plasmid yields of 5% total dry cell weight.
Keywords: DNA vaccine, plasmid, vector, fermentation, Escherichia coli, gene therapy
1. Introduction
Plasmid DNA is a new generation biotechnology product (gene medicines and DNA vaccines) that is just beginning to enter the marketplace. Plasmid DNA vectors may find application as preventive or therapeutic DNA vaccines for viral, bacterial, or parasitic diseases or for other indications such as cancer, or gene therapy products. Four DNA plasmid products are currently licenced for veterinary applications. This includes two infectious disease vaccines for West Nile virus in horses (Ft Dodge Animal Health) and infectious haematopoietic necrosis virus in salmon (Novartis). As well, a melanoma cancer vaccine for dogs (Merial) and a growth hormone releasing factor therapy for pigs (VGX Animal Health) are approved. Numerous human plasmid products are in the pipeline. In a recent review, 72 Phase I, 20 Phase II and 2 phase III human DNA vaccine clinical trials were identified (Kutzler and Weiner, 2008).
The vectors utilized in these ongoing trials often utilize generic CMV promoter containing vectors such as derivatives of the Vical VR1012 vector (Hartikka et al., 1996). However, immunogenicity with such vectors in humans and large animals is generally low. Indeed, the approved veterinary vaccines cited above are with highly immunogenic target antigens. While improved expression is often not critical to vaccine performance in murine models, increased expression correlates with improved immunogenicity in large animals and humans (Kutzler and Weiner, 2008). For general application to large animals with a range of antigens, target gene expression needs to be increased. As well, most vectors such as VR1012 and pVAX1 (Invitrogen, Carlsbad, CA) have not been optimized for production yield. Poor production yield can impose significant additional cost post licensure. As reviewed herein, improved vector design will be critical to ensure safety, efficacy and cost effective manufacture of these new generation vaccines.
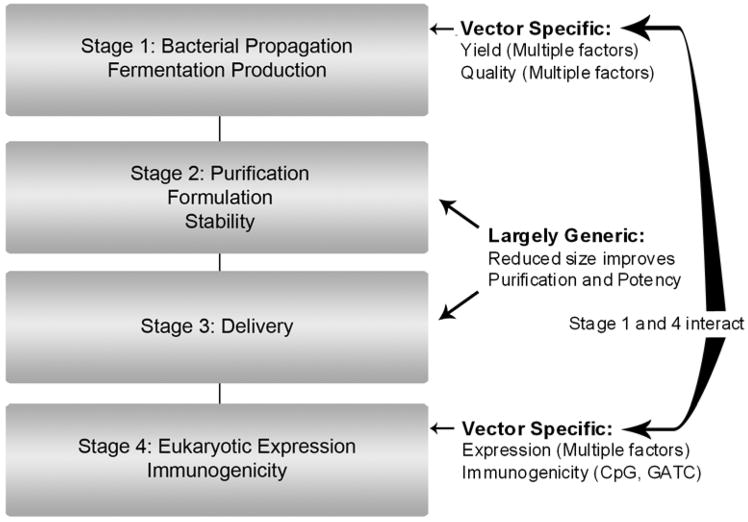
The production and application of plasmid DNA involves four general stages (Figure 1). First, plasmid encoding a gene of interest is transformed into a bacterial cell, typically Escherichia coli (E. coli), propagated to make master and working cell banks, and further propagated in a bioreactor to make production cells that contain high yields of the plasmid (Stage 1). Second, the production cells are lysed, plasmid DNA purified utilizing one of a plurality of purification methods (reviewed in Shamlou, 2003, Carnes and Williams, 2007, Prather et al., 2003) and formulated for delivery (Stage 2). Third, plasmid is delivered to a eukaryotic cell, again using one of a plurality of delivery methods (Stage 3; reviewed in Kutzler and Weiner, 2008). Finally, in Stage 4, the gene of interest is expressed, while the vector backbone stimulates innate immune responses through unmethylated CpG or 5-methyladenosine GATC sequences, or general B DNA mediated Type 1 interferon activation (reviewed in Takeshita and Ishii, 2008; Wagner, 2008).
Figure 1.
DNA Vaccine Vector production and use flowchart.
Stages 2 and 3 are surprisingly generic. For the most part, a single purification process, formulation or delivery method can be applied, without modification, to new unrelated vectors. While smaller vectors are more potent than large vectors (Bloquel et al., 2004), and some purification steps, such as ion exchange membrane separation, do not perform well with large plasmids, this is a generic effect, and is not sequence specific.
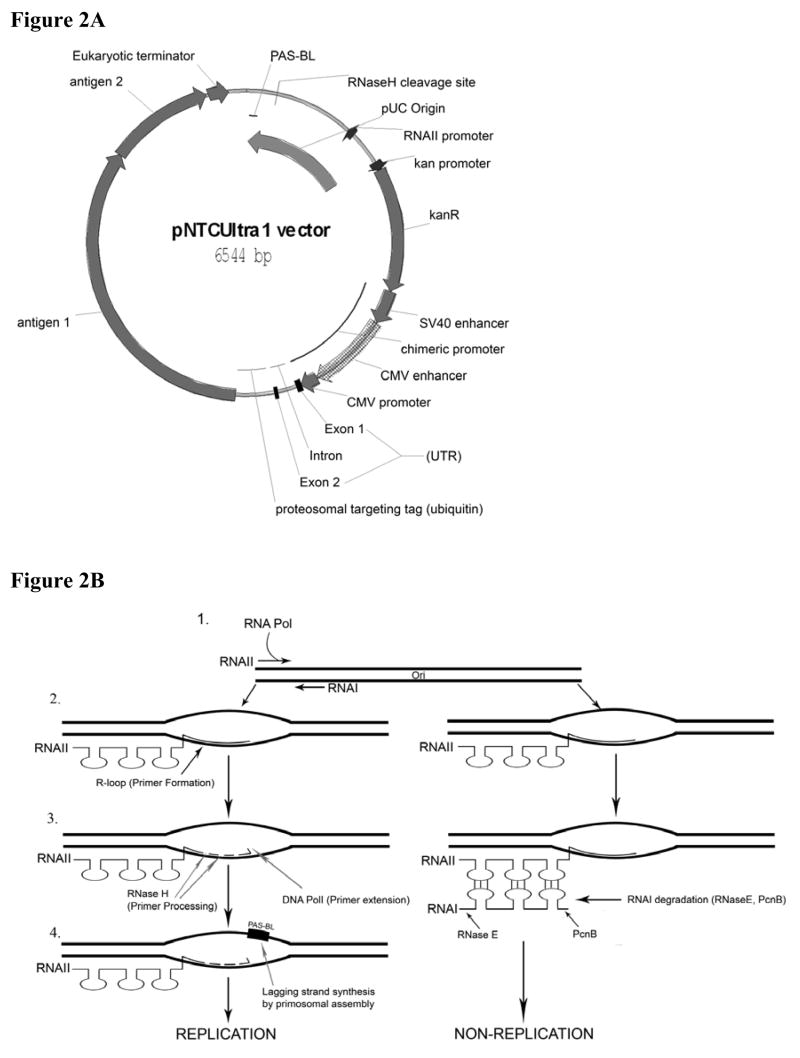
By contrast, Stages 1 and 4 are very sensitive to vector changes. DNA vaccine plasmids contain functional elements that afford propagation (replication origin) and selection (e.g. antibiotic resistance) in a bacterial host organism and elements that drive high level expression in the eukaryotic host (expression cassette comprising eukaryotic enhancer, promoter, terminator/polyadenylation signal; Figure2A ) and activate innate immunity. These components must be carefully arranged, since slight modification of a vector to enhance one parameter can have multiple undesired effects on other parameters.
Figure 2.
A. Annotated pDNAVACCUltra-1 (pNTCUltra-1) restriction map. Critical features such as the mRNA leader (UTR), chimeric SV40-CMV promoter, kanR gene, and functional domains in the origin (PAS-BL, RNAII promoter, RNase H cleavage site) are indicated. This vector targets a fusion antigen (antigens 1 and 2) for proteosomal degradation by fusion to the C-terminus of Ubiquitin A76. B. Plasmid DNA replication. For the leading strand, a RNA polymerase transcript (RNAII) either forms a replication driving R-loop (RNA:DNA hybrid) with the origin (left) or a nonproductive interaction with the antisense RNAI copy number regulator (right). The R-loop is cleaved by RNase H, the resulting 3′ OH serving as a primer for DNA pol I DNA synthesis. Replication of the leading strand exposes a phiX174 type primosome assembly site on the lagging strand (PAS-BL), which binds primosomal proteins (see Section 4.1). Lagging strand replication initiated at PAS-BL terminates at a termination site (ter) within the origin. Leading strand replication proceeds around the plasmid, also stopping at the ter site. DNA Pol I and DNA ligase are required after Pol III mediated leading and lagging strand replication to remove primers and seal nicks, respectively. DNA gyrase then negatively supercoils the resultant covalently closed circular (CCC) plasmid.
Herein, vector design criteria for optimal plasmid performance in both bacteria (Stage 1) and eukaryotic (Stage 4) cells are extensively reviewed. For researchers that wish to clone new genes into existing vectors, a flowchart for design and synthesis of optimal gene inserts is presented. Finally, high yield plasmid fermentation processes are reviewed, with promising new low metabolic burden fed-batch fermentation strategies highlighted.
2. Components affecting gene expression in eukaryotic cells
Vector modifications that improve antigen expression (e.g. codon optimization, inclusion of an intron, a strong promoter) are highly correlative with improved immune responses (reviewed in Manoj et al., 2004). Antigen expression is affected by the following components.
2.1. Promoter
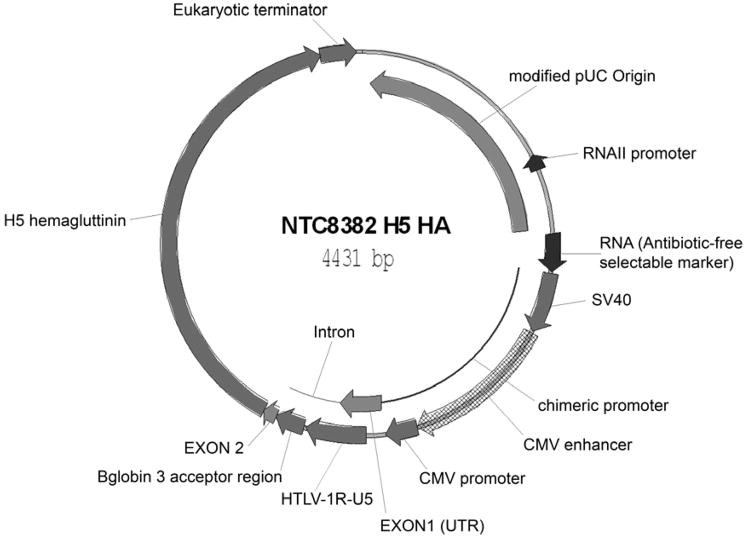
DNA vaccines have traditionally incorporated the human cytomegalovirus/immediate-early or CMV-Chicken-β actin (CAGG) promoters. These promoters drive higher constitutive expression levels than alternative viral promoters (e.g. SV40) or cellular promoters such as the human ubiquitin C (UbC) or human elongation factor factor 1α (EF-1α) promoters. However, improvement in expression beyond that of current CMV or CAGG based vectors may improve immunogenicity in humans. This may be obtained utilizing modified CMV promoters. For example, incorporating the HTLV-1 R-U5 region downstream of the CMV promoter increased expression and improved cellular immune responses to HIV DNA vaccines in mice and nonhuman primates (CMV/R promoter; Barouch et al., 2005) and humoral responses to a influenza H5 hemagluttinin DNA vaccine (Luke et al., 2008; Williams, 2008). Improved expression and/or immunogenicity have also been observed with chimeric SV40-CMV promoters (Li et al., 2001; Williams, 2006; Li et al., 2007; reviewed in Kutzler and Weiner, 2008) (Figures 2A and 3).
Figure 3.
Antibiotic Free Bird Flu DNA vaccine vector. Vector specific features such as the CMV-HTLV-1 R-U5 derived mRNA leader (UTR) chimeric SV40-CMV promoter, and RNA selectable marker are indicated.
2.2. mRNA leader
Expression is generally higher if an intron is present. Typically, this is included in the vector backbone downstream of the promoter, as part of an mRNA leader separating exon 1 and exon 2 (Figure 2A). A typical arrangement is to have the Kozak consensus sequence (Kozak, 1997) at the start of exon 2 immediately prior to the ATG start codon. The spliced untranslated leader region (UTR) is typically 50–150 base-pairs (bp) long, and optimized to limit secondary structure (especially affecting the ATG), and to remove upstream ATG start codons and RNaseL substrates (clustered single stranded UU or UA motifs).
2.3. Gene of interest
Expression is typically higher if the gene is codon optimized to match that of the target organism and terminated by a dual stop codon to limit read through translation.
2.4. Terminator/polyadenylation
Transcription terminators/polyadenylation signals derived from the bovine growth hormone, SV40 or rabbit β-globin genes are commonly utilized. DNA between the stop codon and the terminator is limited to reduce the possibility of cryptic peptide expression or unintended microRNA-mediated expression alteration.
2.5. Prokaryotic sequences affecting expression
During development of pDNAVACCUltra (Williams et al., 2006) and VR1012 (Hartikka et al., 1996) it was determined that gene expression in eukaryotic cells from the CMV promoter was dramatically affected by alterations to vector sequences in close proximity to the CMV enhancer. A number of prokaryotic sequences have been shown to negatively affect gene expression in eukaryotic cells (Leite et al., 1989a and b; Peterson et al., 1987) or bind eukaryotic transcription factors (Tully and Cidlowski, 1987; Ghersa et al., 1994; Kushner et al., 1994). This may account for why expression levels are dependent on the composition and orientation of elements within the prokaryotic region, especially in the immediate vicinity of the eukaryotic expression cassette. All sequence changes made to a plasmid backbone need to be carefully evaluated to detect unexpected detrimental effects of such modifications on the intended purpose of the vector.
2.6. Sequences causing DNA helical instability
Certain sequences promote DNA unwinding that separate the stands (cruciforms). Cruciforms confer sensitivity to endonucleases, such as mung bean endonuclease, and nucleases present in the eukaryotic lysosomal compartment. Such regions reduce the stability of plasmids when trafficked to the nucleus through the endosome, due to nicking by acidic endonucleases. Cruciforms occur in promoter and terminator regions, inverted repeats (e.g. replication origins), homopurine segments and other sequences promoting secondary structures. Computer programs can be utilized to identify potential cruciform cleavage hot spots (e.g. WebSIDD, Bi and Betham, 2004). Interestingly, in the pVAX1 plasmid (Invitrogen, Carlsbad, CA) the bovine growth hormone terminator region contains a homopurine region that is nuclease sensitive. While an alternative polyA sequence significantly improved plasmid nuclease stability, expression after transfection was reduced indicating polyadenylation efficiency is epistatic to nuclease resistance (Azzoni et al., 2007). Interestingly in a study of supercoiled plasmid stability, thermal degradation rates were not increased with a plasmid containing this nuclease sensitive homopurine region (Ribeiro et al., 2008; see Middaugh et al., 1997 for a review of plasmid degradation pathways and testing methods).
The replication origin contains a strong cruciform and is the most sensitive region to endonuclease attack in most plasmids. The elimination of this region in minicircle vectors may in part account for the improved expression with these vectors. Minicircle vectors incorporate target sequences for a site specific recombinase flanking the replication origin and selectable marker in the vector backbone; expression of the recombinase post propagation recombines the vector into two minicircle vectors. The eukaryotic expression cassette minicircle is then purified away from the bacterial origin and selectable marker containing minicirle (reviewed in Darquet et al., 1997). There are significant production issues that need to be overcome to cost effectively manufacture minicircle vectors using recombinase enzymes (reviewed in Mairhofer and Grabherr, 2008).
In summary, minor variations in the vector backbone can alter expression levels, alter target protein intracellular localization (Ramanathan et al., 2001) and ultimately modify immune responses (Zinckgraf and Silbart, 2002).
3. Components affecting immunogenicity
3.1. Antigen targeting
Adaptive immune responses can be enhanced by improving antigen processing for MHCI and or MHCII presentation (reviewed in Leifert et al., 2004). This can be accomplished by targeting heterologous proteins to various intracellular destinations. For example, using the tissue plasminogen activator signal sequence (TPA) to direct antigen secretion often enhances immune responses (Zhongming et al., 1999). As well, membrane-anchoring using human alkaline phosphatase (PLAP) (Gerber et al., 1992), endosomal targeting using human Lamp1 (Wu et al., 1995; August et al., 1997; Weiss et al., 2000), proteosomal targeting using murine Ubiquitin A76 (Rodriguez et al., 1998; Delogu et al., 2000), or endoplasmic reticulum targeting (Xu et al., 2005) may alter or enhance immune responses. Endosomal targeting promotes a MHC class II response, while the destabilizing ubiquitin molecule (UbiquitinA76 versus native UbiquitinG76) is utilized to enhance entry into proteosomal degradation pathway, increase MHC class I presentation, and promote T-helper 1 (TH1) biased immune responses. Endosomal targeting (De Arruda et al., 2004) and secretion (Qiu et al., 2000) has been shown to enhance the immune response against HIV-1 gag protein. Dendritic or immune cell targeting with a variety of leader peptide tags can dramatically improve antibody responses. For example, dendritic cell targeting with CTLA-4 has been shown to improve antibody responses to plasmid DNA delivered by gene gun in pigs (Tachedjian et al., 2000). Most of these tags are extracellular domains of human or highly related proteins (e.g. CTLA-4, CD154, IgG-Fc, C3d, L-Selectin; reviewed in Manoj et al., 2004) which could potentially promote autoimmune responses.
3.2. Unmethylated CpG
Immunostimulatory sequences such as unmethylated CpG can be utilized to enhance T lymphocyte recruitment or expansion (innate immune responses). CpG motifs are 10–20 fold underrepresented in mammalian genomic DNA and, where present, are often methylated. Unmethylated CpG from bacterial DNA or plasmids stimulates TH1 biased immune responses through toll like receptor 9 (TLR9), and may contribute both to the potency of plasmid based DNA vaccines and to the limited duration of expression. Unmethylated CpG potency is affected by flanking dinucleotide context (Krieg, 1999). The immunostimulatory CpG content of a plasmid can be increased by increasing CpG content throughout the plasmid (Krieg et al., 2002) or by adding a block of CpG to a non essential region.
For some applications, such as gene replacement therapy or shRNA mediated gene expression knockdown, immune stimulation and short term expression is undesirable. Promoters and protein selectable marker genes can be modified to reduce or eliminate CpG motifs. Although the pMB1 origin (and derivative pUC origin) contains multiple CpG motifs that are difficult to remove without disrupting origin function, CpG reduced ColE1 plasmids have been created for therapeutic use (Krieg et al., 2002; Yew et al., 2002). For applications where any unmethylated CpG is undesirable, an alternative origin or modified host strain overproducing CpG methylase (Crouzet and Cameron, 2002) should be considered. A CpG free plasmid vector, incorporating CpG free versions of the R6K origin, EM2K bacterial promoter expressed Zeomycin resistance gene, mammalian promoter, leader and terminator is commercially available (pCpG; Invivogen, San Diego CA). For therapeutic use, zeomycin could be replaced with a codon-optimized CpG free version of the kanamycin resistance marker or an alternative marker.
However, the innate immune response to a plasmid cannot be eliminated by removing CpG; cytoplasmically localized B-DNA also induces innate pattern receptors through TBK1 and has been implicated as a key component in activating immune responses to plasmid DNA vaccines (Ishii et al., 2008; reviewed in Takeshita and Ishii, 2008). Recently, Wagner (2008) proposed activation of TLR9 by CpG containing synthetic DNA is phosphothioate base dependent and that TLR9 recognition of unmodified DNA may be sequence independent. Considering that minor alterations in vector sequences can dramatically affect expression (see Sections 2.5, 2.6, and 3.3.1) and immunogenicity (see Section 3.3.2), immunostimulatory alterations resulting from vector backbone CpG alteration need to be carefully interpreted to ensure the result is not due to a secondary effect of expression or methylation alteration.
3.3. dam and dcm methylation
The dcm gene encodes a DNA methylase that methylates the internal cytosine residues in the recognition sequence 5′-CC*AGG-3′ or 5′-CC*TGG-3′ to 5-methyl-cytosine (5mC). The dam methylase encodes a DNA methylase that methylates the internal adenosine in the recognition sequence 5′-GA*TC-3′ to N6-methyladenosine (6mA). As well, E. coli K12 and E. coli B encode type 1 restriction enzymes (HsdRMS) that have methylase enzymes that methylate the internal adenosine in the recognition sequence 5′-AA*CNNNNNNGTGC-3′ or 5′-TGA*CNNNNNNNNTGCT-3′ respectively to 6mA. This is important since these methylation patterns are not found in vertebrates and confer bacterial signatures to DNA prepared in E. coli. This may affect expression ormake it prone to recognition as foreign DNA by the host innate immune system as described below.
3.3.1. Expression
Methylation of plasmid DNA at dam and dcm sites alters expression in a promoter and study specific manner. Expression in mammalian cells was equivalent with dam+ dcm+ versus dam- dcm-plasmid DNA using either CMV promoter vectors, or CMV promoter vectors driving expression of methylated downstream genes (Lai et al., 2008). This contrasts the results from a separate study wherein CMV promoter expression was 2-fold higher after in vivo electroporation of mice with dam+ dcm+ methylated plasmid compared to dam- dcm- unmethylated plasmid (Allamane et al., 2000). CMV promoter driven expression was also slightly lower with unmethylated plasmid (versus dam+ dcm+ methylated) after IV hydrodynamic lipofection delivery to mice (Ochiai et al., 2005). The CMV promoter contains only 2 dcm and no dam sites; increased expression in mice with methylated DNA may be a secondary effect, perhaps promoter stimulation due to dam methylation-mediated cytokine induction (see Section 3.3.2). In contrast, Lai et al. (2008) demonstrated that expression with alternative promoters-enhancers containing multiple dam or dcm sites was reduced in mammalian cells with dam+ dcm+ methylated plasmids compared to the unmethylated versions. This may be a direct effect of methylation-mediated inhibition of transcription factor binding to these promoters. It will be interesting to determine the individual contributions of dam+ and dcm+ methylation to these promoter specific effects. Finally, no effect of dam or dcm methylation status on plasmid transgene silencing after liver delivery was observed (Chen et al., 2008).
3.3.2. Immunogenicity
No difference between dam- dcm- unmethylated or dam+dcm+ methylated plasmid was observed for macrophage activation (Roberts et al., 2005) or TLR9 independent activation of human neutophils (Trevani et al., 2003). In contrast, oligonucleotides or plasmid DNA containing N6-methyladenosine increased cytokine induction (IL-12) in mice compared to unmethylated control DNA (Tsuchiya et al., 2005). IL12 induction was 2 fold lower with unmethylated plasmid (versus dam+ dcm+ methylated) after IV hydrodynamic lipofection delivery to mice (Ochiai et al., 2005). Dam and K12 methylation product N6-methyladenosine is a wide spread bacterial signature not present in mammalian cells, so it may be a pattern recognized by a component of the innate immune system. This is less likely for dcm methylation which is restricted to E. coli and other closely related gram negative organisms (Gomez-Eichelman et al., 1991), and creates 5-methyl-cytosine, a common mammalian pattern (on CpG sites). However, dcm methylated cytosine is in a different context in bacteria compared to mammals (on CT and CA sequences rather than CG).
In summary, it is important to determine the optimal methylation pattern for eukaryotic cell function, and maintain it during preclinical and clinical development.
3.4. Immunostimulatory RNA
Cytoplasmic pattern receptors retinoic acid-inducible gene I (RIG-I) double-stranded RNA-dependent protein kinase (PKR), and melanoma differentiation-associated gene 5 (MDA-5) can activate interferon α and β after activation by double stranded RNA (dsRNA). An RNA polymerase III promoter may be integrated into the vector backbone, and used to create cytoplasmically exported uncapped 5′ tri-phosphate double stranded RNA (dsRNA). 5′ triphosphate dsRNA is a ligand of retinoic acid-inducible gene I (RIG-I) and double-stranded RNA-dependent protein kinase (PKR), the activation of which can affect plasmid immunostimulation (Williams, 2006; Williams, 2008).
4. Sequences necessary for bacterial function: Replication origins
Circular bacterial plasmids replicate by a theta, strand displacement or rolling circle mechanisms (see Del Solar, et al., 1998 for a comprehensive review). To date, all gene therapy plasmids utilize theta replication origins. Theta replication requires synthesis of a primer RNA, and DNA synthesis is initiated by extension of the RNA primer. This type of replication is characterized by the separation of the DNA strands at the origin creating a characteristic theta-shaped replication bubble.
Most theta plasmids such as R6K or R1 (but not pMB1 or ColE1 derived; see below) require a plasmid-encoded Rep initiator protein which binds to specific sequences (often tandem direct repeats called iterons) in the replication origin and recruit host DnaA initiator protein to flanking dnaA box to prime replication.
4.1. ColE1 type origin
The vast majority of therapeutic plasmids currently in use are derived from pBR322 or pUC plasmids, and use high copy derivatives of the pMB1 origin (closely related to the ColE1 origin). For ColE1 replication, plasmid DNA synthesis is unidirectional (Figure 2B) and does not require a plasmid borne initiator protein. For the leading strand, a RNA polymerase transcript (RNAII) forms a R-loop (RNA:DNA hybrid) that is cleaved by RNase H, the resulting 3′ OH serving as a primer for DNA pol I directed DNA synthesis. DNA synthesis then converts to DNA pol III. Replication of the leading strand exposes a phiX174 type primosome assembly site on the lagging strand (PAS-BL; Figure 2A) that binds priA, which recruits priB, dnaT, dnaB, and dnaC to form the preprimosome. Primase (dnaG) then makes a short RNA substrate for DNA pol III. PriC may also be involved in this pathway. Lagging strand replication initiated at PAS-BL terminates at a termination site (ter) within the origin. Leading strand synthesis, with dnaB and dnaC bound to polIII generating lagging strand Okazaki fragments every 1–2 kb, proceeds around the plasmid, stopping at the ter site (reviewed in Masai and Arai, 1996).
ColE1 plasmid copy number is controlled by RNAI, an antisense RNA (reviewed in Del Solar et al., 1998, Prather et al., 2003, Wang et al., 2004). RNAI forms a tRNA like structure, with three stem loops. The 5′ end of the primer, RNAII, is complementary to RNAI and forms a three loop structure antisense to RNAI. The RNAI interaction initiates at the complementary unpaired loops in RNAII (kissing interaction; reviewed in Wagner and Brantl, 1998) preventing its maturation into the replication primer (Figure 2B).
Antisense regulation of copy number by uncharged tRNAs has been observed. Amino acid starvation leads to expression of the stringent (wild type) or relaxed (relA) response. In relA strains, uncharged tRNAs are elevated under conditions of amino acid limitation such as in starved stationary phase cells. Binding of the uncharged tRNAs to loops in RNAI and/or RNAII prevents RNAI interaction with RNAII, and increases plasmid copy number. This can increase copy number up to 10 fold in amino acid starved stationary phase cells versus log phase cells (pBR322 is amplified from 50 to 340; pUC9 is amplified from 90 to 940; Schroeter et al., 1988). This amplification has been exploited to increase pBR322 yield in fermentation culture (Hofmann et al., 1990; Wang et al., 2004).
5. High copy number replicons
Table 1 lists replication origins, with high copy number modifications, that may have utility in therapeutic or DNA vaccine plasmids.
Table 1.
High copy replication origins
| Parent Origin | Regulation | High copy Derivation | Therapeutic plasmids |
|---|---|---|---|
| pMB1 | Antisense RNAI binds RNAII. Rop accessory protein stabilizes interaction | pUC origin (Rop deletion and second site mutation that alters RNAI/II interaction at 37–42°C, not 30°C) | pVAX1 (Invitrogen); V1JNS (Merck); pNTCUltra, (Nature Technology); pPJV7563 (PowderMed); VR1012, (Vical) |
| Rop deletion | pCMVKm2 (Chiron)+ | ||
| ColEI | Same as pMB1 | pMM1, pMM7 (Rop deletion and second site mutation that alters RNAI/RNAII interaction at 37–42°C, not 30°C) | pVC0396 (Valentis)++ |
| R6K (ori α, ori β ori β) | π rep protein binds iteron, copy number dependent activation (low) or repression (high) | Host strain pir-116 mutant (π rep protein copy-up mutation in oligomerization domain removed from plasmid and provided in trans from chromosome) | pCOR (Aventis); pCpG (Invivogen) |
| R1 | RepA initiator protein binds non repeated target. Antisense CopA repressor binds RepA leader (CopT). Auxiliary CopB protein represses RepA expression | RepA controlled by temperature inducible promoter lambda PR (temperature sensitive lambda repressor) | pCWH24-6 (Genecare Development APS) |
pVC0396 is an optimized vector backbone, for insertion of eukaryotic expression cassettes (Eastman and Durland, 2000).
5.1. R6K
The pCpG (Invivogen, San Diego, CA) and pCOR vectors (Soubrier et al., 1999) contain the core R6K origin (400 bp ori γ) on the plasmid, and the pir116 π protein (π mutation that increases copy number) integrated in the host chromosome (Soubrier et al., 1999) (Table 1). This design restricts the host range of the plasmid to the specific production strain. Plasmid multimerization of the pCOR vectors occurs at these constitutively high copy numbers; this is resolved by inclusion of the ColE1 cer resolution of multimer site on the plasmid.
5.2. ColEI type origin
Commonly utilized copy up derivatives of the pMB1 origin (e.g. pUC19; Lin-Chao et al., 1992) or ColE1 origin (pMM1, pMM7; Wong et al., 1982) are approximately 1 kb, and delete the accessory ROP (rom) protein and have an additional temperature sensitive alteration (e.g. pUC, pMM1, pMM7) that destabilizes the RNAI/RNAII interaction. Shifting of a culture containing these origins from 30 to 42°C leads to 30–40 fold increase in plasmid copy number. Perhaps due to the additional regulation by uncharged tRNA binding under amino acid starvation conditions, many of these derivatives are maximally induced by both temperature and entry into stationary phase (pMM1, pMM7, pUC; Lin-Chao et al., 1992; Wong et al., 1982). Inducible plasmid copy number replication origins are preferred for plasmid production since constitutive high plasmid levels increases metabolic burden on the cells, and may result in instability, multimerization, lower productivity, or toxicity to cells (see Section 15). Most existing DNA vaccine plasmids incorporate either the pMB1 based rop- pBR322 origin, the rop- pUC origin, or the ColE1 based pMM1 origin (Table 1).
5.3. R1
Runaway temperature sensitive R1 derivatives have been isolated spontaneously, as well as rationally designed, and include the highest copy number runaway replication vectors. A DNA vaccine vector, pCWH24-6, incorporating an inducible high copy R-plasmid origin has been developed (Hamann et al., 2000; Table 1). However, R-plasmids are large for therapeutic use. For example, the basic R1 replicon is 2.5 kb, while the temperature inducible lambda PR promoter controlled R1 origin in pCWH24-6 is 3.5 kb.
6. Preventing plasmid dimerization during production
Therapeutic plasmids should be stably monomeric (>90%) and resistant to multimerization. In recombination proficient hosts, plasmid multimerization may be controlled by inclusion of a multimer resolution system (MRS) that utilizes a site specific recombinase to resolve plasmid multimers into monomers (reviewed in Summers, 1998).
Standard pUC derived DNA vaccine vectors do not contain a MRS sequence so strict control of multimerization is required. This is accomplished by propagation in a recA- host since plasmid recombination requires the RecF pathway, that itself requires RecA. However, oligomers can be formed by RecA independent pathways, for example, the RecE pathway in sbcA mutants (Fishel et al., 1981). The degree of oligomerization is higher if the plasmid contains large inserts (Berg et al., 1989) or direct repeats (Ribeiro et al., 2008). Once a multimer is formed, it will be stably inherited and out-compete the monomer (dimer catastrophe hypothesis; Summers et al., 1993) although runaway multimerization is balanced with dimer mediated reduced growth rate of the bacteria. Formation of seed stocks at low temperature has been demonstrated to reduce pUC plasmid multimerization in subsequent bioreactor production (Williams et al., 2008; Carnes et al., 2008), potentially by reducing metabolic burden and stationary phase SOS induction (Lee et al., 2002).
The high copy pUC-derived DNA vaccine plasmids also do not have active partitioning functions, and are not precisely distributed during cell division. Consequently, plasmid-free cells arise continuously during culture. The R1 and RK2 plasmids encode an active partition function to ensure plasmid stability (reviewed in Zielenkiewicz and Ceglowsik, 2001). However, inclusion of a partition function on a therapeutic plasmid would require considerable additional DNA. For example, two proteins (ParA and ParB) and a cis-active site (par) are required for partitioning RK2. As well, partitioning plasmids are generally only moderately high copy number. Consequently, with therapeutic plasmids, a selectable marker is included on the plasmid to ensure plasmid retention.
7. Sequences necessary for bacterial retention: Selectable markers
Antibiotic resistance markers are the most commonly utilized selectable markers. Alternative selection strategies have been designed, to address concerns regarding dissemination of antibiotic resistance genes to a patient’s enteric bacteria, as well as to reduce the size of the prokaryotic portion of the therapeutic plasmid. For example, replacement of the 1 kb kanamycin resistance (kanR) gene will potentially increase bioavailability by decreasing plasmid size. Various selection systems are discussed below (also see Mairhofer and Grabherr, 2008).
7.1. Antibiotic resistance
KanR is the most commonly utilized antibiotic resistance marker. Versions of this gene without CpG motifs can be made synthetically or purchased (e.g. pCpGvitro-neo-mcs vector; Invivogen, San Diego CA). Ampicillin resistance is not acceptable due to concerns with hyper reactivity of some patients to β lactam antibiotics. The tetracycline resistance marker is toxic to the E. coli host at high copy number and/or stationary phase (Chiang and Bremer, 1988; Valenzuela et al., 1996a, b).
7.2. Auxotrophy complementation
In these systems, an essential, or conditionally essential gene, is present in the plasmid backbone. Such plasmids can be selected and maintained in bacterial host strains that contain a corresponding chromosomal deletion or suppressible mutation of the essential gene. Numerous examples exist; these systems eliminate the need for antibiotic resistance markers on the plasmid, and are often very small (i.e <100 bp for tRNA genes). For example, the pCOR vectors use a suppressor tRNA selectable marker to suppress a host chromosomal arg gene mutation, required for growth in minimal media (Soubrier et al., 1999).
7.3. Repressor titration
In this case, an operator sequence, placed on a multicopy plasmid, derepresses a chromosomal gene. A number of repressor-operator systems that could be utilized are available (reviewed in Eastman and Durland, 2000; Mairhofer and Grabherr, 2008). For example, the lac operator or tet operator may be on the plasmid, and an antibiotic gene, regulated by the relevant operator is on the chromosome. Titration of the repressor by the operator leads to expression of the chromosomal gene, and antibiotic resistance. Therapeutic plasmids incorporating the pMM1 origin of replication and kanR marker (e.g. pVC0396) and derivatives that can be utilized for repressor titration selection (e.g. pGM509) have been developed (Eastman and Durland, 2000). An alternative system (Cranenburgh et al., 2001) has the dapD essential chromosomal gene under the control of the lac operator/promoter. Three copies of the lac operator on the plasmid titrates the lac repressor, allowing expression of dapD. In the absence of the multicopy plasmid, dapD expression is repressed, and the cell dies. The advantage of such systems is small size (only three copies of the lac operator), elimination of antibiotic resistance (in the case of dapD) or other expressed factors that may have safety concerns (Sherratt et al., 1999). A potential disadvantage of this system is that promoter/operator complexes have been shown to interfere with DNA replication (McGlynn and Guy, 2008); fermentation yields with operator titration plasmids have not been reported.
7.4. Protein based antidote/poison selection schemes
A plasmid stabilization system wherein the F-plasmid ccd antidote-poison operon was modified for plasmid selection has been reported (Szpirer and Milinkovitch, 2005). The antidote protein gene (ccdA) was incorporated onto the plasmid, and the ccdB poison gene was expressed from the chromosome; this scheme allowed plasmid stabilization. This system does contain a protein based selection marker (ccdA) and has not been evaluated in high yield plasmid fermentation.
7.5. RNA based selectable markers
Two groups have developed RNA based selection systems that utilize the endogenous RNAI/RNAII antisense regulators of the pMB1 origin. In both examples, chromosomal genes were engineered to contain an RNAII sequence within the untranslated region of the mRNA. In the presence of a ColE1 type plasmid, the abundantly expressed plasmid encoded RNAI repressor binds both the plasmid encoded RNAII and the ectopic chromosomally expressed RNAII sequence. This latter RNAI:RNAII binding suppresses the translation of the chromosomal gene by RNA-RNA antisense regulation. The regulated gene can be an antibiotic resistance marker, transcriptional repressor (controlling expression of a second gene) or a lethal gene (Mairhofer et al., 2008; Grabherr and Pfaffenzeller, 2005; Cranenburgh, 2005). These systems are limited to existing ColE1 type origin containing plasmid vectors only. Recently a new RNA based antibiotic-free selection system has been reported (Luke et al., 2008; Williams, 2008). Vectors incorporate and express a 150 bp antisense RNA that represses expression of a chromosomally integrated constitutively expressed counter-selectable marker (Bacillus subtilis levansucrase, sacB), allowing plasmid selection on sucrose. These sucrose selectable DNA vaccine vectors (Figure 3) combine antibiotic-free selection, chimeric eukaryotic promoter (SV40-CMV), and optimized HTLV-1 R-U5 containing mRNA leader with highly productive fermentation manufacturing (>1 g/L plasmid DNA yields).
8. Vector design for optimal performance and regulatory compliance
A therapeutic plasmid vector must combine the bacterial and eukaryotic components described above into a vector that combines high mammalian expression with high copy number, high % supercoiling, and effective multimer resolution to meet manufacturing, efficacy and quality requirements for pharmaceuticals. Vectors are difficult to design, since as discussed above, numerous factors affect these critical attributes. For example, plasmid copy number is probably set by: 1) the efficiency of the replication origin (e.g. pUC is higher than pBR322 at 37–42°C); and 2) the percent of initiated replication cycles that are completed. Plasmid copy number will be reduced by sequences that cause inhibition of DNA polymerase III replication, such as protein-DNA complexes assembled on replication termini (Ter) or repressor binding sequences (McGlynn and Guy, 2008). As well, stable RNA-DNA hybrids, unusual DNA structures (e.g. internal-ribosome entry sites; Levy, 2003), and strong convergent promoters leading to head on transcription-replication collisions reduce copy number (reviewed in Mirkin and Mirkin, 2007). Optimal vectors will have a higher copy number due to reduction of these factors. For example, the pDNAVACCultra1 plasmid (Figure 2A) with a chimeric SV40-CMV promoter increases plasmid copy number 2 fold versus alternative vectors such as gWIZ, with fermentation yields up to 2.2 g/L in an inducible fed-batch process (Williams et al., 2008)(see Section 14). This vector has high eukaryotic expression, and does not produce replication intermediates, which is a problem with a number of minimalized vectors such as pVAX1 (see Section 9.1 below).
In Sections 9–11, DNA vaccine vector design issues affecting quality, yield or regulatory compliance are discussed. In Section 12, these design criteria are compiled into an flowchart to design synthetic gene inserts.
9. Sequences detrimental to plasmid quality
Nicking is associated with AT rich regions or cruciforms that ‘breathe’ and are susceptible to endogenous single stranded nucleases. This results in a high percentage of open circular plasmid in the bacterial host, and endosome mediated nuclease digestion in mammalian cells (Azzoni et al., 2007). Palindrome sequences are unstable, as are direct or inverted repeats. Direct repeats are mutational hot spots, especially if cells are grown to stationary phase during propagation (Ribeiro et al., 2008). Unusual DNA structures are formed by runs of potentially Z DNA-forming alternating pyrimidine/purine sequences (such as CpG repeat sequences; Bichara et al., 1995), G-rich sequences that may form tetraplex structures, and oligopyrimidine or oligopurine sequences that may form triplex DNA. Plasmids containing Z DNA forming regions are unstable while intra-molecular triplex regions reduce plasmid supercoiling in batch fermentation (Cooke et al., 2004) presumably due to endogenous nuclease mediated nicking of triplex-formed single stranded DNA regions. If such structures must be retained in a vector, reversing the orientation of these elements, relative to the replication origin, may alleviate propagation problems. For example, a GC rich TGG repeat inhibits plasmid replication only where the TGG strand forms the template for lagging strand synthesis, possibly due to tetraplex structures formed by the G-rich strand (Pan and Leach, 2000).
9.1. Replication intermediates
Reducing plasmid size by elimination of extraneous DNA to increase potency, remove antigenic peptides, and meet regulatory guidance can cause new problems. Replication tends to terminate in internal ribosome entry sites (IRES’s) and eukaryotic enhancer repeat regions (e.g. CMV promoter). Termination is enhanced when the orientation of the origin is close to and parallel with the CMV promoter, resulting in production of linear fragments called replication intermediates. Interference is perhaps due to secondary structure, or fortuitous binding of bacterial proteins (Levy, 2003). This phenomenon is only seen in high copy plasmids; the same arrangement on low copy plasmids does not make detectable intermediates. New minimized DNA vaccine vectors such as pVAX1 (Invitrogen, Carlsbad, CA) contain the CMV promoter and replication origin in close proximity due to size reduction; this vector produces replication intermediates, thus reducing the quality of plasmid produced in the bacterial host (Levy, 2003). The presence of the replication intermediate is readily observed as an extra small band (which is difficult to remove during downstream processing) in purified plasmid preparations. Generally, replication intermediates are not a problem with larger vectors such as gWIZ (Genlantis, San Diego, CA) or VR1012 (Hartikka et al., 1996) which have not been optimized to reduce size (Williams and Carnes, unpublished observations). Configurations for minimal vectors that do not form replication intermediates are shown in Figures 2A and 3 (Williams et al., 2006; Williams, 2006).
9.2. Dimerization sequences
Most plasmid recombination is independent of RecBCD, and is mediated by the RecA-dependent RecF pathway. RecBCD is the major enzyme responsible for degradation of linear DNA in E. coli. RecBCD degradation is arrested at Chi sites (5′ GCTGGTGG-3′). RecBCD directed recruitment of RecA to chi sites stimulates homologous recombination. The presence of chi sites in plasmids promotes RecF independent recombination by the RecBCD pathway in recA+ sbcB+ (wild type) cells (Zaman and Boles, 1996). Oligo-pyrimidine or oligo-purine sequences in a plasmid increases dimerization, presumably through formation of unusual DNA structures (Kato, 1993).
9.3. dcm methylation
dcm is an E. coli DNA methylase that methylates the internal cytosine residues in the recognition sequence 5′-CC*A/TGG-3′. The methylated cytosine is mutagenic to thymine in certain genetic backgrounds. Mutation of the dcm gene or elimination of CCA/TGG sequences will reduce methylation, and possibly reduce mutation rate, of plasmid DNA. The effect of dcm methylation on expression and immunogenicity is unknown (see Section 3.3).
10. Sequences detrimental to plasmid yield
As discussed below, elements that limit plasmid yield and quality will result in plasmids that are carried at lower copy number in the cell.
10.1. Cryptic bacterial promoters
In general, the replication origin needs to be protected from read-through transcription from adjacent genes, for example from cryptic promoters in an antigen gene insert, to prevent plasmid destabilization or reduced copy number (Engels and Meyer, 1993). This may be accomplished by inclusion of shielding transcriptional terminators, optimizing the orientation of composite elements, or both (Eastman and Durland, 2000; Brosius, 1984; Williams et al., 2005). For example, transcriptional terminators shielding the prokaryotic replication origin from insert mediated transcriptional read-through reduce insert toxicity due to eukaryotic gene transcription (Brosius, 1984; Chen et al., 1993; Chen and Morrison, 1987).
10.2. Toxic peptides
A gene insert may contain proteins or peptides that are toxic to the host cell. Hydrophobic membrane spanning peptides are particularly toxic to bacteria. Translation of an insert encoded toxic peptide may be prevented by introduction of a reverse oriented promoter after the insert, to generate translation-disrupting antisense RNA (Weiner et al., 2005; reviewed by Saida et al., 2006). However, not all insert borne toxic peptides are expressed from the sense strand; for example, bacterial toxicity of an influenza H1 serotype antigen gene is mediated by expression of the antisense strand (Williams, unpublished observations). Alternatively, plasmid stabilization and improved production has been achieved through insertion of an intron to disrupt bacterial production of a known toxic protein (Boyd et al., 1999).
10.3. Ter sites
Bacterial replication is blocked at defined terminator or ‘ter’ sites. These sites contain a core 5′-GTTGTAAC-3′ conserved sequence, with less conserved 5′ and 3′ flanks (reviewed in Neylon et al., 2005). While ter sites within the replication origin are essential for normal replication termination, additional sites may be detrimental.
11. Regulatory considerations
The introduction of plasmid DNA into humans requires special considerations which have been addressed in several recent World Heath Organization (WHO), US Food and Drug Administration (FDA), or European Agency for the Evaluation of Medicinal Products (EMEA) regulatory draft guidance’s (EMEA, 2006; EMEA, 2001; FDA, 2007; FDA, 1998; WHO, 2007). The content of these guidance’s has been reviewed (Schalk et al., 2006; Glenting and Wessels, 2005). Key issues to consider in plasmid design are discussed below.
11.1. Homology to target organism or natural pathogens
To reduce chances of chromosomal integration, the plasmid should contain no significant homology to the target organism genome (Wang et al., 2004). This can be determined by BLAST search by specifying to search for short, nearly exact matches against the relevant genome. Recombination with natural viruses inside the vaccinated person is also possible, for example, between the CMV promoter and infecting CMV virus. However, this is considered to be a lower risk with DNA vaccines than conventional vaccines (Schalk et al., 2006) and is not raised as a specific concern in FDA, WHO or EMEA regulatory guidance’s, with the exception of recombination events that would restore viral virulence.
Vectors should not contain bacterial DNA uptake sequences that preferentially transform Pasteurellaceae or Neisseriaceae species (Schalk et al., 2006; see Section 12 below).
11.2. Nonfunctional sequences
Spacer or junk sequences that do not encode a defined function should be removed. This is difficult to accomplish, since minor variations in the vector backbone can have dramatic effects on bacterial yield and quality and eukaryotic expression and immunogenicity (see Sections 2.5, 2.6 and 9.1).
11.3. Immunogenic sequences
Regions encoding antigenic peptides should not be present. These include cryptic open reading frames (ORFs) in bacterial or eukaryotic sequences that may be expressed in eukaryotic cells (van Hall et al., 1998; Schirmbeck et al., 2005). The removal of spacer and junk sequences and use of RNA-based selectable markers (i.e. to eliminate the kanR ORF) reduces this risk.
Of particular concern is use of hepatitis viral enhancers. Hepatitis genomes are highly compacted such that enhancer functions overlap antigenic protein coding regions (Yuh and Ting, 1990). For example, pPJV7563 and derivatives (Fuller, 2006) contain a single copy of the HBV posttranscriptional regulatory element (PRE) immediately downstream of the stop codon prior to the transcriptional terminator. This element is included to increase mRNA nuclear export. However, the PRE also encodes an 178 amino acid fragment of the hepatitis C virus (HCV) polymerase gene which, by design, is within the eukaryotic expression cassette; this type of design has been demonstrated to generate T-cells reactive to MHC I epitopes in the cryptic ORF (Schirmbeck et al., 2005). Such antigenic regions might significantly alter immune responses in individuals with prior exposure and memory T-cells to Hepatitis.
11.4. Host range restriction
The pMB1 origin is host restricted to E. coli and closely related gram negative bacteria. In the case of the R6K origin, the replication initiator protein may be deleted from the plasmid, and integrated into the genome of the host. This is advantageous, since it both reduces the size of the plasmid origin, and restricts the host range of the plasmid to production strains containing the activator. The R6K expression plasmid systems have this configuration (Table 1). This greatly reduces the concern that plasmid containing therapeutic genes or antibiotic resistance markers (see below) will be taken up and propagated in enteric bacteria in the patient. The dissemination of some therapeutic genes (e.g. growth factors) or resistance markers (see below) in this manner would be undesirable.
11.5. Antibiotic resistance markers
The use of antibiotic resistance markers is generally discouraged, and if used the in vivo effect needs to be evaluated. For example, the EMEA, 2001 guidance indicates ‘lack of expression in mammalian cells should be verified due to regulatory concerns’. Ampicillin resistance is not acceptable due to concerns with hyper reactivity of some patients to β lactam antibiotics. Resistance to kanamycin encoded by the gene neomycin phosphotransferase III [npt-III, aph(3′)-III] should be avoided, since this also confers resistance to amikacin, a reserve antibiotic (EMEA, 2008). Most vectors utilize either npt-II (e.g. pVAX1) or npt-Ia (e.g. VR1012, pDNAVACCUltra). The npt-II [aph(3′)-IIa] gene from TN5 confers resistance to neomycin B, kanamycin A and neamine. Geneticin (G418) and amikacin are inefficient substrates (Siregar et al., 1994). The npt-Ia [aph(3′)-Ia] gene from TN903 confers resistance to kanamycin A, neomycin B, neamine, lividomycin A and geneticin. Amikacin and butirosin A are only weakly inhibited and the enzyme has only weak phosphatase activity towards them (Siregar et al., 1995). Alternative vectors with the more specific ANT(4′)1a kanR marker have also been developed (Snyder and Satishchandran, 1998).
11.6. Enzymatic activities
For antigen genes having enzymatic activity or virulence issues, incorporation of inactivating or attenuating mutations, respectively, should be considered (WHO, 2007). For example, the multiple basic amino acid motifs at the influenza H5 hemagluttinin cleavage site (PQRERRRKKRGL) correlative with pathogenicity can be replaced with the corresponding region (PQRESRGL) from several nonpathogenic influenza strains, to protect against virulence factor gene transfer during immunization to infecting non pathogenic viruses.
Transient antibody responses against DNA have been observed with DNA binding proteins such as HIV-nef or Hepatitis C virus core. This raises concern that immune responses against DNA may be induced by protein/nucleic acid complexes (reviewed in Schalk et al., 2006). Incorporation of mutations that inactivate DNA binding, or eliminate nuclear localization may decrease risk of DNA protein complexes; as well, cytoplasmic proteins should have improved proteosomal processing, and presentation by MHCI, compared to nuclear localized proteins.
12. Insert design flowchart
Once an optimized vector is validated, new versions are created by substitution of new genes into the backbone. Vectors designed by the criteria described above typically allow cloning of the gene of interest into an existing intron containing transcript (introns increase mRNA export and subsequent expression levels). Immediately downstream of the intron in exon 2 is an optimized Kozak sequence conforming to the consensus gccRccAUGG where R =G or A, and AUG is the gene start codon. SalI (GTCGAC) functions as a kozak sequence, and is often used as the 5′ restriction site to clone genes into vectors as kozak-ATG (GTCGAC-ATG) fusions. The gene is cloned starting with the ATG codon immediately downstream of the kozak sequence. A non-native amino acid may be included at codon two, since highly expressed genes have an ATGG start. Alternatively, the gene is cloned with or without the ATG codon downstream of a signal sequence such as TPA to direct the protein to the endoplasmic reticulum for secretion. Secretion often improves immune responses.
Using a validated vector backbone may reduce the extent of required preclinical testing for versions incorporating new genes. For example the FDA (2007) guidance indicates “biodistribution studies may be waived for DNA vaccines produced by inserting a novel gene into a plasmid vector previously documented to have an acceptable biodistribution/integration profile” while the WHO (2007) guidance indicates “biodistribution and persistence studies are required, unless substantial experience is already gained with an almost identical or similar product” and the EMEA (2006) guidance indicates “previous experience with the vector system will allow optimization and focusing of non-clinical studies”. Safety studies using DNA vaccine vectors with different inserts have demonstrated similar biodistribution (Sheets et al., 2006a) and toxicology (Sheets et al., 2006b). It is therefore critical that new antigen genes are well designed, to avoid unnecessary insert-specific regulatory issues.
Therapeutic or antigen genes traditionally are transferred from a genomic DNA construct directly into a DNA vaccine vectors. However, expression of unmodified gene inserts, especially those with dramatically different codon usage patterns compared to the target organism, is often suboptimal. Gene synthesis technology (e.g. Geneart, Regensburg Germany; DNA2.0, Menlo Park, CA) has become highly affordable (reviewed in Wu et al., 2007). We recommend, for DNA vaccines intended for potential licensure, that new gene inserts should be designed de novo for compatibility, regulatory compliance and improved eukaryotic expression, and then made synthetically.
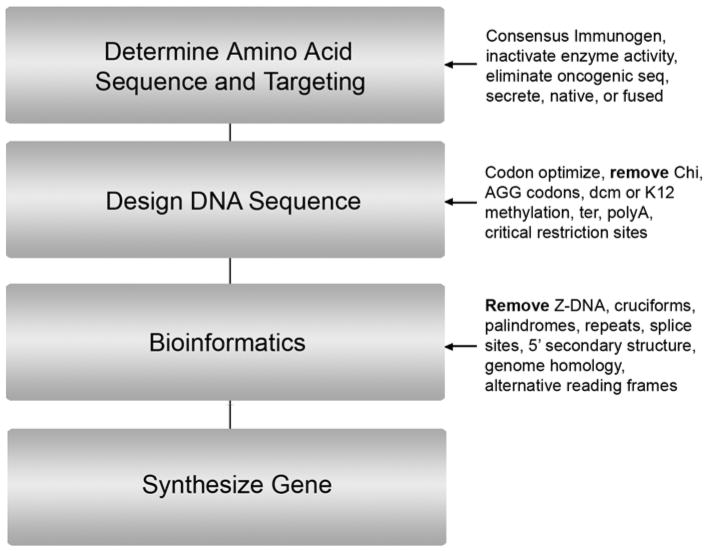
A flowchart for designing new inserts (Figure 4) is discussed below. Many of the factors relating to vector design are also important for creating new inserts in existing vectors.
Figure 4.
Insert design flowchart. See Section 12 for a detailed discussion.
Determine desired protein amino acid sequence: For pathogens containing various serotypes or amino acid variations, this may involve defining a “consensus immunogen” (Laddy et al., 2007). Highly expressed genes start with ATGG sequence, which includes Met-Gly, Met-Ala, Met-Val, Met-Glu and Met-Asp dipeptide amino termini. Bioinformatics should be utilized to ensure no oncogenic sequences exist in the predicted protein (EMEA, 2006). A fusion protein containing two antigens (e.g. Figure 2A) can be utilized in advanced designs to co-immunize against one or more target organisms.
Determine targeting: For example, if the gene is a secreted antigen determine if secretion will be retained or removed. If retained, determine if a native or heterologous secretion sequence (e.g. TPA) will be utilized. The advantage of TPA is extensive prior clinical experience and demonstrated performance for high yield production of secreted protein with a variety of target genes. Inactivation of nuclear localization sequences should be considered for DNA binding proteins.
-
Design synthetic gene: Use codon optimization software such as OPTIMIZER (Puigbo et al. 2007) or gene design software described in Villalobos et al. (2006).
During design, define sequences to be eliminated from the final gene sequence: Sequences to exclude include AGG codons (underrepresented in highly expressed human genes; Huang et al., 2008), Chi sites (5′ GCTGGTGG-3′), ‘ter’ sites (core 5′-GTTGTAAC-3′), dcm sites (CCAGG or CCTGG), K12 methylation sites (AACNNNNNNGTGC), polyadenylation sites (AATAAA or ATTAAA), TATA boxes (TATAa/tAa/t), immunosuppressive telomeric TTAGGG motif (Gursel et al., 2003), Pasteurellaceae or Neisseriaceae DNA uptake sequences (GCCGTCTGAA, AAGTGCGGT or ACAAGCGGTC; reviewed in Bakkali, 2007) and flanking restriction sites that will be utilized for cloning into the vector.
During design, the CpG content and immunostimulatory context (Krieg, 1999) can be modified up or down as desired.
When designing, consider that protein secondary structure information may be encoded in mRNA through codon usage bias (Gu et al., 2003).
Bioinformatics screen putative synthetic gene sequences to identify undesired sequences for bacterial propagation: AT rich regions (TT or TA clusters also encode UU or UA eukaryotic RNase L cleavage sites; these regions are unlikely in codon optimized GC rich genes for mammalian expression), hairpins, Z-DNA forming alternating purine-pyrimidine repeats, oligo-pyrimidine or oligo-purine sequences (WEB based software can be utilized to screen for these structures: ZHUNT for Z-DNA; Ho et al., 1986; EMBOSS programs einverted, palindrome, equicktandem or etandem, to detect inverted repeats, palindromes, or tandem repeats, respectively; Rice et al., 2000) or DNA cruciforms (WebSIDD; Bi and Betham, 2004).
Bioinformatics screen putative synthetic genes to identify undesired sequences for eukaryotic expression: cryptic splice sites (acceptor, donor, or branch sites) that could lead to aberrant splicing; >16 bp dsRNA from inverted repeats (identified in step 4 above) that could lead to RNAi mediated gene silencing; cryptic microRNA targets that could lead to inadvertent cell specific regulation. Such sequences can be identified by bioinformatics; for example internal splice sites (Pertea et al., 2001), and microRNA (see Ioshikhes et al., 2007 for a review of available programs).
Determine secondary structure at 5′ end of gene. Screen using a program such as Mfold (Zucker, 2003) for RNA folding patterns at the 5′ 100 bp of the gene, as it would be present in RNA produced from the final vector (i.e. fused to the vector encoded spliced leader RNA). Select optimized version that does not have substantial secondary structure in region of the Kozak-ATG start site.
Blast search target organism genome (specify search for short homology regions) and modify sequences to remove regions with significant identity.
Translate entire insert in all 6 reading frames (3 forward and 3 reverse) and search for potential cryptic open reading frames in the 5 alternative frames. If necessary, modify to create stop codons in alternative frames by making synonymous codon changes as necessary.
Incorporate two tandem stop codons (combinations of TGA or TAA) to ensure correct termination and prevent read through translation. 10) Have gene synthesized, and clone into vector.
Occasionally, a plasmid insert that meets these design criteria is low yield or toxic to the bacterial host. Since bioinformatics effectively identifies alternative DNA structures, it is likely that the problem is due to expression of toxic peptides from cryptic bacterial promoters. However, a correlation between weak bacterial promoters in the insert predicted by various bioinformatics programs and plasmid productivity has not been observed (Williams, unpublished observations). This may be due to the limitation of the search algorithms. Such vectors often can be produced using low metabolic burden seed-stock and fermentation processes (Williams et al., 2008; see Section 15 below).
Cryptic antisense eukaryotic promoters may also be detrimental, by making antisense RNA that leads to RNA interference and gene silencing. Utilization of improved bioinformatics programs for detection of bacterial promoters (reviewed in Wang and Benham, 2006) and human promoters (reviewed in Abeel et al., 2008) may increase insert design success rate in the future.
13. Host cell lines for plasmid manufacture
DNA vaccine plasmids typically are manufactured using NIH automatic exempt attenuated E. coli K12 strains such as DH5α (Carnes et al., 2006), DH5 (Listner et al., 2006), DH1 (Cooke et al., 2004), JM108 (Huber et al., 2005), SCS1-L (Singer et al., 2008) or DH10B (Lahijani et al., 1996). BL21 (an E. coli B strain) has recently been demonstrated to be a high yielding plasmid production host (Phue et al., 2008). The genotypes of these cell lines are shown in Table 2.
Table 2.
Plasmid methylation profile from various E. coli plasmid production hosts
| Genotype + | Dam | Dcm | K12 Methylase | |
|---|---|---|---|---|
| DH1 | F- endA1 hsdR17(rK-mK+) supE44 thi-1 recA1 gyrA96 (Na1r) relA1 | + | + | + |
| DH5 | F- deoR endA1 hsdR17(rK-mK+) supE44 thi-1 recA1 gyrA96 (Na1r) relA1 DeoR derivative of DH1 | + | + | + |
| DH5α | F- endA1 hsdR17(rK-mK+) supE44 thi-1 recA1 gyrA96 (Na1r) relA1 Δ (lacZYA-argF)U169(m80lacZDM15) | + | + | + |
| JM108 | F-. recA1 endA1 gyrA96 thi-1 hsdR17(rK-mK+) supE44 relA1 Δ(lac-proAB) | + | + | + |
| SCS1-L | F- recA1 endA1 gyrA96 thi-1 hsdR17(rK-mK+) supE44 relA1 | + | + | + |
| DH10B | F- mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 recA1 endA1 araD139 Δ(ara, leu)7697 galU galK λ-rpsL nupG | + | + | − |
| BL21 | F- ompT hsdSB (rB- mB-) gal dcm | + | − | − |
| GT115++ | F̄ mcrA Δ(mrr-hsdRMS-mcrBC) F80lacZ ΔM15 ΔlacX74 recA1 endA1 Δdcm uidA(DMluI)::pir-116 ΔsbcC-sbcD | + | − | − |
+endA1: increases plasmid quality. EndA is a periplasmic nonspecific nuclease that protects cells from invading DNA. However, upon lysis, EndA will degrade plasmid DNA. recA: reduces plasmid recombination. relA: Eliminates stringent response, allowing very slow growth. sbcC and sbcD Operon encoded nuclease that creates double stranded breaks at hairpins that are then repaired using recombination pathways (Connelly and Leach, 1996). SbcC and sbcD mutants stabilize plasmids with palindromes (8–16 minimum stem loop; Chalker et al., 1988; Connelly et al., 1999) or a TGG24 trinucleotide repeat (Pan and Leach, 2000). In sbcC mutant, RecA (or RecQ helicase) is required for palindrome viability, perhaps to allow replicative bypass of secondary structure (Cromie et al., 2000).
++ Pir-116 π protein mutation increases copy number of plasmids harboring the R6K gamma origin of replication. GT116 is a related cell line without uidA(DMluI)::pir-116.
Auxotrophic strains are suitable for growth in semi defined media but not for production in minimal media since they require expensive supplementation. DH5 and JM108 have been utilized for defined media plasmid production (Huber et al., 2005; Huber et al., 2005b; Listner et al., 2006). For defined media growth, it is critical that strains are not mucoid. For example, gal+ strains produces additional capsule through colonic acid production in minimal media. Mucoid cultures are difficult to harvest and foul downstream processing. Colonic acid production can be blocked by mutagenesis of the galE gene.
For therapeutic production of plasmid, strains that contain F′ conjugation plasmids are undesirable from a regulatory perspective due to the potential for conjugative enhanced transfer of genetic material to environmental organisms. As well, blue white selection (the reason for the inclusion of the F plasmid in several strains) is not required for production. This problem was addressed in an innovative way with the pCOR vector host by inactivation of the F factor traD conjugative coupling protein (Soubrier et al., 1999).
As illustrated in Table 2, the various production hosts differ in their methylation pattern. Since, as described above, methylation can effect eukaryotic expression and immunogenicity, it is important to determine the optimal methylation pattern, and maintain plasmid production in hosts with the same pattern during subsequent preclinical and clinical development (EMEA, 2001).
While dcm function is dispensable, dam- strains have not been used for large scale plasmid production; Dam methylation is involved in multiple cellular processes including chromosomal and ColE1 replication (reviewed in Lobner-Olesen et al., 2005) and dam- strains are mutagenic (Glickman and Radman 1980; Marinus et al., 1983). Dam- strains such as SCS110 (Stratagene, La Jolla, CA) are available for small scale plasmid production to evaluate dam methylation effects on function, to determine if dam sites should be removed from critical regions of the vector.
13.1. Strain specific shake flask productivity differences are not predictive of fermentation performance
Several studies have been performed to identify optimal strains for plasmid production. For example, JM108 was selected as the best strain for fermentation production after evaluation of a panel of 7 strains (DH1, DH5, DH5α, JM83, JM101, HB101, and JM108) for production of three different plasmids, using LB shake flask media. One of the plasmids in these strains was then evaluated in 37°C semi defined batch fermentation. Significantly, plasmid productivity in shake flask was poorly predictive of fermentation productivity. DH5α was one of the poorer fermentation producers in this study (1.3 mg/OD550/L; 65% supercoiled), with DH1 intermediate (3.0 mg/OD550/L; 84% supercoiled) while JM108 was the highest (8.2 mg/OD550/L; 91% supercoiled) (Huber et al., 2005). By contrast, in an inducible 30–42°C semi defined batch fermentation process, DH1 and DH5α were both high producing strains (Carnes et al., 2006). An extensive shake flask evaluation of plasmid specific yield and % supercoiling in 17 host strains with three different pUC origin based plasmids (5.8 kb gWIZ-GFP, 6.9 kb pSVβ, 20 kb pQR150) has been reported (Yau et al., 2008). HB101 and DH5α were appropriate for production of all 3 plasmids; these strains had near 100% supercoiling with all 3 plasmids and high specific yields with 2/3 plasmids. By contrast, the other 15 strains were found to have poor supercoiling or production yields with one or more plasmids. For example while BL21(DE3) plasmid yields were high, % supercoiling was inconsistent (gWIZ-GFP=98%, pSVβ= 82% and pQR150=45%). Recently, BL21 recAendA has been demonstrated to produce high yields (1.9 g/L) of predominantly supercoiled plasmid with a gWIZ derived plasmid in a 30°C to 42°C inducible process (Phue et al., 2008). In contrast to the Yau et al., 2008 shake flask results, BL21 was a poor producer; this was attributed to the RecA+ gene product (Phue et al., 2008). These studies demonstrate strain specific plasmid productivity in shake flask is poorly predictive of fermentation productivity, and strains performance varies between different fermentation processes.
13.2. Cell stocks for manufacturing pharmaceutical grade plasmids
Cell lines for production should be created by transformation of the relevant cell line with diluted preparations of plasmid, to ensure cells are transformed with only a single plasmid molecule. Individual colonies are screened for plasmid quality (i.e. % monomer) and quantity (plasmid yield) prior to creating Master and Working Cell banks. Care should be taken with high copy pUC origin plasmids to avoid stationary phase at high temperatures (e.g. 37°C) when using relA mutant strains such as DH1, DH5, DH5α and JM108. Such conditions create very high plasmid copy numbers, increased metabolic burden, and induction of mutagenic repair pathways. For example, high copy pUC8 plasmids reduce stationary phase viability; the toxicity correlated with the size of DNA insert, but not transcription of the insert (Cheah et al., 1987). Growth of transformed cells for seed stock creation at lower temperature (e.g. 30°C) results in reduced plasmid dimerization and improved downstream fermentation production yields with traditionally poor yielding plasmids (Williams et al., 2008; Carnes et al., 2008).
13.3. Insertion elements
Insertion of transposable elements into plasmids during production may be an issue with some processes and plasmids. Prather et al. (2006) report high frequency transposition of insertion element 1 (IS1) into the promoter of the neo selectable marker during cell line creation in defined media while Posfai et al. (2006) identified IS1 transposition into plasmids propagated in standard E. coli strains. Thus, the cell bank-fermentation process must be carefully engineered to limit insertion sequence activation, especially with defined media processes.
14. Fermentation Processes for Plasmid Production
Plasmid fermentation processes ideally maximize both the volumetric yield (mg/L) and specific yield (mg/g DCW or mg/OD600/L) of high quality supercoiled plasmid. High volumetric yields facilitate smaller and more economical fermentations, while high specific yield drastically improves plasmid purity and yield in downstream processing. The fermentation process should also be optimized to retain a high percentage of supercoiled plasmid since other plasmid forms (e.g. nicked) are difficult to remove during purification and are considered undesirable isoforms by regulatory agencies (FDA, 1996). Media optimization needs to be carefully performed, since differences in growth media carbon:nitrogen ratio can dramatically affect cell harvest and lysis characteristics (O’Kennedy et al., 2000) as can cell storage conditions (Kong et al., 2008). Thus, fermentation media and processes needs to be carefully optimized for plasmid yield, plasmid quality (% supercoiling), and compatibility of the resultant cells for harvest and lysis.
Temperature can be employed to selectively induce temperature sensitive plasmid amplification. Of known origin mutations, the pUC mutation is well suited for this application since its copy number can be shifted from a low level (30°C) to a very high level (42°C). Higher copy numbers of pUC plasmid are obtained by temperature shift to 42°C rather than 37°C in restricted growth fed-batch fermentation (Carnes et al., 2006). The mutated RNAII primer in pUC may be totally resistant to RNAI inhibition at 42°C (runaway replication), but only partially resistant at 37°C. It is possibly that uncharged tRNA-mediated RNAI inhibition (see Section 5.2) in restricted grown amino acid starved cells contributes to runaway replication. Indeed, uncharged tRNA-mediated RNAI inhibition increased stationary phase plasmid yields at 37°C with lower copy pBR322 based origins (Carnes et al., 2006) and media and process designs to increase levels of uncharged tRNAs may have application to improve 37°C pUC fermentations (reviewed in Wang et al., 2004). Transcription initiated from the weak RNAII primer promoter may be a key rate limiting factor affecting pUC plasmid copy number and production yields. However, unregulated RNAII promoter up mutations or alternative promoters may increase copy number at 30°C, increasing metabolic burden during the critical early growth phase, ultimately reducing productivity (see Section 16).
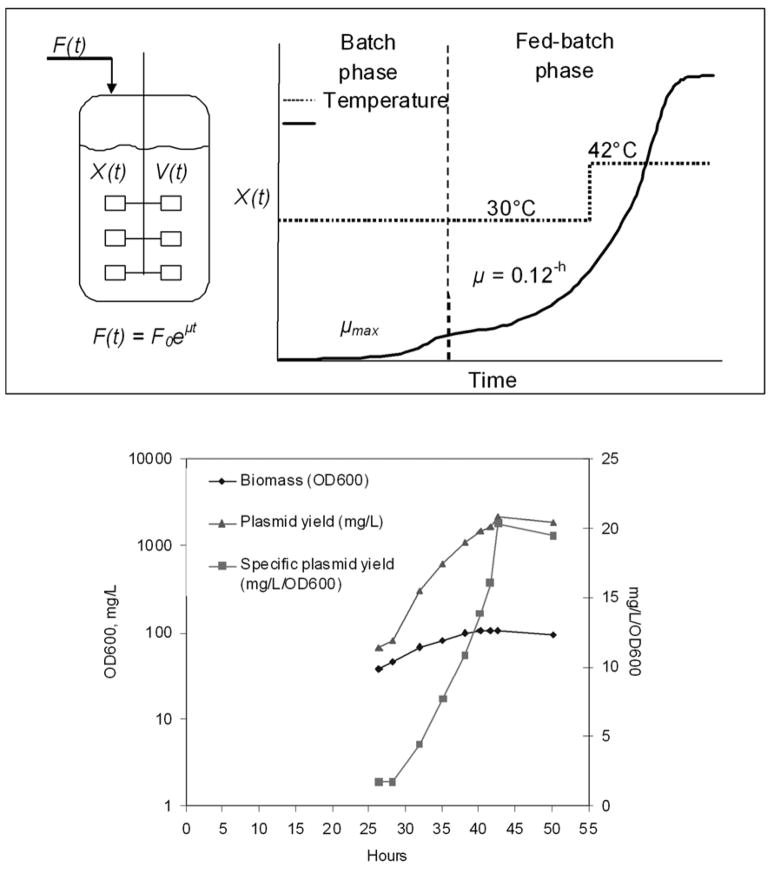
14.1. Fed-batch fermentation processes
For fed-batch fermentation (Figure 5A), cells are inoculated into an initial volume of medium that contains all non-limiting nutrients and an initial concentration of the limiting substrate. After this batch growth phase, controlled feeding of the limiting nutrient begins once the cells have consumed the initial amount of substrate allowing control of growth rate < μmax during the fed-batch phase. Feeding strategies are either feedback controlled (e.g. DO-stat, pH stat, metabolic activity, biomass concentration, substrate concentration), or predetermined (e.g. constant, linear, stepwise, or exponential feeding). Metabolic overflow from excess substrate is reduced, avoiding excessive formation of inhibitory acetate (reviewed in Carnes, 2005; Carnes and Williams, 2007).
Figure 5.
A. Inducible fed-batch plasmid production fermentation process. B. E. coli strain DH5α + pNTCUltra1 plasmid inducible fed-batch fermentation growth and plasmid yield profiles. Fermentation was shifted from 30 to 42°C at 29hrs to induce plasmid production, plasmid yield reached 2130 mg/L.
14.2. R6K origin
Fermentation production yields are an issue with R6K origin vectors: moderate yields of 100mg/L yield in fed-batch fermentation using minimal media are reported for the pCOR vector system. Production of an alternative R6K origin containing vector, derived from pCpG (Invitrogen, Carlsbad, CA) in either E. coli strain GT115 (Table 2; Invivogen, San Diego, CA) or DH10B:pir116, also had similarly low fermentation yields of up to 250 mg/L and low specific yields of up to 1.4 mg plasmid/g dry cell weight (DCW) (Hebel et al., 2008). Mutagenesis of the pir-116 replication protein and selection for increased copy number has been used to make new production strains (TEX2pir42) with reported yields of 205 mg/L (7.6 mg plasmid/g DCW; Soubrier, 2004) or 421 mg/L (116 OD600; Soubrier et al., 2005; this is 7.3 mg plasmid/g DCW assuming 50 g DCW/100 OD600/L). These yields are 5–10 fold reduced compared to that obtained with optimal pUC origin vectors that can reach 2.2 g/L and 5% total dry cell weight (50 mg plasmid/g DCW) (see below).
14.3. pUC origin
Several batch and fed-batch plasmid fermentation processes have been described that provide yields between 100–250 mg/L plasmid (1–17 mg plasmid/g DCW) with pUC origin plasmids (Lahijani et al., 1996; Schmidt et al., 2003; reviewed in Carnes and Williams, 2007; Carnes, 2005; Hoare et al., 2006; Prather et al., 2003; Shamlou, 2003). A few high yield fed-batch processes (0.5–2.2 g/L, up to 51 mg plasmid/g DCW) have been described that enable cost effective plasmid manufacture. These processes all couple reduced growth rate during plasmid production with high copy replication origins in cells grown at 37°C (Listner et al., 2006; Huber et al., 2005b)or 42°C (Singer et al., 2009; Carnes et al., 2006; Carnes and Williams, 2006; reviewed in Carnes and Williams, 2007). Reduced growth rate increases copy number (Lin-Chao and Bremer, 1986; Seo and Bailey, 1985), % supercoiling (O’Kennedy et al., 2003) and plasmid retention. Plasmid yields at harvest from the Listner et al. (2006) and Carnes et al. (2006) processes are compared in Table 3. The Huber et al. (2005b) process is not compared, since maximum specific plasmid yields occur relatively early in the process and is lower during later biomass accumulation. Productivity yields of 2200 mg/L (25 mg plasmid/L/OD600, 51 mg/g DCW =5% DCW) with the pUC based pNTCUltra plasmid were obtained with the Carnes et al. (2006) in fed-batch fermentation process. The plasmid DNA from the process is predominantly supercoiled monomer (Williams et al., 2008). A 37°C −42°C shifted DH5α fed-batch fermentation of pUC19 (which was growth arrested at 22 OD600) has been reported that also reached 5% DCW specific yield (Danquah and Forde, 2008). Thus, the highest specific yield of plasmid/biomass obtained to date is about 5% DCW, and this has been obtained only with optimized fed-batch processes.
Table 3.
Plasmid Specific yields from Fed Batch Fermentation
| Media + | Strain | Plasmid | Fermentation Process | Cell Density (OD600 ) | Volumetric plasmid yield (mg/L) | Specific plasmid yield (mg/L/OD600) |
|---|---|---|---|---|---|---|
| SD | DH5α | pNTCUltra1 | 30°C growth 42°C induction1 |
88 | 2220 ++ | 25 ++ (51 mg/gDCW) |
| SD | DH5α | gWIZ-derived (kanR) | 30°C growth 42°C induction1 |
97 | 1070 ++ | 11 ++ |
| SD | DH5α | gWIZ-derived (ampR) | 30°C growth 42°C induction2 |
141 | 991 | 7 |
| SD | BL21 | gWIZ-derived (ampR) | 30°C growth 42°C induction2 |
187 | 1923 | 10 |
| D | DH5 | pV1JNS3 derived | 37°C throughout3 | 90 | 1600 | 18 (39 mg/gDCW) |
+ SD = semidefined, D = defined
++ pNTCUltra1 volumetric and specific yields are approximately 2 fold higher than gWIZ
= Listner et al. (2005)
14.4. Theoretical plasmid yields
Studies on replication machinery overproduction of chromosomal DNA (i.e. when driven from an integrated plasmid R1 runaway replicon; Bernander et al., 1989) showed that the upper limit for chromosomal DNA content tolerated in the cell may be a 3–4 fold increase. Studies with runaway R plasmids have achieved 75–80% total DNA as plasmid (up to 3x relative to chromosomal; >2 fold increase in total DNA, standardized to protein). Thus, utilizing host replication systems in current strains, if all the additional replication capacity was directed to plasmid production, plasmid yields of 300–400% of chromosomal (75–80% total DNA; Uhlin et al., 1979) may be theoretically possible. This may represent an upper limit for cell tolerance, as this is the maximum obtained with a variety of different sized plasmids, and is associated with viability loss and altered cell morphology (Uhlin and Nordstrom, 1978, Nordstrom and Uhlin, 1992). Projecting these yields to % DCW is difficult, since the number of genomes per cell and cell size is dependent on strain, growth rate, and media composition. For example, while unrestricted logarithmic growing cultures can contain 2, 4 or even 8 copies of the chromosome (Skarstad et al., 1985), rapidly growing cells are much larger than growth-restricted cells (Akerlund et al., 1996; Churchward et al., 1982) such that smaller slow growing cells can contain 5% chromosomal DNA/DCW while logarithmically growing larger cells contain 2% chromosomal DNA/DCW (reviewed in Bremer and Dennis, 1996). The R plasmid runaway replication studies were generally performed with exponentially growing cultures, so this would predict 6–8% plasmid DNA/DCW is the maximum limit (i.e. 300–400% of chromosomal, assuming 2% chromosomal DNA/DCW). This of course assumes the capacity of cells for DNA does not vary depending on strain, cell size or growth rate.
15. Metabolic burden limits plasmid productivity
Generally, lower growth rate favors reduced selection against plasmid containing cells because plasmid presence reduces the maximum growth rate. While the optimal temperature for E. coli growth is 37°C, lower temperatures (such as 30°C) may be used in the batch phase of fed-batch fermentation to both cause reduced maximum specific growth rate and reduced plasmid copy number (Carnes et al., 2006; Carnes and Williams, 2006). This combination reduces metabolic burden and plasmid loss during the batch phase by minimizing growth difference between plasmid-containing and plasmid-free cells. This will also prevent the accumulation of uncharged tRNAs, and unwanted early plasmid amplification by antisense inhibition of RNAI. Plasmid production is then induced by temperature shifting to 42°C after biomass production (Figure 5A, B; Carnes et al., 2006; Carnes and Williams, 2006).
Alternative strategies developed to reduce plasmid mediated metabolic stress aim to alleviate growth rate inhibition at 37°C. For example, engineering a strain to overproduce intermediaries of the pentose-phosphate pathway (required for maintaining nucleotide precursor pools, plasmid replication and protein production) by overexpression of glucose-6-phosphate dehydrogenase (zwf) partially alleviated plasmid mediated growth rate reduction (Flores et al., 2004). The ribose-5-phosphate isomerase A (rpiA) gene product is downregulated in BL21 cells containing a high copy plasmid; overexpression of rpiA improved plasmid copy number (Wang et al., 2006). However, while overexpression of zwf-rpiA in the inducible fed-batch process of Carnes et al.(2006) increased the rate of plasmid induction, it did not affect final plasmid yield, indicating nucleotide precursor pools do not limit plasmid copy number at 42°C (Williams et al., 2008). Inactivation of a global regulator, FruR, in DH5α increased both growth rate of plasmid bearing cells at 37°C (Ow et al., 2007) and copy number by 21% to 19.2 mg/gDCW (Ow et al., 2009). The effect of the fruR knockout on plasmid production in a low metabolic burden high yielding process, if any, is unknown.
Protein overexpression from plasmids also places a metabolic load on the host cell that increases oxygen demand (Kay et al., 2003) and may limit plasmid productivity (reviewed in Ricci and Hernandez, 2000; Glick, 1995). For example, fermentation of a kanamycin resistant (kanR) plasmid at 37°C in a batch defined media fermentation process resulted in reduced growth rate and biomass, and overflow metabolism; overexpression of the kanR gene product to 18% of total protein was speculated as the cause of these effects (Rozkov et al., 2004). Plasmid protein expression is reduced by growth at 30°C during biomass accumulation, by lowering both copy number and copy number-dependent kanR production (Carnes et al., 2006).
15.1. Low metabolic burden process improves yield
As described above, growth at reduced temperature (e.g. 30°C) during the batch phase eliminates the need for engineering cells to reduce metabolic burden during the biomass accumulation phase, since metabolic burden is inherently reduced or eliminated by the process (Williams et al., 2008). High metabolic burden during early growth may account for the generally poor plasmid yields with most fermentation processes using a variety of media, since these typically are performed at 37°C throughout (reviewed in Carnes and Williams 2007; Prather et al., 2003). Consistent with this, a recent report compared 35°C versus 37°C growth during the batch phase, with subsequent fed-batch production at 37°C under restricted growth rate. Production yields with 35°C batch phase growth were dramatically improved (250 mg/L versus 50 mg/L; OMahony et al., 2007). Interestingly, creation of seed stocks from transformations performed at 30°C, rather than 37°C, also dramatically increased plasmid yield, productivity and seed stock viability with traditionally low yield plasmids (Williams et al., 2008; Carnes et al., 2008) indicating a general benefit on plasmid productivity of reduced temperature growth in all steps prior to the growth controlled restricted fed-batch stage. Low metabolic burden seed stock-fermentation processes allow generic production of a spectrum of vectors of both good and bad design; this is critical for contract manufacturers with no control of incoming vector design.
16. Conclusions
Methods to design licensable DNA vaccine vectors that combine high yield and integrity during bacterial production with increased expression in mammalian cells are presented. While all new vectors will require extensive preclinical testing to validate safety and performance prior to clinical use, regulatory testing burden for follow-on products can be reduced by combining carefully designed synthetic genes with existing validated vector backbones. Utilizing recently developed low metabolic burden seed stock and fermentation strategies, even poorly designed vectors can now be produced in usable yields, while optimized vectors can be manufactured in high yields exceeding 2 g/L, with specific plasmid yields of 5% total DCW. While specific yield may be approaching a maximum, combining high-yield low metabolic stress processes with alternative cell lines capable of producing 2–3 fold more biomass in fed-batch fermentation (reviewed in Shiloach and Fass, 2005) may further improve plasmid yields to 4–6 g/L. Collectively, DNA vaccination efficacy and cost of goods will be improved using new vectors and processes that combine improved potency with more economical manufacture.
Acknowledgments
The authors would like to thank Justin Vincent for preparation of Figures.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abeel T, et al. Generic eukaryotic core promoter prediction using structural features of DNA. Genome Res. 2008;18:310–23. doi: 10.1101/gr.6991408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akerlund T, Nordstrom K, Bernander R. Analysis of cell size and DNA content in exponentially growing and stationary-phase batch cultures of Escherichia coli. J Bacteriol. 1995;177:6791–7. doi: 10.1128/jb.177.23.6791-6797.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allamane S, Jourdes P, Ratel D, Vicat JM, Dupre I, Laine M, Berger F, Benabid AL, Wion D. Bacterial DNA methylation and gene transfer efficiency. Biochem Biophys Res Commun. 2000;276:1261–4. doi: 10.1006/bbrc.2000.3603. [DOI] [PubMed] [Google Scholar]
- August JT, Pardoll DM, Guarnieri FG. Lysosomal targeting of immunogens. 5633234. 1997
- Azzoni AR, Ribeiro SC, Monteiro GA, Prazeres DMF. The impact of polyadenylation signals on plasmid nuclease-resistance and transgene expression. J Gene Med. 2007;9:392–402. doi: 10.1002/jgm.1031. [DOI] [PubMed] [Google Scholar]
- Bakkali M. Genome dynamics of short oligonucleotides: the example of bacterial DNA uptake enhancing sequences. PLoS ONE. 2007;2:e741. doi: 10.1371/journal.pone.0000741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barouch DH, et al. A human T-cell leukemia virus type 1 regulatory element enhances the immunogenicity of human immunodeficiency virus type 1 DNA vaccines in mice and nonhuman primates. J Virol. 2005;79:8828–34. doi: 10.1128/JVI.79.14.8828-8834.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barros De Arruda L, Chickhlikar PR, August JT, Marques ETA. DNA vaccine encoding human immunodeficiency virus-1 Gag, targeted to the major histocompatibility complex II compartment by lysosomal-associated membrane protein, elicits long-term memory response. Immunol. 2004;112:126–33. doi: 10.1111/j.1365-2567.2004.01823.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg CM, et al. pBR322-derived multicopy plasmids harboring large inserts are often dimers in Escherichia coli K-12. Plasmid. 1989;21:138–41. doi: 10.1016/0147-619x(89)90057-7. [DOI] [PubMed] [Google Scholar]
- Bernander R, Merryweather A, Nordstrom K. Overinitiation of replication of the Escherichia coli chromosome from an integrated runaway-replication derivative of Plasmid R1. J Bacteriol. 1989;171:674–83. doi: 10.1128/jb.171.2.674-683.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi CP, Benham CJ. WebSIDD: Server for Prediction of the Stress-induced Duplex Destabilized Sites in Superhelical DNA. Bioinformatics. 2004;20:1477–79. doi: 10.1093/bioinformatics/bth304. [DOI] [PubMed] [Google Scholar]
- Bichara M, Schumacher S, Fuchs RP. Genetic instability within monotonous runs of CpG sequences in Escherichia coli. Genetics. 1995;140:897–907. doi: 10.1093/genetics/140.3.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloquel C, Fabre E, Bureau MF, Scherman D. Plasmid DNA electrotransfer for intracellular and secreted proteins expression: new methodological developments and applications. J Gene Med. 2004;(Suppl 1):S11–23. doi: 10.1002/jgm.508. [DOI] [PubMed] [Google Scholar]
- Boyd AC, Popp F, Michaelis U, Davidson H, Davidson-Smith H, Doherty A, McLachlan G, Porteous DJ, Seeber S. Insertion of natural intron 6a–6b into a human cDNA-derived gene therapy vector for cystic fibrosis improves plasmid stability and permits facile RNA/DNA discrimination. J Gene Med. 1999;1:312–21. doi: 10.1002/(SICI)1521-2254(199909/10)1:5<312::AID-JGM55>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- Bremer H, Dennis PP. Modulation of chemical composition and other parameters of the cell by growth rate. In: Neidhardt FC, Curtiss R, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE, editors. Escherichia coli and Salmonella: Cellular and molecular biology. Vol. 2. Washington: ASM Press; 1996. pp. 1553–69. [Google Scholar]
- Brosius J. Toxicity of an overproduced foreign gene product in Escherichia coli and its use in plasmid vectors for the selection of transcription terminators. Gene. 1984;27:161–72. doi: 10.1016/0378-1119(84)90137-9. [DOI] [PubMed] [Google Scholar]
- Carnes AE. Fermentation design for the manufacture of therapeutic plasmid DNA. BioProcess Intl. 2005;3:36–44. [Google Scholar]
- Carnes AE, Hodgson CP, Williams JA. Inducible Escherichia coli fermentation for increased plasmid DNA production. Biotechnol Appl Biochem. 2006;45:155–66. doi: 10.1042/BA20050223. [DOI] [PubMed] [Google Scholar]
- Carnes AE, Hodgson CP, Williams JA. Improved E. coli plasmid DNA production. 60931465. 2008
- Carnes AE, Williams JA. Process for plasmid DNA fermentation. World Patent Application WO2006023546. 2006
- Carnes AE, Williams JA. Plasmid DNA manufacturing Technology. Recent Patent Biotechnol. 2007;1:151–6. doi: 10.2174/187220807780809436. [DOI] [PubMed] [Google Scholar]
- Chalker AF, Leach DR, Lloyd RG. Escherichia coli sbcC mutants permit stable propagation of DNA replicons containing a long palindrome. Gene. 1988;71:201–5. doi: 10.1016/0378-1119(88)90092-3. [DOI] [PubMed] [Google Scholar]
- Chapman BS, Thayer RM, Vincent KA, Haigwood NL. Effect of intron A from human cytomegalovirus (Towne) immediate-early gene on heterologous expression in mammalian cells. Nucleic Acids Res. 1991;19:2979–86. doi: 10.1093/nar/19.14.3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheah UE, Weigand WA, Stark BC. Effects of recombinant plasmid size on cellular processes in Escherichia coli. Plasmid. 1987;18:127–34. doi: 10.1016/0147-619x(87)90040-0. [DOI] [PubMed] [Google Scholar]
- Chen JD, Morrision DA. Cloning of Streptococcus pneumoniae DNA fragments in Escherichia coli requires vectors protected by strong transcriptional terminators. Gene. 1987;55:179–87. doi: 10.1016/0378-1119(87)90278-2. [DOI] [PubMed] [Google Scholar]
- Chen W, Kallio PT, Bailey JE. Construction and characterization of a novel cross-regulation system for regulating cloned gene expression in Escherichia coli. Gene. 1993;130:15–22. doi: 10.1016/0378-1119(93)90341-y. [DOI] [PubMed] [Google Scholar]
- Chen ZY, Riu E, He CY, Xu H, Kay MA. Silencing of episomal transgene expression in liver by plasmid bacterial backbone DNA is independent of CpG methylation. Mol Ther. 2008;16:548–56. doi: 10.1038/sj.mt.6300399. [DOI] [PubMed] [Google Scholar]
- Chiang CS, Bremer H. Stability of pBR322-derived plasmids. Plasmid. 1988;20:207–20. doi: 10.1016/0147-619x(88)90027-3. [DOI] [PubMed] [Google Scholar]
- Churchward G, Bremer H, Young R. Macromolecular composition of bacteria. J Theor Biol. 1982;94:651–70. doi: 10.1016/0022-5193(82)90305-8. [DOI] [PubMed] [Google Scholar]
- Connelly JC, de Leau ES, Leach DR. DNA cleavage and degradation by the SbcCD protein complex from Escherichia coli. Nucleic Acids Res. 1999;27:1039–46. doi: 10.1093/nar/27.4.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connelly JC, Leach DR. The sbcC and sbcD genes of Escherichia coli encode a nuclease involved in palindrome inviability and genetic recombination. Genes Cells. 1996;1:285–91. doi: 10.1046/j.1365-2443.1996.23024.x. [DOI] [PubMed] [Google Scholar]
- Cooke JR, McKie EA, Ward JM, Keshavarz-Moore E. Impact of intrinsic DNA structure on processing of plasmids for gene therapy and DNA vaccines. J Biotechnol. 2004;114:239–54. doi: 10.1016/j.jbiotec.2004.06.011. [DOI] [PubMed] [Google Scholar]
- Cranenburgh RM. Plasmid Maintenance. World Patent Application WO2005052167. 2005
- Cranenburgh RM, Hanak JA, Williams SG, Sherratt DJ. Escherichia coli strains that allow antibiotic-free plasmid selection and maintenance by repressor titration. Nucleic Acids Res. 2001;29:E26. doi: 10.1093/nar/29.5.e26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cromie GA, Millar CB, Schmidt KH, Leach DR. Palindromes as substrates for multiple pathways of recombination in Escherichia coli. Genetics. 2000;154:513–22. doi: 10.1093/genetics/154.2.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crouzet J, Cameron B. Method for producing therapeutic DNA. 20020187527. 2002
- Danquah MK, Forde GM. Development of a pilot-scale bacterial fermentation for plasmid based biopharmaceutical production using stoichiometric medium. Biotech Bioprocess Engineering. 2008;13:158–67. [Google Scholar]
- Darquet AM, Cameron B, Wils P, Scherman D, Crouzet J. A new DNA vehicle for nonviral gene delivery: supercoiled minicircles. Gene Ther. 1997;4:1341–9. doi: 10.1038/sj.gt.3300540. [DOI] [PubMed] [Google Scholar]
- Delogu G, Howard A, Collins FM, Morris SL. DNA vaccination against tuberculosis: Expression of a Ubiquitin-conjugated tuberculosis protein enhances antimycobacterial immunity. Infect Immun. 2000;68:3097–102. doi: 10.1128/iai.68.6.3097-3102.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Solar G, Giraldo R, Ruiz-Echevarria MJ, Espinosa M, Diaz-Orejas R. Replication and control of circular bacterial plasmids. Microbiol Molec Biol Reviews. 1998;62:434–64. doi: 10.1128/mmbr.62.2.434-464.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durland RH, Eastman EM. Manufacturing and quality control of plasmid-based gene expression systems. Adv Drug Deliver Rev. 1998;30:33–48. doi: 10.1016/s0169-409x(97)00105-1. [DOI] [PubMed] [Google Scholar]
- EMEA. Note for guidance on the quality, preclinical and clinical aspects of gene transfer medicinal products. European Medicines Agency Doc. Ref. CPMP/BWP/3088/99, 2001
- EMEA. Guideline on non-clinical studies required before first clinical use of gene therapy medicinal product. European Medicines Agency Doc. Ref. EMEA/CHMP/GTWP/125459/2006, 2006
- EMEA. Overview of comments received on guideline on non-clinical studies required before first clinical use of gene therapy medicinal products. European Medicines Agency Doc. Ref. EMEA/CHMP/GTWP/65260/2008, 2008
- Engels P, Meyer P. Inactivation of the transcriptional-dependent inhibition of plasmid replication: a selection method for cloning large DNA fragments. Biotechniques. 1993;14:324–5. [PubMed] [Google Scholar]
- FDA. Guidance for industry: Considerations for Plasmid DNA Vaccines for Infectious Disease Indications. US Food and Drug Administration, 2007
- FDA. Guidance for industry: Guidance for human somatic cell therapy and gene therapy. US Food and Drug Administration, 1998
- FDA. Points to Consider on Plasmid DNA Vaccines for Preventive Infectious Disease Indications. US Food and Drug Administration, 1996
- Fishel RA, James AA, Kolodner R. recA-independent general genetic recombination of plasmids. Nature. 1981;294:184–6. doi: 10.1038/294184a0. [DOI] [PubMed] [Google Scholar]
- Flores S, Anda-Herrera R, Gosset G, Bolivar FG. Growth-rate recovery of Escherichia coli cultures carrying a multicopy plasmid, by engineering of the pentose-phosphate pathway. Biotech Bioeng. 2004;87:485–94. doi: 10.1002/bit.20137. [DOI] [PubMed] [Google Scholar]
- Fuller J. Nucleic Acid Constructs. World Patent Application WO2006082398. 2006
- Gerber L, Kodukula K, Udenfriend S. Phosphatidylinositol Glycan (PI-G) anchored Membrane Proteins. J Biol Chem. 1992;267:12168–73. [PubMed] [Google Scholar]
- Ghersa P, Whelan J, Pescini R, DeLamarter JF, Hooft van Huijsduijnen R. Commonly used cat reporter vectors contain a cAMP-inducible, cryptic enhancer that co-operates with NF-kappa B-sites. Gene. 1994;151:331–2. doi: 10.1016/0378-1119(94)90681-5. [DOI] [PubMed] [Google Scholar]
- Glenting J, Wessels S. Ensuring safety of DNA vaccines. Microbial Cell Factories. 2005;4:26–31. doi: 10.1186/1475-2859-4-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glick BR. Metabolic load and heterologous gene expression. Biotechnol Adv. 1995;13:247–61. doi: 10.1016/0734-9750(95)00004-a. [DOI] [PubMed] [Google Scholar]
- Glickman BW, Radman M. Escherichia coli mutator mutants deficient in methylation-instructed DNA mismatch correction. Proc Natl Acad Sci USA. 1980;77:1063–7. doi: 10.1073/pnas.77.2.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Eichelmann MC, Levy-Mustri A, Ramirez-Santos J. Presence of 5-methylcytosine in CC(A/T)GG sequences (Dcm methylation) in DNAs from different bacteria. J Bacteriol. 1991;173:7692–4. doi: 10.1128/jb.173.23.7692-7694.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr R, Pfaffenzeller I. Host-vector system for antibiotic-free ColE1 plasmid propagation. World Patent Application WO2006029985. 2006
- Gu W, Zhou T, Ma J, Sun X, Lu Z. Folding type specific secondary structure propensities of synonymous codons. IEEEE Trans Nanobioscience. 2003;2:150–7. doi: 10.1109/tnb.2003.817024. [DOI] [PubMed] [Google Scholar]
- Gursel I, Gursel M, Yamada H, Ishii KJ, Takeshita F, Klinman DM. Repetitive elements in mammalian telomeres suppress bacterial DNA-induced immune activation. J Immunol. 2003;171:1393–400. doi: 10.4049/jimmunol.171.3.1393. [DOI] [PubMed] [Google Scholar]
- Hamann CW, Nielsen J, Ingersle E. Novel Plasmids for use in medicine and method of producing same. World Patent Application WO0028048. 2002
- Hartikka J, et al. An improved plasmid DNA expression vector for direct injection into skeletal muscle. Hum Gene Ther. 1996;7:1205–17. doi: 10.1089/hum.1996.7.10-1205. [DOI] [PubMed] [Google Scholar]
- Hebel HL, Cai Y, Davies LA, Hyde SC, Pringle IA, Gill DR. Challenges in the process development of a novel Zero-CpG CFTR plasmid for human clinical use. ASGT. 2008 [Google Scholar]
- Ho PS, Ellison MJ, Quigley GJ, Rich A. A computer aided thermodynamic approach for predicting the formation of Z-DNA in naturally occurring sequences. EMBO J. 1986;5:2737–44. doi: 10.1002/j.1460-2075.1986.tb04558.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoare M, Levy S, Bracewell DG, Doig SD, Kong S, Titchener-Hooker N, Ward JM, Dunnill P. Bioprocess engineering issues that would be faced in producing a DNA vaccine at up to 100 m3 fermentation scale for an influenza pandemic. Biotechnol. Prog. 2005;21:1577–92. doi: 10.1021/bp050190n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann KH, Neubauer P, Riethdorf S, Hecker M. Amplification of pBR322 plasmid DNA in Escherichia coli relA strains during batch and fed-batch fermentation. J Basic Microbiol. 1990;30:37–41. doi: 10.1002/jobm.3620300111. [DOI] [PubMed] [Google Scholar]
- Huang Y, et al. A recoding method to improve the humoral response to an HIV DNA vaccine. PLoS ONE. 2008;3:e3214. doi: 10.1371/journal.pone.0003214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber H, Pacher C, Necina R, Kollmann F, Reinisch C. Method for producing plasmid DNA on a manufacturing scale by fermentation of the Escherichia coli K-12 strain JM108. World Patent Application WO2005098002. 2005
- Huber H, Weigl G, Buchinger W. Fed-batch fermentation process and culture medium for the production of plasmid DNA in E. coli on a manufacturing scale. World Patent Application WO2005097990. 2005
- Ioshiknhes I, Roy S, Sen CK. Algorithms for mapping of mRNA targets for MicroRNA. DNA Cell Biology. 2007;26:265–72. doi: 10.1089/dna.2006.0566. [DOI] [PubMed] [Google Scholar]
- Ishii KJ, et al. TANK-binding kinase-1 delineates innate and adaptive immune responses to DNA vaccines. Nature. 2008;451:725–9. doi: 10.1038/nature06537. [DOI] [PubMed] [Google Scholar]
- Kato M. Polypyrimidine/polypurine sequence in plasmid DNA enhances formation of dimer molecules in Escherichia coli. Dimerization of plasmid DNA in Escherichia coli. Mol Biol Rep. 1993;18:183–7. doi: 10.1007/BF01674429. [DOI] [PubMed] [Google Scholar]
- Kay A, O’Kennedy RD, Ward J, Keshavarz-Moore E. Impact of plasmid size on cellular oxygen demand in Escherichia coli. Biotechnol Appl Biochem. 2003;38:1–7. doi: 10.1042/BA20030022. [DOI] [PubMed] [Google Scholar]
- Kong S, Rock CF, Booth A, Willoughby N, O’Kennedy RD, Relton J, Ward JM, Hoare M, Levy MS. Large-scale plasmid DNA processing: evidence that cell harvesting and storage methods affect yield of supercoiled plasmid DNA. Biotechnol Appl Biochem. 2008;51:43–51. doi: 10.1042/BA20070174. [DOI] [PubMed] [Google Scholar]
- Kozak M. Recognition of AUG and alternative initiator codons is augmented by G in position +4 but is not generally affected by the nucleotides in position +5 and +6. EMBO J. 1997;16:2482–92. doi: 10.1093/emboj/16.9.2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieg AM. Direct immunologic activities of CpG DNA and implications for gene therapy. J Gene Med. 1999;1:56–63. doi: 10.1002/(sici)1521-2254(199901/02)1:1<56::aid-jgm5>3.3.co;2-y. [DOI] [PubMed] [Google Scholar]
- Krieg AM, Davis HL, Wu T, Schorr J. Vectors and methods for immunization or therapeutic protocols. 6339068. 2002
- Kushner PJ, Baxter JD, Duncan KG, Lopez GN, Schaufele F, Uht RM, Webb P, West BL. Eukaryotic regulatory elements lurking in plasmid DNA: the activator protein-1 site in pUC. Mol Endocrinol. 1994;8:405–7. doi: 10.1210/mend.8.4.8052261. [DOI] [PubMed] [Google Scholar]
- Kutzler MA, Weiner DB. DNA vaccines: ready for prime time? Nature Reviews Genetics. 2008;9:776–88. doi: 10.1038/nrg2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laddy DJ, Yan J, Corbitt N, Kobasa D, Kobinger GP, Weiner DB. Immunogenicity of novel consensus-based DNA vaccines against avian influenza. Vaccine. 2007;25:2984–9. doi: 10.1016/j.vaccine.2007.01.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahijani R, Hulley G, Soriano G, Horn NA, Marquet M. High-yield production of pBR322-derived plasmids intended for human gene therapy by employing a temperature-controllable point mutation. Hum Gene Ther. 1996;7:1971–80. doi: 10.1089/hum.1996.7.16-1971. [DOI] [PubMed] [Google Scholar]
- Lai YF, Tseng YJ, Yang FY, Au LC. Mammalian cis-reporting plasmid may alter activities due to the derivation of host Escherichia coli strains. Anal Biochem. 2008;376:103–7. doi: 10.1016/j.ab.2008.01.030. [DOI] [PubMed] [Google Scholar]
- Lee J, Him HC, Kim SW, Hong SI, Park YH. Interplay of SOS induction, recombinant gene expression, and multimerization of plasmid vectors in Escherichia coli. Biotechnol Bioeng. 2002;80:84–92. doi: 10.1002/bit.10354. [DOI] [PubMed] [Google Scholar]
- Leifert JA, Rodriguez-Carreno MP, Rodriguez F, Whitton JL. Targeting plasmid-encoded proteins to the antigen presentation pathways. Immunological Review. 2004;199:40–53. doi: 10.1111/j.0105-2896.2004.0135.x. [DOI] [PubMed] [Google Scholar]
- Leite JP, Cousin C, Heysen A, D’Halluin JC. Negative effect of a cis-acting pBR322 element on adenovirus E1a gene expression. Gene. 1989;82:351–6. doi: 10.1016/0378-1119(89)90062-0. [DOI] [PubMed] [Google Scholar]
- Levy J. Avoidance of undesirable replication intermediates in plasmid propagation. 7390654. 2003
- Li HS, et al. Enhancement of DNA vaccine-induced immune responses by a 72-bp element from SV40 enhancer. Chin Med J. 2007;120:496–502. [PubMed] [Google Scholar]
- Li S, MacLaughlin FC, Fewell JG, Gondo M, Wang J, Nicol F, Dean DA, Smith LC. Muscle-specific enhancement of gene expression by incorporation of SV40 enhancer in the expression plasmid. Gene Ther. 2001;8:494–7. doi: 10.1038/sj.gt.3301419. [DOI] [PubMed] [Google Scholar]
- Lin-Chao S, Bremer H. Effect of the bacterial growth rate on replication control of plasmid pBR322 in Escherichia coli. Mol Gen Genet. 1986;203:143–9. doi: 10.1007/BF00330395. [DOI] [PubMed] [Google Scholar]
- Lin-Chao S, Chen W, Wong T. High copy number of the pUC plasmid results from a Rom/Rop-suppressible point mutation in RNAII. Mol Micro. 1992;6:3385–93. doi: 10.1111/j.1365-2958.1992.tb02206.x. [DOI] [PubMed] [Google Scholar]
- Listner K, et al. Development of a highly productive and scalable plasmid DNA production platform. Biotechnol Prog. 2006;22:1335–45. doi: 10.1021/bp060046h. [DOI] [PubMed] [Google Scholar]
- Lobner-Olesen A, Skovgaard O, Marinus MG. Dam methylation: coordinating cellular processes. Current Opin Microbiol. 2005;8:154–60. doi: 10.1016/j.mib.2005.02.009. [DOI] [PubMed] [Google Scholar]
- Luke J, Carnes AE, Hodgson CP, Williams JA. Improved antibiotic-free DNA vaccine vectors utilizing a novel RNA based plasmid selection system. Vaccine. 2008 doi: 10.1016/j.vaccine.2009.06.017. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mairhofer J, Grabherr R. Rational vector design for efficient non-viral gene delivery: challenges facing the use of plasmid DNA. Mol Biotechnol. 2008;39:97–104. doi: 10.1007/s12033-008-9046-7. [DOI] [PubMed] [Google Scholar]
- Mairhofer J, Pfaffenzeller I, Merz D, Grabherr R. A novel antibiotic free plasmid selection system: Advances in safe and efficient DNA therapy. Biotechnol J. 2008;3:83–9. doi: 10.1002/biot.200700141. [DOI] [PubMed] [Google Scholar]
- Manoj S, Babiuk LA, van Drunen Littel-van den Hurk S. Approaches to enhance the efficacy of DNA vaccines. Critical Rev Clin Lab Sci. 2004;41:1–39. doi: 10.1080/10408360490269251. [DOI] [PubMed] [Google Scholar]
- Marinus MG, Carraway M, Frey AZ, Brown L, Arraj AJ. Insertion mutants in the dam gene of Escherichia coli K-12. Mol Gen Genet. 1983;192:288–9. doi: 10.1007/BF00327681. [DOI] [PubMed] [Google Scholar]
- Masai H, Arai K. Mechanisms of primer RNA synthesis and D-loop/R-loop-dependent DNA replication in Escherichia coli. Biochimie. 1996;78:1109–17. doi: 10.1016/s0300-9084(97)86737-5. [DOI] [PubMed] [Google Scholar]
- McGlynn P, Guy CP. Replication forks blocked by protein-DNA complexes have limited stability in vitro. J Mol Biol. 2008;381:249–55. doi: 10.1016/j.jmb.2008.05.053. [DOI] [PubMed] [Google Scholar]
- Middaugh CR, Evans RK, Montgomery DL, Casimiro DR. Analysis of plasmid DNA from a pharmaceutical perspective. J Pharmaceutical Sciences. 1997;87:130–46. doi: 10.1021/js970367a. [DOI] [PubMed] [Google Scholar]
- Mirkin EV, Mirkin SM. Replication fork stalling at natural impediments. Microbiol Molec Biol Reviews. 2007;71:13–35. doi: 10.1128/MMBR.00030-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neylon C, Kralicek AV, Hill TM, Dixon NE. Replication termination in E. coli: Structure and antihelicase activity of the Tus-Ter complex. Microbiol Molec Biol Reviews. 2005;69:501–526. doi: 10.1128/MMBR.69.3.501-526.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordstrom K, Uhlin BE. Runway replication plasmids as tools to produce large quantities of proteins from cloned genes in bacteria. Bio/Technology. 1992;10:661–6. doi: 10.1038/nbt0692-661. [DOI] [PubMed] [Google Scholar]
- Ochiai H, Harashima H, Kamiya H. Effect of methylated adenine in plasmid DNA on transgene expression in mice. Biol Pharm Bull. 2005;28:2019–22. doi: 10.1248/bpb.28.2019. [DOI] [PubMed] [Google Scholar]
- O’Kennedy RD, Baldwin C, Keshavarz-Moore E. Effects of growth medium selection on plasmid DNA production and initial processing steps. J Biotechnol. 2000;76:175–83. doi: 10.1016/s0168-1656(99)00187-x. [DOI] [PubMed] [Google Scholar]
- O’Kennedy RD, Ward JM, Keshavarz-Moore E. Effects of fermentation strategy on the characteristics of plasmid DNA production. Biotechnol Appl Biochem. 2003;37:83–90. doi: 10.1042/ba20020099. [DOI] [PubMed] [Google Scholar]
- O’Mahony K, Freitag R, Hilbrig F, Muller P, Schumacher I. Strategies for high titre plasmid DNA production in Escherichia coli DH5α. Process Biochem. 2007;42:1039–49. [Google Scholar]
- Ow DS, Lee RM, Nissom PM, Philp R, Oh SK, Yap MG. Inactivating FruR global regulator in plasmid-bearing Escherichia coli alters metabolic gene expression and improves growth rate. J Biotechnol. 2007;131:261–9. doi: 10.1016/j.jbiotec.2007.07.508. [DOI] [PubMed] [Google Scholar]
- Ow DS, Yap MG, Oh SK. Enhancement of plasmid DNA yields during fed-batch culture with a fruR knockout Escherichia coli strain. Biotechnol Appl Biochem. 2009;52:53–9. doi: 10.1042/BA20070260. [DOI] [PubMed] [Google Scholar]
- Pan X, Leach DR. The roles of mutS, sbcCD and recA in the propagation of TGG repeats in Escherichia coli. Nucleic Acids Res. 2000;28:3178–84. doi: 10.1093/nar/28.16.3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M, Lin X, Salzberg SL. GeneSplicer: a new computational method for splice site prediction. Nucleic Acids Res. 2001;29:1185–90. doi: 10.1093/nar/29.5.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson DO, Beifuss KK, Morley KL. Context-Dependent Gene expression: cis-acting negative effects of specific prokaryotic plasmid sequences on eukaryotic genes. Molec Cell Biol. 1987;7:1563–7. doi: 10.1128/mcb.7.4.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phue JN, Lee SJ, Trinh L, Shiloach J. Modified Escherichia coli B (BL21), a superior producer of plasmid DNA compared with Escherichia coli K (DH5α) Biotechnol Bioeng. 2008;101:831–6. doi: 10.1002/bit.21973. [DOI] [PubMed] [Google Scholar]
- Posfai G, et al. Emergent properties of reduced-genome Escherichia coli. Science. 2006;312:1044–6. doi: 10.1126/science.1126439. [DOI] [PubMed] [Google Scholar]
- Prather KL, Edmonds MC, Herod JW. Identification and characterization of IS1 transposition in plasmid amplification mutants of E. coli clones producing DNA vaccines. Appl Microbiol Biotechnol. 2006;73:815–26. doi: 10.1007/s00253-006-0532-1. [DOI] [PubMed] [Google Scholar]
- Prather KJ, Sagar S, Murphy J, Chartrain M. Industrial scale production of plasmid DNA for vaccine and gene therapy: plasmid design, production and purification. Enzyme Microbial Technol. 2003;33:865–83. [Google Scholar]
- Puigbo P, Guzman E, Romeu A, Garcia-Vallve S. Optimizer: a web server for optimizing the codon usage of DNA sequences. Nucleic Acids Res. 2007;35:W126–W131. doi: 10.1093/nar/gkm219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu JT, Liu B, Tian C, Pavlakis G, Yu XF. Enhancement of primary and secondary cellular immune responses against human immunodeficiency virus type 1 gag by using DNA expression vectors that target gag antigen to the secretory pathway. J Virol. 2000;74:5997–6005. doi: 10.1128/jvi.74.13.5997-6005.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanathan MP, Ayyavoo V, Weiner DB. Choice of expression vector alters the localization of a human cellular protein. DNA Cell Biol. 2001;20:101–5. doi: 10.1089/104454901750070300. [DOI] [PubMed] [Google Scholar]
- Ribeiro SC, Monteiro GA, Prazeres DMF. Evaluation of the effect of Non-B DNA structures on plasmid integrity via accelerated stability studies. J Pharmaceutical Sciences. 2008 doi: 10.1002/jps.21503. Epub. [DOI] [PubMed] [Google Scholar]
- Ribeiro SC, Oliveira PH, Prazeres DM, Monteiro GA. High frequency plasmid recombination mediated by 28 bp direct repeats. Mol Biotechnol. 2008;40:252–60. doi: 10.1007/s12033-008-9082-3. [DOI] [PubMed] [Google Scholar]
- Ricci JCD, Hernandez ME. Plasmid effects on Escherichia coli metabolism. Crit Rev Biotechnol. 2000;20:79–108. doi: 10.1080/07388550008984167. [DOI] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genetics. 2000;16:276–7. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- Roberts TL, Dunn JA, Terry TD, Jennings MP, Hume DA, Sweet MJ, Stacey KJ. Differences in macrophage activation by bacterial DNA and CpG-containing oligonucleotides. J Immunol. 2005;175:3569–76. doi: 10.4049/jimmunol.175.6.3569. [DOI] [PubMed] [Google Scholar]
- Rodriguez F, et al. DNA immunization with Minigenes: Low frequency of memory cytotoxic T lymphocytes and inefficient antiviral protection are rectified by ubiquitination. J Virol. 1998;72:5174–81. doi: 10.1128/jvi.72.6.5174-5181.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozkov A, Avignone-Rossa CA, Ertl PF, Jones P, O’Kennedy RD, Smith JJ, Dale JW, Bushell ME. Characterization of the metabolic burden on Escherichia coli DH1 cells imposed by the presence of a plasmid containing a gene therapy sequence. Biotechnol Bioeng. 2004;88:909–15. doi: 10.1002/bit.20327. [DOI] [PubMed] [Google Scholar]
- Saida F, Uzan M, Odaert B, Bontems F. Expression of highly toxic genes in E. coli: Special strategies and genetic tools Current Protein Peptide. Science. 2006;7:47–56. doi: 10.2174/138920306775474095. [DOI] [PubMed] [Google Scholar]
- Schalk JA, Mooi FR, Berbers GA, van Aerts LA, Ovelgonne H, Kimman TG. Preclinical and clinical safety studies on DNA vaccines. Human Vaccines. 2006;2:45–53. doi: 10.4161/hv.2.2.2620. [DOI] [PubMed] [Google Scholar]
- Schirmbeck R, Riedl P, Fissolo N, Lemonnier FA, Bertoleti A, Reimann J. Translation from cryptic reading frames of DNA vaccines generates an extended repertoire of immunogenic, MHC Class I-restricted epitopes. J Immunol. 2005;174:4647–56. doi: 10.4049/jimmunol.174.8.4647. [DOI] [PubMed] [Google Scholar]
- Schmidt T, Friehs K, Flaschel E, Schleef M. Method for the isolation of ccc plasmid DNA. 6664078. 2003
- Schroeter A, Riethdorf S, Hecker M. Amplification of different ColE1 plasmids in an Escherichia coli relA strain. J Basic Microbiol. 1988;28:553–5. doi: 10.1002/jobm.3620280818. [DOI] [PubMed] [Google Scholar]
- Seo JH, Bailey JE. Effects of recombinant plasmid content on growth properties and cloned gene product formation in Escherichia coli. Biotechnol Bioeng. 1985;27:1668–74. doi: 10.1002/bit.260271207. [DOI] [PubMed] [Google Scholar]
- Shamlou PA. Scaleable processes for the manufacture of therapeutic quantities of plasmid DNA. Biotechnol Appl Biochem. 2003;37:207–18. doi: 10.1042/BA20030011. [DOI] [PubMed] [Google Scholar]
- Sheets RL, Stein J, Manetz TS, Andrews C, Bailer R, Rathmann J, Gomez PL. Toxicological safety evaluation of DNA plasmid vaccines against HIV-1, Ebola, Severe Acute Respiratory Syndrome, or West Nile virus is similar despite differing plasmid backbones or gene-inserts. Toxicol Sci. 2006;91:620–30. doi: 10.1093/toxsci/kfj170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheets RL, Stein J, Manetz TS, Duffy C, Nason M, Andrews C, Kong WP, Nabel GJ, Gomez PL. Biodistribution of DNA plasmid vaccines against HIV-1, Ebola, Severe Acute Respiratory Syndrome, or West Nile virus is similar, without integration, despite differing plasmid backbones or gene insert. Toxicol Sci. 2006;91:610–9. doi: 10.1093/toxsci/kfj169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherratt DJ, Williams SG, Hanak JAJ. Plasmid stabilization. 5972708. 1999
- Shiloach J, Fass R. Growing E. coli to high cell density – A historical perspective on method development. Biotechnol Adv. 2005;23:345–57. doi: 10.1016/j.biotechadv.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Singer A, Eiteman MA, Altman E. DNA plasmid production in different host strains of Escherichia coli. J. Ind Microbiol Biotechnol. 2009 doi: 10.1007/s10295-008-0522-7. epub. [DOI] [PubMed] [Google Scholar]
- Siregar JJ, Lerner SA, Mobashery S. Purification and characterization of aminoglycoside 3′-phosphotransferase Type IIa and kinetic comparison with a new mutant enzyme. Antimicrobial Agents Chemotherapy. 1994;38:641–7. doi: 10.1128/aac.38.4.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siregar JJ, Miroshnikov K, Mobashery S. Purification, characterization, and investigation of the mechanism of aminoglycoside 3′-phosphotransferase Type Ia. Biochemistry. 1995;34:12641–88. doi: 10.1021/bi00039a026. [DOI] [PubMed] [Google Scholar]
- Skarstad K, Steen HB, Boye E. Escherichia coli DNA distributions measured by flow cytometry and compared with theoretical computer simulations. J Bacteriol. 1985;163:661–8. doi: 10.1128/jb.163.2.661-668.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder LA, Satishchandran C. Chimeric kanamycin resistance gene. 5851804. 1998
- Soubrier F. Circular DNA molecule having a conditional origin of replication, process for their preparation and their use in gene therapy. World Patent Application WO2004033664. 2004
- Soubrier F, et al. pCOR: a new design of plasmid vectors for nonviral gene therapy. Gene Ther. 1999;6:1482–8. doi: 10.1038/sj.gt.3300968. [DOI] [PubMed] [Google Scholar]
- Soubrier F, Laborderie B, Cameron B. Improvement of pCOR plasmid copy number for pharmaceutical applications. Appl Microbiol Biotechnol. 2005;66:683–8. doi: 10.1007/s00253-004-1729-9. [DOI] [PubMed] [Google Scholar]
- Summers D. Timing, self-control and a sense of direction are the secrets of multicopy plasmid stability. Mol Microbiol. 1998;29:1137–45. doi: 10.1046/j.1365-2958.1998.01012.x. [DOI] [PubMed] [Google Scholar]
- Summers DK, Beton CW, Withers HL. Multicopy plasmid instability: the dimer catastrophe hypothesis. Mol Microbiol. 1993;8:1031–8. doi: 10.1111/j.1365-2958.1993.tb01648.x. [DOI] [PubMed] [Google Scholar]
- Szpirer CY, Milinkovitch MC. Separate-component-stabilization system for protein and DNA production without the use of antibiotics. Biotechniques. 2005;38:775–81. doi: 10.2144/05385RR02. [DOI] [PubMed] [Google Scholar]
- Tachedjian M, Boyle JS, Lew AM, Horvatic B, Scheerlinck JP, Tennent JM, Andrew ME. Gene gun immunization in a preclinical model is enhanced by B7 targeting. Vaccine. 2003;21:2900–5. doi: 10.1016/s0264-410x(03)00162-2. [DOI] [PubMed] [Google Scholar]
- Takeshita F, Ishii KJ. Intracellular DNA sensors in immunity. Current Opinion Immunology. 2008;20:383–8. doi: 10.1016/j.coi.2008.05.009. [DOI] [PubMed] [Google Scholar]
- Trevani AS, et al. Bacterial DNA activates human neutrophils by a CpG-independent pathway. Eur J Immunol. 2003;33:3164–74. doi: 10.1002/eji.200324334. [DOI] [PubMed] [Google Scholar]
- Tsuchiya H, Matsuda T, Harashima H, Kamiya H. Cytokine induction by a bacterial DNA-specific modified base. Biochem Biophys Res Commun. 2005;326:777–81. doi: 10.1016/j.bbrc.2004.11.115. [DOI] [PubMed] [Google Scholar]
- Tully DB, Cidlowski JA. pBR322 contains glucocorticoid regulatory element DNA consensus sequences. Biochem Biophys Res Commun. 1987;144:1–10. doi: 10.1016/s0006-291x(87)80467-9. [DOI] [PubMed] [Google Scholar]
- Uhlin BE, Molin S, Gustafsson P, Nordstrom K. Plasmids with temperature-dependent copy number for amplification of cloned genes and their products. Gene. 1979;6:91–106. doi: 10.1016/0378-1119(79)90065-9. [DOI] [PubMed] [Google Scholar]
- Uhlin BE, Nordstrom K. A runaway-replication mutant of plasmid R1drd-19: Temperature-dependent loss of copy number control. Molec Gen Genet. 1978;165:167–79. doi: 10.1007/BF00269904. [DOI] [PubMed] [Google Scholar]
- Valenzuela MS, Ikpeazu EV, Siddiqui KA. E. coli growth inhibition by a high copy number derivative of plasmid pBR322. Biochem Biophys Res Commun. 1996;219:876–83. doi: 10.1006/bbrc.1996.0339. [DOI] [PubMed] [Google Scholar]
- Valenzuela MS, Siddiqui KA, Sarkar BL. High expression of plasmid-encoded tetracycline resistance gene in E. coli causes a decrease in membrane-bound ATPase activity. Plasmid. 1996;36:19–25. doi: 10.1006/plas.1996.0027. [DOI] [PubMed] [Google Scholar]
- van Hall T, van de Rhee NE, Schoenberger SP, Vierboom MP, Verreck FA, Melief CJ, Offringa R. Cryptic open reading frames in plasmid vector backbone sequences can provide highly immunogenic cytotoxic T-lymphocyte epitopes. Cancer Res. 1998;58:3087–93. [PubMed] [Google Scholar]
- Villalobos A, Ness JE, Gustafsson C, Minshull J, Govindarajan S. Gene Designer: a synthetic biology tool for constructing artificial DNA segments. BMC Bioinformatics. 2006;7:285. doi: 10.1186/1471-2105-7-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner EG, Brantl S. Kissing and RNA stability in antisense control of plasmid replication. TIBS. 1998;23:451–4. doi: 10.1016/s0968-0004(98)01322-x. [DOI] [PubMed] [Google Scholar]
- Wagner H. The sweetness of the DNA backbone drives toll-like receptor 9. Curr Opin Immunol. 2008;20:396–400. doi: 10.1016/j.coi.2008.06.013. [DOI] [PubMed] [Google Scholar]
- Wang H, Benham CJ. Promoter prediction and annotation of microbial genomes based on DNA sequence and structural responses to superhelical stress. BMC Bioinformatics. 2006;7:248. doi: 10.1186/1471-2105-7-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, et al. Detection of integration of plasmid DNA into host genomic DNA following intramuscular injection and electroporation. Gene Ther. 2004;11:711–21. doi: 10.1038/sj.gt.3302213. [DOI] [PubMed] [Google Scholar]
- Wang Z, Xiang L, Shao J, Wegrzyn A, Wegrzyn G. Effects of the presence of ColE1 plasmid DNA in Escherichia coli on the host cell metabolism. Microbial Cell Factories. 2006;5:34. doi: 10.1186/1475-2859-5-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Yuan Z, Hengge UR. Processing of plasmid DNA with ColE1-like replication origin. Plasmid. 2004;51:149–61. doi: 10.1016/j.plasmid.2003.12.002. [DOI] [PubMed] [Google Scholar]
- Weiner DB, Zhang D, Cohen A. Expression system for cloning toxic genes. 6881558. 2005
- Weiss R, et al. Improvement of the immune response against plasmid DNA encoding OspC of Borrelia by an ER-targeting leader sequence. Vaccine. 2000;18:815–24. doi: 10.1016/s0264-410x(99)00338-2. [DOI] [PubMed] [Google Scholar]
- WHO. Annex 1: Guidelines for assuring the quality and nonclinical safety evaluation of DNA vaccines. World Heath Organization Technical Report Series No 941, 2007
- Williams JA. Vectors and methods for genetic immunization. World Patent Application WO2006078979. 2006
- Williams JA. Vectors and methods for genetic immunization. World Patent Application WO2008153733. 2008
- Williams JA, Luke J, Johnson L, Hodgson C. pDNAVACCultra vector family: high throughput intracellular targeting DNA vaccine plasmids. Vaccine. 2006;24:4671–6. doi: 10.1016/j.vaccine.2005.08.033. [DOI] [PubMed] [Google Scholar]
- Williams JA, Luke J, Langtry S, Anderson S, Carnes AE, Hodgson CP. Generic plasmid DNA production platform incorporating low metabolic burden seed-stock and fed-batch fermentation processes. Biotechnol Bioeng. 2008 doi: 10.1002/bit.22347. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong EM, Muesing MA, Polisky B. Temperature-sensitive copy number mutants of ColE1 are located in an untranslated region of the plasmid genome. Proc Natl Acad Sci USA. 1982;79:3570–4. doi: 10.1073/pnas.79.11.3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Dress L, Freeland SJ. Optimal encoding rules for synthetic genes: the need for a community effort. Molecular Systems Biology. 2007;3:134. doi: 10.1038/msb4100176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T, et al. Engineering an intracellular pathway for major histocompatibility complex class II presentation of antigens. Proc Natl Acad Sci USA. 1995;92:11671–5. doi: 10.1073/pnas.92.25.11671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Chu Y, Zhang R, Xu H, Wang Y, Xiong S. Endoplasmic reticulum targeting sequence enhances HBV-specific cytotoxic T lyphocytes induced by a CTL epitope-based DNA vaccine. Virology. 2005;334:255–63. doi: 10.1016/j.virol.2005.01.040. [DOI] [PubMed] [Google Scholar]
- Yau SY, Keshavarz-Moore E, Ward J. Host strain influences on supercoiled plasmid DNA production in Escherichia coli: Implications for efficient design of large-scale processes. Biotechnol Bioeng. 2008;101:529–44. doi: 10.1002/bit.21915. [DOI] [PubMed] [Google Scholar]
- Yew NS, Zhao H, Przybylska M, Wu IH, Tousignant JD, Scheule RK, Cheng SH. CpG-depleted plasmid DNA vectors with enhanced safety and long-term gene expression in vivo. Mol Ther. 2002;5:731–8. doi: 10.1006/mthe.2002.0598. [DOI] [PubMed] [Google Scholar]
- Yuh CH, Ting LP. The genome of Hepatitis B virus contains a second enhancer: cooperation of two elements within this enhancer is required for its function. J Virol. 1990;64:4281–7. doi: 10.1128/jvi.64.9.4281-4287.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaman MM, Boles TC. Plasmid recombination by the RecBCD pathway of Escherichia coli. J Bacteriol. 1996;178:3840–45. doi: 10.1128/jb.178.13.3840-3845.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhongming L, Howard A, Kelley C, Delogu G, Collins F, Morris S. Immunogenicity of DNA vaccines expressing tuberculosis proteins fused to tissue plasminogen activator signal sequences. Infect Immun. 1999;67:4780–6. doi: 10.1128/iai.67.9.4780-4786.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielenkiewicz U, Ceglowsik P. Mechanism of plasmid stable maintenance with special focus on plasmid addiction systems. Acta Biochimica Polonica. 2001;48:1003–23. [PubMed] [Google Scholar]
- Zinckgraf JW, Silbart LK. Modulating gene expression using DNA vaccines with different 3′-UTRs influence antibody titer, seroconversion and cytokine profiles. Vaccine. 2002;21:1640–9. doi: 10.1016/s0264-410x(02)00740-5. [DOI] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]