Abstract
The structure of the 21-30 fragment of the amyloid β-protein (Aβ) was investigated by ion-mobility mass spectrometry and replica exchange dynamics simulations. Mutations associated with familial Alzheimer’s disease (E22G, E22Q, E22K, and D23N) of Aβ(21-30) were also studied, in order to understand any structural changes that might occur with these substitutions. The structure of the WT peptide shows a bend and a perpendicular turn in the backbone which is maintained by a network of D23 hydrogen bonding. Results for the mutants show that substitutions at E22 do little to alter the overall structure of the fragment. A substitution at D23 resulted in a change of structure for Aβ(21-30). A comparison of these gas-phase studies to previous solution-phase studies reveals that the peptide can fold in the absence of solvent to a structure also seen in solution, highlighting the important role of the D23 hydrogen bonding network in stabilizing the fragment’s folded structure.
Keywords: amyloid β-protein, Alzheimer’s disease, protein folding, ion-mobility mass spectrometry, molecular dynamics
INTRODUCTION
A growing body of evidence closely links the amyloid β-protein (Aβ) to Alzheimer’s disease (AD). Aβ is the primary component of amyloid plaques found in the brain tissue of Alzheimer’s patients. Originally the amyloid plaques themselves were believed to be the causative agents of AD, as outlined by the Amyloid Cascade Hypothesis[1, 2]. However, recent clinical and experimental studies have shown that soluble oligomers of Aβ are neurotoxic[3]. The Aβ peptide typically exists as either a 40 or 42 amino acid residue peptide (Aβ40 and Aβ42, respectively). While Aβ40 normally exists in the plasma at a concentration ∼10-fold higher than that of Aβ42, the latter peptide is the more neurotoxic[4] and is the primary component of amyloid plaques. A small number of familial forms of AD exist that result in early onset of the disease. These forms typically arise from single amino acid substitutions of the primary structure of the peptide, all of which are known to occur at either the 22nd or 23rd residue of the Aβ protein[5-10].
Limited proteolysis/mass spectrometry experiments[11] showed that both Aβ(1-40) and Aβ(1-42) contain a 10-residue trypsin-resistant segment, Ala21-Ala30. The homologous decapeptide exhibited the same resistance to proteolysis, suggesting that the segment is particularly stable. Solution NMR studies with molecular dynamics simulations showed that the segment forms a turn-like structure at the center of the peptide, resides V24-K28. The stability of this region suggests that it may be involved in the folding and subsequent aggregation of the full-length monomer peptide.
Proteolysis and solution state NMR have also been employed to probe the stability and structure of the Aβ(21-30) mutants associated with familial Alzheimer’s disease (FAD)[12]. The results showed that the FAD-associated substitutions at Glu-22 and Asp-23 have an increased susceptibility to proteolysis, compared to wild-type (WT) Aβ(21-30). NMR experiments revealed that the increased susceptibility to proteolysis is caused by destabilization of the peptide’s structure. This destabilization of the folding nucleus may explain the acceleration of Aβ aggregation that is associated with FAD.
Here, we use nano-electrospray (ESI) mass spectrometry (MS) and ion mobility spectrometry (IMS) to provide a structural characterization of wild-type (WT) Aβ(21-30) as well as the mutant forms of the peptide associated with each type of hereditary AD. Ion mobility mass spectrometry (IM-MS) is a technique that has been successfully employed to study the full length Aβ peptide[13, 14]. These experiments are coupled with modeling in order to provide an accurate understanding of the molecular interactions that help to maintain the peptide’s integrity as well as investigate possible conformational changes that may occur as a result of various FAD mutations.
METHODS
Sample preparation
The Aβ(21-30) fragment, A21E22D23VGSNKGA30 as well as the mutant forms associated with FAD: Italian (E22K)[10], Arctic (E22G)[7, 8], Dutch (E22Q)[9], and Iowa (D23N)[6], were synthesized by FMOC (N-(9-fluorenyl)methoxycarbonyl) chemistry. All peptides were synthesized in their N-terminal Nα-acetyl and C-terminal carboxyamide (blocked) forms so as to mimic the structure of the segment within the full-length peptide. The samples were purified by reversed-phase HPLC, characterized by mass spectrometry and amino analysis[15] and lyophilized. The lyophilized peptide was dissolved in 25 mM ammonium acetate pH 7.4 for a final concentration of 400μM.
IM-MS Experiments
Mass spectra were obtained using an instrument built in-house with a nanoelectrospray ionization (nano-ESI) source, ion funnel, temperature-controlled drift cell, and quadrupole mass filter[16]. To record mass spectra, 5 μL of sample solution is introduced into the instrument from gold-coated borosilicate capillaries (0.1 mm o.d./0.78 mm i.d.). The capillaries are made with a Sutter Instrument Co. P-97 Flaming/brown micropipette puller and coated with gold by an Emitech K550X sputter coater. The ions are generated continuously at atmospheric pressure in the nano-ESI source and guided through the ion funnel to the analysis region of the instrument. The ion beam is passed through the drift cell, analyzed by the quadrupole mass filter, and detected.
Ion mobility measurements were recorded using the instrument described above[16]. In ion mobility experiments, the ions are stored at the end of the ion funnel and pulsed into the drift cell filled with 5 Torr helium gas. The injection energy of the ions pulsed into the drift cell can be varied from near 0 to 150 eV. The ions in the cell are quickly thermalized by collisions with the helium buffer gas and are drawn through the cell under the influence of a weak electric field.
The force of the weak electric field and the frictional drag of the collisions with the He gas produce a constant drift velocity vD. The drift velocity is proportional to the applied field E where the proportionality constant K is termed the ion mobility:
| (1) |
As the ions exit the drift cell, they are detected as a function of time producing an arrival time distribution (ATD). Kinetic theory[17] relates the ion mobility to the ion-He collision cross-section (σ), which can in turn be related to the arrival time at the detector (tA):
| (2) |
where q is the ion charge, T is the temperature, μ is the reduced mass of the ion-He collision, N is the He number density at STP, l is the drift cell length (4.503 cm), kB is the Boltzmann constant, and t0 is the time the ion spends outside of the cell before reaching the detector. All of the quantities on the right hand side of eq 2 are either measured or know for each experiment, so that a very precise value of σ can be determined.
Simulations
In all our simulations the AMBER94 force field [18] was used to describe the peptide. The temperature was maintained by the Nosé-Hoover algorithm [19] with a coupling constant of 0.05 ps. The LINCS [20] algorithm was used to constrain covalent bonds involving hydrogens. The Lennard-Jones potential decays smoothly to zero between 1.5nm and 2.5nm. Neighbor lists for the nonbonded interactions were updated every 1000 simulation steps. Short-range neighbors were defined within a cutoff of 4.0nm. Generous twin-range cut-offs of 4.0nm/4.0nm were used to evaluate electrostatic interactions. No periodic boundary conditions are used. Simulations were performed using GROMACS software [21-23].
The initial structure for each peptide is taken from the central structure of the most populated cluster in the solution phase simulations[24]. All-atom models constructed for the various ten-residue peptides are capped by a neutral acetyl and amide group at the N and C terminus, respectively. The species modeled here corresponds to the peptide at neutral pH. Here, Aβ(21-30) has an overall -1 charge and contains two negatively charged sites (E22, D23) and one positively charged site (K28). The Arctic mutant (E22G) and the Dutch mutants (E22Q) have no overall charge and both contain one negatively charge site (D23) and one positively charged site (K28). The Italian mutant (E22K) has an overall +1 charge and has one negatively charge site (D23) and two positively charged (K22, K28). The Iowa mutant (D23N) has no overall charge and one negatively charged site (E22) and one positively charged site (K28). The replica-exchange algorithm [25] (REMD) was used to enhance equilibrium sampling. 40 replicas of the original system were exponentially spaced between 300K and 2200K. Every 250 simulation steps (0.125ps) exchanges were attempted among neighboring replicas and atomic coordinates were saved. Acceptance ratios for exchange ranged from 16% to 24%. Simulations were run for 28-30 ns and all but the first 1ns of the trajectories were analyzed.
Snapshots of the 300 K replica taken every 5 ps (∼5800 structures) were clustered into families of structures. The GROMOS clustering method [26] is employed in which the root mean square deviations (RMSD) among conformations provide a measure of structural similarity. For each structure, the number of other structural neighbors, i.e. those with RMSD <1 Å over Cα of resides 22-28, is calculated. The structure with the highest number of neighbors is taken as the central structure of a cluster (C1), which comprises this structure and all of its neighbors. All of the C1 structures are eliminated from the pool of available structures and the procedure is repeated until all structures are eliminated.
Hydrogen bonds are considered present when the donor and acceptor are within 3.5Å and the donor-acceptor-hydrogen angle is <60°. Salt bridges are defined as an interaction between oppositely charged groups in which the charged atoms are separated by ≤ 4.5Å. Cross sections of the model structures are calculated using a modified projection method[27, 28].
RESULTS
IM-MS
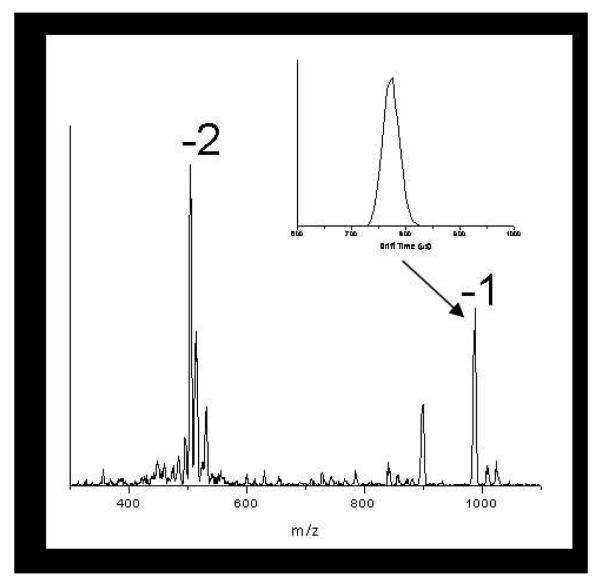
Figure 1 shows a typical mass spectrum for WT Aβ(21-30). The spectrum has two groups of peaks corresponding to the -1 (m/z=988) and -2 (m/z=495) charge states with the addition of up to two sodium atoms (m/z=1011 and 1034) for the -1 charge state and one sodium for the -2 (m/z=506) charge state. Although the -1 charge state is the solution charge state at pH 7.4, the -2 charge state is the dominant peak in the mass spectrum. Mass spectra were recorded for each mutant of Aβ(21-30) and shared similar characteristics as those for the WT peptide. The mass spectra were recorded in positive or negative ion mode, according to the charge state of each species in solution. In the case of peptides with a neutral charge at pH 7, data were recorded for both positive and negative ions.
Figure 1.
A typical mass spectrum of WT Aβ(21-30). The ATD for the z/n= 1 charge state is included as an inset (z is the charge and n is the oligomer number: n=1 is a monomer, n=2 is a dimer, etc).
Ion mobility data was recorded for each charge state observed in the mass spectrum. A typical ATD of WT Aβ(21-30) is given as an inset of figure 1. The ATD consists of one narrow and symmetric peak, suggesting that only one conformation of the peptide is present (or if multiple conformers are present, they either interconvert on the time scale of the experiment or have very similar cross sections). Therefore, the ATD of the z=-1 peak represents the peak for the monomer with a -1 charge. The ATDs for each Aβ mutant show a similar single, symmetric peak for the monomer. The experimental cross sections for each species are listed in Table 1.
Table 1.
Experimental and theoretical cross-sections for WT Aβ(21-30) and its mutant forms. Theoretical cross-sections are given for the central structure of the most populated cluster (see Table 2 for spread of values).
| Mutant | Charge (pH 7) |
Cross Sections (Å2) |
||
|---|---|---|---|---|
| Experimental* | Theoretical** | % Deviation | ||
| WT | -1 | 227 | 231 | 1.73% |
| D23N | 0 | 222.5 | 220 | 1.12% |
| E22G | 0 | 210 | 208 | 0.95% |
| E22K | +1 | 230 | 224 | 2.61% |
| E22Q | 0 | 226 | 225 | 0.44% |
| S26I | -1 | 229 | 233 | 1.75% |
| D23G | 0 | 216 | 217 | 0.46% |
Experimental cross sections correspond to the protein at pH 7. For proteins with a neutral charge, cross sections were taken for the +1 and -1 charge states and averaged.
Theoretical cross sections correspond to the protein at pH 7.
Molecular Modeling
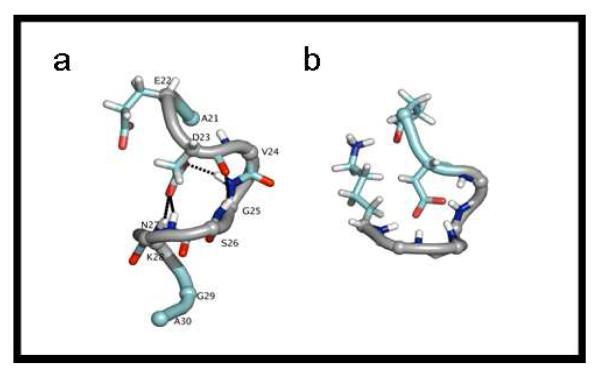
As shown in Table 2, most of the mutants of Aβ(21-30) spend a majority of time in a preferred family of structures (C1). The actual structures shown here are the central structures from this most populated cluster, C1. Figure 2a shows the central structure from the most populated cluster for the WT Aβ(21-30) at 300K. The structure contains both a bend* at residues G25-K28 and a turn** at D23-S26. The plane in which the turn lies is perpendicular to the bend at G25-K28. The bend at G25-K28 is stabilized by D23 Oδ side chain hydrogen bonds to G25, N27, and K28 amide hydrogen atoms (present in 96.6%, 99.0% and 83.0% of the C1 structures, respectively) The turn in the peptide is stabilized by an O-N backbone hydrogen bond between residues D23(O) and S26(N) which is present in 82.8% of the structures in C1. There is a salt bridge at E22-K28 present in 100% of the C1 structures of the WT and a salt bridge between D23 and K28 in 99.2% of the structures.
Table 2.
The most populated cluster of structures obtained from replica exchange simulations at 300K is analyzed. The percentage of all structures belonging to the most populated cluster is given. The average cross section and standard deviation are calculated over 10 structures within C1. The root mean square deviation of Cα distances from the central structure of the WT C1 cluster is calculated for the Cα atoms of residues 22-28 only. The percentage of structures within cluster C1 that contain salt bridges among residues E22, D23 and K28 are given.
| % of structures in C1 |
σ (Å2) | RMSD from WT (Å) |
E22-K28 salt bridge population (%) |
D23-K28 salt bridge population (%) |
|
|---|---|---|---|---|---|
| WT | 99 | 231 ±6.1 | - | 100 | 99.2 |
| D23N | 61 | 222 ±4.2 | 2.67 | 99.9 | - |
| E22G | 79 | 210 ±6.0 | 1.1 | - | 98.7 |
| E22K | 99 | 225 ±2.2 | 3.13 | - | 100* |
| E22Q | 89 | 225 ±3.9 | 0.943 | - | 99.9 |
In the E22K mutant, a salt bridge exists between K22 and D23 in 100% of C1 structures.
Figure 2.
The central structures of the C1 cluster for WT Aβ(21-30) from gas phase simulations (a) and solution phase simulations (b).
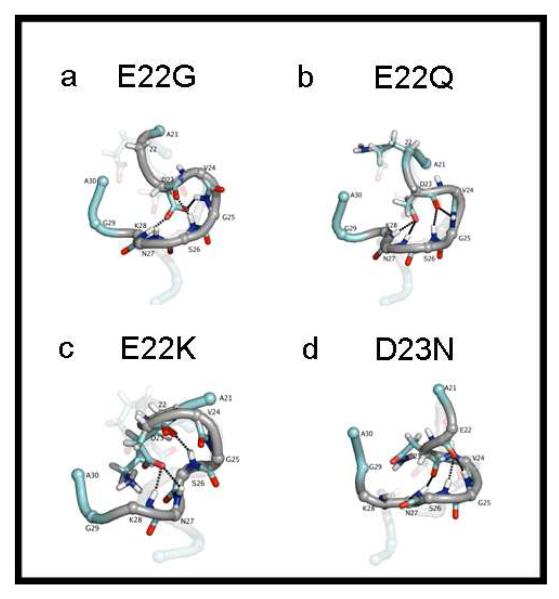
Figures 3a and 3b show the structures of the Arctic (E22G) and Dutch (E22Q) mutants respectively, with the structure of the WT faded in the background. Both mutants show similar structural elements and intermolecular interactions to those of the WT. For all mutants, the positions of the termini do not match those of the wild type. The termini of the peptides were not included in the statistical analysis of the simulations because they are not present within full-length Aβ. The turn and bend that are present in WT Aβ(21-30) are maintained in the Arctic (E22G) and Dutch (D22Q) mutants. The RMSD from WT of E22Q is 0.943 Å and for E22G is 1.10 Å (Table 2). In E22G the amount of bonding between residues D23 and G25 and between D23 and N27 is noticeably lower (48.6% and 55.4% respectively) and the percentage of hydrogen bonds formed with K28 is the same as in the WT (84.3%). Bonding between D23 Oδ side chain atoms and amide hydrogen of G25 is not present in E22Q but the amount of D23 side chain bonding remains similar to that of the WT for D23 bonds with N27 and K28. The hydrogen bond between backbone atoms at D23(O) and S26(NH) are still present at high percentages for both E22G (71.9%) and E22Q (77.6%) (Table 2). A salt bridge formed between D23 and K28 in 98.7% and 99.9% of the C1 structures for E22G and E22Q respectively.
Figure 3.
Gas phase structures for Aβ(21-30) mutants: Arctic E22G (a), Dutch E22Q (b) Italian E22K (c) and Iowa D23N (d). All structures are the central structure of cluster C1. The structure of the WT peptide is shown as a transparent image behind each mutant. For all the structures, the bend region at residues 23-28 is shown in gray and dotted lines denote key hydrogen bonds.
The Italian (E22K) mutant has a higher RMSD from wild type of 2.67 Å although it still has similar structural elements to the WT (figure 3c). This structure shows both the turn (K22-S26) and bend (G25-K28) that are present in the WT, however the bonding motif is different. The turn at K22-S26 is stabilized by a K22(O) backbone hydrogen bond to S26(N) and G25(N) atoms (present in 74.2% and 98.4% of the structures, respectively). These bonds are not present in the WT, the Arctic mutant, nor the Dutch mutant. The bend is stabilized by D23O sidechain hydrogen bonds to N28 amide hydrogen atoms (present in 90.0% of the structures). A salt bridge at D23-K28 was present in 100% of the all of the C1 structures.
The structure of the Iowa form of the peptide (D23N) deviates the most from the WT, with a RMSD of 3.13Å. The structure of Aβ(21-30) D23N shows two turns at N23-N27 and at E22-S26 (figure 3d). The first turn is stabilized by a backbone hydrogen bond between N23(O) and N27(N) atoms. The second turn (E22-S26) is maintained by another backbone hydrogen bond between E22(O) and S26(N) atoms, found in 90.7% of the structures. Backbone hydrogen bonds were calculated for the Nδ and Oδ atoms of the Asn side-chain for this mutant, but these populated less than 25% of the C1 structures. For D23N, a salt bridge between E22 and K28 was present in 99.9% of the structures.
Solution-phase v. gas-phase structure
In solution-phase molecular dynamics studies[24], the WT structures in cluster C1 contain a single backbone bend, while the WT structures in the second most highly populated structural cluster C2 have a single turn. In solution, these two structures are independent and in equilibrium with each other. This is in contrast to the results in the gas phase, where the bend and turn exist simultaneously, perpendicular to each other (Fig 2).
In solution, only a single salt bridge exists, located between residues E22 and K28. Substitutions at E22 remove this salt bridge although the backbone structure remains intact, suggesting that D23 hydrogen bonds are the primary forces stabilizing the structure of the fragment’s backbone. In the gas phase however, two salt bridges occur: one between E22-K28 and another between D23-K28. Further details of the solution structures of the Aβ(21-30) fragment and the related FAD mutants can be found elsewhere[24].
DISCUSSION AND CONCLUSIONS
A comparison between experimental cross-sections and theoretical cross-sections (Table 1) shows excellent agreement between experiment and theory. Deviations fall below 3% in all cases and below 2% in almost all cases. Substitutions at E22 do little to alter the turn-bend structure of the decapeptide, with subtle changes in the factors that stabilize this structure, as discussed here. Both E22Q and E22G show the same D23 Oδ backbone NH hydrogen bonds, which act to preserve the bend in residues 23-28. Still, these bonds are not as highly populated as in the wild type. E22Q has the lowest RMSD from WT, consistent with the very similar resistance to proteolysis displayed by the two peptides[12]. Hydrogen bonding between D23 and N27 is present in E22Q and WT but not present in E22G. Conversely E22G maintains a hydrogen bond between D23 and G25, which is not present in E22Q. Since both mutations have similar RMSD to the WT we reiterate that these bonding differences have little impact on the core structure. The structure of E22K contains a dissimilar network of backbone hydrogen bonds to the WT but, perhaps surprisingly, has a similar structure.
The Iowa substitution, D23N, on the other hand, does result in a change of backbone structure. Here, a negatively charged aspartic acid residue is replaced with a neutral asparagine residue. With this substitution, the hydrogen bonding between residue 23 and the surrounding residues is disrupted, suggesting that the D23 network of hydrogen bonds is primarily responsible for maintaining the backbone structure in the WT peptides, as well as in the E22X mutants. This is further emphasized by the decreased population of the C1 cluster of the D23N mutant, as compared to the E22X mutants (Table 2). With a mutation at residue 23, the peptide able to access slightly more conformational space than the WT and other E22X mutants.
These results confirm the presence of a turn in the 24-28 region of the peptide, as previously reported in NMR experiments11,12. Proteolysis experiments on the mutant forms of Aβ(21-30) by Grant et al.[12] showed that substitutions at E22 and D23 increase the peptide’s susceptibility to digestion, suggesting that these changes act to destabilize the core structure, perhaps encouraging higher order Aβ assembly. Our results for the Iowa form support this hypothesis, showing a change from a bend at G25-K28 with a turn at D23-S26 for the wild-type Aβ(21-30) to two turns between residues N23-N27 and E22-S26 in the mutant. On the other hand, our calculations on substitutions at E22 show that none of these FAD associated mutations significantly alter the backbone structure of WT Aβ(21-30). Regardless, subtle changes in structure for the various E22X mutants may either alter the intramolecular interactions between the Aβ(21-30) segment and the rest of the full-length peptide or the intermolecular interactions between the Aβ monomers. Consequently, the kinetics of monomer folding and the ensuing peptide assembly processes may be altered by these mutations.
This hypothesis however, does not explain the differences between the results presented here and a previous proteolysis study on Aβ(21-30)[12]. In those studies, the E22G, E22Q and E22K mutants all showed decreased resistance to digestion relative to the WT. There can be several reasons for the differences. First, the proteolysis occurs at K28, which is at the C-terminal end of the conserved core structure of the peptide. Hence, other factors may dominate the trypsin-induced proteolysis. Second, the proteolysis is a relatively slow process and may occur from minor structural components that are not considered in our structural analysis of Fig. 3. In fact, a central feature of the kinetic analysis of the proteolysis experiments was the equilibrium among protease-sensitive and protease-insensitive conformers. Finally, proteolysis occurs in solution and solution structures23 are more variable than our solvent free structures, where the vast majority of the structures fall in a single cluster family. For example, the population of cluster C1 for the gas phase structure of WT Aβ(21-30) is 99%, while the population in solution is 44%.
Overall, however, the above conclusions are consistent with those drawn from our solution phase molecular dynamics simulations[24, 30] even though some important differences result from the loss of solvent. The small size of the peptide ensures that it is completely desolvated during the IM-MS experiments. Without the presence of solvent, electrostatic interactions and salt bridges play a significant structural role. The presence of stable salt bridges in the gas phase is primarily responsible for the simultaneous existence of the bend and turn that is found in the gas phase for the WT and the E22X mutants. In solvent, the D23-K28 salt bridge is not present. As a consequence, the fraction of structures containing a backbone bend is substantially reduced, although the bend remains the dominant structural element for the WT and E22X mutants.
Conversely, the D23N structure consists of a single turn in both the gas and solution phase. This provides further evidence that the D23-K28 salt bridge is the cause of the structural difference upon transitioning from solution to gas phase for the WT and E22X mutants.
Simulations in explicit solvent[24] show that a network of D23 hydrogen bonds serves as the primary source of structural stabilization for the bend in the peptide backbone of the WT and E22X mutants. This same network of D23 hydrogen bonds still exists in the gas phase even in the presence of the D23-K28 salt bridge. Even though the gas phase structure is completely desolvated, the important interactions from the biologically-relevant solution phase structure remain intact. These results speak to the persistence of the fundamental intramolecular interactions in these peptides and why they are so resistant to proteolysis.
Table 3.
The percentages of C1 structures containing a hydrogen bond between one or both D23 sidechain Oδ atoms and the given amide hydrogen atom are calculated. Analogous hydrogen bonds were also calculated for the Nδ and Oδ atoms of the Asn sidechain in the D23N mutant and no hydrogen bonds are populated in more than 25% of the C1 structures for these cases.
| V24(NH) | G25(NH) | S26(NH) | N27(NH) | K28(NH) | |
|---|---|---|---|---|---|
| WT | 3.9 | 96.6 | 16.7 | 99 | 83.9 |
| E22G | 1.46 | 48.6 | 18.3 | 55.4 | 84.3 |
| E22K | 0 | 0 | 4.07 | 12.4 | 90 |
| E22Q | 0.235 | 1.29 | 36.1 | 95.9 | 71.9 |
Table 4.
The percentage of C1 structures containing a hydrogen bond between two backbone atoms is calculated.
| 23(O)-G25(NH) | 23(O)- S26(NH) |
23(O)- N27(NH) |
22(O)- G25(NH) |
22(O)- S26(NH) |
|
|---|---|---|---|---|---|
| WT | 3.02 | 82.8 | 0 | 0 | 0 |
| D23N | 0 | 0.166 | 0 | 17.9 | 90.7 |
| E22G | 78.3 | 71.9 | 0 | 0 | 0 |
| E22K | 0.425 | 2.01 | 45.5 | 98.4 | 74.2 |
| E22Q | 34.9 | 77.6 | 13.1 | 0 | 0 |
ACKNOWLEDGMENTS
The support of the National Institute of Health under grant AG027818, the David and Lucile Packard Foundation and the NSF (MCB # 0642086), is gratefully acknowledged.
Footnotes
A bend is defined as a region of curvature such that the bond angle formed by the three Cα atoms of residues i-2, i, and i+2 is at least 70°29.
A turn is defined as a region where a hydrogen bond exists between CO of residue i and NH of residue i+n29.
REFERENCES
- 1.Hardy JA, Higgins GA. Alzheimers-Disease - the Amyloid Cascade Hypothesis. Science. 1992;256(5054):184–185. doi: 10.1126/science.1566067. [DOI] [PubMed] [Google Scholar]
- 2.Hardy J, Selkoe DJ. Medicine - The amyloid hypothesis of Alzheimer’s disease: Progress and problems on the road to therapeutics. Science. 2002;297(5580):353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- 3.Roychaudhuri R, Yang M, Teplow DB. J. Biol. Chem. 2008 doi: 10.1074/jbc.R800036200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dahlgren KN, et al. Oligomeric and fibrillar species of amyloid-beta peptides differentially affect neuronal viability. Journal of Biological Chemistry. 2002;277(35):32046–32053. doi: 10.1074/jbc.M201750200. [DOI] [PubMed] [Google Scholar]
- 5.Hendriks L, et al. Presenile-Dementia and Cerebral-Hemorrhage Linked to a Mutation at Codon-692 of the Beta-Amyloid Precursor Protein Gene. Nature Genetics. 1992;1(3):218–221. doi: 10.1038/ng0692-218. [DOI] [PubMed] [Google Scholar]
- 6.Grabowski TJ, et al. Novel amyloid precursor protein mutation in an Iowa family with dementia and severe cerebral amyloid angiopathy. Annals of Neurology. 2001;49(6):697–705. doi: 10.1002/ana.1009. [DOI] [PubMed] [Google Scholar]
- 7.Kamino K, et al. Linkage and Mutational Analysis of Familial Alzheimer-Disease Kindreds for the App Gene Region. American Journal of Human Genetics. 1992;51(5):998–1014. [PMC free article] [PubMed] [Google Scholar]
- 8.Nilsberth C, et al. The ‘Arctic’ APP mutation (E693G) causes Alzheimer’s disease by enhanced A beta protofibril formation. Nature Neuroscience. 2001;4(9):887–893. doi: 10.1038/nn0901-887. [DOI] [PubMed] [Google Scholar]
- 9.Levy E, et al. Mutation of the Alzheimers-Disease Amyloid Gene in Hereditary Cerebral-Hemorrhage, Dutch Type. Science. 1990;248(4959):1124–1126. doi: 10.1126/science.2111584. [DOI] [PubMed] [Google Scholar]
- 10.Tagliavini F, et al. Alzheimer’s Rep. Suppl. Vol. 2. 1999. p. 528. [Google Scholar]
- 11.Lazo ND, et al. On the nucleation of amyloid beta-protein monomer folding. Protein Science. 2005;14(6):1581–1596. doi: 10.1110/ps.041292205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grant MA, et al. Familial Alzheimer’s disease mutations alter the stability of the amyloid beta-protein monomer folding nucleus. PNAS. 2007;104(42):16522–16527. doi: 10.1073/pnas.0705197104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baumketner A, et al. Amyloid Beta-Protein Monomer Strucutre: A Computational and Experimental Study. Protein Science. 2006;15(3):420–428. doi: 10.1110/ps.051762406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bernstein SL, et al. Amyloid beta-protein: Monomer structure and early aggregation states of A beta 42 and its Pro(19) alloform. Journal of the American Chemical Society. 2005;127(7):2075–2084. doi: 10.1021/ja044531p. [DOI] [PubMed] [Google Scholar]
- 15.Lomakin A, et al. On the nucleation and growth of amyloid beta-protein fibrils: Detection of nuclei and quantitation of rate constants. Proceedings of the National Academy of Sciences of the United States of America. 1996;93(3):1125–1129. doi: 10.1073/pnas.93.3.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wyttenbach T, Kemper PR, Bowers MT. Design of a new electrospray ion mobility mass spectrometer. International Journal of Mass Spectrometry. 2001;212(13):13–23. [Google Scholar]
- 17.Mason EA, McDaniel EW. Transport Properties of ions in gases. Wiley; New York: 1988. [Google Scholar]
- 18.Cornell WD, et al. A 2nd Generation Force-Field for the Simulation of Proteins, Nucleic-Acids, and Organic-Molecules. Journal of the American Chemical Society. 1995;117(19):5179–5197. [Google Scholar]
- 19.Nose S. Constant Temperature Molecular-Dynamics Methods. Progress of Theoretical Physics Supplement. 1991;(103):1–46. [Google Scholar]
- 20.Hess B, et al. LINCS: A linear constraint solver for molecular simulations. Journal of Computational Chemistry. 1997;18(12):1463–1472. [Google Scholar]
- 21.Berendsen HJC, Vanderspoel D, Vandrunen R. Gromacs -a Message-Passing Parallel Molecular-Dynamics Implementation. Computer Physics Communications. 1995;91(13):43–56. [Google Scholar]
- 22.Lindahl E, Hess B, van der Spoel D. GROMACS 3.0: a package for molecular simulation and trajectory analysis. Journal of Molecular Modeling. 2001;7(8):306–317. [Google Scholar]
- 23.Sorin EJ, Pande VS. Exploring the helix-coil transition via all-atom equilibrium ensemble simulations. Biophysical Journal. 2005;88(4):2472–2493. doi: 10.1529/biophysj.104.051938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krone MG, et al. Effects of Familial Alzheimer’s Disease Mutations on the Folding Nucleation of the Amyloid Beta-Protein. Journal of Molecular Biology. 2008;381:221–228. doi: 10.1016/j.jmb.2008.05.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sugita Y, Okamoto Y. Replica-exchange molecular dynamics method for protein folding. Chemical Physics Letters. 1999;314(12):141–151. [Google Scholar]
- 26.Daura X, et al. Peptide folding: When simulation meets experiment. Angewandte Chemie-International Edition. 1999;38(12):236–240. [Google Scholar]
- 27.Wyttenbach T, et al. Effect of the Long-Range Potential on Ion Mobility Measurements. Journal of the American Society for Mass Spectrometry. 1997;8:275–282. [Google Scholar]
- 28.Wyttenbach T, von Helden G, Bowers MT. Conformations of alkali ion cationized polyethers in the gas phase: polyethylene glycol and bis[(benzo-15-crown-5)-15-ylmethyl] pimelate. International Journal of Mass Spectrometry. 1997;165/166:363–375. [Google Scholar]
- 29.Kabsch W, Sander C. Dictionary of Protein Secondary Structure - Pattern-Recognition of Hydrogen-Bonded and Geometrical Features. Biopolymers. 1983;22(12):2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]