Summary
In multicellular organisms, telomerase is required to maintain telomere length in the germline but is dispensable in the soma. Mice, for example, express telomerase in somatic and germline tissues, while humans express telomerase almost exclusively in the germline. As a result, when telomeres of human somatic cells reach a critical length the cells enter irreversible growth arrest called replicative senescence. Replicative senescence is believed to be an anticancer mechanism that limits cell proliferation. The difference between mice and humans led to the hypothesis that repression of telomerase in somatic cells has evolved as a tumor-suppressor adaptation in large, long-lived organisms. We tested whether regulation of telomerase activity coevolves with lifespan and body mass using comparative analysis of 15 rodent species with highly diverse lifespans and body masses. Here we show that telomerase activity does not coevolve with lifespan but instead coevolves with body mass: larger rodents repress telomerase activity in somatic cells. These results suggest that large body mass presents a greater risk of cancer than long lifespan, and large animals evolve repression of telomerase activity to mitigate that risk.
Keywords: body mass, cancer, evolution, lifespan, rodents, telomerase
Introduction
During replication of linear chromosomes, the leading strands are synthesized completely to their ends, while lagging strand synthesis leaves nascent DNAs incomplete at their 5′ ends — the so-called ‘end-replication problem’ (Olovnikov, 1973). This problem arises because of the inability of the most distal RNA primer to be replaced by DNA and by inefficient initiation of DNA synthesis by DNA polymerase α-primase from the very end of linear DNA. To prevent progressive telomere shortening most organisms use telomerase, a ribonucleoprotein that uses an RNA molecule as a template for telomeric DNA synthesis (Chan & Blackburn, 2004). In unicellular organisms, such as yeasts and ciliates, telomerase is uniformly expressed. In multicellular organisms, however, telomerase is only required in the germline provided that telomeres in somatic cells are sufficiently long to allow the appropriate number of cell divisions for development and daily function of an organism.
In humans, telomerase is expressed in early embryos but is progressively shut off in somatic tissues (Bekaert et al., 2004). Normal human somatic cells, with the exception of stem cells, have no detectable telomerase activity, and their telomeres shorten with every division (Harley et al., 1990). After approximately 60 population doublings, human fibroblasts in culture enter replicative senescence (Hayflick & Moorhead, 1961) when telomeres reach a critical length (Shay & Wright, 2000). Replicative senescence is believed to have evolved as an adaptive mechanism to protect organisms from uncontrolled cell proliferation and cancer (Campisi, 2001). Indeed, telomerase is activated in most human tumors (Kim et al., 1994). Replicative senescence is not, however, a universal phenomenon, even among mammals. Mice, for instance, express telomerase in most of their somatic tissues (Prowse & Greider, 1995), and mouse fibroblasts in culture at physiological oxygen concentration (2–5%) do not experience replicative senescence (Parrinello et al., 2003).
The dramatic difference between human and mouse telomerase regulation is generally explained by their differences in lifespan and body mass (Wright & Shay, 2000; Forsyth et al., 2002). Longer-lived organisms experience more cell divisions and thus a greater risk of multistage carcinogenesis. Under this hypothesis, mice are short lived, presumably succumbing to predation or disease in the wild, and therefore require fewer anticancer mechanisms. Notably, up to 90% of captive mice die of cancer (Lipman et al., 2004). In contrast, humans are long lived, and have therefore evolved genetic tumor-suppressor systems that set limits on cell proliferation. Similarly, telomerase activity is thought to coevolve with body mass: larger animals, having more cells than smaller ones, could experience a greater susceptibility to cancer (Nunney, 1999). Repressed telomerase activity in human somatic cells is thus thought to be one such antitumor adaptation. This hypothesis, based primarily on humans and laboratory mice, suggests that telomerase activity should coevolve with lifespan and body mass. Limited data from other species (primarily farm animals) show that telomerase activity is repressed in somatic tissues of cows (Thomas et al., 2000), sheep (Davis et al., 2005), horses (Argyle et al., 2003), cats (McKevitt et al., 2003), and primates (Steinert et al., 2002), but the situation is less clear in dogs (Nasir et al., 2001) and pigs (Pathak et al., 2000; Fradiani et al., 2004) as conflicting results have been reported. Most of the farm animals are relatively large and long lived, and do not provide enough variability to study the contributions of body mass and lifespan in evolution of telomerase activity.
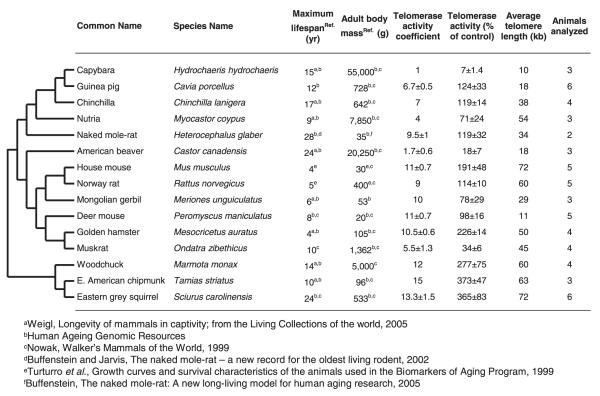
Here we test the hypothesis that telomerase activity coevolves with lifespan and body mass using a collection of 15 rodent species. These species have diverse lifespans: short-lived rodents, such as house mouse and Norway rat, have a maximum lifespan of 4–5 years; long-lived rodents, such as grey squirrel and naked mole rat, have lifespans that exceed 20 years (Buffenstein, 2005; Buffenstein & Jarvis, 2002; Human Ageing Genomic Resources, 2006; Nowak, 1999; Turturro et al., 1999; Weigl, 2005). Body mass is also very diverse among rodents: capybara, the largest rodent, has an adult body mass of 55 kg while deer mouse has an adult body mass of 20 g (Human Ageing Genomic Resources, 2006; Nowak, 1999). This diversity provides an excellent opportunity to test for coevolution between telomerase activity, lifespan, and body mass.
Results
Telomerase activity
To test the hypothesis that telomerase activity coevolves with lifespan and body mass we examined telomerase activity in a collection of 15 rodent species (Fig. 1). We chose phylogenetically diverse species that differ dramatically in their lifespans and body masses providing an excellent model to study evolution of telomerase regulation. To obtain a representative picture of telomerase activity status in these rodents, we measured telomerase activity in a panel of seven tissues using the telomeric repeat amplification protocol (TRAP). The TRAP assay measures the extension of telomeric sequence by telomerase in whole cell extract (Fig. 2). We used three controls in each assay. First, because telomerase activity is present in the germ cells of all mammals, we used testicular tissue as a positive control for the assay and specimen quality. However, the level of telomerase activity in the testes is developmentally regulated, and mature sperm is telomerase negative (Prowse & Greider, 1995). We therefore included a second control: a telomerase-positive human cancer cell line was used in each reaction set as a reference for quantification. Third, each reaction contains two primers that produce a 36-bp product that serves as an internal control for polymerase chain reaction (PCR).
Fig. 1.
Phylogeny and phenotypic data for 15 rodent species. The tree topology is based on molecular phylogenies inferred from (Martin et al., 2000; Michaux et al., 2001; Murphy et al., 2001; Montgelard et al., 2002; Adkins et al., 2003; Steppan et al., 2004). The telomerase activity coefficients, and percentage of telomerase activity for each species are species averages ± SD.
Fig. 2.
Telomerase activity assays. Lanes are labeled as follows: +, telomerase positive human cancer cell line; H, heart; Li, liver, Sp, spleen; K, kidney; Sk, skin; Lu, lung; T, testis. The number below each lane corresponds to relative units of telomerase activity calculated as described in the text. Each gel represents one animal. The reaction includes internal positive control for the polymerase chain reaction (PCR) amplification step (indicated by arrow). Some telomerase negative samples show nonspecific bands of wrong molecular weight caused by unspecific PCR amplification.
To quantify telomerase activity we measured the total intensity of the bands (excluding the internal positive control) in each lane using ImageQuant software. Total telomerase activity rather than processivity was used, as processivity is more dependent on reaction conditions and may vary across species (Chen & Greider, 2003). Telomerase activity for each tissue was calculated as a percentage of telomerase activity of the human cancer cell line. We then used two methods to calculate species-specific telomerase activity.
First, we calculated a telomerase activity index from 0 to 3 for each tissue: if a tissue had no detectable telomerase activity it was assigned ‘0’; telomerase activity of up to 10% of the human cancer cell line was assigned ‘1’; activity from 10% to 50% of the human control was assigned ‘2’; and activity greater than 50% of the positive control cell line was assigned ‘3’. For each individual, the six index values, corresponding to the tissues assayed excluding testis (in which telomerase is constitutively active), were summed to yield an overall telomerase activity coefficient that, in principle, could vary from 0 to 18 (Fig. 1). Telomerase activity coefficients for each species were calculated as averages between two and six animals (Fig. 1). Representative TRAP assays are shown in Fig. 2.
The index was used for the following reasons. Telomerase activity varied dramatically between tissues, from no detectable activity to 257% of the human cancer cell line. Thus, if the raw values for telomerase activity were used, a single tissue with very high activity would determine the total value. Since we do not have a quantitative estimate of a contribution of each tissue to aging and tumorigenesis, we assumed equal contribution of each tissue. Furthermore, when telomerase activity is higher than in human cancer cells the TRAP reaction reaches a plateau and may not be quantitative. The indexing addresses these potential distortions by equalizing contribution of each tissue and setting an upper limit for telomerase activity.
The second method to calculate telomerase activity for the species was to use raw telomerase activity values expressed as percent of the activity in the human cancer cell line. Total telomerase activity summed across the six tissues was calculated for each animal and the species value was calculated as an average between two and six animals (Fig. 1).
Most species showed high telomerase activity in multiple somatic tissues, even the longest-lived rodents such as naked mole rat and grey squirrel (Fig. 2). This is a surprising observation: if repression of telomerase activity serves as a tumor-suppressor mechanism, then long-lived animals would be expected to repress telomerase in somatic tissues. The only two rodents that showed nearly complete somatic repression of telomerase activity, similar to humans, were the largest rodents: beaver and capybara.
We statistically evaluated whether telomerase activity coefficient correlates with body mass or lifespan across rodent species. These analyses reveal that telomerase activity shows a significant negative correlation with body mass (F1,13 = 14.9, r2 = 0.534, P = 0.002) but not with maximum lifespan (F1,13 = 1.2, r2 = 0.085, P = 0.293) (Fig. 3). The latter probability value should be treated with caution as the regression analysis of maximum lifespan violates the heterogeneity of variances assumption; we therefore also present nonparametric Spearman rank correlation, which remains nonsignificant: rs = -0.052, P = 0.856. To correct these analyses for phylogenetic non-independence, we analyzed phylogenetically independent contrasts (Felsenstein, 1985; Harvey & Pagel, 1991; Purvis et al., 1994). These analyses confirm that evolutionary change in telomerase activity coefficient is negatively correlated with change in body mass (F1,13 = 42.1, r2 = 0.76, P < 0.0001), not maximum lifespan (F1,13 = 6.5, r2 = 0.024, P = 0.584). Qualitatively the same result was obtained for the raw values of telomerase activity without the use of indexing. Evolutionary change in telomerase activity is negatively correlated with change in body mass (F1,13 = 6.5, r2 = 0.34, P = 0.024), not maximum lifespan (F1,13 = 1.9, r2 = 0.13, P = 0.187).
Fig. 3.
Relationships of telomerase activity with (A) body mass (y = -2.8x + 16.1), and (B) maximum lifespan. Plots use species data uncorrected for phylogeny.
We then examined whether average telomerase activity in individual tissues correlates with body mass or lifespan. We present the analysis of phylogenetically independent contrasts. In liver, spleen, and kidney, telomerase activity shows a significant negative correlation with body mass (liver: F1,13 = 11.6, r2 = 0.47, P = 0.005; spleen: F1,13 = 8.7, r2 = 0.4, P = 0.01; kidney: F1,13 = 9.9, r2 = 0.43, P = 0.008). In heart and lung telomerase activity is not significantly correlated with body mass (heart: F1,13 = 1.8, r2 = 0.12, P = 0.208; lung: F1,13 = 1.3, r2 = 0.09, P = 0.268). The analysis did not detect significant correlation between telomerase activity in heart, liver, spleen, kidney, or lung and maximum lifespan. We were unable to analyze telomerase activity in the skin because an evolutionary assumption was violated.
Body mass was previously shown to correlate positively with longevity (Harvey et al., 1989; Austad & Fischer, 1991; Austad, 2005). To control for the effects of possible correlations between body mass and lifespan, we studied the independent contribution of each to the telomerase activity coefficient by analyzing independent contrasts with multiple regression (Harvey & Pagel, 1991). Only body mass was significantly related to telomerase activity (F2,12 = 22.9, r2 = 0.79, P < 0.0001; body mass, t = -2.6, P < 0.0001; maximum lifespan, t = -2.64, P = 0.228). Thus, all three analyses — of uncorrected data, of phylogenetically corrected data, and of phylogenetically corrected data controlling for a possibly confounding third variable — confirmed a significant negative correlation of telomerase activity with body mass but not with lifespan. Reduced telomerase activity thus appears to have evolved in larger, but not in longer-lived, rodents.
Telomere length
We examined whether telomere length in these species correlates with body mass or lifespan because: (i) telomerase is responsible for telomere length maintenance; and (ii) we found that telomerase activity correlates with body mass. We measured telomere length using the terminal restriction fragment (TRF) method. TRF length was determined using pulse-field gel electrophoresis followed by Southern blot hybridization with telomere-specific probes (Fig. 4). Average telomere lengths are shown in Fig. 1. The two largest rodents capybara and beaver that had the lowest telomerase activity had relatively short telomeres (10 and 18 kb, respectively), which is similar to telomere length in humans. However, statistical analysis of the 15 species showed that telomere length is not significantly related to body mass (uncorrected species data: F1,13 = 0.6, r2 = 0.04, P = 0.455; independent contrasts: F1,13 = 0.02, r2 = 0.001, P = 0.894), or to maximum lifespan (uncorrected species data: F1,13 = 0.5, r2 = 0.035, P = 0.504; independent contrasts: F1,13 = 3.9, r2 = 0.23, P = 0.071). Thus, telomere length does not coevolve with either body mass or lifespan.
Fig. 4.
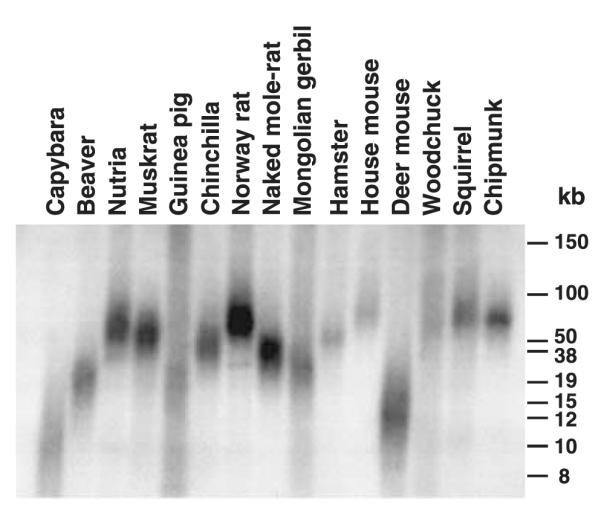
Telomere length measurement. Telomere length was analyzed using the terminal restriction fragment (TRF) assay as described in Experimental procedures.
Discussion
Our study shows a strong negative correlation between telomerase activity and body mass. The correlation was significant both for the composite telomerase activity coefficient for six tissues and for the total raw telomerase activity. Analysis of individual tissues showed that telomerase activity in spleen, liver, and kidney negatively correlates with body mass. We propose the following model to explain coevolution of telomerase activity and body mass (Fig. 5). Evolutionary increases in body mass increase cancer risk, as larger animals contain more cells in their bodies and malignant transformation may occur in any single cell. The increased cancer mortality rate drives the adaptive evolution of tumor-suppressor mechanisms (Nunney, 1999; Leroi et al., 2003). Our results provide evidence that such an adaptive tumor-suppressor mechanism — somatic repression of telomerase activity — has evolved with body mass in animals. Other tumor-suppressor mechanisms such as more efficient DNA repair may also evolve with body mass (Promislow, 1994).
Fig. 5.
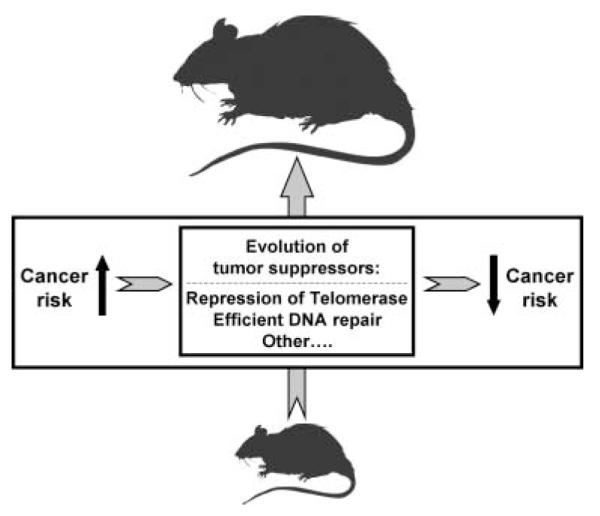
Coevolution of telomerase activity and body mass. Increase in body mass leads to increased cancer risk. To counteract this risk large species evolve additional tumor-suppressor mechanisms, such as repression of somatic telomerase activity.
Previous studies did not allow differentiation between the contributions of body mass and lifespan on evolution of somatic repression of telomerase activity, as the existing data on telomerase activity among mammals was derived from a limited number of species, primarily mice and rats that are small and short lived, and on large long-lived species such as humans and cattle. The importance of considering body mass when analyzing evolution of longevity has been previously emphasized. By controlling for the effect of body mass and phylogeny Promislow (1994) concluded that DNA repair rates correlate with body mass rather than with longevity in mammals. Similarly, Lorenzini et al. (2005) found that replicative capacity of fibroblasts positively correlates with body mass. The latter result may appear inconsistent with our finding of negative correlation between telomerase activity and body mass. This apparent contradiction can be easily explained by considering the definition of replicative capacity employed by Lorenzini et al. (2005). In that study fibroblasts were cultured in 20% oxygen and out of 59 cultures 21 spontaneously immortalized after a period of reduced growth rate. It has been demonstrated by Parrinello et al. (2003) that senescence of mouse fibroblasts in 20% oxygen is caused by oxidative stress rather than by telomere shortening, while at physiological oxygen concentration mouse fibroblasts are immortal. Since half of the species used by Lorenzini et al. were rodents, the observed positive correlation between replicative capacity and body mass is likely to reflect the positive correlation between oxidative stress resistance and body mass, but has no direct relation to telomerase activity. The largest species (cattle, gorilla, and human) examined by Lorenzini et al. display telomere-mediated senescence, and do not express telomerase activity in the soma (Thomas et al., 2000; Steinert et al., 2002), which is in agreement with our finding of negative correlation between body mass and telomerase activity.
In contrast to telomerase activity, we did not find correlations between telomere length and body mass or lifespan. The majority of rodents in this study had telomeres longer than 30 kb. Notable exceptions are beaver and capybara that also had the lowest telomerase activity. Short telomeres in beaver and capybara suggest the presence of replicative senescence in these two species. It is likely that the species with high telomerase activity do not have replicative senescence, thus telomere length in these species does not affect cancer rates, and therefore does not coevolve with body mass or lifespan. Previous studies have shown telomere length to be highly variable even within closely related inbred mouse strains, and found no correlation of telomere length with lifespan (Hemann & Greider, 2000). The importance of telomerase activity level but not telomere length in the species with high somatic telomerase activity can be explained as follows. Multiple studies suggest that telomerase has oncogenic and growth promoting functions independent of its role in telomere maintenance (reviewed in Chang & DePinho, 2002; Gorbunova & Seluanov, 2003). Thus it is possible that in the species at the high end of telomerase activity spectrum the level of telomerase activity affects cancer rates independently of telomere length.
We found that several long-lived rodent species such as grey squirrel and naked mole rat have high telomerase activity in somatic cells, and are therefore unlikely to use replicative senescence as an anticancer mechanism. The presence of telomerase activity in somatic cells may provide benefits such as better wound healing and stronger immune response. It is important to note, however, that the regulation of telomerase activity is one of many anticancer adaptations. Thus small long-lived rodents may maintain high telomerase activity in the soma and rely on other tumor suppressor mechanisms. Since human cancer is believed to originate from stem cells (Reya et al., 2001), and stem cells have telomerase activity, understanding anticancer mechanisms employed by long-lived rodents with high telomerase activity may provide valuable information to fight cancer in humans.
Our inability to detect an effect of lifespan on the evolution of telomerase activity is puzzling, because any increase in lifetime cell divisions should increase the opportunity for tumorigenic somatic mutation. The lack of correlation between lifespan and a tumor-suppressor mechanism may suggest that large body mass presents greater risk for cancer development than long lifespan. Lifetime cancer risk is thought to depend on the number of cell divisions that occur during development and the number of cell divisions needed for maintenance during lifespan. Thus, a speculative explanation for why body mass but not lifespan coevolves with tumor-suppressor mechanisms could be that initial events leading to cancer occur during growth and development. It is important to note, however, that tumor-suppressor mechanisms other than telomerase repression may coevolve with lifespan. Future studies aimed at understanding why telomerase coevolves with body mass but not lifespan may shed new light on the mechanisms of cancer development and evolution of longevity.
Experimental procedures
Animal samples
Capybaras were farm raised at Profauna Farm at Iguape (São Paulo, Brazil). Outbred multicolored guinea pigs were purchased from Elmhill Labs. Chinchillas were from the colony of R. Salvi (University at Buffalo, Buffalo, NY, USA). Nutrias were wild caught in Maryland as a part of US Department of Agriculture Nutria Eradication Program. Naked mole rats were from the colony of Kenneth C. Catania. at Vanderbilt University, Nashville, TN, USA. Beavers were wild caught. One beaver was from Washington State, and two beavers were obtained locally at Montezuma National Wildlife refuge, Seneca County, NY, USA. Three house mice had mixed genetic background, and two mice were inbred 129/Sv. Inbred rats were purchased from Charles River Laboratories (Wilmington, MA, USA). Three rats were Brown-Norway (BN) and two rats were Fischer 344. Mongolian gerbils of Crl:Mon(Tum) strain were purchased from Charles River Laboratories. Hamsters of Crl:LVG(Syr) strain were from Charles River Laboratories. Deer mice, muskrats, woodchucks, chipmunks, and squirrels were wild trapped in New York State. All animals used in this study were young adults. Exact age was known for laboratory animals and was estimated for wild-caught animals from body measurements and color. Live animals were euthanized according to the University of Rochester Animal Care and Use Committee guidelines. Care was taken to minimize pain and discomfort to the animals. All carcasses were placed on ice immediately. Organs were processed for analysis and frozen in liquid nitrogen within 48 h. In pilot experiments with mice and woodchucks we saw no decline in telomerase activity between the tissues stored at 4 °C for up to 72 h prior to freezing (Supplementary Fig. S1). We therefore used 48 h as a cut-off point for storage and shipping time.
Telomeric repeat amplification protocol
Telomeric repeat amplification protocol assay was performed using TRAPeze kit (Chemicon, Temecula, CA, USA) according to manufacturer’s instructions. Briefly, in the first step of the TRAP assay, radiolabeled substrate oligonucleotide is added to 0.5 μg of protein extract. If telomerase is present and active, telomeric repeats (GGTTAG) are added to the 3′ end of the oligonucleotide. In the second step, extended products are amplified by PCR. Telomerase extends the oligonucleotide by multiples of 6 bp, generating a ladder of products of increasing length. For each species, telomerase activity was analyzed in seven tissues from two to six animals. The assays were repeated at least two times for each individual animal to ensure reproducibility. A human cancer cell line overexpressing telomerase was used as a reference in each assay.
Telomere lengths
Telomere length was analyzed by Southern blot using the TRF method. Genomic DNA was extracted from liver, digested with a mixture of AluI, HaeIII, RsaI, and HinfI restriction enzymes that do not cut within telomeric repeat sequence, separated using pulse field gel electrophoresis, and hybridized with radiolabeled oligonucleotide containing telomeric sequence (TTAGGG)4. Pulse field gels were run using a CHEF-DR II apparatus (Bio-Rad, Hercules, CA, USA) for 24 h at constant 150 V, using ramped pulse times from 10 to 30 s.
Statistical analysis
Phylogenetic relationships among the 15 species studied here were inferred by combining the information from six studies (Martin et al., 2000; Michaux et al., 2001; Murphy et al., 2001; Montgelard et al., 2002; Adkins et al., 2003; Steppan et al., 2004), which were based on sequence analysis of 22 genes. Felsenstein’s method (1985) of analyzing phylogenetically independent contrasts was used to test for correlated evolution between telomerase activity, telomere length, maximum lifespan, and body mass. Standardized independent contrasts were generated for phenotypic values using the CRUNCH algorithm of the comparative analysis by independent contrasts (CAIC) program (Purvis & Rambaut, 1995). We were unable to infer branch lengths for the tree as no common molecular phylogeny of all 15 species exists. We therefore assumed equal branch lengths, which is equivalent to assuming a punctuational model of evolution (Purvis et al., 1994). To test for correlated evolution, linear and multiple regression models forced through the origin were fit to independent contrasts (Harvey & Pagel, 1991). For all analyses, we confirmed that the statistical and evolutionary assumptions of the independent contrasts methods were met (Purvis & Rambaut, 1995). Normality was tested using the Shapiro-Wilk test implemented in R. In analyses of the uncorrected data, body mass was log transformed to meet the assumption of normality. In analyses of phylogenetically corrected data all variables were log transformed prior to generating independent contrasts. All statistical tests are two tailed.
Supplementary Material
Fig. S1 Telomerase activity is not affected by short-term storage of carcass at 4 °C. Woodchuck and mouse carcasses were stored at 4 °C and skin and liver biopsies were taken at indicated times after death.
Acknowledgments
We are grateful to Hiram Lyon, Linn Sajdak, Lidza Kalifa, Ingrid Sarelius, Coeli Lopes, and Rulang Jiang (University of Rochester), Stephen Kendrot (USDA Nutria Eradication program), George Brady (Cascade Biosupply), John Ryan (NY State Department of Environmental Conservation) and Richard Salvi (University of Buffalo) for providing animal tissue and carcasses. We thank David Mittelman for help at the beginning stages of the project, and Steven Rosenblatt for help with quantification of TRAP data. We thank H. Allen Orr and Kurt A. McKean for comments on the manuscript. This work was supported by grants from US National Institutes of Health, Ellison Medical Foundation, and American Federation for Aging Research, to V.G.
References
- Adkins RM, Walton AH, Honeycutt RL. Higher-level systematics of rodents and divergence time estimates based on two congruent nuclear genes. Mol. Phylogenet. Evol. 2003;26:409–420. doi: 10.1016/s1055-7903(02)00304-4. [DOI] [PubMed] [Google Scholar]
- Argyle D, Ellsmore V, Gault EA, Munro AF, Nasir L. Equine telomeres and telomerase in cellular immortalisation and ageing. Mech. Ageing Dev. 2003;124:759–764. doi: 10.1016/s0047-6374(03)00104-0. [DOI] [PubMed] [Google Scholar]
- Austad SN. Diverse aging rates in metazoans: targets for functional genomics. Mech. Ageing Dev. 2005;126:43–49. doi: 10.1016/j.mad.2004.09.022. [DOI] [PubMed] [Google Scholar]
- Austad SN, Fischer KE. Mammalian aging, metabolism, and ecology: evidence from the bats and marsupials. J. Gerontol. 1991;46:B47–B53. doi: 10.1093/geronj/46.2.b47. [DOI] [PubMed] [Google Scholar]
- Bekaert S, Derradji H, Baatout S. Telomere biology in mammalian germ cells and during development. Dev. Biol. 2004;274:15–30. doi: 10.1016/j.ydbio.2004.06.023. [DOI] [PubMed] [Google Scholar]
- Buffenstein R. The naked mole-rat: a new long-living model for human aging research. J. Gerontol. A Biol. Sci. Med. Sci. 2005;60:1369–1377. doi: 10.1093/gerona/60.11.1369. [DOI] [PubMed] [Google Scholar]
- Buffenstein R, Jarvis JU. The naked mole rat — a new record for the oldest living rodent. Sci. Aging Knowl. Environ. 2002:pe7. doi: 10.1126/sageke.2002.21.pe7. 2002. [DOI] [PubMed] [Google Scholar]
- Campisi J. Cellular senescence as a tumor-suppressor mechanism. Trends Cell Biol. 2001;11:S27–S31. doi: 10.1016/s0962-8924(01)02151-1. [DOI] [PubMed] [Google Scholar]
- Chan SR, Blackburn EH. Telomeres and telomerase. Philos. Trans. R. Soc. Lond., B, Biol. Sci. 2004;359:109–121. doi: 10.1098/rstb.2003.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S, DePinho RA. Telomerase extracurricular activities. Proc. Natl Acad. Sci. USA. 2002;99:12520–12522. doi: 10.1073/pnas.212514699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JL, Greider CW. Determinants in mammalian telomerase RNA that mediate enzyme processivity and cross-species incompatibility. EMBO J. 2003;22:304–314. doi: 10.1093/emboj/cdg024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis T, Skinner JW, Faragher RG, Jones CJ, Kipling D. Replicative senescence in sheep fibroblasts is a p53 dependent process. Exp. Gerontol. 2005;40:17–26. doi: 10.1016/j.exger.2004.09.004. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Phylogenies and the comparative method. Am. Nat. 1985;125:1–15. [Google Scholar]
- Forsyth NR, Wright WE, Shay JW. Telomerase and differentiation in multicellular organisms: turn it off, turn it on, and turn it off again. Differentiation. 2002;69:188–197. doi: 10.1046/j.1432-0436.2002.690412.x. [DOI] [PubMed] [Google Scholar]
- Fradiani PA, Ascenzioni F, Lavitrano M, Donini P. Telomeres and telomerase activity in pig tissues. Biochimie. 2004;86:7–12. doi: 10.1016/j.biochi.2003.11.009. [DOI] [PubMed] [Google Scholar]
- Gorbunova V, Seluanov A. Telomerase as a growth-promoting factor. Cell Cycle. 2003;2:534–537. doi: 10.4161/cc.2.6.515. [DOI] [PubMed] [Google Scholar]
- Harley CB, Futcher AB, Greider CW. Telomeres shorten during ageing of human fibroblasts. Nature. 1990;345:458–460. doi: 10.1038/345458a0. [DOI] [PubMed] [Google Scholar]
- Harvey PH, Pagel MD. The Comparative Method in Evolutionary Biology. Oxford University Press; Oxford: 1991. [Google Scholar]
- Harvey PH, Read AF, Promislow DEL. Life history variation in placental mammals: unifying the data with theory. Oxf. Surv. Evol. Biol. 1989;6:13–31. [Google Scholar]
- Hayflick L, Moorhead PS. The serial cultivation of human diploid strains. Exp. Cell Res. 1961;25:585–621. doi: 10.1016/0014-4827(61)90192-6. [DOI] [PubMed] [Google Scholar]
- Hemann MT, Greider CW. Wild-derived inbred mouse strains have short telomeres. Nucl. Acids Res. 2000;28:4474–4478. doi: 10.1093/nar/28.22.4474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Human Ageing Genomic Resources. 2006 http://genomics.senescence.info/species/
- Kim NW, Piatyszek MA, Prowse KR, Harley CB, West MD, Ho PL, Coviello GM, Wright WE, Weinrich SL, Shay JW. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- Leroi AM, Koufopanou V, Burt A. Cancer selection. Nat. Rev. Cancer. 2003;3:226–231. doi: 10.1038/nrc1016. [DOI] [PubMed] [Google Scholar]
- Lipman R, Galecki A, Burke DT, Miller RA. Genetic loci that influence cause of death in a heterogeneous mouse stock. J. Gerontol. A Biol. Sci. Med. Sci. 2004;59:977–983. doi: 10.1093/gerona/59.10.B977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzini A, Tresini M, Austad SN, Cristofalo VJ. Cellular replicative capacity correlates primarily with species body mass not longevity. Mech. Ageing Dev. 2005;126:1130–1133. doi: 10.1016/j.mad.2005.05.004. [DOI] [PubMed] [Google Scholar]
- Martin Y, Gerlach G, Schlotterer C, Meyer A. Molecular phylogeny of European muroid rodents based on complete cytochrome b sequences. Mol. Phylogenet. Evol. 2000;16:37–47. doi: 10.1006/mpev.1999.0760. [DOI] [PubMed] [Google Scholar]
- McKevitt TP, Nasir L, Wallis CV, Argyle DJ. A cohort study of telomere and telomerase biology in cats. Am. J. Vet. Res. 2003;64:1496–1499. doi: 10.2460/ajvr.2003.64.1496. [DOI] [PubMed] [Google Scholar]
- Michaux J, Reyes A, Catzeflis F. Evolutionary history of the most speciose mammals: molecular phylogeny of muroid rodents. Mol. Biol. Evol. 2001;18:2017–2031. doi: 10.1093/oxfordjournals.molbev.a003743. [DOI] [PubMed] [Google Scholar]
- Montgelard C, Bentz S, Tirard C, Verneau O, Catzeflis FM. Molecular systematics of sciurognathi (rodentia): the mitochondrial cytochrome b and 12S rRNA genes support the Anomaluroidea (Pedetidae and Anomaluridae) Mol. Phylogenet. Evol. 2002;22:220–233. doi: 10.1006/mpev.2001.1056. [DOI] [PubMed] [Google Scholar]
- Murphy WJ, Eizirik E, Johnson WE, Zhang YP, Ryder OA, O’Brien SJ. Molecular phylogenetics and the origins of placental mammals. Nature. 2001;409:614–618. doi: 10.1038/35054550. [DOI] [PubMed] [Google Scholar]
- Nasir L, Devlin P, McKevitt T, Rutteman G, Argyle DJ. Telomere lengths and telomerase activity in dog tissues: a potential model system to study human telomere and telomerase biology. Neoplasia. 2001;3:351–359. doi: 10.1038/sj.neo.7900173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak R. Walker’s Mammals of the World. John Hopkins University Press; Baltimore: 1999. [Google Scholar]
- Nunney L. Lineage selection and the evolution of multistage carcinogenesis. Proc. Biol. Sci. 1999;266:493–498. doi: 10.1098/rspb.1999.0664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olovnikov AM. A theory of marginotomy. The incomplete copying of template margin in enzymic synthesis of polynucleotides and biological significance of the phenomenon. J. Theor. Biol. 1973;41:181–190. doi: 10.1016/0022-5193(73)90198-7. [DOI] [PubMed] [Google Scholar]
- Parrinello S, Samper E, Krtolica A, Goldstein J, Melov S, Campisi J. Oxygen sensitivity severely limits the replicative lifespan of murine fibroblasts. Nat. Cell Biol. 2003;5:741–747. doi: 10.1038/ncb1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak S, Multani AS, McConkey DJ, Imam AS, Amoss MS., Jr Spontaneous regression of cutaneous melanoma in sinclair swine is associated with defective telomerase activity and extensive telomere erosion. Int. J. Oncol. 2000;17:1219–1224. doi: 10.3892/ijo.17.6.1219. [DOI] [PubMed] [Google Scholar]
- Promislow DE. DNA repair and the evolution of longevity: a critical analysis. J. Theor. Biol. 1994;170:291–300. doi: 10.1006/jtbi.1994.1190. [DOI] [PubMed] [Google Scholar]
- Prowse KR, Greider CW. Developmental and tissue-specific regulation of mouse telomerase and telomere length. Proc. Natl Acad. Sci. USA. 1995;92:4818–4822. doi: 10.1073/pnas.92.11.4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purvis A, Gittleman JL, Luh H-K. Truth or consequences: effects of phylogenetic accuracy on two comparative methods. J. Theor. Biol. 1994;167:293–300. [Google Scholar]
- Purvis A, Rambaut A. Comparative analysis by independent contrast (CAIC): an Apple Macintosh application for analysing comparative data. Comp. Appl. Biosci. 1995;11:247–251. doi: 10.1093/bioinformatics/11.3.247. [DOI] [PubMed] [Google Scholar]
- Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- Shay JW, Wright WE. Hayflick, his limit, and cellular ageing. Nat. Rev. Mol. Cell Biol. 2000;1:72–76. doi: 10.1038/35036093. [DOI] [PubMed] [Google Scholar]
- Steinert S, White DM, Zou Y, Shay JW, Wright WE. Telomere biology and cellular aging in nonhuman primate cells. Exp. Cell Res. 2002;272:146–152. doi: 10.1006/excr.2001.5409. [DOI] [PubMed] [Google Scholar]
- Steppan S, Adkins R, Anderson J. Phylogeny and divergence-date estimates of rapid radiations in muroid rodents based on multiple nuclear genes. Syst. Biol. 2004;53:533–553. doi: 10.1080/10635150490468701. [DOI] [PubMed] [Google Scholar]
- Thomas M, Yang L, Hornsby PJ. Formation of functional tissue from transplanted adrenocortical cells expressing telomerase reverse transcriptase. Nat. Biotechnol. 2000;18:39–42. doi: 10.1038/71894. [DOI] [PubMed] [Google Scholar]
- Turturro A, Witt WW, Lewis S, Hass BS, Lipman RD, Hart RW. Growth curves and survival characteristics of the animals used in the Biomarkers of Aging Program. J. Gerontol. A Biol. Sci. Med. Sci. 1999;54:B492–B501. doi: 10.1093/gerona/54.11.b492. [DOI] [PubMed] [Google Scholar]
- Weigl R. Longevity of Mammals in Captivity; from the Living Collections of the World. Schweizerbart; Stuttgart: 2005. [Google Scholar]
- Wright W, Shay J. Telomere dynamics in cancer progression and prevention: fundamental differences in human and mouse telomere biology. Nat. Med. 2000;6:849–851. doi: 10.1038/78592. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Telomerase activity is not affected by short-term storage of carcass at 4 °C. Woodchuck and mouse carcasses were stored at 4 °C and skin and liver biopsies were taken at indicated times after death.