Abstract
In the major pathway of homologous DNA recombination in prokaryotic cells, the Holliday junction intermediate is processed through its association with RuvA, RuvB, and RuvC proteins. Specific binding of the RuvA tetramer to the Holliday junction is required for the RuvB motor protein to be loaded onto the junction DNA, and the RuvAB complex drives the ATP-dependent branch migration. We solved the crystal structure of the Holliday junction bound to a single Escherichia coli RuvA tetramer at 3.1-Å resolution. In this complex, one side of DNA is accessible for cleavage by RuvC resolvase at the junction center. The refined junction DNA structure revealed an open concave architecture with a four-fold symmetry. Each arm, with B-form DNA, in the Holliday junction is predominantly recognized in the minor groove through hydrogen bonds with two repeated helix-hairpin-helix motifs of each RuvA subunit. The local conformation near the crossover point, where two base pairs are disrupted, suggests a possible scheme for successive base pair rearrangements, which may account for smooth Holliday junction movement without segmental unwinding.
In all living organisms, DNA homologous recombination is a crucial process not only for generating the genomic diversity but also for repairing damaged chromosomes. At the molecular level, the key events in homologous recombination are the formation of a universal DNA intermediate, the Holliday junction (1), and the processing of this intermediate into mature recombinant DNA products through branch migration of the junction followed by resolution. In the late stage of the Escherichia coli recombination process, the RuvA, RuvB, and RuvC proteins are involved in the processing of Holliday junction DNA (2–4). Specific binding of the RuvA tetramer to a Holliday junction is followed by loading of the RuvB hexameric rings and the formation of a tripartite structure, in which the RuvA tetramer is flanked by the two RuvB rings on opposite sides (5). The RuvAB complex facilitates the migration of the junction point and expands the heteroduplex region in an ATP-dependent manner. Recent studies have suggested that the RuvA, RuvB, and RuvC proteins assemble to form a transient complex, before resolution of the Holliday junction by RuvC (6–8).
Crystallographic and biochemical studies revealed that RuvA adopts a unique tetrameric architecture formed by identical subunits with three distinct domains (9, 10). Proteolytic and mutational analyses demonstrated that domain III plays a major role in the ATP-dependent branch migration through direct contact with RuvB whereas the remaining major core (I and II) is responsible for Holliday junction binding (10, 11).
The RuvA tetramer forms two types of complexes, termed complex I and complex II (12–14). They both contain a single junction DNA but different numbers of the RuvA tetramer, one tetramer for complex I and two tetramers for complex II. The crystal structure of the E. coli RuvA-Holliday junction complex in the complex I form was solved at 6-Å resolution, and an overall structural view of the complex was reported (15). More recently, the crystal structure of octameric complex II from Mycobacterium leprae has been determined at 3.0-Å resolution (16). However, the internal DNA structure appeared to be so substantially disordered, and the critical junction DNA conformation was not described in detail. We report here the crystal structure of the E. coli RuvA-Holliday junction complex in the complex I form. This analysis allowed us to refine both structures, the protein and the Holliday junction, at 3.1-Å resolution. The atomic model of the complex provides insights into specific recognition between the protein and the junction DNA.
Materials and Methods
Purification of the RuvA-Holliday Junction Complex.
The RuvA protein (203 amino acids) was purified as reported (10). The DNA oligonucleotides, which were designed to form immobile four-way junctions, were obtained commercially (BEX, Tokyo). Each set of the four oligonucleotides was mixed at an equimolar ratio, and the immobile four-way junctions were prepared by annealing, as described (17). The RuvA tetramer and the synthetic four-way junction were combined in a 2:1 molar ratio and were dialyzed against a buffer containing 20 mM Tris⋅HCl buffer at pH 7.5, 150 mM NaCl, 5% glycerol, and 1 mM EDTA at 4°C. The complex was fractionated by gel filtration on a Superdex 200 10/30 column (Amersham Pharmacia).
Crystallization and Data Collection.
Crystallization was carried out by using 15 kinds of synthetic junctions with various arm lengths. Among the various crystal forms produced from polyethylene glycol or ammonium sulfate solutions by the vapor diffusion or microdialysis method, only one crystal form diffracted to 3.0-Å resolution. This crystal form was grown at 20°C by the hanging-drop vapor diffusion method from a solution containing 0.1 M Mes⋅NaOH, 2.0–2.2 M ammonium sulfate, and 5% glycerol (pH 7.5). The diffraction pattern showed unit cell dimensions of a = b = c = 158.65 Å with the I cubic space group. A careful examination of the intensity data identified the space group as I432. A data set from a native crystal (Native1) was collected on beam line BL41XU at SPring8 (Hyogo, Japan). A second native (Native2) and derivative data sets were measured on beam line BL6B of the Photon Factory at KEK (Tsukuba, Japan). All data sets were collected under liquid nitrogen cryo-conditions at a temperature of 100 K. Data were reduced with the hkl program suite (18) (Table 1).
Table 1.
Statistics of crystallographic analysis
| Data collection | Diffraction data | Native 1* | Native 2* | Bakers Hg |
|---|---|---|---|---|
| Wavelength, Å | 0.708 | 1.00 | 1.00 | |
| Resolution, Å | 80.0–3.2 | 80.0–3.0 | 80.0–3.5 | |
| Total observations | 147,496 | 143,269 | 75,377 | |
| Unique reflections | 5,956 | 7,136 | 4,582 | |
| Completeness, % | 99.9 (100) | 99.9 (100) | 99.8 (100) | |
| Rmerge, % | 6.4 (17.9) | 5.4 (27.5) | 7.1 (33.4) | |
| Rderiv, % | 27.8 | |||
| SIRAS phasing, 20–3.5 Å | Heavy atom site | 1 | ||
| Phasing power; acentric/centric | 1.44/0.95 | |||
| Rcullis; acentric/centric, % | 0.79/0.74 | |||
| Refinement, 50.0–3.1 Å | No. of reflections | 6,491 | ||
| Rc %/Rfree, % | 25.0 (33.6)/27.9 (36.9) | |||
| rmsd bond length, Å/rmsd bond angles, degrees | 0.011/1.5 | |||
| No. of atoms (average B value, Å2); protein/DNA† | 1,528 (45.6)/2,038 (94.8) | |||
Rmerge = Σ|I − 〈I〉|/ΣI; Rderiv = Σh∥Fderiv(h)| − |Fnative2(h)∥/Σh|Fnative2(h)|. Phasing power = 〈|FH|〉/E, where |FH| is the structure factor amplitude for the heavy atom and E is the estimated lack-of-closure error. Rcullis = Σ∥FPH − FP| − |FH(calc)∥/Σ|FPH − FP|; Rc and Rfree = Σh∥F(h)obs| − |F(h)calc∥/Σh|F(h)obs| for reflections in the working and test (9.5%) sets, respectively. Numbers in parentheses are for the highest shells.
Native1 was used for initial phasing by the molecular replacement method whereas SIRAS phasing and crystallographic refinements were carried out with Native2.
DNA atoms include four sets of 25 nucleotides at 0.25 occupancy.
Phasing.
Initial phasing of the native data were attempted by using the molecular replacement technique. The structure of the NH2 fragment alone (residues 1–138), extracted from the crystal structure of E. coli RuvA (10), was used as a search model. For the calculations, the program amore (19) and intensity data from 15- to 4-Å resolution were used. The molecular replacement solution resulted in a crystallographic R factor of 43.0% and a correlation factor of 55.5% after rigid body refinement. The electron density map showed domain III of RuvA omitted from the search model whereas the DNA molecule could not be identified.
These phases were improved by combining them with those obtained from the single isomorphous replacement and anomalous scattering (SIRAS) of a mercury derivative (Table 1); the mercury derivative was prepared by soaking the crystal in 1 mM Baker's dimercurial for 1 h. The mercury atom, bound to Cys-34, was clearly located in a difference Fourier map, which was calculated by using the phases from the molecular replacement. Refinements of the heavy atom parameters and phase calculations were carried out by using the program mlphare (20). The SIRAS phases were modified by solvent flattening and histogram matching by using dm (21). The resultant 4-Å resolution electron-density map clearly showed the junction DNA. The model of the NH2 fragment, obtained from the solution of the molecular replacement, was fitted to the solvent-flattened map, and the phases calculated from this model were combined with the SIRAS phases, using the program sigmaa (22). These phases provided a map good enough to reveal the remaining portion of the RuvA protein and the DNA phosphodiester backbone.
Crystallographic Refinement.
The four-fold axis of the RuvA tetramer in the complex coincides with the crystallographic four-fold axis. The synthetic Holliday junction has no internal four-fold symmetry in the sequence and has one overhanging nucleotide on each 5′ end of the two arms (Fig. 4C). Thus, the structural analysis based on I432 produces an averaged density for the junction DNA, and hence the identification of bases would be hampered even in the final Fourier map.
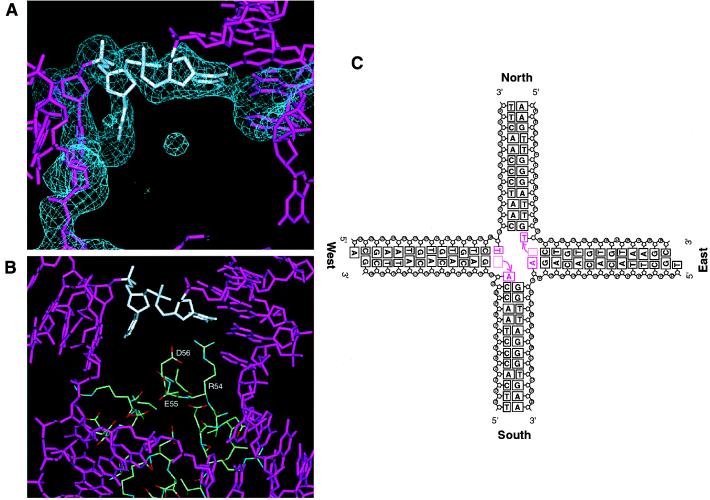
Figure 4.
Structure of the Holliday junction center. (A) Fo-Fc annealed omit electron density map (>2.5 σ) showing a DNA moiety within the junction center. The two bases closest to the junction center, indicated by a white stick model, were omitted from the map calculation. (B) Environments around unpaired bases in the tetrameric RuvA center. Arg-54, Glu-55, and Asp-56 of each RuvA subunit, which form the acidic pin, are shown by a ball-and-stick representation. (C) Schematic drawing of the Holliday junction structure. The synthetic Holliday junction was designed to form two pairs of opposite arms with different lengths: the north and south arms of a 12-bp DNA duplex and the east and west arms of a 13-bp DNA duplex with a single base overhang at the 5′ end. Two AT base pairs disrupted at the crossover are colored by magenta. The topological features of the unpaired bases may reflect a scene during branch migration, in which the base pair rearrangements are in progress and the new base pairs will be subsequently formed.
Data from 50- to 3.1-Å resolution (no σ cut off) were used for the crystallographic refinement. Temperature factors, grouped into main chain and side chain atoms of each protein residue and into sugar-phosphate and base atoms of each nucleotide, were refined, and a bulk solvent correction was applied. The present structure, refined by simulated annealing with cns (23), has yielded an R factor of 0.250 and a free R factor of 0.279. Four kinds of DNA sequences in the complex were averaged in the refinement, and hence the asymmetric unit of the crystal structure contains one RuvA subunit, which includes 199 amino acid residues (residues 1–150 and 155–203), and four DNA duplex arms (100 nucleotides) at 0.25 occupancy. Thus, the final model of the complex consists of four identical RuvA subunits and four DNA strands of 25 nucleotides.
Figs. 1B, 3A, B, and D, and 4A and B were drawn in quanta98. Figs. 2 A and B and 5 were produced with the program grasp (24).
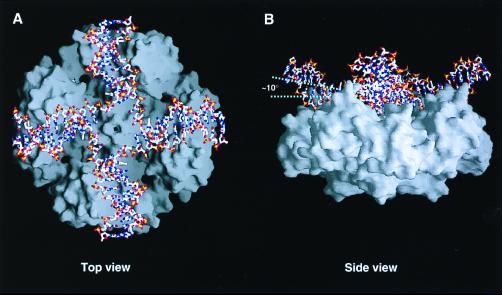
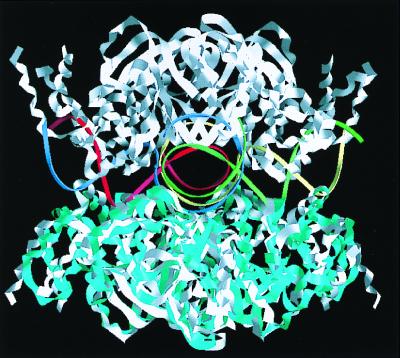
Figure 1.
Entire view of the RuvA-Holliday junction complex. (A) Top view of the Holliday junction bound to the RuvA tetramer, looking down the protein-DNA interface along the four-fold axis. The DNA molecule is shown in a stick representation colored with the atoms by type: oxygen atoms in red, phosphorus atoms in yellow, nitrogen atoms in blue, and carbon atoms in white. The molecular surface representation shows the RuvA tetramer. (B) Side view of the complex, rotated about 90° from A. The four-fold axis lies perpendicular to the plane at the center of the complex.
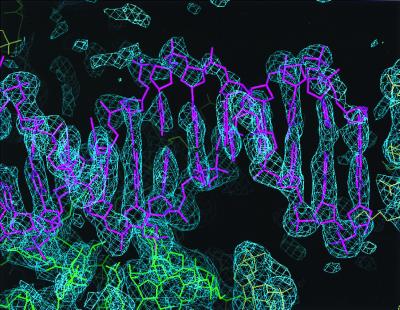
Figure 3.
2Fo-Fc electron density map (>1.0 σ) showing a DNA duplex, corresponding to one of the four arms of Holliday junction. The DNA molecule is shown in a magenta-colored stick representation. The green wire model indicates the RuvA protein in the complex whereas the yellow ones denote the symmetry-related complex molecules.
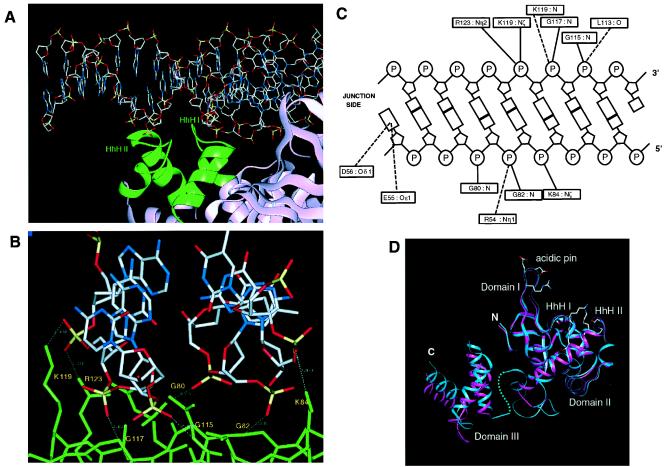
Figure 2.
Representative protein-Holliday junction interaction. (A) A DNA arm (stick representation) is recognized on the minor groove side by the two HhH motifs (green ribbon representation) of RuvA. The view of the complex is the same as that in Fig. 1B. The junction center is located at the right end of the figure. (B) Close-up view showing the interactions between RuvA and DNA. RuvA is shown in a green-colored stick representation. Hydrogen bonds formed between the protein and the DNA phosphate backbone are indicated by white dotted lines. (C) Schematic representation of protein-DNA interactions. Solid lines indicate polar interactions between the protein and DNA atoms at a distance of less than 3.2 Å. Dotted lines represent candidates for water-mediated interactions within a distance of less than 6.0 Å. (D) Ribbon representations of the single subunit of E. coli RuvA. The subunit of the free form (magenta) is superimposed onto that of the complex with the junction DNA (blue). The blue dot line indicates the structurally disordered connection between the flexible loop and domain III in the complex whereas the connection in the free form structure is not shown. The residues, involved in DNA binding through direct (Lys-84, Gly-117, Lys-119, and Arg-123) or putative indirect (Arg-54 and Leu-113) polar interactions, are indicated on the complex model by their side chains. The side chains of Glu-55 and Asp-56, which form the acidic pin, are also indicated.
Figure 5.
Structural comparison between complex I and complex II (16). Octameric complex II is superimposed onto tetrameric complex I, which contains the refined Holliday junction structure. The RuvA tetramer in complex I is represented by the blue ribbon whereas the white ribbons indicate the two tetramers in complex II. Each strand of the junction DNA in complex I is drawn with different ribbon color. The DNA structure in complex II is not shown here because its coordinates are unavailable from the Protein Data Bank.
Results
Overall structure of the RuvA tetramer-Holliday junction complex. For cocrystallization, the RuvA tetramer was mixed with a synthetic Holliday junction, which consists of four immobile arms with non-homologous sequences (Fig. 4C). We purified octameric complex II by size exclusion chromatography. The fraction containing the complex was concentrated and then immediately subjected to crystallization. The subsequent x-ray analysis revealed that the crystal actually contained complex I. We found that increasing the salt concentration converted complex II to complex I at 0.5 M NaCl in neutral solutions (data not shown). It is thus likely that the high ammonium sulfate concentration in the crystallization drops caused the conversion of complex II into complex I.
The RuvA-Holliday junction complex retains the four-fold symmetry (Fig. 1 A and B), as previously revealed by the 6-Å resolution structural analysis of a different complex with shorter DNA arms (15). The junction DNA conforms well to the positively charged concave surface of the RuvA tetramer. One RuvA subunit covers eight base pairs of a single DNA arm starting at the crossover point. The junction DNA bound to RuvA adopts a remarkable conformation, which substantially deviates from the strict square-planar arrangement, as observed in the previous lower resolution structure (15). The Holliday junction is most depressed in close vicinity of the crossover point, to allow intimate contact between the DNA interface and the RuvA tetramer. The helical axis of each junction arm is inclined by about 10 degrees from the ideal plane (Fig. 1B). On the extensive basic surface of the RuvA tetramer, eight residues, corresponding to Glu-55 and Asp-56 in each subunit, form an acidic central pin (9) (Fig. 2D). This acidic pin appears to repel the DNA backbone away from the junction center (Fig. 1A).
DNA binding produces only minor conformational changes of RuvA, except for an approximately 4-Å relocation of domain III toward the protein-DNA interface (Fig. 2D). The average root-mean-square displacement for 132 Cα atoms of domains I and II is at 0.76 Å between the complex and the E. coli DNA-free form. To explicitly detect a difference in the intersubunit configuration over the entire tetramer, the local superposition was also made with respect to only domains I and II of a single subunit. This comparison revealed that DNA binding induced a significant alteration in the intersubunit orientations, so that the RuvA tetramer twisted slightly about the central four-fold axis.
The average B-factor of the main chain atoms for domain III in the complex (39.0 Å2) is much lower than that in the DNA-free form [94.6 Å2 (10)] and is comparable to those in the other two domains. In the DNA free crystal, the entire linker region between domains II and III (residues 141–157) adopted a disordered structure. In contrast, the same region in the complex showed a well defined electron density, which allowed us to refine the structure, except for four residues (residues 151–154). It contains a short α helix between Ala-144 and Thr-149 (Fig. 2D), which was not found in the DNA-free form. As the junction DNA makes no interaction with domain III, it is likely that the packing effects in the complex crystal account for the movement of domain III and the well defined structure of the linker region.
Each end of the DNA arms, furthest from the crossover point, is in contact with the fifth α-helix in an adjacent RuvA molecule, through hydrophobic interactions between the bases and the side chains of Val-107 and Val-111 and through nonpolar interactions between the sugar and the aliphatic moiety of the Lys-118 side chain. Another crystal contact was observed between symmetry-related protein molecules. No interaction was found between DNA molecules.
Protein-DNA Interaction.
Domain II plays a major role in the junction DNA binding whereas domains I and III hardly contribute to DNA recognition. Each RuvA subunit is responsible for fixing one of the four DNA arms. The two helix-hairpin-helix (HhH) motifs (25) in domain II (Fig. 2D) are exposed to the DNA binding interface and play a major role in the recognition of the Holliday junction (Fig. 2A). The two hairpin loops in the repeated HhH motifs, with a helical insertion, contact with each phosphodiester backbone of a DNA duplex on the minor groove side (Fig. 2A). Both phosphates of two adjacent nucleotides in each strand are recognized by two peptide groups (Fig. 2 B and C); the phosphate oxygens in one DNA strand are hydrogen-bonded with the nitrogen atoms of the main chain amides of Gly-80 and Gly-82, which lie on the hairpin loop between the α2 and α3 helices in HhH I. In the complementary strand, the amide nitrogens of Gly-115 and Gly-117 on the hairpin between the α5 and α6 helices in HhH II form hydrogen-bonds with the phosphate oxygens. The basic side chains of Lys-119 and Arg-123 near the second HhH motif are located close enough to make polar interactions with the phosphate oxygens of one strand. The Lys-84 side chain also forms a hydrogen bond with the other strand (Fig. 2 B and C). Despite these strong polar interactions, no significant conformational change was observed in the HhH motifs.
Holliday Junction Structure.
Overall, each of the four DNA arms in the junction adopts the B-DNA conformation, except for the junction center. The final map showed sequential and well defined electron densities, which fit to one strand of B-form DNA on the side of the protein-DNA interface whereas the other DNA strand, facing the solvent, shows partially poor densities, particularly for the sugar phosphate groups (Fig. 3). Each end of the arms, in contact with the protein of an adjacent complex, represents the base pair formation, implying that the structures of the overhanging nucleotides at the eastern and western termini (Fig. 4C) are disordered. The refinement of the complex model, using the I23 symmetry, excluded the possibility that this structural disorder was derived from the higher symmetry of I432.
We identified 12 base pairs for each arm of the four-way junction. The information of the bases is lost because the electron density was smeared owing to averaging among the four non-homologous arms. However, the refined structure clearly revealed that the two AT base pairs nearest to the junction center in the east-west arms are disrupted (Fig. 4A). As indicated in the refined electron density map, the unpaired adenine or thymine base on each strand at the junction is stacked to the inner end of each arm as a nucleotide 3′ extension (Fig. 4 A and C). Consequently, in the final model, each arm is composed of the DNA duplex with 12 base pairs and a single unpaired nucleotide at the junction center. Each of four DNA single strands, consisting of 25 nucleotides, is distributed into two adjacent DNA duplex arms, with 13 nucleotides from the 5′ end and 12 nucleotides from the 3′ end. The unpaired nucleotides do not directly interact with the protein. They are located at a distance of about 6 Å from the carboxyl side chains of Glu-55 and Asp-56 in the central acidic pin (Fig. 4B), implying that water-mediated hydrogen bonds may be formed between these acidic side chains and unpaired nucleotides. The side chain of Arg-54 approaches the phosphate group of the DNA backbone (Figs. 2C and 4B), in such a way that water-mediated polar interactions might be possible between them.
Discussion
The present structural analysis of the RuvA-Holliday junction complex allowed the successful refinements of both the structures of the RuvA tetramer and of the Holliday junctions at 3.1-Å resolution. The present structure of the complex, with a much larger junction DNA than those in the other complex crystals, also has provided insights into the mechanisms of RuvA-Holliday junction recognition and the RuvAB-mediated branch migration.
The refined structure of complex I shows that the Holliday junction contains four unpaired bases at the crossover point (Fig. 4 A and C), although the previous structural analysis at 6-Å resolution suggested that the base pairs in the corresponding region were maintained (15). The configurations of these unpaired bases seem to hint at a possible intermediate DNA structure during branch migration, where two base pairs, located on opposite sides across the junction center, are disrupted and subsequently reformed with other partners in the adjacent arms (Fig. 4C). This putative action within the RuvA-Holliday junction complex implies more positive and important roles of RuvA in branch migration driven by the RuvAB complex. Recent studies have suggested that the RuvAB complex promotes migration of the junction point, without the extensive separation of DNA duplexes as found classically for hexameric helicases (26, 27). This agrees with the idea that the RuvB motor functions as a pump to pull DNA duplexes along the junction arms on the two opposite sides (28). Thus, RuvA, which executes rearrangements of base pairs, would smoothly translate the crossover point on the protein platform in concert with the ATP-dependent RuvB pumping behavior. This model is more consistent with the electron microscopic observation that double-stranded DNA, rather than single-stranded DNA, passes through the central hole of the RuvB ring (5, 28).
The direct hydrogen bonds of a pair of HhH motifs in domain II with phosphate backbones appear to be a predominant factor for RuvA to make specific recognition of junction DNA (Fig. 2). On the other hand, protein-DNA recognition generally involves multiple water-mediated interactions (29). In addition, the structural features of the protein-DNA interface imply the formation of many water-mediated hydrogen bonds, despite the insufficient resolution to identify them convincingly. In the vicinity of the junction center, the side chains of Arg-54, Glu-55, and Asp-56 are situated close enough to contact the phosphate backbone or the unpaired nucleotide base through water-mediated interactions and may contribute to maintaining a unique DNA conformation. Efficient base pair rearrangements, coupled with the migration of DNA arms, may rely on water-mediated interactions between the central acidic pin and the unpaired nucleotides.
The internal architecture of each RuvA domain is essentially identical between complexes I and II, although domain III is shifted by several angstroms. On the other hand, the conformation of the Holliday junction and the DNA-protein interface in our complex I structure appear to differ substantially from those in complex II (16). In contrast with the concave junction DNA architecture in complex I (Fig. 1B), a doublet of the RuvA tetramers in complex II sandwiches a flat junction DNA with a central four-fold axis. The complex II crystal structure also showed a distinct scheme for holding the DNA backbones on the RuvA tetramer by the HhH motifs. Actually, both of the RuvA tetramers, the upper and the lower, simultaneously contact each strand of a DNA duplex arm (16). By contrast, in complex I, the two repeated HhH motifs bridge the two phosphodiester backbones across the minor groove of the B-form DNA (Fig. 2A). This implies that the arrangement of a junction DNA arm relative to each RuvA subunit may differ between the two complexes. When the RuvA tetramer of complex I is superimposed on the lower tetramer of complex II (Fig. 5), there are two serious steric conflicts, observed between the junction DNA of complex I and the upper RuvA tetramer in complex II, because of the large deviation of the Holliday junction from the planar conformation. Each domain III of the upper tetramer in complex II collides with the extension of each DNA arm (Fig. 5), and the other conflict is observed between the junction DNA and the acidic pin of the upper tetramer. We suspect that the RuvB motor could act as a molecular switch to alter the conformation of RuvA and the Holliday junction between the two forms. Thus, complexes I and II may reflect two of various stages in the dynamic and consecutive action during branch migration.
It has been proposed that the octameric complex in form II provides a stable anchor for the RuvB motor, which would drive branch migration by rotating the DNA (16). On the other hand, the RuvC dimer was found to more efficiently resolve the junction DNA in the presence of both RuvA and RuvB, implying the formation of a higher order complex, termed the RuvABC resolvasome (7, 8, 30). However, the Holliday junction covered on both sides by RuvA, as in form II, could not be cleaved by RuvC resolvase. Actually, the addition of RuvA was found to inhibit the cleavage of the junction DNA (13). By contrast, the junction DNA in complex I is open to contact RuvC, and, hence, this complex would be more favorable for the formation of RuvABC resolvasome, where RuvC is bound to RuvB (7). However, docking examination showed obvious steric conflicts between the crystal structure of RuvC (31) and the Holliday junction in complex I. As the rigid dimeric structure of RuvC is unlikely to be deformed (32), we presume that the junction DNA would be at least partly disjoined from RuvA so as to contact RuvC. RuvB in complex with RuvC may play an unknown role in adjusting the junction DNA conformation to the interface of RuvC. A full understanding of the specific interactions among the RuvA, RuvB, and RuvC proteins and the Holliday junction awaits more detailed structural studies of the higher order complex at the atomic level.
Acknowledgments
We thank Drs. N. Kamiya and M. Kawamoto for their help with data collection at Spring8. Drs. S. E. Tsutakawa and A. Paler are acknowledged for critical reading of the manuscript. This research was supported by Grants-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Science, Sports and Culture of Japan (Grants 08280102 and 0828010) to H.S.
Abbreviation
- SIRAS
single isomorphous replacement and anomalous scattering
Footnotes
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 1C7Y).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.140212997.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.140212997
References
- 1.Holliday R. Genet Res. 1964;5:282–304. [Google Scholar]
- 2.Shinagawa H, Iwasaki H. Trends Biochem Sci. 1996;21:107–111. [PubMed] [Google Scholar]
- 3.West S C. Annu Rev Genet. 1997;31:213–244. doi: 10.1146/annurev.genet.31.1.213. [DOI] [PubMed] [Google Scholar]
- 4.Sharples G J, Ingleston S M, Lloyd R G. J Bacteriol. 1999;181:5543–5550. doi: 10.1128/jb.181.18.5543-5550.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu X, West S C, Egelman E H. J Mol Biol. 1997;266:217–222. doi: 10.1006/jmbi.1996.0799. [DOI] [PubMed] [Google Scholar]
- 6.Davies A A, West S C. Curr Biol. 1998;8:725–727. doi: 10.1016/s0960-9822(98)70282-9. [DOI] [PubMed] [Google Scholar]
- 7.van Gool A J, Shah R, Mezard C, West S C. EMBO J. 1998;17:1838–1845. doi: 10.1093/emboj/17.6.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Gool A J, Hajibagheri N M, Stasiak A, West S C. Genes Dev. 1999;13:1861–70. doi: 10.1101/gad.13.14.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rafferty J B, Sedelnikova S E, Hargreaves D, Artymiuk P J, Baker P J, Sharples G J, Mahdi A A, Lloyd R G, Rice D W. Science. 1996;274:415–421. doi: 10.1126/science.274.5286.415. [DOI] [PubMed] [Google Scholar]
- 10.Nishino T, Ariyoshi M, Iwasaki H, Shinagawa H, Morikawa K. Structure (London) 1998;6:11–21. doi: 10.1016/s0969-2126(98)00003-3. [DOI] [PubMed] [Google Scholar]
- 11.Nishino T, Iwasaki H, Kataoka M, Ariyoshi M, Fujita T, Shinagawa H, Morikawa K. J Mol Biol. 2000;298:407–416. doi: 10.1006/jmbi.2000.3675. [DOI] [PubMed] [Google Scholar]
- 12.Parsons C A, Tsaneva I, Lloyd R G, West S C. Proc Natl Acad Sci USA. 1992;89:5452–6. doi: 10.1073/pnas.89.12.5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Whitby M C, Bolt E L, Chan S N, Lloyd R G. J Mol Biol. 1996;264:878–890. doi: 10.1006/jmbi.1996.0684. [DOI] [PubMed] [Google Scholar]
- 14.Rice D W, Rafferty J B, Artymiuk P J, Lloyd R G. Curr Opin Struct Biol. 1997;7:798–803. doi: 10.1016/s0959-440x(97)80149-2. [DOI] [PubMed] [Google Scholar]
- 15.Hargreaves D, Rice D W, Sedelnikova S E, Artymiuk P J, Lloyd R G, Rafferty J B. Nat Struct Biol. 1998;5:441–446. doi: 10.1038/nsb0698-441. [DOI] [PubMed] [Google Scholar]
- 16.Roe S M, Barlow T, Brown T, Oram M, Keeley A, Tsaneva I R, Pearl L H. Mol Cell. 1998;2:361–372. doi: 10.1016/s1097-2765(00)80280-4. [DOI] [PubMed] [Google Scholar]
- 17.Iwasaki H, Takahagi M, Nakata A, Shinagawa H. Genes Dev. 1992;6:2214–20. doi: 10.1101/gad.6.11.2214. [DOI] [PubMed] [Google Scholar]
- 18.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 19.Navaza J. Acta Crystallogr A. 1994;50:157–163. [Google Scholar]
- 20.Otwinowski Z. In: In Proceedings of the CCP4 Study Weekend. Wolf W, Evans P R, Leslie A G W, editors. Warringtom, U.K.: SERC Daresbury Laboratory; 1991. pp. 80–88. [Google Scholar]
- 21.Collaborative Computational Project, Number 4. Acta Crystallogr D. 1994;50:760–763. [Google Scholar]
- 22.Read R J. Acta Crystallogr A. 1986;42:140–149. [Google Scholar]
- 23.Brunger A T, Adams P D, Clore G M, Delano W L, Gros P, Grosse-Kunstleve R W, Jiang J-S, Kuszewski J, Nilges M, Pannu N S, et al. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 24.Nicholls A. grasp. New York: Columbia Univ.; 1993. [Google Scholar]
- 25.Doherty A J, Serpell L C, Ponting C P. Nucleic Acids Res. 1996;24:2488–2497. doi: 10.1093/nar/24.13.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.George H, Mezard C, Stasiak A, West S C. J Mol Biol. 1999;293:505–519. doi: 10.1006/jmbi.1999.3187. [DOI] [PubMed] [Google Scholar]
- 27.George H, Kuraoka I, Nauman D A, Kobertz W R, Wood R, West S C. Curr Biol. 2000;10:103–106. doi: 10.1016/s0960-9822(00)00296-7. [DOI] [PubMed] [Google Scholar]
- 28.Egelman E H. J Struct Biol. 1998;124:123–128. doi: 10.1006/jsbi.1998.4050. [DOI] [PubMed] [Google Scholar]
- 29.Janin J. Structure (London) 1999;7:R277–R279. doi: 10.1016/s0969-2126(00)88333-1. [DOI] [PubMed] [Google Scholar]
- 30.Zerbib D, Mezard C, George H, West S C. J Mol Biol. 1998;281:621–630. doi: 10.1006/jmbi.1998.1959. [DOI] [PubMed] [Google Scholar]
- 31.Ariyoshi M, Vassylyev D G, Iwasaki H, Nakamura H, Shinagawa H, Morikawa K. Cell. 1994;78:1063–1072. doi: 10.1016/0092-8674(94)90280-1. [DOI] [PubMed] [Google Scholar]
- 32.Morikawa K. Nucleic Acids Mol Biol. 1998;12:275–299. [Google Scholar]