Abstract
The insect compound eye is one of the most precise and highly ordered patterns in the living world. It develops from an unpatterned simple epithelium by a series of cell fate decisions and complex morphogenetic movements. In the first days of metamorphosis, this interplay is particularly noticeable. Recent insights have revealed how interactions between neighboring cells drive the process. Interaction between Delta on cone cells and Notch proteins on the surface of neighbors induces the first pigment cells to differentiate. The primary pigment cells then express a Nephrin protein, Hibris, that interacts with a different Nephrin, Roughest, on their neighbors. Heterophilic adhesion between Hibris and Roughest result in remodeling contacts between cells to favour their contact with the pigment cells. In conjunction, the primary pigment cells signal to neighbors through the EGF Receptor to survive rather than undergo apoptosis. This sorting and culling process results in a sculpted pattern with a precise number and position of cells that is repeated hundreds of times in each compound eye.
Introduction
The development of organ systems in the body requires both the determination of diverse cell types and the organization of these cells into an elaborate pattern. Most of our current understanding of organogenesis comes from work on a small number of species whose characteristics make them suitable for analysis. In this review, I shall consider our current understanding about pattern formation of the Drosophila visual organ, the compound eye.
Compound eyes develop over a period of about 10 days. Beginning in mid-embryogenesis, two anterior clusters of ectodermal cells are fated as the eye anlagen [1]. Head involution folds the clusters internally to form the eye-antennal discs. After the embryo has hatched, an initial phase of eye development occurs for the first three days of larval life. The eye-antennal discs grow by cell division until the middle of the third instar, when each eye contains ~10,000 cells [2]. Beginning early in the third instar, cells at the posterior of the disc begin to differentiate. This transition from a proliferative phase to a differentiation phase is marked by a dorsoventral groove in the eye epithelium, called the morphogenetic furrow. The furrow moves across the disc until it reaches the anterior margin after approximately two days [3]. Cells arrest their proliferation and begin to differentiate as they enter the furrow. Differentiation occurs in a highly prescribed sequence beginning with differentiation of the R8 photoreceptor neurons at uniformly spaced positions. These neurons then signal to neighboring cells to develop incipient ommatidia (unit eyes). Sequential differentiation of the other seven photoreceptor types (R1–R7) occurs in four steps that are timed approximately four hours apart [4]. Each R8 neuron recruits one cell of each type, such that seven photoreceptors cluster around each R8 neuron [5]. Subsequently, four non-neuronal cells per ommatidium are recruited to differentiate– these are called cone cells.
By the end of the larval stage, ~26 rows of ommatidia have emerged. The remaining rows are generated during the first 10 hours of pupation [6]. Shortly thereafter, the eye discs reverse the process of involution to re-expose the eyes to the outside of the animal. Upon eye eversion, the cellular architecture of an ommatidium is crudely formed, and ommatidia are separated by a matrix of undifferentiated cells (Fig. 1A). However two days later, by mid-pupa, the cellular architecture of the eye is essentially complete with each ommatidium neatly juxtaposed to other ommatidia (Fig. 1B–E). The subsequent stages of pupal life are predominated by development of the cell specializations that make the ommatidium a functional simple visual unit (Fig. 2). I will focus my review on the remarkable pattern formation that occurs over the first two days of pupation.
Figure 1. Pattern formation in the pupal eye.
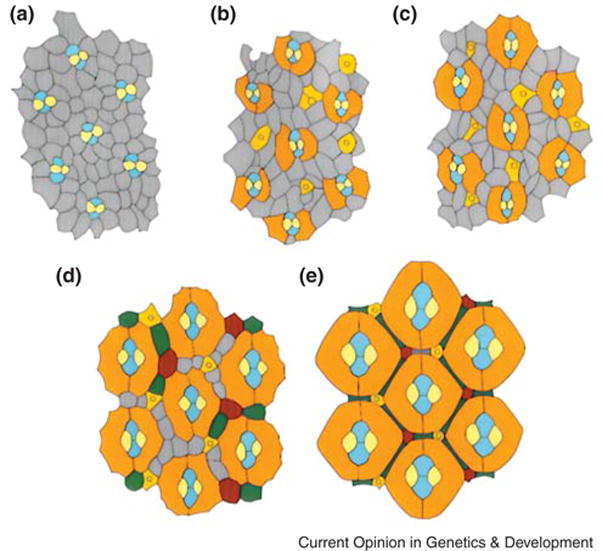
The apical surface of eyes at different stages of pupal life. Grey cells are uncommitted IPCs and coloured cells are determined. (A) At the beginning of pupation, the cone cells (blue and yellow) are embedded within a mosaic of IPCs. (B) After 20 hours pupation, PP cells (orange) are enlarging and surrounding the cone cells. (C) After 30 hours pupation, PP cells are contacting each other. (D) After 40 hours pupation, cone and PP cells are enlarging, and IPCs sort into single rows between them. IPCs positioned in certain niches differentiate into SP (green) and TP cells (red). IPCs which are not committed undergo apoptosis. (E) After 60 hours pupation, pattern formation is complete. Figure is modified after [2].
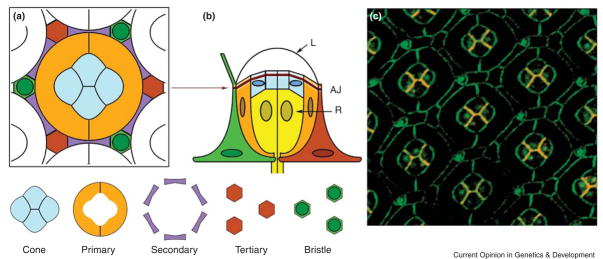
Figure 2. Cell topologies and geometries in the eye.
(A,B) Schematic ommatidium at 60 hours of pupal life. A cross-section view (A) is at the level of the adherens junction (AJ), and a side view (B) is equatorial to the midplane. A central group of cone cells are laterally surrounded by the two PP cells. The cone cell group sits over a cluster of eight photoreceptor cells (R) and under the lens (L). A lattice of SP and TP cells, and bristles are indicated. Nuclei in their defined positions are indicated as ovals. The schematics do not contain the small apical and basal processes of photoreceptor and cone cells that project from top to bottom. (C) Cell outlines of a retina at 55 hours of pupal life, as stained for DE-cadherin (green) and DN-cadherin (red). Colocalization of the cadherins results in orange staining.
Primary Pigment Cell Determination
For the first part of pupal life, new cell types continue to be specified. The primary pigment (PP) cells become distinct from neighboring cells as evident when two cells enlarge and displace other cells from contact with the cone cells [2]. The two PP cells then reach around and contact each other to form a concentric ring around the cone cells (Fig. 1B–D). The Banerjee group has shown that a signal from the cone cells triggers this phenomenon [7]. During pupation, cone cells begin to express high levels of Delta, a ligand for the Notch receptor. Since Delta only activates Notch on neighboring cells, it means that adjacent undetermined cells receive the signal. Interestingly, the level of Delta expression is not uniform in all four cone cells but appears higher in the anterior-posterior pair than the equatorial-polar pair. Correlated with this, it is the uncommitted cells contacting the anterior-posterior pair of cone cells that appear to enlarge and differentiate into PP cells ([2] and R.W.C. unpublished observations). An attractive idea is that Delta-Notch signaling from the anterior-posterior cone cells to uncommitted neighbors triggers their determination into PP cells. However, the Delta-Notch signal is not sufficient to do the task. These presumptive PP cells must also contain Lozenge, a RUNX transcription factor, for their determination [7].
Yet more inputs also play a role in PP cell determination. The retinal determination factors Eyes Absent (Eya) and Sine oculis (So) make a protein complex together to control the transcription of downstream target genes [8]. Eya/So by themselves are not essential for PP cell differentiation [9]. However, they act redundantly with an intercellular signal that is used reiteratively to instruct all cell fates in the eye. Signals mediated through the EGF Receptor (EGFR) and Ras transduction pathway are critical for eye development [10]. Although knockout of either EGFR or the transduction pathway have no effect on PP determination, loss of Eya/So and EGFR signaling together blocks PP determination [9]. This redundancy between such dissimilar molecular mechanisms is intriguing, particularly in light of the fact that So/Eya and EGFR are individually essential for determination of photoreceptors and cone cells. The exact mechanism behind their activities in PP cell remains a mystery.
The formation of the pigment lattice pattern
As the PP cells occupy their positions, all remaining cells are pushed into the space between ommatidia (Fig. 1B–C). Following closure of the PP cell ring, the cone and PP cells expand in size at the apical surface, constraining the interommatidium precursor cells (IPCs) into a lattice array [2]. From these IPCs arise the secondary pigment (SP) and tertiary pigment (TP) cells. SP cells have the unique topology of occupying the horizontal and oblique faces of an ommatidium; TP cells occupy the vertices of ommatidia (Fig. 2). The process of selection of SP and TP cells involves two morphogenetic events that occur in an approximate order: 1) remodeling of cell contacts, 2) apoptosis.
Remodeling of adhesive contacts between neighboring cells is the first event in IPC pattern formation. The IPCs are initially arranged in double or triple rows between the forming ommatidia. These cells then sort themselves to form a single row of cells, aligned head-to-tail [6,11]. Cell sorting is primarily mediated by cell contact remodeling. IPCs rearrange contact with each other to favor contact with PP cells from adjacent ommatidia, thus creating a single row of cells[12–14]. Further cell sorting by remodeling is seen at each vertex of an ommatidium, where three IPCs initially occupy that position. One of these IPCs reaches past the other two to contact a third PP cell – this IPC displaces the others and by singly occupying this niche, becomes the TP cell.
Recently, insights into this patterning process have been made by analysis of two genes: roughest and hibris. Loss of either gene results in a number of sorting defects. IPCs fail to rearrange into single rows, and three cells co-occupy the vertex niche where only one cell should reside [11–13]. Both genes encode members of the Neph1/Nephrin protein family [12,15]. Neph1 and Nephrin proteins are members of the immunoglobulin superfamily, which includes transmembrane proteins that mediate calcium-independent cell-cell adhesion [16,17]. Roughest protein is localized within IPCs specifically at the interfaces between PP cells and IPCs [11]. Roughest is less abundant along IPC-IPC interfaces. Moreover, localization is restricted to the adherens junction, where Roughest colocalizes with DE-cadherin, which is also enriched at IPC-PP cell interfaces [12,13].
In contrast, hibris is expressed in PP cells [12]. Although the precise localization of the Hibris protein is not yet known, several lines of evidence indicate that it is likely localized to the adherens junction of PP cells where it directly interacts with Roughest protein localized to IPCs. First, both heterophilic and homophilic adhesive interactions of Neph1/Nephrin proteins have been suggested [18,19]. Second, Roughest coimmunoprecipitates with Hibris when both proteins are expressed in co-cultured Drosophila cells [12]. Third, reduced Hibris in single PP cells results in reduced Roughest protein along IPC interfaces with those cells. Conversely, increased Hibris in PP cells results in increased Roughest along IPC interfaces with those cells. All these data suggest that Hibris in PP cells and Roughest in IPCs directly interact with each other in a heterophilic manner. This interaction is functionally important for IPC sorting since roughest and hibris show strong genetic interactions on the sorting process [12].
How does the interaction mediate cell sorting? One possibility is that heterophilic adhesion between Roughest and Hibris drives cell sorting akin to the differential sorting seen in aggregates of cells with different adhesivities (Fig. 3). Steinberg suggested that cells sort into layers based upon their relative adhesiveness: highly adhesive cells nucleate a core layer and exclude weakly adhesive cells [20]. This mechanism would account for the cell sorting seen with IPCs also. Moreover, the importance of differential adhesion for patterning the pupal eye has already been demonstrated. Cone cells segregate and assemble into their simple pattern by minimizing surface contact with PP cells [21]. It is a process analogous to the way in which soap bubbles minimize surface contact with a less adhesive environment. This process is mediated at least in part by DN-cadherin, which is specifically expressed in cone cells, and provides differential adhesion between cone cells [21,22]. Perhaps the strongest evidence for differential adhesion driving IPC sorting is from experiments in which IPCs were given more or less Roughest than their neighbors [12]. Cells with less Roughest were outcompeted to become SP and TP cells whereas cells with more Roughest became supercompetitors.
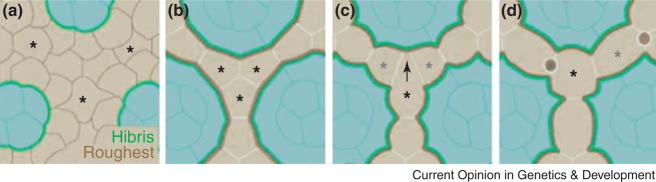
Figure 3. IPC Sorting and Apoptosis.
IPCs are marked in beige, and PPs and cone cells are marked in blue. Roughest protein is indicated by brown lines in IPCs while Hibris protein is indicated by green lines in cones and PPs. (A) At 10 hours pupal life, low uniform levels of Roughest in IPCs result in uniform contact between cells. (B) At 30 hours pupal life, heterophilic interaction between Hibris and Roughest (thick green and brown lines) along IPC:PP contacts result in sorting such that the size of IPC:IPC contacts is reduced in favour of IPC:PP contacts. (C) One IPC establishes contact with a third PP and becomes (D) a TP cell. The other two competing IPCs then must compete for a SP cell fate. Some IPCs which fail to outcompete undergo apoptosis (circles). Figure is modeled after [12].
The mechanism does not occur independently of other adhesion events, however. Loss of Notch function causes uniform distribution of both Roughest and DE-cadherin around the adherens junction of each IPC, suggesting that the localization of the two molecules could be related [13,23]. Indeed, the Hibris-Roughest interaction profoundly promotes the specific distribution of DE-cadherin along the IPC-PP interfaces [12,13]. Conversely, DE-cadherin influences Roughest distribution. Elimination or disruption of DE-cadherin leads to redistribution of Roughest throughout all interfaces [13]. These results imply co-dependence between Roughest-Hibris and DE-cadherin that might be exerted directly or possibly indirectly through establishment and/or maintenance of a functional adherens junction.
Culling of the Pattern
After IPC sorting is complete, two to three surplus IPCs are eliminated by apoptosis, suggesting that the cell-sorting process might be a prerequisite [6,11,24,25]. Indeed, roughest mutants possess extra SP and TP cells due to decreased apoptosis [25]. Ectopic localization of Roughest protein throughout IPCs rather than at IPC-PP interfaces also blocks apoptosis of IPCs [11,23]. These results suggest that the differential adhesion mediated by Roughest is a critical determinant to triggering apoptosis. How does this occur? One idea is that IPCs compete for a limiting survival factor whose distribution is dependent upon differential adhesion. If this factor is unevenly distributed, then cells with less of the factor die while cells with more of the factor survive.
Using laser ablation, Miller and Cagan observed that cone and PP cells are required for IPCs to avoid apoptosis [26]. Ablation of cone/PP cells result in ectopic apoptosis. This effect was blocked, however, if IPCs received a constitutive EGFR signal, indicating that cone/PP cells send a survival signal to IPCs via the EGFR signal transduction pathway. Indeed, cone and PP cells express an EGFR ligand called Spitz at the time of IPC apoptosis, consistent with this model [26]. Furthermore, loss of EGFR activity results in ectopic cell death [27,28]. EGFR-dependent signaling in IPCs inhibits the Head Involution Defective (Hid) protein, an inhibitor of DIAP, which is itself an inhibitor of the apoptotic caspase pathway [29]. Surviving cells appear not to activate the caspase pathway, and instead adopt either a SP or TP identity.
Interestingly, the EGFR survival signal is counterbalanced by antagonistic signals. Argos, a secreted repressor of EGFR signaling, is expressed in cone, SP, and TP cells [30]. Overexpression of Argos results in enhanced apoptosis [28,30,31]. One attractive model is that Argos diffuses from cone cells and limits the influence of Spitz to promote survival to only those IPCs closest to the Spitz source. Another model, not exclusive with the first, is that IPCs that are fated to survive send out an anti-survival (Argos) signal to other IPCs, thereby ensuring their destruction. A second antagonistic signal is mediated by the Notch receptor. Mutations in the Notch gene result in more IPCs escaping death [32]. Epistasis experiments suggest that Notch acts upstream of EGFR [29], which is difficult to simply reconcile with a model of direct PP-to-IPC signaling. Moreover, Notch appears to act downstream of cone/PP cells, the presumed sole source of EGFR ligand [26]. Either other cells such as photoreceptors also secrete a pro-survival EGFR ligand or cone/PP cells also produce a Notch inhibitor. The precise source and agent of Notch signaling is not yet known. However, a consequence of Notch activation in IPCs is the restricted distribution of Roughest protein; loss of Notch causes uniform localization of Roughest around IPC cells [23]. Since this also would lead to a block in differential cell-sorting, it is tempting to speculate that Notch organizes Hibris-Roughest interactions that drive cell competition for the pro-apoptotic EGFR signal.
Conclusions
Pattern formation of the Drosophila eye during the early stages of pupal development is a complex interplay of intercellular signals of two basic varieties. One variety informs cells about their fates in a manner consistent with differential gene expression. Another variety re-orders cell neighbor relationships in a manner consistent with differential cell adhesion. What is fascinating is how these two types of signals cascade in a precise temporal order to achieve what is ultimately one of the most precise and highly ordered patterns in the living world.
Acknowledgments
This work was supported by the NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Green P, Hartenstein AY, Hartenstein V. The embryonic development of the Drosophila visual system. Cell Tissue Res. 1993;273:583–598. doi: 10.1007/BF00333712. [DOI] [PubMed] [Google Scholar]
- 2.Wolff T, Ready D. Pattern formation in the Drosophila retina. In: Bate M, Martinez-Arias A, editors. The Development of Drosophila melanogaster. Vol. 2 Cold Spring Harbor Press; 1993. pp. 1277–1325. [Google Scholar]
- 3.Ready DF, Hanson TE, Benzer S. Development of the Drosophila retina, a neurocrystaline lattice. Developmental Biology. 1976;53:217–240. doi: 10.1016/0012-1606(76)90225-6. [DOI] [PubMed] [Google Scholar]
- 4.Tomlinson A. Cellular interactions in the developing Drosophila eye. Development. 1988:104. doi: 10.1242/dev.104.2.183. [DOI] [PubMed] [Google Scholar]
- 5.Morante J, Desplan C, Celik A. Generating patterned arrays of photoreceptors. Curr Opin Genet Dev. 2007:14. doi: 10.1016/j.gde.2007.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cagan RL, Ready DF. The emergence of order in the Drosophila pupal retina. Developmental Biology. 1989;136:346–362. doi: 10.1016/0012-1606(89)90261-3. [DOI] [PubMed] [Google Scholar]
- 7.Nagaraj R, Banerjee U. Combinatorial signaling in the specification of primary pigment cells in the Drosophila eye. Development. 2007;134:825–831. doi: 10.1242/dev.02788. [DOI] [PubMed] [Google Scholar]
- 8.Silver SJ, Rebay I. Signaling circuitries in development: insights from the retinal determination gene network. Development. 2005;132:3–13. doi: 10.1242/dev.01539. [DOI] [PubMed] [Google Scholar]
- 9.Hayashi T, Carthew RW. The role of retinal determination genes in directing cell differentiation in the Drosophila eye. Development. 2007 (submitted) [Google Scholar]
- 10.Voas MG, Rebay I. Signal integration during development: insights from the Drosophila eye. Dev Dyn. 2004;229:162–175. doi: 10.1002/dvdy.10449. [DOI] [PubMed] [Google Scholar]
- 11.Reiter C, Schimansky T, Nie Z, Fischbach KF. Reorganization of membrane contacts prior to apoptosis in the Drosophila retina: the role of the IrreC-rst protein. Development. 1996;122:1931–1940. doi: 10.1242/dev.122.6.1931. [DOI] [PubMed] [Google Scholar]
- 12.Bao S, Cagan R. Preferential adhesion mediated by Hibris and Roughest regulates morphogenesis and patterning in the Drosophila eye. Dev Cell. 2005;8:925–935. doi: 10.1016/j.devcel.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 13.Grzeschik NA, Knust E. IrreC/rst-mediated cell sorting during Drosophila pupal eye development depends on proper localisation of DE-cadherin. Development. 2005;132:2035–2045. doi: 10.1242/dev.01800. [DOI] [PubMed] [Google Scholar]
- 14.Vidal M, Larson DE, Cagan RL. Csk-deficient boundary cells are eliminated from normal Drosophila epithelia by exclusion, migration, and apoptosis. Dev Cell. 2006;10:33–44. doi: 10.1016/j.devcel.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 15.Ramos RG, Igloi GL, Lichte B, Baumann U, Maier D, Schneider T, Brandstatter JH, Frohlich A, Fischbach KF. The irregular chiasm C-roughest locus of Drosophila, which affects axonal projections and programmed cell death, encodes a novel immunoglobulin-like protein. Genes Dev. 1993;7:2533–2547. doi: 10.1101/gad.7.12b.2533. [DOI] [PubMed] [Google Scholar]
- 16.Barclay AN. Membrane proteins with immunoglobulin-like domains--a master superfamily of interaction molecules. Semin Immunol. 2003;15:215–223. doi: 10.1016/s1044-5323(03)00047-2. [DOI] [PubMed] [Google Scholar]
- 17.Hynes RO, Lander AD. Contact and adhesive specificities in the associations, migrations, and targeting of cells and axons. Cell. 1992;68:303–322. doi: 10.1016/0092-8674(92)90472-o. [DOI] [PubMed] [Google Scholar]
- 18.Barletta GM, Kovari IA, Verma RK, Kerjaschki D, Holzman LB. Nephrin and Neph1 co-localize at the podocyte foot process intercellular junction and form cis hetero-oligomers. J Biol Chem. 2003;278:19266–19271. doi: 10.1074/jbc.M301279200. [DOI] [PubMed] [Google Scholar]
- 19.Gerke P, Huber TB, Sellin L, Benzing T, Walz G. Homodimerization and heterodimerization of the glomerular podocyte proteins nephrin and NEPH1. J Am Soc Nephrol. 2003;14:918–926. doi: 10.1097/01.asn.0000057853.05686.89. [DOI] [PubMed] [Google Scholar]
- 20.Steinberg MS. Reconstruction of tissues by dissociated cells. Some morphogenetic tissue movements and the sorting out of embryonic cells may have a common explanation. Science. 1963;141:401–408. doi: 10.1126/science.141.3579.401. [DOI] [PubMed] [Google Scholar]
- 21.Hayashi T, Carthew RW. Surface mechanics mediate pattern formation in the developing retina. Nature. 2004;431:647–652. doi: 10.1038/nature02952. [DOI] [PubMed] [Google Scholar]
- 22.Nern A, Nguyen LV, Herman T, Prakash S, Clandinin TR, Zipursky SL. An isoform-specific allele of Drosophila N-cadherin disrupts a late step of R7 targeting. Proc Natl Acad Sci U S A. 2005;102:12944–12949. doi: 10.1073/pnas.0502888102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gorski SM, Brachmann CB, Tanenbaum SB, Cagan RL. Delta and notch promote correct localization of irreC-rst. Cell Death Differ. 2000;7:1011–1013. doi: 10.1038/sj.cdd.4400742. [DOI] [PubMed] [Google Scholar]
- 24.Brachmann CB, Cagan RL. Patterning the fly eye: the role of apoptosis. Trends Genet. 2003;19:91–96. doi: 10.1016/S0168-9525(02)00041-0. [DOI] [PubMed] [Google Scholar]
- 25.Wolff T, Ready DF. Cell death in normal and rough eye mutants of Drosophila. Development. 1991;113:825–839. doi: 10.1242/dev.113.3.825. [DOI] [PubMed] [Google Scholar]
- 26.Miller DT, Cagan RL. Local induction of patterning and programmed cell death in the developing Drosophila retina. Development. 1998;125:2327–2335. doi: 10.1242/dev.125.12.2327. [DOI] [PubMed] [Google Scholar]
- 27.Baker NE, Yu SY. The EGF receptor defines domains of cell cycle progression and survival to regulate cell number in the developing Drosophila eye. Cell. 2001;104:699–708. doi: 10.1016/s0092-8674(01)00266-5. [DOI] [PubMed] [Google Scholar]
- 28.Sawamoto K, Taguchi A, Hirota Y, Yamada C, Jin MH, Okano H. Argos induces programmed cell death in the developing Drosophila eye by inhibition of the Ras pathway. Cell Death Differ. 1998;5:262–270. doi: 10.1038/sj.cdd.4400342. [DOI] [PubMed] [Google Scholar]
- 29.Yu SY, Yoo SJ, Yang L, Zapata C, Srinivasan A, Hay BA, Baker NE. A pathway of signals regulating effector and initiator caspases in the developing Drosophila eye. Development. 2002;129:3269–3278. doi: 10.1242/dev.129.13.3269. [DOI] [PubMed] [Google Scholar]
- 30.Wildonger J, Sosinsky A, Honig B, Mann RS. Lozenge directly activates argos and klumpfuss to regulate programmed cell death. Genes Dev. 2005;19:1034–1039. doi: 10.1101/gad.1298105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Freeman M. Misexpression of the Drosophila argos gene, a secreted regulator of cell determination. Development. 1994;120:2297–2304. doi: 10.1242/dev.120.8.2297. [DOI] [PubMed] [Google Scholar]
- 32.Cagan RL, Ready DF. Notch is required for successive cell decisions in the developing Drosophila retina. Genes Dev. 1989;3:1099–1112. doi: 10.1101/gad.3.8.1099. [DOI] [PubMed] [Google Scholar]