Abstract
An acetyl-histone peptide library was used to determine the thermodynamic parameters that define acetylation-dependent bromodomain—histone interactions. Bromodomains interact with histones by binding acetylated lysines. The bromodomain used in this study, BrD3, is derived from the polybromo-1 protein, which is a subunit of the PBAF chromatin remodeling complex. Steady-state fluorescence anisotropy was used to examine the variations in specificity and affinity that drive molecular recognition. Temperature and salt concentration dependence studies demonstrate that the hydrophobic effect is the primary driving force, consistent with lysine acetylation being required for binding. An electrostatic effect was observed in only two complexes where the acetyl-lysine was adjacent to an arginine. The large change in heat capacity determined for the specific complex suggests that the dehydrated BrD3—histone interface forms a tightly bound, high-affinity complex with the target site. These explorations into the thermodynamic driving forces that confer acetylation site-dependent BrD3—histone interactions improve our understanding of how individual bromodomains work in isolation. Furthermore, this work will permit the development of hypotheses regarding how the native Pb1, and the broader class of bromodomain proteins, directs multisubunit chromatin remodeling complexes to specific acetyl-nucleosome sites in vivo.
Keywords: Acetyl-lysine, Histone acetylation, Polybromo, Histone code, Protein—protein interactions, Thermodynamics
Histone acetylation is a key regulator of transcription; however, it is still a matter of debate as to whether the regulatory features are influenced by modification at specific lysine sites or it is the cumulative electrostatic consequences of random lysine acetylation. Mounting evidence indicates that large multiprotein chromatin remodeling complexes regulate transcription by localizing to certain chromatin sites through the interaction of bromodomain (BrD)1 proteins with acetylated nucleosomes [1-7]. The BrD is a protein motif, approximately 100 amino acids in length, primarily associated with targeting subunits of chromatin remodeling complexes [8]. The biological role of the BrD centers on its ability to discriminate the acetylation state of lysine residues in histone proteins. In fact, there is growing evidence that the position of the acetyl-lysine is not random, but instead, the BrD requires acetylation at specific sites for high-affinity binding [9]. For example, fluorescence resonance energy transfer assays were used to show that the bromodomain protein Brd2 requires the interaction between its bromodomain and an acetyl-histone to amplify transcription in vivo [10]. To activate transcription after SAGA (Spt-Ada-Gcn5 acetyl-transferase) complex directed acetylation at specific nucleosomal locations, BrD proteins of the Swi2/Snf2 complex are required to displace acetyl-histones [11]. Likewise, the yeast RSC (remodels the structure of chromatin) complex requires multiple BrD proteins to recognize acetylated nucleosomes so as to facilitate movement of RNA polymerase II, possibly regulating movement through the coding region [12]. This correlation between histone acetylation state and gene activity led to speculation that these molecular tags are involved in regulating gene function [13,14].
The human polybromo-1 (Pb1) protein was recently identified as a subunit of the polybromo, BRG1-associated factors (PBAF) chromatin remodeling complex required for kinetochore localization and regulation of certain genes [15-17]. Computational analysis of the 1634-amino acid Pb1 protein revealed the presence of six tandem bromodomains [18], indicating that Pb1 is capable of binding multiply acetylated histone proteins. Recent in vitro studies indicate that Pb1 bromodomains discriminate both the acetylation state and the location of the lysine side chains within the histone tail regions [9]. The work investigated the acetylation site specificity of all six bromodomains binding to acetylated histones using steady-state fluorescence anisotropy. It was shown that Pb1 bromodomains bind acetylated lysine side chains with high affinity at specific positions within the N-terminal tail of histone H3. The specific and nonspecific binding activities for a given BrD were shown to differ by more than two orders of magnitude in some cases. Interestingly, duplicate target sites were identified for two of the bromodomains, suggesting that the Pb1 protein may interact with both copies of histone H3 on nucleosome assembly. It is possible that the multiple bromodomains from Pb1 cooperatively bind a discreet histone acetylation array to localize the PBAF complex to specific chromatin locations.
The thermodynamic features of many protein complexes have been studied, but such investigations into the acetylation dependence of BrD—histone interactions, central to transcriptional regulation and epigenetic phenomena, have not been explored prior to this work. Because many biological processes are mediated by highly selective noncovalent interactions, an analysis of the thermodynamic parameters that define specificity will yield a quantitative description of BrD—histone interactions and provide insights into the so-called histone code hypothesis. Here the detailed thermodynamic parameters associated with site-specific and nonspecific binding events are described for acetylation-dependent BrD—histone interactions. The range of binding affinities for different AcK positions in the histone H3 tail makes the third amino-terminal bromodomain (BrD3) an interesting model for the study of the thermodynamic features driving acetylation-dependent protein—protein binding events. The results of this work are a strong first step in characterizing the driving forces that confer the molecular recognition features of this important system. Furthermore, this work will permit the development of hypotheses regarding how the native Pb1, and the broader class of bromodomain proteins, directs multisubunit chromatin remodeling complexes to specific acetyl-nucleosome sites in vivo.
Materials and methods
Materials
Enzymes were purchased from New England Biolabs (Ipswich, MA, USA). All reagents used for cloning and standard molecular biology procedures were obtained from Fisher (Hanover Park, IL, USA). Kanamycin, chloram-phenicol, and isopropyl-β-D-thiogalactopyranoside (IPTG) were ordered from Acros Organics (Hampton, NH, USA). Nickel—nitrotriloacetic acid (Ni2+—NTA) agarose used in protein purification was ordered from Qiagen (Valencia, CA, USA). Fmoc amino acids and peptide synthesis reagents were purchased from Advanced Chemtech (Louisville, KY, USA) and synthesized at the protein core facility at Arizona State University.
Instrumentation
PCR was performed using the Mastercycler Gradient Thermocycler from Eppendorf (Westbury, NY, USA). Histone H3 peptides were synthesized on a 9050 peptide synthesizer (Millipore, Bedford, MA, USA), purified using high-performance liquid chromatography, and characterized using matrix-assisted laser desorption ionization time-of-flight (MALDI—TOF) mass spectrometry. Steady-state fluorescence anisotropy measurements were performed on a Quantamaster-6/2003 spectrofluorometer in T-format with a 75-W Xenon lamp and were analyzed with FeliX32 fluorescence analysis software (PTI, London, ON, Canada). A USB2000 UV—Vis spectrometer from Ocean Optics (Dunedin, FL, USA) was used to measure absorbance. A Gel Logic 200 imaging system from Kodak (New Haven, CT, USA) was used to quantify DNA and protein gels.
Expression and purification of single bromodomains
Cloning, expression, and purification of BrD3 from the PB1 gene (GenBank accession no. AF225871) were performed using a previously described method [19]. BrD3 corresponds to amino acids 315 to 473 in the native Pb1 sequence. In general, BrD3 was PCR amplified, inserted into a pET30(b) expression vector using NdeI/KpnI, and transformed into DH5α cells for subcloning. The construct positions a carboxy-terminal protease cleavable hexa-histidine tag for effective one-step purification. Rosetta (DE3) cells (Novagen, San Diego, CA, USA) were used for protein expression due to the high number of rare tRNAs in the polybromo region of the PB1 gene. Cells were grown in Luria—Bertani (LB) media supplemented with kanamycin (50 μg/ml) and chloramphenicol (34 μg/ml) and induced with IPTG. The high purity of each BrD was achieved by one-step nickel affinity chromatography (Ni2+—NTA), as visualized by Coomassie-stained SDS—PAGE analysis [19]. Dialyzed samples examined by SDS—PAGE showed only one band at approximately 22 kDa, confirming the purity of these preparations. The concentration of purified BrD3 was determined by Bradford assay using a bovine serum albumin (BSA) standard.
Synthesis, purification, and characterization of fluorescein-labeled histone H3 peptides
All peptides were synthesized on PAL-PEG-PS resin by automated solid-phase peptide synthesis using Fmoc chemistry as described previously [19]. Fluorescein-labeled H3 peptides (Fig. 1) were derived from the amino-terminal 25 amino acids (the tail region) of human histone H3 (GenBank accession no. AAN10051). Acetylated lysines were incorporated at one of the indicated lysine positions, and fluorescein was coupled to the α-amine of the N-terminal end using O-(7-azabenzotriazol-1-yl)-N,N,N’,N’-tetramethyluronium hexafluorophosphate (HATU)-activated coupling chemistry. The peptides were removed from the resin and deprotected by cleavage with trifluoroacetic acid (TFA) methods for 4 h. Fluorescein is stable to standard deprotection and cleavage conditions. The peptides were purified by a two-step process using Sephadex G-25 size exclusion chromatography in 10 mM Tris (pH 7.5), followed by reversed-phase HPLC on a Zorbax C8 column (9.4 mm × 25 cm) using a water (0.1% TFA)-to-acetonitrile (0.1% TFA) gradient. Identities and purities of the H3 peptides were confirmed by MALDI—TOF using α-cyano-hydroxycinnamic acid as matrix. The expected mass of each monoacetylated peptide was 3011.4, and multiple peaks centered at m/z 3012.0 in each mass spectrum were observed for the singly charged species (data not shown). Each H3 peptide had the same mass and yielded nearly identical mass spectra. The expected mass of the unmodified peptide was 2969.2 Da, and a single peak was observed at m/z 2968.8 Da in the mass spectrum [9]. Concentrations of purified H3 peptides were determined spectrophotometrically based on the extinction coefficient of ε491nm = 88,000 M-1 cm-1 for fluorescein [20].
Fig. 1.

Monoacetylated histone H3 peptides. The primary sequence of the H3 peptide is derived from the amino-terminal 25 amino acids of the human histone H3 protein. The name for each respective histone H3 peptide is based on the position of the underlined acetyl-lysine (AcK). Fluorescein was introduced at the α-amine of the peptide backbone (not shown).
Fluorescence anisotropy measurements
Steady-state fluorescence anisotropy measurements were performed as forward titrations as described previously [19,21]. In general, 10 to 50 nM of a given fluorescein-labeled histone peptide was titrated with 0 to 0.1 mM (final concentration) of BrD3 until the maximum anisotropy was attained. Fluorescence anisotropy measurements used vertically polarized excitation at 488 nm and detection in the vertical and horizontal planes at 520 nm, corresponding to the spectral properties of fluorescein. Anisotropy is calculated as A =(Ivv — gIvh) / (Ivv + 2gIvh), where Ivv is the fluorescence intensity with vertically polarized excitation and vertically polarized emission, Ivh is the fluorescence intensity with vertically polarized excitation and horizontally polarized emission, and g = (Ihv/Ihh) is a factor correcting for the polarization dependence of the spectrometer. Steady-state fluorescence measurements were performed in 20 mM Tris (pH 7.8), 100 mM NaCl, and 5% glycerol except in the case of the salt titrations, where the concentration of NaCl was as given in the figures and tables. Salt dependence measurements were performed by the addition of the appropriate amount of 1 M NaCl in the same Tris buffer to yield the indicated final salt concentration. Temperature dependence measurements were performed by incubating each sample at the given temperature. The data are the averages of at least three independent measurements and fit using a nonlinear least squares algorithm (SigmaPlot). A thermally controlled sample compartment was used to maintain a constant temperature for each set of measurements. To minimize sample evaporation during experiments, the cuvettes were capped during titrations at higher temperatures. The change in the total fluorescence signal, after being adjusted at each titration point for dilution, varied by less than 2% over the course of these measurements, suggesting that evaporation did not adversely affect these measurements.
Determination of thermodynamic functions
The contribution of the hydrophobic effect and the electrolyte effect to the stability of each BrD3—H3 complex was determined by analyzing the temperature and salt concentration dependence of the observed association equilibrium constant Kobs. For binding to a specific DNA sequence, Kobs is given by:
| (1) |
where [BrD]eq is the concentration of unbound BrD3, [H3]eq is the concentration of a given H3 peptide, and [BrD — H3]eq is the concentration of bound complex at equilibrium. This analysis is based on the assumption that site-specific binding can be described as a two-state system. Considering that there is only one acetyl-lysine per peptide and that bromodomains show negligible binding to unmodified lysines, the two-state model is an appropriate description of the unbound-to-bound transition. Here the stability of the bound complex is determined by the differences in the noncovalent interactions between the BrD3 and the H3 peptide as solution conditions are varied. Equilibrium titration data were evaluated by fitting to Eq. (1) using a nonlinear least squares algorithm as described previously [9,21]. Each equilibrium binding constant value reported is the average of at least three independent titrations. The experimental error in determining equilibrium binding constant values was approximately ±15%.
Results
Specific binding of BrD3 to acetylated K9 of histone H3
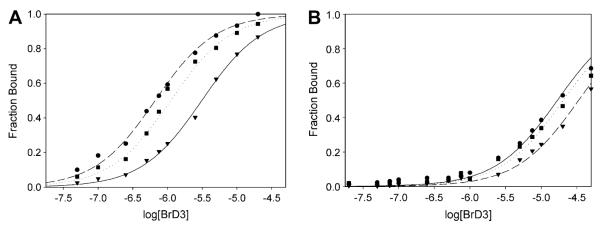
The specific binding affinity of BrD3 was determined for different solution conditions by monitoring the anisotropy of a fluorescein-labeled histone H3 peptide displaying an acetyl-lysine at position 9 (AcK9) as a function of the BrD3 concentration. Fig. 2 shows representative binding curves at a few selected temperatures and salt concentrations for the association of BrD3 with H3—AcK9, the previously identified high-affinity target site for BrD3 [9]. Titrations were performed to attain an equilibrium constant for each sample condition based on a two-state binding model. Plots of the fluorescence anisotropy titrations are represented as a fraction of the indicated monoacetylated H3 peptide bound as a function of BrD3 concentration (Ai — Amin/Amax — Amin). Raw anisotropy data are rescaled along the y axis to represent the fraction of H3 peptide complexed with BrD3 simply by linearly rescaling the measured anisotropies from 0 (experimentally measured lower baseline) to 1 (measured upper baseline). The titration midpoint for individual determinations represents the equilibrium dissociation constant. The binding constants (Kobs) and the corresponding binding free energies (ΔG°) determined for the complete range of salt concentrations (50–250 mM NaCl) and temperatures (283–313 K) are shown in Table 1.
Fig. 2.
Representative binding curves for BrD3 binding to the fluorescein-labeled AcK9 (A) and AcK14 (B) histone H3 peptides. The fluorescence anisotropy of 10 nM fluorescein-labeled H3 peptide is plotted as a function of the log of the nonlabeled BrD3 concentration. (A) BrD3 binding to AcK9 at several salt concentrations and temperatures: ●, 298 K, 0.050 M Na+; ▾, 308 K, 0.050 M Na+; ∎, 298 K, 0.250 M Na+. (B) BrD3 binding to AcK14 at several salt concentrations and temperatures: ●, 298 K, 0.050 M Na+; ▾, 308 K, 0.050 M Na+; ∎, 298 K, 0.250 M Na+. All plots were fit to a two-state binding model.
Table 1.
Equilibrium association constants (Kobs) and free energy (ΔG°) for site-specific binding of BrD3 with the five acetylated histone H3 peptides
| Temperature (K) |
Na+ (M) |
AcK4 |
AcK9 |
AcK14 |
AcK18 |
AcK23 |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
Kobs (105 M-1) |
ΔG (kcal/mol) |
Kobs (105 M-1) |
ΔG (kcal/mol) |
Kobs (105 M-1) |
ΔG (kcal/mol) |
Kobs (105 M-1) |
ΔG (kcal/mol) |
Kobs (105 M-1) |
ΔG (kcal/mol) |
||
| 283 | 0.10 | 0.3 | -5.6 | 4.6 | -7.3 | 0.3 | -5.9 | 0.7 | -6.3 | 0.6 | -6.2 |
| 288 | 0.10 | 0.3 | -5.9 | 7.4 | -7.7 | 0.3 | -5.9 | 0.8 | -6.4 | 1.0 | -6.6 |
| 298 | 0.10 | 0.4 | -6.3 | 14.1 | -8.4 | 0.4 | -6.2 | 1.0 | -6.8 | 1.1 | -6.9 |
| 303 | 0.10 | 0.5 | -6.5 | 11.1 | -8.4 | 0.4 | -6.4 | 0.8 | -6.8 | 1.0 | -6.9 |
| 308 | 0.10 | 0.4 | -6.5 | 5.2 | -8.1 | 0.3 | -6.3 | 0.6 | -7.1 | 0.7 | -6.9 |
| 313 | 0.10 | 0.3 | -6.2 | 2.9 | -7.8 | 0.2 | -6.3 | 0.6 | -6.4 | 0.4 | -6.6 |
| 298 | 0.05 | 0.4 | -6.3 | 13.7 | -8.2 | 0.3 | -6.1 | 1.1 | -6.9 | 1.1 | -6.9 |
| 298 | 0.15 | 0.4 | -6.3 | 13.9 | -8.4 | 0.4 | -6.2 | 0.9 | -6.8 | 1.1 | -6.9 |
| 298 | 0.20 | 0.4 | -6.3 | 10.4 | -8.2 | 0.2 | -5.9 | 0.8 | -6.7 | 1.0 | -6.8 |
| 298 | 0.25 | 0.3 | -6.2 | 8.3 | -8.1 | 0.3 | -6.1 | 0.5 | -6.4 | 1.0 | -6.8 |
To understand the role of the electrostatic effect in site-specific BrD—acetyl-histone recognition, binding experiments were performed using buffer conditions with increasing ionic strength. As the NaCl concentration is increased from 50 to 250 mM, the Kobs for the interaction of BrD3 and AcK9 shows a decrease from 13.7 × 105 M-1 to 8.3 × 105 M-1 (Table 1). This nearly twofold reduction in the binding constant represents a modest, yet detectible, electrostatic contribution to the binding event. In contrast, the observed equilibrium constant for BrD3 binding to AcK9 shows a strong dependence on temperature. The increase in binding strength as temperature is increased from 283 to 300 K is followed by a decrease in the binding strength as the temperature increases beyond 300 K (Table 1). This is due to an unfavorable binding enthalpy at lower temperatures that becomes favorable at higher temperatures, as described in more detail below. BrD3 interacts with the peptide displaying AcK9 with high affinity from 298 to 303 K with Kobs values of 14.1 × 105 M-1 and 11.1 × 105 M-1, respectively. However, binding at the temperature extremes is weaker (Kobs ∼ 3 × 105 M-1), representing nearly a fivefold difference in the association constant. Overall, equilibrium binding to the AcK9 site occurs with association constants in the 2.9 × 105 M-1 to 14.1 × 105 M-1 range for the temperatures examined here.
Nonspecific acetyl-lysine binding by BrD3
The salt and temperature dependence data for the interaction of BrD3 with each of the low-affinity acetylation sites are shown in Table 1. The equilibrium constant is lower for nonspecific binding than for the specific binding for all samples and solution conditions. The Kobs values for the nonspecific acetyl-lysine sites (AcK4, AcK14, and AcK23) from 50 to 250 mM NaCl show a 10 to 20% decrease in the binding constant over the NaCl concentration range examined. The Kobs values for BrD3 binding to AcK4 and AcK23 are roughly 0.4 × 105 M-1 and 1.0 × 105 M-1, respectively, showing a decrease in binding only when NaCl concentrations exceed 200 mM. The AcK18 peptide exhibits the greatest NaCl dependence of all BrD—acetyl-histone complexes, and this is attributed to the arginine flanking the acetyl-lysine at that position. At the highest salt concentrations, BrD3 binds the nonspecific acetylation sites with an affinity in the range of 8- to 27-fold lower than it binds the AcK9 target site (Table 1). In comparison, at the lowest salt concentrations, BrD3 binds nonspecific acetyl-histone sites with a 12- to 43-fold lower association constant than is observed for the specific AcK9 site. In cases where a strong electrostatic component exists, nonspecific interactions between biomolecules compete less effectively than binding of ions in solution as salt concentration is increased.
The temperature dependence of Kobs for the interaction of BrD3 with the nontarget acetyl-lysine positions (AcK4, AcK14, AcK18, AcK23) was also examined to better understand the thermodynamic driving forces for nonspecific BrD—histone interactions. Parallel measurements of BrD3 binding at these sites occur at affinities 10- to 40-fold less than those observed for the high-affinity AcK9 target site and show smaller relative temperature-dependent changes. This is evident when comparing the data as a function of temperature for specific binding (BrD3—AcK9) with the nonspecific binding of BrD3 to AcK4, AcK14, AcK18, and AcK23 (Fig. 3). As the temperature is increased from 283 to 303 K, the nonspecific binding shows a 1.3- to 1.8-fold (less than 2-fold) increase in the association constant. This weaker dependence on temperature exhibited by nonspecific binding reflects a smaller negative change in heat capacity than is observed for the specific BrD3—AcK9 complex.
Fig. 3.
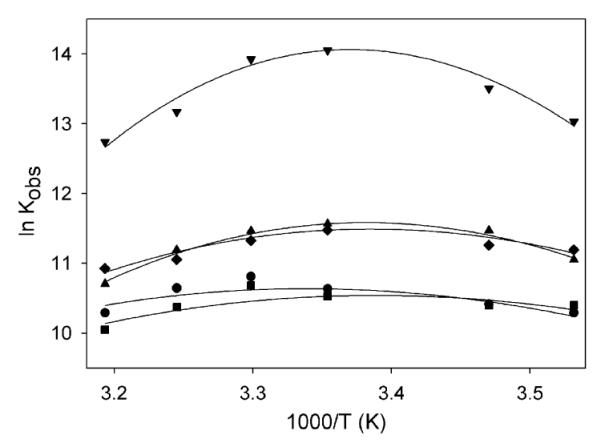
Dependence of ln Kobs on T-1 for binding interaction between BrD3 and AcK4 (●), AcK9 (▾), AcK14 (∎), AcK18 (◆), and AcK23 (▴). ln Kobs versus reciprocal temperature (1000/T) is plotted over a range of temperatures (283–313 K). Lines are fit to Eq. (2) as described in Discussion.
Titrations of BrD3 with unmodified histone H3 show negligible anisotropy change in the concentration range and solution conditions examined here, indicating that BrD3 does not interact with peptides in the absence of acetyl-lysine. The hydrophobic binding pocket of the BrD is unable to form a stable thermodynamic complex with unmodified (charged) lysines. The KD values previously were estimated to be in the millimolar range [19], beyond the titration range of the current experiments. Effectively, the Pb1 bromodomains show only modest binding to unmodified histone H3 peptide even at the highest concentrations examined. Because BrD3 shows only residual anisotropy change in the absence of acetyl-lysine, the salt and temperature dependence studies show no condition-dependent changes in binding. Rigorous controls were carried out to ensure that anisotropy changes resulted from the formation of the BrD3—histone complex. Titrations without BrD using the buffer itself resulted in no anisotropy change. Parallel titrations substituting a BrD with BSA were performed to rule out nonspecific binding to the fluorescein-labeled H3 peptide because BSA is not expected to interact with histone peptides. BSA concentrations up to 10 mM caused no change in anisotropy (data not shown), confirming that the anisotropy change was a function of BrD—acetyl-histone complex formation.
Examination of hydrophobic effect for specific and nonspecific binding
Large negative changes in heat capacity occurring on the removal of hydrophobic surfaces from water exposure are characteristic of protein folding and conformational changes [22,23]. More recently, such changes in heat capacity were shown to be proportional to the change in water-exposed hydrophobic surface area, yielding important information about the binding interface [24-28]. To understand this hydrophobic effect, the temperature dependence of Kobs (Fig. 3 and Table 1) was fit to Eq. (2) and the standard heat capacity change (ΔCp) was determined for each BrD3—acetyl-histone complex:
| (2) |
Here lnKobs and are values at T* = 298 K. The choice of reference temperature is arbitrary and does not affect the results of the fit. This equation assumes that is independent of temperature. The errors from fitting the equilibrium constants for each temperature to Eq. (2) are within 20% of the stated values for ΔCp (Table 2). Eq. (2) assumes that ΔCp is independent of temperature within experimental uncertainty [29]. It was suggested [30,31] that the thermodynamics of such processes (where heat capacity is large and temperature is independent) can be characterized by the change in heat capacity and two temperatures: TH and TS. Using this approach, the temperature dependence of the enthalpic and entropic contributions to the binding free energy can be calculated with the above parameters using the following two equations:
| (3) |
| (4) |
where TH is the temperature at which ΔH° = 0 and occurs at the maxima of the van’t Hoff plot (Kobs is maximum) and TS is the temperature at which ΔS° = 0 and occurs at the minima of a plot of ΔG° versus temperature (ΔG° is minimum).
Table 2.
Thermodynamic parameters
| Parameter | AcK4 | AcK9 | AcK14 | AcK18 | AcK23 |
|---|---|---|---|---|---|
| ΔCp | -0.4 | -1.6 | -0.4 | -0.7 | -0.9 |
| TH | 299.9 | 296.6 | 294.8 | 295.4 | 295.7 |
| TS | 314.5 | 301.6 | 309.0 | 304.0 | 303.4 |
Association constants for site-specific protein interactions typically exhibit maxima at temperatures in the physiological range [32,33]. This is observed, for example, in Fig. 3, which plots the ln Kobs as a function of 1/T for the binding of BrD3 to each H3 peptide. For AcK9, the natural log of the equilibrium constant shows a maximum at approximately 297 K. Furthermore, because aqueous solutions of aliphatic and aromatic molecules have positive heat capacities [34], binding of the ligand should be accompanied by a negative change in heat capacity resulting from the reduced solvent exposure of the acetyl-lysine and regions of the hydrophobic pocket. Analysis of these data using Eq. (2) reveals a negative heat capacity change on the association for binding of BrD3 to AcK9 (ΔCp = -1.6 kcal/K · mol). The plots indicate that the interaction enthalpies exhibit strong temperature dependencies, and this is commonly observed for specific binding interactions involving proteins [35].
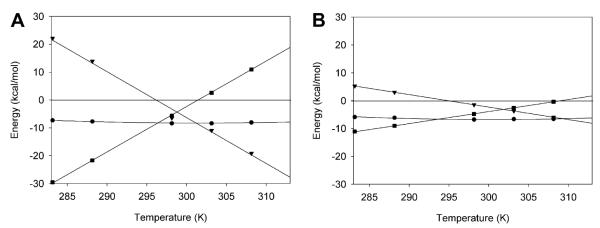
The temperature dependence of the enthalpic and entropic contributions to the binding free energy was further evaluated using Eqs. (3) and (4). The resulting thermodynamic profiles of the histone H3—BrD3 interaction are shown in Fig. 4, which plots the free energy, entropy, and enthalpy as functions of temperature. At 298 K, binding is both enthalpically and entropically favorable for BrD3 binding to AcK9 (ΔH = -6.5 kcal/mol and ΔS = 15.8 cal/K·mol). The TH and TS values for AcK9 binding are 296.6 and 301.6 K, respectively, whereas the nonspecific complexes range from 294.8 to 299.9 K for TH and from 301.6 to 314.5 K for TS (Table 2). Site-specific binding for BrD3 becomes enthalpically favorable and entropically unfavorable as temperature increases, such that at 308 K association is entirely enthalpically driven for AcK9 (ΔH = -26.3 kcal/mol and ΔS = -58.6 cal/K·mol). Each of the nonspecific sites shows weaker dependencies on temperature than BrD3 binding to the target AcK9 site as indicated by the slope of the enthalpy and entropy plots (Fig. 4).
Fig. 4.
Thermodynamic profiles of specific and nonspecific interactions of BrD3. Plots of free energy (●), enthalpy (▾), and entropy (∎) of binding versus temperature are given for AcK9 (A) and AcK14 (B). It should be noted that the entropy is given as —TΔS to present the data with enthalpy and free energy in the same units.
Discussion
The condition-dependent interaction between BrD3 and acetyl-histone H3 was examined using steady-state fluorescence anisotropy to yield a thermodynamic description of the molecular-level features that confer binding selectivity and affinity. The bromodomain used in this study, BrD3, is derived from the Pb1 protein, which is a subunit of the PBAF chromatin remodeling complex. This BrD has been shown to interact with different acetyl-lysines on histone H3 as high-affinity (specific) or low-affinity (nonspecific) complexes that differ by nearly two orders of magnitude. The BrD3 binds the target site (AcK9) with high affinity, having an equilibrium constant in the nanomolar range, and binds the nontarget sites with affinities in the micromolar range. The dependence of Kobs on NaCl concentration and temperature indicates that the overall binding event is driven predominantly by the hydrophobic effect, consistent with binding occurring only after lysine acetylation (loss of positive charge). These explorations into acetylation-dependent BrD—histone interactions provide a clear understanding of how individual domains work in isolation, thereby permitting the development of hypotheses regarding the larger histone acetylation pattern that acts as the binding target of multi-Brd-containing chromatin remodeling complexes.
The polyampholytic character of proteins makes their dependence on salt more complicated than a structurally rigid polyanion such as DNA, which has a high axial charge density [36]. Equilibrium constants for specific and nonspecific interactions are weak functions of the salt concentration for the acetylation-dependent protein—protein interactions examined here. A decrease in the equilibrium association constant with increasing ionic strength, characteristic of the competition between the charged ligands and ions for available binding sites at the protein interface, is not observed for most acetylated peptides and is only weakly observed for AcK9 and AcK18. A comparison of the Kobs as a function of salt concentration, for the interaction of BrD3 with the high-affinity AcK9 and low-affinity AcK18 peptides, shows a modest electrostatic effect (Table 1). Although BrD3 binds AcK18 with nearly 20-fold lower affinity under optimal conditions, the observed equilibrium association constant for both complexes reveals the largest ionic contribution to the binding stabilization for all of the complexes examined here. Inspection of both AcK9 and AcK18 peptides shows an arginine to the amino side of the acetyl-lysine (Fig. 1), suggesting that a salt bridge forms at the histone—BrD3 interface. Computational analysis of Pb1—BrD3 reveals a glutamic acid within the binding region capable of forming a salt bridge with the arginine of the histone tail [37-39]. In contrast, the nonspecific sites AcK4, AcK14, and AcK23, which do not have charged side chains flanking the acetyl-lysine, show a negligible electrostatic effect. Interestingly, AcK4 has an amino acid separating the acetyl-lysine from the arginine, yet it exhibits negligible salt concentration dependence. This suggests that the glutamic acid might be too constrained to form a stable salt bridge with the arginine over a larger separation distance. The selective nature of the observed electro-static effect in peptides having charged groups near the acetyl-lysine supports the observations from structural data that the BrD—histone interface encompasses multiple amino acids in addition to the acetyl-lysine [37,39-46].
The variation in the ΔCp values between specific and nonspecific BrD3—acetyl-histone complexes determined experimentally was compared with the values calculated based on the change in solvent-exposed surface area [47-49]:
| (5) |
where ΔAnp and ΔAp are the changes in nonpolar and polar surface area, respectively. The ΔCp values from nonspecific to specific range from -400 to -1600 J/K · mol, indicating significant variation in the hydration of the BrD—histone interface. In the case of the specific BrD3—AcK9 complex, the experimental ΔCp shows a greater effect than can be attributed solely to the change in solvent-exposed surface area on complex formation. Here the relationship between the change in heat capacity and solvent-exposed surface area suggests that the acetyl-lysine flanking side chains exert an effect on the structure of the bound complexes that is greater than simply the additional change in heat capacity contributed by AcK alone. Such differences previously were linked to ligand-induced folding or conformational changes associated with the binding event. For comparison, structural data for the Brg1 bromodomain complexed to histone H3 [39] were used to determine the total surface area for the interface, where the Brg1 engulfs the acetyl-lysine and two amino acids to each side of the AcK. The surface area of the Brg1—histone interface is 292.4 Å2. Using Eq. (5), the ΔCp was calculated to be 850 ± 50 J/K · mol for the Brg1—H3 complex. Assuming that the homologous BrD3—H3 complex has a comparable surface area, the additional contributions to heat capacity change may be due to an induced folding mechanism.
In general, nonspecific interactions are stabilized by the formation of the hydrophobic binding pocket around the AcK. The ΔCp of approximately 500 ± 100 J/K · mol observed for the nonspecific complexes most likely represents the baseline surface area dehydrated on binding the acetyl-lysine within the binding pocket. The additional 1000 J/K·mol observed in the specific complex is a significantly more dehydrated interface formed by optimal contacts from the flanking side chains of the histone and optimized folding by the BrD. Specific interactions further exert contact ion pairs between the flanking side chains of the histone and the BrD loop region as well as the added hydrophobic contribution by the greater complementary contact interface formed by the flanking side chains with the BrD. The difference in each case likely is a result of the larger solvent-exposed surface area in the nonspecific complex, which does not form a tight fit at the macromolecular interface, compared with the specifically bound complex. The site-specific binding of BrD3 to histone H3 displaying AcK9 may be due to multiple features inherent to an induced fit mechanism, including the folding of the BrD around the acetyl-lysine and dehydration of the binding interface that encompasses one or more amino acids to the amino and carboxy sides of the acetyl-lysine.
A comparison of the loop regions of Brg1, P/CAF, and Gcn5 shows significant conformational differences to accommodate peptide binding [37,39,41]. Furthermore, the BrD—acetyl-lysine interface in the hydrophobic pocket is further maximized by contacting amino acids that flank the acetyl-lysine. The position variability of acetyl-lysine within the hydrophobic pocket is roughly 8 Å, with larger effects on spatial orientation of flanking side chains of the peptide ligand. Because the histone tail sequences are nearly invariant from yeast to human, this variation in sequence and structure of BrD loop regions is likely to participate in the recognition mechanism. A key observation from sequence alignments between bromodomains is the lack of sequence variation at positions that comprise the interface with the acetyl-lysine. Conversely, BrD regions that make contact with side chains surrounding the acetyl-lysine of the histone tail are highly variable. As the data here show, there are clear biophysical consequences to these sequence variations that contribute to thermodynamic stabilization of complex formation and, by extension, site-specific binding by bromodomains.
Acknowledgments
We thank Dan Brune (Arizona State University) for synthesizing the histone H3 peptides. We also thank the Thompson laboratory for helpful discussions and contributions.
Footnotes
- BrD
- bromodomain
- SAGA
- Spt-Ada-Gcn5 acetyl-transferase
- RSC
- remodels the structure of chromatin
- Pb1
- polybromo-1
- BrD3
- third amino-terminal bromodomain
- IPTG
- isopropyl-β-D-thioga-lactopyranoside
- Ni2+—NTA
- nickel—nitrotriloacetic acid
- MALDI—TOF
- matrix-assisted laser desorption ionization time-of-flight
- LB
- Luria—Bertani
- BSA
- bovine serum albumin
- HATU
- O-(7-azabenzotriazol-1-yl)-N,N,N’,N’-tetramethyluronium hexafluorophosphate
- TFA
- trifluoroacetic acid
- AcK
- acetyl-lysine
References
- [1].Denis GV, McComb ME, Faller DV, Sinha A, Romesser PB, Costello CE. Identification of transcription complexes that contain the double bromodomain protein Brd2 and chromatin remodeling machines. J. Proteome Res. 2006;5:502–511. doi: 10.1021/pr050430u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Jang MK, Mochizuki K, Zhou MS, Jeong HS, Brady JN, Ozato K. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol. Cell. 2005;19:523–534. doi: 10.1016/j.molcel.2005.06.027. [DOI] [PubMed] [Google Scholar]
- [3].Kzhyshkwska J, Rusch A, Wolf H, Dobner T. Regulation of transcription by the heterogeneous nuclear ribonucleoprotein E1B—AP5 is mediated by complex formation with the novel bromodomain-containing protein Brd7. Biochem. J. 2003;371:385–393. doi: 10.1042/BJ20021281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Deng Z, Chen CJ, Chamberlin M, Lu F, Blobel GA, Speicher D, Cirillo LA, Zaret KS, Lieberman PM. The CBP bromodomain and nucleosome targeting are required for Zta-directed nucleosome acetylation and transcription activation. Mol. Cell. Biol. 2003;23:2633–2644. doi: 10.1128/MCB.23.8.2633-2644.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Staal A, Enserink JM, Stein JL, Stein GS, Van Wijnen AJ. Molecular characterization of Celtix-1, a bromodomain protein interacting with the transcription factor interferon regulatory factor 2. J. Cell. Physiol. 2000;185:269–279. doi: 10.1002/1097-4652(200011)185:2<269::AID-JCP12>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- [6].Jones MH, Hamana N, Shimane M. Identification and characterization of BPTF, a novel bromodomain transcription factor. Genomics. 2000;63:35–39. doi: 10.1006/geno.1999.6070. [DOI] [PubMed] [Google Scholar]
- [7].Nielsen MS, Petersen CM, Gliemann J, Madsen P. Cloning and sequencing of a human cDNA encoding a putative transcription factor containing a bromodomain. Biochim. Biophys. Acta. 1996;1306:14–16. doi: 10.1016/0167-4781(95)00239-1. [DOI] [PubMed] [Google Scholar]
- [8].Zeng L, Zhou M. Bromodomain: An acetyl-lysine binding domain. FEBS Lett. 2002;513:124–128. doi: 10.1016/s0014-5793(01)03309-9. [DOI] [PubMed] [Google Scholar]
- [9].Chandrasekaran R, Thompson M. Polybromo-1 bromodomains bind histone H3 at specific acetyl-lysine positions. Biochem. Biophys. Res. Commun. 2007;355:661–666. doi: 10.1016/j.bbrc.2007.01.193. [DOI] [PubMed] [Google Scholar]
- [10].Kanno T, Kanno Y, Siegel RM, Jang MK, Lenardo MJ, Ozato K. Selective recognition of acetylated histones by bromodomain proteins visualized in living cells. Mol. Cell. 2004;13:33–43. doi: 10.1016/s1097-2765(03)00482-9. [DOI] [PubMed] [Google Scholar]
- [11].Chandy M, Gutierrez JL, Prochasson P, Workman JL. SWI/SNF displaces SAGA-acetylated nucleosomes. Euk. Cell. 2006;5:1738–1747. doi: 10.1128/EC.00165-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Carey M, Li B, Workman JL. RSC exploits histone acetylation to abrogate the nucleosomal block to RNA polymerase II elongation. Mol. Cell. 2006;24:481–487. doi: 10.1016/j.molcel.2006.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Eberharter A, Becker PB. Histone acetylation: A switch between repressive and permissive chromatin. EMBO Rep. 2002;3:224–229. doi: 10.1093/embo-reports/kvf053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Kimura A, Umehara T, Horikoshi M. Chromosomal gradient of histone acetylation established by Sas2p and Sir2p functions as a shield against gene silencing. Nat. Genet. 2002;32:370–377. doi: 10.1038/ng993. [DOI] [PubMed] [Google Scholar]
- [15].Xue Y, Canman J, Lee C, Nie Z, Yang D, Moreno G, Young M, Salmon E, Wang W. The human SWI/SNF-B chromatin-remodeling complex is related to yeast Rsc and localizes at kinetochores of mitotic chromosomes. Proc. Natl. Acad. Sci. USA. 2000;97:13015–13020. doi: 10.1073/pnas.240208597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Wang Z, Zhai WG, Richardson JA, Olson EN, Meneses JJ, Firpo MT, Kang CH, Skarnes WC, Tjian R. Polybromo protein BAF180 functions in mammalian cardiac chamber maturation. Genes Dev. 2004;18:3106–3116. doi: 10.1101/gad.1238104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].DeCristofaro MF, Betz BL, Rorie CJ, Reisman DN, Wang WD, Weissman BE. Characterization of SWI/SNF protein expression in human breast cancer cell lines and other malignancies. J. Cell. Physiol. 2001;186:136–145. doi: 10.1002/1097-4652(200101)186:1<136::AID-JCP1010>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- [18].Horikawa I, Barrett J. cDNA cloning of the human polybromo-1 gene on chromosome 3p21. DNA Seq. 2002;13:211–215. doi: 10.1080/1042517021000021590. [DOI] [PubMed] [Google Scholar]
- [19].Chandrasekaran R, Thompson M. Expression, purification, and characterization of individual bromodomains from human polybromo-1. Prot. Expr. Purif. 2006;50:111–117. doi: 10.1016/j.pep.2006.07.004. [DOI] [PubMed] [Google Scholar]
- [20].Haugland RP. In: Handbook of Fluorescent Probes and Research Chemicals. 6th ed Spence MTZ, editor. Molecular Probes; Eugene, OR: 1996. pp. 144–156. [Google Scholar]
- [21].Thompson M, Woodbury N. Thermodynamics of specific and nonspecific DNA-binding by cyanine dye labeled DNA-binding domains. Biophys. J. 2001;81:1793–1804. doi: 10.1016/S0006-3495(01)75830-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Privalov PL, Gill SJ. Stability of protein—structure and hydrophobic interaction. Adv. Prot. Chem. 1988;39:191–234. doi: 10.1016/s0065-3233(08)60377-0. [DOI] [PubMed] [Google Scholar]
- [23].Record MT, Ha JH, Fisher MA. Analysis of equilibrium and kinetic measurements to determine thermodynamic origins of stability and specificity and mechanism of formation of site-specific complexes between proteins and helical DNA. Methods Enzymol. 1991;208:291–343. doi: 10.1016/0076-6879(91)08018-d. [DOI] [PubMed] [Google Scholar]
- [24].Brenowitz M, Jamison E, Majumdar E, Adhya S. Interaction of the Escherichia coli Gal repressor protein with its DNA operators in vitro. Biochemistry. 1990;29:3374–3383. doi: 10.1021/bi00465a033. [DOI] [PubMed] [Google Scholar]
- [25].Ladbury J, Wright J, Sturtevant J, Stigler P. A thermodynamic study of the trp repressor—operator interaction. J. Mol. Biol. 1994;238:669–681. doi: 10.1006/jmbi.1994.1328. [DOI] [PubMed] [Google Scholar]
- [26].Jin L, Yang J, Carey J. Thermodynamics of ligand binding to trp repressor. Biochemistry. 1993;32:7302–7309. doi: 10.1021/bi00079a029. [DOI] [PubMed] [Google Scholar]
- [27].Merabet E, Ackers GK. Calorimetric analysis of (cl repressor binding to DNA operator sites. J. Mol. Biol. 1995;34:8554–8563. doi: 10.1021/bi00027a005. [DOI] [PubMed] [Google Scholar]
- [28].Takeda Y, Ross P, Mudd C. Thermodynamics of Cro protein DNA interactions. Proc. Natl. Acad. Sci. USA. 1992;89:8180–8184. doi: 10.1073/pnas.89.17.8180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Frank D, Saeker R, Bond J, Capp M, Tsoidkov O, Melcher S, Levandoski M, Record MT., Jr. Thermodynamics of the interactions of Lac repressor with variants of the symmetric Lac operator: Effects of converting a consensus site to a non-specific site. J. Mol. Biol. 1997;267:1186–1206. doi: 10.1006/jmbi.1997.0920. [DOI] [PubMed] [Google Scholar]
- [30].Baldwin RL. Temperature-dependence of the hydrophobic interaction in protein folding. Proc. Natl. Acad. Sci. USA. 1986;83:8069–8072. doi: 10.1073/pnas.83.21.8069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Ha J-H, Spolar RS, Record MT. Role of the hydrophobic effect in stability of site-specific protein—DNA complexes. J. Mol. Biol. 1989;209:801–816. doi: 10.1016/0022-2836(89)90608-6. [DOI] [PubMed] [Google Scholar]
- [32].deHaseth PL, Lohman TM, Record MT. Nonspecific interaction of Lac repressor with DNA: An association reaction driven by counterion release. Biochemistry. 1977;16:4783–4790. doi: 10.1021/bi00641a004. [DOI] [PubMed] [Google Scholar]
- [33].Revzin A. Analyzing DNA—protein interactions by gel-electrophoresis. J. Chem. Educ. 1990;67:749–753. [Google Scholar]
- [34].Tanford C. The Hydrophobic Effect. 2nd ed John Wiley; New York: 1980. [Google Scholar]
- [35].Sturtevant JM. Heat capacity and entropy changes in processes involving proteins. Proc. Natl. Acad. Sci. USA. 1977;74:2236–2240. doi: 10.1073/pnas.74.6.2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Olmsted MC, Anderson CF, Record MT. Monte-Carlo description of oligoelectrolyte properties of DNA oligomers: Range of the end effect and the approach of molecular and thermodynamic properties to the poly-electrolyte limits. Proc. Natl. Acad. Sci. USA. 1989;86:7766–7770. doi: 10.1073/pnas.86.20.7766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Hudson B, Martinez-Yamout M, Dyson H, Wright P. Solution structure and acetyl-lysine binding activity of the GCN5 bromodomain. J. Mol. Biol. 2000;304:355–370. doi: 10.1006/jmbi.2000.4207. [DOI] [PubMed] [Google Scholar]
- [38].Pizzitutti F, Giansanti A, Ballario P, Ornaghi P, Torreri P, Ciccotti G, Filetici P. The role of loop ZA and Pro371 in the function of yeast Gcn5p bromodomain revealed through molecular dynamics and experiment. J. Mol. Recogn. 2006;19:1–9. doi: 10.1002/jmr.748. [DOI] [PubMed] [Google Scholar]
- [39].Shen WQ, Xu C, Huang W, Zhang JH, Carlson JE, Tu XM, Wu JH, Shi YY. Solution structure of human Brg1 bromodomain and its specific binding to acetylated histone tails. Biochemistry. 2007;46:2100–2110. doi: 10.1021/bi0611208. [DOI] [PubMed] [Google Scholar]
- [40].Pantano S, Marcello A, Ferrari A, Gaudiosi D, Sabo A, Pellegrini V, Beltram F, Giacca M, Carloni P. Insights on HIV-1 Tat:P/CAF bromodomain molecular recognition from in vivo experiments and molecular dynamics simulations. Prot.—Struct. Funct. Bioinf. 2006;62:1062–1073. doi: 10.1002/prot.20805. [DOI] [PubMed] [Google Scholar]
- [41].Mujtaba S, He Y, Zeng L, Farooq A, Carlson J, Ott M, Verdin E, Zhou M. Structural basis of lysine-acetylated HIV-1 Tat recognition by PCAF bromodomain. Mol. Cell. 2002;9:575–586. doi: 10.1016/s1097-2765(02)00483-5. [DOI] [PubMed] [Google Scholar]
- [42].Jacobson R, Ladurner A, King D, Tjian R. Structure and function of a human TAFII250 double bromodomain module. Science. 2000;288:1422–1427. doi: 10.1126/science.288.5470.1422. [DOI] [PubMed] [Google Scholar]
- [43].Mujtaba S, He Y, Zeng L, Yan S, Plotnikova O, Sachchidanand, Sanchez R, Zeleznik-Le N, Ronai Z, Zhou M. Structural mechanism of the bromodomain of the coactivator CBP in p53 transcriptional activation. Mol. Cell. 2004;13:251–263. doi: 10.1016/s1097-2765(03)00528-8. [DOI] [PubMed] [Google Scholar]
- [44].Mujtaba S, Zhou MM. Chromatin and Chromatin Remodeling Enzymes. 2004. pp. 119–130.
- [45].Dhalluin C, Carlson JE, Zeng L, He C, Aggarwal AK, Zhou MM. Structure and ligand of a histone acetyltransferase bromodomain. Nature. 1999;399:491–496. doi: 10.1038/20974. [DOI] [PubMed] [Google Scholar]
- [46].Nakamura Y, Umehara T, Nakano K, Jang MK, Shirouzu M, Morita S, Uda-Tochio H, Hamana H, Terada T, Adachi N, Matsumoto T, Tanaka A, Horikoshi M, Ozato K, Padmanabhan B, Yokoyama S. Crystal structure of the human Brd2 bromodomain: Insights into dimerization and recognition of acetylated histone H4. J. Biol. Chem. 2007;282:4193–4201. doi: 10.1074/jbc.M605971200. [DOI] [PubMed] [Google Scholar]
- [47].Freire E, Murphy KP. Molecular-basis of cooperativity in protein folding. J. Mol. Biol. 1991;222:687–698. doi: 10.1016/0022-2836(91)90505-z. [DOI] [PubMed] [Google Scholar]
- [48].Murphy KP, Gill SJ. Solid model compounds and the thermodynamics of protein unfolding. J. Mol. Biol. 1991;222:699–709. doi: 10.1016/0022-2836(91)90506-2. [DOI] [PubMed] [Google Scholar]
- [49].Spolar RS, Record MT. Coupling of local folding to site-specific binding of proteins to DNA. Science. 1994;263:777–784. doi: 10.1126/science.8303294. [DOI] [PubMed] [Google Scholar]