Abstract
Background & Aims
Variants in the cytochrome P450 2C9 (CYP2C9) gene are associated with impaired metabolism of celecoxib. We examined the influence of CYP2C9*2 (R144C) and CYP2C9*3 (I359L) variants on dose-related response or toxicity in a randomized trial of celecoxib.
Methods
We identified individuals with CYP2C9*2 and CYP2C9*3 genotypes (≥1 variant allele) in the Adenoma Prevention with Celecoxib trial. Following adenoma removal, patients were assigned randomly to groups given placebo or low-dose (200 mg, twice-daily) or high-dose (400 mg, twice-daily) celecoxib and underwent follow-up colonoscopies at 1 and/or 3 years.
Results
Among 1660 patients, 21% were CYP2C9*2 and 12% were CYP2C9*3 genotypes. Overall, celecoxib was associated with a dose-dependent reduction in adenoma, compared with placebo, with relative risks (RR) of 0.65 (0.56–0.76) for the low-dose and 0.54 (0.46–0.63) for the high-dose groups. However, the additional protective effect of the high dose, compared with the low dose, was observed only in those with CYP2C9*3 genotypes (RR, 0.51; 0.30–0.87). The high dose, compared with low dose, was not associated with significant risk reduction among those with CYP2C9*2 (RR, 0.83; 0.57–1.21) or wild-type (RR, 0.89; 0.72–1.11) genotypes. Compared with placebo, a higher incidence of cardiovascular events was associated with both doses among patients with wild-type genotypes, but only with the high dose among patients with variant genotypes.
Conclusions
The greater efficacy of high-dose celecoxib, compared with the low dose, in preventing colorectal adenoma appears confined to individuals with slow-metabolizer (CYP2C9*3) genotypes. Genetic variability influences susceptibility to the potential benefits and hazards of celecoxib.
Introduction
Although colorectal cancer can be effectively prevented through early detection and removal of precursor adenomatous polyps, suboptimal rates of population screening highlight the importance of investigating chemopreventative strategies.1 Recently, two randomized, placebo-controlled, clinical trials have offered compelling evidence that the cyclooxygenase-2 (COX-2) selective inhibitor celecoxib reduces the risk of sporadic colorectal adenoma among patients with a prior history of adenoma.2, 3 In the Adenoma Prevention with Celecoxib (APC) trial, patients who had recently undergone colonoscopic removal of an adenoma were randomly assigned to receive placebo, low-dose (200 mg twice daily) of celecoxib, or high-dose (400 mg twice daily) celecoxib and underwent follow-up colonoscopies at 1 and 3 years. The estimated cumulative incidence of the detection of one or more new adenomas by year 3 was 60.7 percent for patients receiving placebo, as compared with 43.2 percent for those receiving low-dose celecoxib (relative risk [RR], 0.67; 95% confidence interval [CI] 0.59–0.77; P<.001) and 37.5 perecent for those receiving high-dose celecoxib (RR, 0.55; 95% CI, 0.48–0.64; P<.001).2 Unfortunately, in a separate, adjudicated safety analysis, the APC trial also revealed unexpected dose-related cardiovascular toxicity.4
The highly polymorphic cytochrome p450 enzyme isoform 2C9 (CYP2C9) is the principal enzyme responsible for the metabolism of several drugs, including some non-selective, non-aspirin NSAIDs, celecoxib, and warfarin.5 A substantial body of in vitro and in vivo evidence demonstrated that the variant CYP2C9*3 allele (I359L), with an estimated prevalence among Causasians of 6%, is associated with impaired metabolism of these drugs.5–13 The more common CYP2C9*2 (R144C) allele, with an estimated prevalence of 10% among Causasians, has also been associated with a less substantial reduction in enzyme activity in some,5, 10, 11, 14, 15 but not all studies.9, 12
Because inter-individual variability in metabolism may influence both drug efficacy and safety, we examined the influence of CYP2C9 genetic variants on the effect of celecoxib in patients enrolled in the APC trial. Because the APC trial was a placebo-controlled, randomized trial examining both low-dose and high-dose celecoxib, we had a unique opportunity to examine the influence of CYP2C9 variant genotypes on celecoxib dosing and subsequent risk of both adenoma and adverse events.
Methods
Study Population
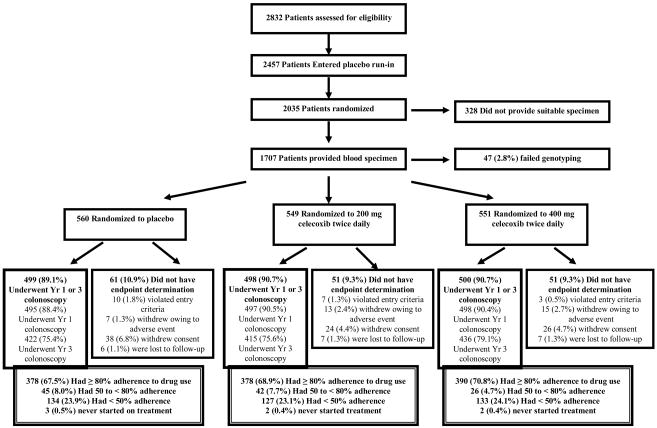
Patients were derived from the Adenoma Prevention with Celecoxib (APC) trial, (ClinicalTrials. gov NCT00005094) for which the methods and main results have been previously described in detail.2 The flow of patients through the study is summarized in the Figure. In brief, the APC trial was a randomized, placebo-controlled trial designed to examine the effect of celecoxib on occurrence of endoscopically detected adenomas among patients who had undergone colonoscopic removal of all colorectal adenomas within 6 months of study entry and had a high risk of recurrent adenomas on the basis of a history of either multiple adenomas or removal of a single adenoma more than 5 mm in diameter. 2,035 patients were randomly assigned to placebo, 200 mg of celecoxib twice daily (low-dose), or 400 mg of celecoxib twice daily (high-dose). Randomization was stratified on the basis of the use or nonuse of low-dose aspirin (325 mg or less every other day or 162.5 mg or less every day) and clinical site. Further details about the trial are available in the supplementary materials.
Figure. Flow of Patients Through the Study.
Patients who violated study entry criteria were those for whom the presence of an adenoma on colonoscopy at baseline could not be confirmed. Patients who withdrew consent for study participation included those who withdrew from the study for medical or nonmedical reasons, those who failed to complete a post-randomization colonoscopy for nonmedical reasons, or those who did not adhere to the protocol for other reasons. Adherence to the use of study medication was calculated as the duration of use in days, divided by 1095. Percentages do not always total 100 due to rounding.
The APC trial was sponsored by the National Cancer Institute and Pfizer Inc through a clinical trials agreement. All patients provided written informed consent before enrollment and the study protocol was approved by the human subjects committee at each study site.
Outcome Ascertainment
A study investigator performed follow-up colonoscopies with endoscopic removal of polyps at one and three years after randomization. A central study pathologist examined, in a blinded fashion, all polyps removed during these colonoscopies. Adverse events were reported by investigators and classified according to criteria from the Medical Dictionary for Regulatory Activities (MedDRA), version 8.1.
Genotyping
Among the 2,035 patients who were enrolled in the study, 1,707 patients at the baseline exam provided a blood specimen suitable for separation into components, which was subsequently stored at −70°C in a central tissue bank. All patients who provided specimens provided separate, informed consent and a genotyping study protocol was approved by the Human Subjects Committee at Partners Healthcare. All genotyping was performed, in a blinded fashion, at the Dana-Farber Harvard Cancer Center High Throughput Polymorphism Detection Core on de-identified patient samples using previously described methods (see supplementary materials for details).16 Among the 1707 patients, genotyping was unsuccessful for the *2 allele in 15 patients, for the *3 allele in 14 patients, and for both alleles in 18 patients. These patients were subsequently excluded from the analysis. We inserted blinded quality control samples equal to 10% of the total number of samples to validate genotype identification procedures; concordance for these samples was 100%. As in our previous study,16 we further validated our genotyping quality by examining Hardy-Weinberg equilibrium among the non-Hispanic White patients who did not develop an adenoma over follow-up. Using the χ2 goodness-of-fit test, the genotype distribution was in Hardy-Weinberg equilibrium for the *2 allele (p=.86) and *3 allele (p=.99).
Statistical Analysis
Consistent with the primary efficacy analyses,2 we used the detection of an adenoma during a post-randomization colonoscopy, regardless of whether the patient adhered to the treatment regimen, as the primary endpoint. We used the Mantel-Cox test, which is a life-table extension of the Mantel-Haenszel statistic, with stratification for aspirin use or nonuse, sex, and age (<65 vs. ≥65 years). The Mantel-Cox procedure also provides a summary risk ratio, which is the weighted average of the relative risk over the two intervals and across strata of aspirin use, sex, and age. Patients with no follow-up colonoscopy (n=163) were excluded from both follow-up intervals. A patient with a colonoscopy at year 3 but with no colonoscopy at year 1 was included in the analysis through year 1, with the assumption that the patient had no adenoma at year 1, and was then included in the analysis through year 3 according to the findings of the colonoscopy at year 3. The analyses at year 3 excluded patients with an adenoma at year 1 colonoscopy and patients with no adenoma at year 1 and no colonoscopy at year 3. According to methods of Kaplan-Meier, three-year cumulative incidences were calculated and patients at risk at each timepoint were those without previously detected adenomas.
Investigator-reported adverse events were analyzed according to prespecified categories to describe cardiovascular and thrombotic disorders, renal and hypertensive disorders, and gastrointestinal ulceration and hemorrhage. The analyses included all events occurring after the first dose of study medication and up to 30 days after the last dose of study medication, including events among patients who continued study medication in the 24 month extension study. The outcome of an adverse event was based on a time-to-event analysis, and a Cox proportional hazards model adjusting for low-dose aspirin use was used to estimate the relative risk.
Among the 1660 patients successfully genotyped, 1102 had no variant *2 or *3 alleles (wild-type/wild-type), 346 had one *2 allele (wild-type/*2), 78 had one *3 allele (wild-type/*3), 17 had one *2 allele and one *3 allele (*2/*3), 11 had two *2 alleles (*2/*2), and 6 had two *3 alleles.(*3/*3) Consistent with prior studies, we defined patients with wild-type genotypes as having no *2 or *3 alleles (wild-type/wild-type), patients with CYP2C9*2 genotypes as having ≥ one *2 allele (wild-type/*2 or *2/*2), and patients with CYP2C9*3 genotypes as having ≥ one *3 allele (wild-type/*3 or *3/3).11, 16, 17 The 17 patients with one *2 allele and one *3 allele were defined as CYP2C9*3 genotypes. The genotype groups were assessed for differences in baseline characteristics using analysis of variance for continuous variables and Chi-squared tests for categorical variables. To test whether the dose effect differed between genotype groups, we used the Cox proportional hazards model with ties handled by the exact method, which assumes that ties arise from grouping continuous, untied data. A linear contrast was constructed from coefficients of this model to assess whether the CYP2C9*2 or CYP2C9*3 genotype was different from wild-type genotype with respect to risk reduction associated with dose. The statistical significance of this contrast was obtained using the Wald method.18, 19 We used the SAS version 9.1 (SAS institute, Cary, NC) for all analyses. All P values are two-sided.
Results
Among the 2,035 patients who were randomly assigned to treatment, 1,707 (84%) provided a suitable blood specimen for genotyping and 1660 (97%) of these patients were successfully genotyped (Figure). Overall, 1,102 (66%) had wild-type genotypes, 357 (21%) had CYP2C9*2 genotypes (≥ one *2 allele) and 201 (12%) had CYP2C9*3 genotypes (≥ one *3 allele). Baseline characteristics were largely similar according to genotype (Table 1). Variant genotypes were more frequently observed in non-Hispanic whites, consistent with other studies,20 as well as in users of low-dose aspirin and patients with a prior history of hypertension.
Table 1.
Baseline Characteristics of the Patients According to CYP2C9 Genotype a
| Characteristics |
All (n =1660) |
Wild-type (n = 1102) |
CYP2C9*2 (n = 357) |
CYP2C9*3 (n = 201) |
P valueb |
|---|---|---|---|---|---|
| Age, median y, (range) | 59 (31, 88) | 59 (31, 84) | 59 (34, 88) | 60 (38, 80) | .09 |
| Women (%) | 31.9 | 32.4 | 29.7 | 33.3 | .57 |
| Race or ethnic group (%)c | |||||
| Non-hispanic White | 92.2 | 89.7 | 96.9 | 98.0 | <.001 |
| Non-hispanic Black | 5.2 | 7.0 | 2.0 | 1.0 | |
| Hispanic | 1.8 | 2.3 | 1.0 | 0.5 | |
| Asian/Pacific Islander/Other | 0.8 | 1.0 | 0.3 | 0.5 | |
| Current cigarette smoker (%) | 16.6 | 17.4 | 16.3 | 12.4 | .25 |
| Body-mass indexd | |||||
| Men | 28.8 (0.1) | 28.8 (0.2) | 28.7 (0.3) | 28.8 (0.4) | .99 |
| Women | 29.0 (0.3) | 28.8 (0.3) | 29.6 (0.7) | 29.0 (0.8) | .56 |
| Colorectal cancer in a parent (%) | 21 | 20.3 | 25.2 | 17.9 | .08 |
| Findings at baseline colonoscopy | |||||
| No. of adenomas | 2.1 (0.04) | 2.1 (0.05) | 2.1 (0.08) | 2.1 (0.11) | .96 |
| At least one adenoma ≥ 1cm (%) | 42.6 | 44.1 | 39.2 | 43.8 | .26 |
| Multiple adenomas (%) | 55.9 | 56.7 | 55.5 | 52.0 | .47 |
| Adenoma burden, cm.e | 1.50 (0.03) | 1.50 (0.04) | 1.45 (0.06) | 1.51 (0.09) | .81 |
| History of cardiovascular events (%)f | 14.2 | 14.0 | 17.1 | 10.5 | .09 |
| History of hypertension (%) | 40.4 | 37.8 | 46.5 | 43.8 | .01 |
| History of diabetes (%) | 9.3 | 9.0 | 8.7 | 12.4 | .27 |
| Use of low-dose aspirin (%)g | 32.0 | 29.7 | 37.0 | 35.8 | .02 |
| Randomized to placebo (%) | 33.7 | 34.5 | 33.9 | 29.4 | -- |
| Randomized to celecoxib, 200 mg twice daily (%) | 33.1 | 32.7 | 32.5 | 36.3 | -- |
| Randomized to celecoxib, 400 mg twice daily (%) | 33.2 | 32.9 | 33.6 | 34.3 | -- |
Data are expressed as mean (SD) unless otherwise indicated. Wild-type genotypes include individuals with no *2 (R144C) or *3 (I359L) alleles. CYP2C9*2 genotypes include individuals with ≥ one *2 allele. CYP2C9*3 genotypes include individuals with ≥ one *3 allele. Individuals with one *2 allele and one *3 allele were classified as having CYP2C9*3 genotypes.
Test of difference between wild-type, CYP2C9*2, and CYP2C9*3 genotype groups was calculated by analysis of variance for continuous variables, χ2 for categorical variables.
Race or ethnic group was determined by the investigator using predefined categories.
Body-mass index is the weight in kilograms divided by the square of the height in meters.
The adenoma burden was defined as the sum of the diameter of all adenomas reported during colonoscopy at baseline.
Cardiovascular events were defined as myocardial infarction, cerebrovascular disease, congestive heart failure, angina, and atherosclerotic heart disease.
Low-dose aspirin was defined as 325 mg or less every other day or 162.5 mg or less every day.
We first examined the main effect of CYP2C9 genotype on adenoma risk. The estimated cumulative incidence through the year 3 colonoscopy of one or more adenomas according to genotype is provided in Table 2. Although there was some evidence of an overall association of the CYP2C9*2 genotype with adenoma risk, this appeared to be confined to the subgroup of patients who were taking low-dose aspirin (RR, 1.44; 95% CI, 1.14–1.81) and was not evident among patients who were randomly assigned to placebo (RR, 1.17; 95% CI, 0.96–1.43). The CYP2C9*3 genotype did not appear to be independently associated with adenoma risk across all subgroups.
Table 2.
Risk of Adenoma According to CYP2C9 Genotypea
| Wild-type |
CYP2C9*2 |
CYP2C9*3 |
|
|---|---|---|---|
| All patients, No. at riskb | 996 | 318 | 183 |
| Cumulative incidence, 3 yrs, % ± SE | 44.8 ± 1.6 | 52.1 ± 2.9 | 46.7 ± 3.8 |
| RR (95% CI)c | 1.0 | 1.16 (1.00–1.35) | 1.05 (0.86–1.28) |
| P valued | -- | .05 | .66 |
|
| |||
| By celecoxib treatment | |||
| Patients randomized to placebo, No. at riskb | 338 | 111 | 50 |
| Cumulative incidence, 3 yrs, % ± SE | 57.2 ± 2.8 | 68.4 ± 4.6 | 64.6 ± 7.0 |
| RR (95% CI)e | 1.0 | 1.17 (0.96–1.43) | 1.06 (0.78–1.42) |
| P valued | -- | .13 | .72 |
| Patients randomized to celecoxib 200 mg twice daily, No. at riskb | 330 | 99 | 69 |
| Cumulative incidence, 3 yrs, % ± SE | 39.9 ± 2.8 | 46.0 ± 5.3 | 49.3 ± 6.2 |
| RR (95% CI)e | 1.0 | 1.20 (0.89–1.62) | 1.25 (0.90–1.72) |
| P valued | -- | .24 | .20 |
| Patients randomized to celecoxib 400 mg twice daily, No. at riskb | 328 | 108 | 64 |
| Cumulative incidence, 3 yrs, % ± SE | 37.0 ± 2.8 | 41.0 ± 4.9 | 29.6 ± 5.9 |
| RR (95% CI)e | 1.0 | 1.11 (0.81–1.53) | 0.79 (0.50–1.25) |
| P valued | -- | .50 | .72 |
|
| |||
| By aspirin strata | |||
| Patients taking aspirin,f No. at riskb | 303 | 116 | 62 |
| Cumulative incidence, 3 yrs, % ± SE | 44.6 ± 3.0 | 60.4 ± 4.7 | 47.1 ± 6.6 |
| RR (95% CI)g | 1.0 | 1.44 (1.14–1.81) | 1.15 (0.82–1.60) |
| P valued | -- | .003 | .43 |
| Patients not taking aspirin,f No. at riskb | 693 | 202 | 121 |
| Cumulative incidence, 3 yrs, % ± SE | 44.9 ± 2.0 | 47.4 ± 3.7 | 46.4 ± 4.7 |
| RR (95% CI)g | 1.0 | 1.02 (0.84–1.24) | 1.00 (0.78–1.28) |
| P valued | -- | .82 | .99 |
Wild-type genotypes include individuals with no *2 (R144C) or *3 (I359L) alleles. CYP2C9*2 genotypes include individuals with ≥ one *2 allele. CYP2C9*3 genotypes include individuals with ≥ one *3 allele. Individuals with one *2 allele and one *3 allele were classified as having CYP2C9*3 genotypes.
No. at risk include patients who underwent a follow-up colonoscopy at year 1 and/or year 3.
Relative risk calculated by the Mantel-Cox test, with stratification for celecoxib treatment, aspirin use, time, age and sex, with the wild-type genotype as the referent group.
The P value is the Cochran-Mantel-Haenszel test of general association compared to the wild-type genotype.
Relative risk calculated by the Mantel-Cox test, with stratification for aspirin use, time, age, and sex, with the wild-type genotype as the referent group.
Patients were stratified at study entry according to the use or nonuse of low-dose aspirin (325 mg or less every other day or 162.5 mg or less every day). Patients not taking aspirin at baseline were required to abstain from taking it during the trial.
Relative risk calculated by the Mantel-Cox test with stratification for celecoxib treatment, time, age, and sex, with the wild-type genotype as the referent group.
Because genotype is associated with celecoxib metabolism, we then investigated the influence of celecoxib on risk of adenoma according to CYP2C9 genotype (Table 3). For patients of all genotypes, the estimated cumulative incidence of one or more adenomas by year 3 was 60.5% for those receiving placebo, as compared with 42.5% for those receiving low-dose (200 mg twice daily) celecoxib (RR, 0.65; 95% CI, 0.56–0.76; P<.001) and 36.9% for those receiving high-dose (400 mg twice daily) celecoxib (RR, 0.54; 95% CI, 0.46–0.63; P<.001), consistent with the primary analysis of the trial.2 Within each subgroup defined by genotype, both low-dose and high-dose celecoxib was associated with a lower risk of adenoma compared with placebo.
Table 3.
Risk of Adenoma According to Celecoxib Dose, Stratified by CYP2C9 Genotypea
| Placebo |
Celecoxib, 200 mg Twice Daily |
Celecoxib, 400 mg Twice Daily |
|
|---|---|---|---|
| All patients, No. at risk.b | 499 | 498 | 500 |
| Cumulative incidence, 3 yrs, % ± SE | 60.5 ± 2.3 | 42.5 ± 2.3 | 36.9 ± 2.2 |
| RR (95% CI) compared with placeboc | 1.0 | 0.65 (0.56–0.76) | 0.54 (0.46–0.63) |
| P valued | -- | <.001 | <.001 |
| RR (95% CI) compared with 200 mge | -- | 1.0 | 0.82 (0.69–0.98) |
| P valuef | -- | -- | .03 |
|
| |||
| By genotype | |||
| Patients with wild-type genotypes, No. at riskb | 338 | 330 | 328 |
| Cumulative incidence, 3 yrs, % ± SE | 57.2 ± 2.8 | 39.9 ± 2.8 | 37.0 ± 2.8 |
| RR (95% CI) compared with placeboc | 1.0 | 0.64 (0.53- 0.77) | 0.56 (0.46–0.68) |
| P valued | -- | <.001 | <.001 |
| RR (95% CI) compared with 200 mge | -- | 1.0 | 0.89 (0.72, 1.11) |
| P valuef | -- | -- | 0.31 |
| Patients with CYP2C9*2 genotypes, No. at riskb | 111 | 99 | 108 |
| Cumulative incidence, 3 yrs, % ± SE | 68.4 ± 4.6 | 46.0 ± 5.3 | 41.0 ± 4.9 |
| RR (95% CI) compared with placeboc | 1.0 | 0.63 (0.47–0.86) | 0.54 (0.40–0.74) |
| P valued | -- | .003 | <.001 |
| RR (95% CI) compared with 200 mge | -- | 1.0 | 0.83 (0.57–1.21) |
| P valuef | -- | -- | .33 |
| Patients with CYP2C9*3 genotypes, No. at riskb | 50 | 69 | 64 |
| Cumulative incidence, 3 yrs, % ± SE | 64.6 ± 7.0 | 49.3 ± 6.2 | 29.6 ± 5.9 |
| RR (95% CI) compared with placeboc | 1.0 | 0.76 (0.51–1.13) | 0.41 (0.24–0.70) |
| P valued | -- | 0.18 | <.001 |
| RR (95% CI) compared with 200 mge | -- | 1.0 | 0.51 (0.30–0.87) |
| P valuef | -- | -- | .01 |
Wild-type genotypes include individuals with no *2 (R144C) or *3 (I359L) alleles. CYP2C9*2 genotypes include individuals with ≥ one *2 allele. CYP2C9*3 genotypes include individuals with ≥ one *3 allele. Individuals with one *2 allele and one *3 allele were classified as having CYP2C9*3 genotypes.
No. at risk include patients who underwent a follow-up colonoscopy at year 1 and/or year 3.
Relative risk calculated by the Mantel-Cox test, with stratification for aspirin use, time, age and sex, with the placebo group as the referent group.
The p value is the Cochran-Mantel-Haenszel test of general association compared to the placebo group.
Relative risk calculated by the Mantel-Cox test, with stratification for aspirin use, time, age and sex, with the celecoxib 200 mg twice daily group as the referent group.
The p value is the Cochran-Mantel-Haenszel test of general association compared to celecoxib 200 mg twice daily group.
To specifically examine the influence of genotype on celecoxib dose and adenoma risk, we compared the cumulative incidence of adenoma among the group receiving high-dose celecoxib with the group receiving low-dose celecoxib (Table 3). Among all genotypes, high-dose celecoxib was associated with a 5.6% greater reduction in 3-year cumulative incidence of adenoma compared with low-dose celecoxib (RR, 0.82; 95% CI, 0.69–0.98; P=.03). However, the effect of dose appeared to be confined to patients with CYP2C9*3 genotypes. Among individuals with CYP2C9*3 genotypes, high-dose celecoxib was associated with a 19.7% greater reduction in cumulative incidence of adenoma compared with low-dose (RR, 0.51; 95% CI, 0.30–0.87; P=.01). In contrast, compared with low-dose celecoxib, high-dose was associated with a 5% reduction in cumulative incidence of adenoma (RR, 0.83; 95% CI, 0.57–1.21, P=.33) among those with CYP2C9*2 genotypes and a 2.9% reduction in cumulative incidence of adenoma (RR, 0.89; 95% CI, 0.72–1.11; P=.31) among those with wild-type genotypes. A formal test of whether the increased risk reduction of higher dose treatment differed between the CYP2C9*3 genotype compared to wildtype genotype approached statistical significance (P=.09).
Although the number of events was limited, we conducted an exploratory analysis examining the influence of CYP2C9 genotype on investigator-reported adverse events associated with celecoxib treatment. We focused on cardiovascular and thrombotic disorders, renal and hypertensive disorders, and gastrointestinal ulceration and hemorrhage separately, consistent with the primary analysis of trial (Table 4). Among patients of any genotype, there was a dose-related increase in cardiovascular and thrombotic disorders with a RR of 1.69 (95% CI, 1.02–2.80) for high-dose compared with placebo. This substantially agreed with the primary analysis of the trial,2 as well as a separate, prespecified analysis of adjudicated cardiovascular events that was previously reported.4 However, the dose relationship between celecoxib and cardiovascular and thrombotic events appeared to vary somewhat according to genotype. Among those with wild-type genotypes, the cumulative incidence of cardiovascular and thrombotic events was 5.7% among those who received placebo, compared with 9.6% for low-dose celecoxib (RR, 1.62; 95% CI, 0.90–2.92) and 8.1% for high-dose (RR, 1.37; 95% CI, 0.75–2.51). However, in the subgroups with variant genotypes, the excess incidence of cardiovascular and thrombotic events appeared primarily among those assigned to high-dose celecoxib, but not low-dose. Among those with either CYP2C9*2 or CYP2C9*3 genotypes, the cumulative incidence of cardiovascular and thrombotic events was 4.0% among those who received placebo, compared with 3.1% for low-dose celecoxib (RR 0.83; 95% CI, 0.25–2.72) and 10.9% for high-dose celecoxib (RR 2.76; 95% CI, 1.08–7.06). These associations appeared consistent when examining the CYP2C9*2 and CYP2C9*3 genotypes separately (Table 4).
Table 4.
Incidence of Adverse Events after Randomization According to Celecoxib Dose, Stratified by CYP2C9 Genotypea
| Placebo |
Celecoxib, 200 mg Twice Daily |
Celecoxib, 400 mg Twice Daily |
|
|---|---|---|---|
| Cardiovascular disordersb | |||
| All patients | |||
| No. with event/No. at risk | 24/557 | 34/547 | 41/549 |
| Cumulative incidence, 3 yrs, % ± SE | 5.1 ± 1.1 | 7.4 ± 1.3 | 9.0 ± 1.4 |
| RR (95% CI) | 1.0 | 1.41 (0.83–2.37) | 1.69 (1.02–2.80) |
| By genotype | |||
| Patients with wild-type genotypes | |||
| No. with event/No. at risk | 18/377 | 29/358 | 25/362 |
| Cumulative incidence, 3 yrs, % ± SE | 5.7 ± 1.4 | 9.6 ± 1.8 | 8.1 ± 1.6 |
| RR (95% CI) | 1.0 | 1.62 (0.90–2.92) | 1.37 (0.75–2.51) |
| Patients with CYP2C9*2 genotypes | |||
| No. with event/No. at risk | 4/121 | 3/116 | 8/118 |
| Cumulative incidence, 3 yrs, % ± | 3.9 ±1.9 | 2.8 ± 1.6 | 9.4 ± 3.2 |
| RR (95% CI) | 1.0 | 0.81 (0.18–3.63) | 2.75 (0.82–9.25) |
| Patients with CYP2C9*3 genotypes | |||
| No. with event/No. at risk | 2/59 | 2/73 | 8/69 |
| Cumulative incidence, 3 yrs, % ± SE | 4.4 ±3.0 | 3.4 ± 2.4 | 13.3 ± 4.4 |
| RR (95% CI) | 1.0 | 0.80 (0.11–5.67) | 2.69 (0.56–12.79) |
|
| |||
| Renal and hypertensive disordersc | |||
| All patients | |||
| No. with event/No. at risk | 97/557 | 126/547 | 99/549 |
| Cumulative incidence, 3 yrs, % ± SE | 19.8 ± 1.9 | 26.2 ± 2.1 | 20.8 ± 2.0 |
| RR (95% CI) | 1.0 | 1.37 (1.05–1.79) | 1.02 (0.77–1.35) |
| By genotype | |||
| Patients with wild-type genotypes | |||
| No. with event/No. at risk | 66/377 | 76/358 | 63/362 |
| Cumulative incidence, 3 yrs, % ± SE | 20.3 ± 2.3 | 23.6 ± 2.5 | 20.5 ± 2.4 |
| RR (95% CI) | 1.0 | 1.23 (0.88–1.71) | 0.99 (0.70–1.40) |
| Patients with CYP2C9*2 genotypes | |||
| No. with event/No. at risk | 22/121 | 25/116 | 20/118 |
| Cumulative incidence, 3 yrs, % ±SE | 19.2 ± 3.9 | 25.0 ± 4.5 | 19.9 ± 4.2 |
| RR (95% CI) | 1.0 | 1.27 (0.71–2.25) | 1.00 (0.55–1.85) |
| Patients with CYP2C9*3 genotypes | |||
| No. with event/No. at risk | 9/59 | 25/73 | 16/69 |
| Cumulative incidence, 3 yrs, % ± SE | 17.2 ± 5.6 | 41.0 ± 6.5 | 23.4 ± 5.5 |
| RR (95% CI) | 1.0 | 2.64 (1.22–5.68) | 1.30 (0.57–2.97) |
|
| |||
| Gastrointestinal ulceration/hemorrhaged | |||
| All patients | |||
| No. with event/No. at risk | 57/557 | 56/547 | 55/549 |
| Cumulative incidence, 3 yrs, % ± SE | 11.9 ± 1.6 | 11.8 ± 1.6 | 11.7 ± 1.5 |
| RR (95% CI) | 1.0 | 0.97 (0.67–1.41) | 0.95 (0.66–1.38) |
| By genotype | |||
| Patients with wild-type genotypes | |||
| No. with event/No. at risk | 41/377 | 35/358 | 38/362 |
| Cumulative incidence, 3 yrs, % ± SE | 12.9 ± 2.0 | 10.9 ± 1.8 | 12.2 ± 1.9 |
| RR (95% CI) | 1.0 | 0.84 (0.54–1.32) | 0.92 (0.59–1.43) |
| Patients with CYP2C9*2 genotypes | |||
| No. with event/No. at risk | 10/121 | 16/116 | 9/118 |
| Cumulative incidence, 3 yrs, % ± SE | 8.5 ± 2.7 | 17.2 ± 4.1 | 9.5 ± 3.1 |
| RR (95% CI) | 1.0 | 1.85 (0.84, 4.07) | 0.99 (0.40, 2.46) |
| Patients with CYP2C9*3 genotypes | |||
| No. with event/No. at risk | 6/59 | 5/73 | 8/69 |
| Cumulative incidence, 3 yrs, % ± SE | 13.7 ±5.3 | 7.9 ± 3.4 | 12.1 ± 4.3 |
| RR (95% CI) | 1.0 | 0.62 (0.19–2.02) | 0.98 (0.34–2.84) |
Wild-type genotypes include individuals with no *2 (R144C) or *3 (I359L) alleles. CYP2C9*2 genotypes include individuals with ≥ one *2 allele. CYP2C9*3 genotypes include individuals with ≥ one *3 allele. Individuals with one *2 allele and one *3 allele were classified as having CYP2C9*3 genotypes. Adverse events include those that were reported during the time after the first dose of the study drug until 30 days after the last dose of study drug. The analysis excludes seven participants with genotype information that were randomized but never initiated treatment: three patients in the placebo group, two assigned to 200 mg of celecoxib twice daily, and two assigned to 400 mg of celecoxib twice daily. Data on adverse events include events reported among 639 patients who continued blinded treatment beyond the 36 month core phase of the study who were enrolled in the 24 month extension study.
Cardiovascular events include cardiovascular death or circulatory collapse, stroke, myocardial infarction, congestive heart failure, venous thrombosis or thromboembolism, cardiovascular therapeutic procedures, vascular therapeutic procedures, cerebrovascular disease, and vascular disease. Four of the cardiovascular events occurred after 36 months. All four of these patients had wild-type genotypes: one patient was randomized to placebo with an event at 38 months; two patients were randomized to 200 mg twice daily with events at 40 and 49 months; one patient was randomized to 400 mg twice daily with an event at 44 months.
Renal and hypertensive events include elevated creatinine, fluid retention or edema, hypertension, proteinuria and renal failure. Eighteen of the renal events occurred after 36 months. Eleven patients had wild-type genotypes: 4 patients were randomized to placebo with events at 36.1, 36.4, 38, and 41 months; four patients were randomized to 200 mg twice daily with events at 36.3, 36.6, 36.8, and 36.9 months; three patients were randomized to 400 mg twice daily with events at 36.2, 36.3, and 41 months. Four patients had CYP2C9*2 genotypes: two patients were randomized to placebo with events at 36.1 and 36.4 months; one patient was randomized to 200 mg twice daily with an event at 36.5 months; one patient was randomized to 400 mg twice daily with an event at 41 months. Three patients had CYP2C9*3 genotypes: one patient was randomized to placebo with an event at 36.3 months; two patients were randomized to 400 mg twice daily with events at 36.6 and 36.8 months.
Gastrointestinal ulceration and hemorrhage events include anemia, gastrointestinal bleeding, gastritis/duodenitis, upper or lower gastrointestinal ulceration, and other hemorrhage. Eleven of the gastrointestinal events occurred after 36 months. Eight patients had wild-type genotypes: three were randomized to placebo with events at 36.3, 36.5, and 37.5 months; three were randomized to 200 mg twice daily with events at 37, 38, and 48 months; two were randomized to 400 mg twice daily with events at 36.7 and 42 months. Two patients had CYP2C9*2 genotypes: one patient was randomized to placebo with an event at 37 months; one patient was randomized to 200 mg twice daily with an event at 36.3 months. One patient had a CYP2C9*3 genotype and was randomized to 400 mg twice daily with an event at 36.6 months.
For all patients of any genotype, there did not appear to be a dose-related increase in either renal and hypertensive disorders or gastrointestinal ulceration and hemorrhage, consistent with the overall results of the trial.2 When examined according to subgroups defined by genotype, there were also no consistent differences in risk. Although there appeared to be, compared with placebo, a higher risk of renal and hypertensive events associated with low-dose celecoxib (RR 2.64; 1.22–5.68) among those with CYP2C9*3 genotypes, this was not observed with the high-dose (RR 1.30; 95% CI, 0.57–2.97).
Discussion
In this large, randomized, placebo-controlled trial, celecoxib was associated with a decrease in the three-year cumulative incidence of adenoma among patients with wild-type, variant CYP2C9*2 (≥ one R144C) or CYP2C9*3 (≥ one I359L allele) genotypes of CYP2C9, the principal enzyme responsible for celecoxib metabolism from the active to inactive state. However, compared with the lower dose, the additional benefit of the higher dose was restricted to those with the CYP2C9*3 genotype. Although statistical power was limited by the small number of events, it also appeared that among patients with variant genotypes, the increased incidence of cardiovascular and thrombotic events was primarily observed with high-dose celecoxib but not low-dose. In contrast, among patients with wild-type genotypes, both doses of celecoxib appeared associated with risk. There did not appear to be a significant independent main effect of genotype on risk of adenoma or cardiovascular events.
Our results regarding a differential effect of celecoxib on adenoma recurrence according to CYP2C9 genotype are consistent with our present understanding of the role of CYP2C9 in phase 1 drug metabolism and the functional alterations associated with variant genotypes.5–13 The CYP2C9 enzyme metabolizes celecoxib from its active state to an inactive form; the CYP2C9*3 genotype is associated with significantly greater impairment of this metabolism than either the CYP2C9*2 or wild-type genotype. Thus, high-dose celecoxib may overwhelm a diminished capacity to inactivate drug among individuals with CYP2C9*3 genotypes. This may lead to substantially increased accumulation of active celecoxib, in turn potentiating greater anti-cancer efficacy. In contrast, the metabolic capacity of patients with wild-type or CYP2C9*2 genotypes may be sufficient to inactivate high-dose and low-dose celecoxib with equivalent efficiency. Thus, for individuals with these genotypes, high and low doses of celecoxib may yield comparable levels of bioavailability, resulting in no clinically apparent difference in therapeutic effectiveness.
To our knowledge, our study is the first to examine the relationship between CYP2C9 genetic variants and randomly assigned celecoxib use with clinical outcomes. In support of our findings, previous work demonstrated the relevance of CYP2C9 genetic variation with other drug substrates and endpoints. Several studies have shown that CYP2C9 genetic variants are associated with decreased elimination of warfarin, resulting in an increased risk of overanticoagulation and complications from bleeding.11, 21–23 Previous case-control studies have also demonstrated that compared with wild-type, individuals with CYP2C9 variant genotypes have a lower risk of colorectal cancer associated with ibuprofen,24 as well as an increased risk of gastroduodenal bleeding related to NSAIDs.17
Interestingly, although there did not appear to be an overall independent effect of CYP2C9 genotype on risk of adenoma, patients in our study with the CYP2C9*2 genotype appeared to have a somewhat higher risk of adenoma in the subgroup using low-dose aspirin. Previous studies have suggested that CYP2C9 variant genotype may be independently associated with adenoma risk through differential metabolism of carcinogens and endogenous prostanoids or induction of COX-2.13, 16, 25–27 This finding may also be explained by an effect of the *3 variant allele in CYP2C8, which is in strong linkage with the CYP2C9 *2 allele.28 In support of this hypothesis, we did not observe any increase risk of adenoma related to the CYP2C9*3 variant, which is not strongly linked to the *3 allele of CYP2C8. The *3 allele of CYP2C8 has been associated with impaired metabolism of arachidonic acid, which influences adenoma risk as well as vascular tone, potentially also explaining the higher prevalence of baseline hypertension among patients in this trial with CYP2C9*2 genotypes.29
Given the importance of the APC trial in identifying unexpected toxicity associated with celecoxib,4 we also examined the influence of CYP2C9 genetic variation on risk of adverse events. For both renal and hypertensive events, there did not appear to be a consistent differential effect of celecoxib according to genotype. We also did not find a relationship between variant genotype and gastrointestinal bleeding associated with celecoxib. This finding contrasts with a small case-control study which did observe an association between CYP2C9 variant genotype and short-term risk of NSAID-related gastroduodenal bleeding.17 However, most patients in this case-control study were exposed to COX-2 non-selective NSAIDs (e.g. diclofenac) which may be associated with a greater risk of bleeding than celecoxib. In fact, in the main analysis of the APC trial, an overall association between celecoxib and gastrointestinal bleeding was not observed.2
Interestingly, among individuals with either CYP2C9*2 or CYP2C9*3 genotypes, the excess number of cardiovascular and thrombotic events associated with celecoxib was restricted to those randomized to high-dose treatment. Overall, these data highlight the potential that individuals with impaired metabolism may be at particularly high risk of dose-related cardiovascular toxicity. Although it is unclear why low-dose celecoxib was not strongly related to cardiovascular and thrombotic events among individuals with variant genotypes, it is important to note that other randomized trials of celecoxib also did not demonstrate an increased risk of cardiovascular events associated with low-dose (200 mg twice daily or 400 mg once daily).3, 30–32 Nonetheless, because we had a limited number of events, our findings should be viewed as exploratory and support the importance of examining CYP2C9 genotype within ongoing clinical trials focused on adverse events associated with celecoxib.32
Our study had several strengths. First, celecoxib treatment was randomly assigned, dosing was strictly defined, treatment duration was long-term, and use of other NSAIDs (except low-dose aspirin) was not permitted.2 Thus, it is unlikely that our findings are related to differential dosing, heterogeneity in treatment duration, or exposure to other CYP2C9-metabolized NSAIDs. Second, patients were randomized to two different doses of celecoxib, permitting a detailed assessment of the effect of genotype on dose. Other studies examining single doses of celecoxib or exposure irrespective of dose would be unable to uncover a specific effect of genotype on dose-related outcomes. Third, as all patients were enrolled in a randomized trial, treatment and colonoscopic surveillance for outcomes, as well as reporting of adverse events, were uniform and standardized.
Several limitations of this study deserve comment. First, we had a limited number of patients with the CYP2C9*3 genotype although our study is consistent with the prevalence of this variant in other cohorts not specifically selected according to genotype.11, 16, 33 Second, as we have previously mentioned, we had a limited number of adverse events for analysis, especially among patients with variant genotypes. Nonetheless, our study represents the largest cohort of patients who have been randomized to long-term treatment with celecoxib for whom toxicity data are available. Third, although two copies of the I359L allele (CYP2C9*3 homozygotes) may be associated with greater impairment of drug metabolism than one copy (CYP2C9*3 heterozygotes),9, 12, 34 we did not have enough patients with two copies of I359L alleles to analyze outcomes according to gene dosage. Fourth, we restricted our analysis of CYP2C9 variants to CYP2C9*2 and CYP2C9*3 polymorphisms, which are the most prevalent polymorphisms among non-Hispanic whites.5, 20, 35, 36 However, because non-Hispanic Whites composed more than 90% of our study population, it is unlikely that misclassification of genotypes which are more prevalent among non-Whites would influence our results. Importantly, our results were also not materially altered when we restricted our analyses to non-Hispanic White patients (data not shown). Given potential differences in the prevalence of genetic variants as well as their associated metabolizer phenotypes in non-White populations, further studies are warranted to examine CYP2C9 variants and celecoxib use in other populations.
At baseline, there were differences in low-dose aspirin use according to genotype. Unlike many non-aspirin NSAIDs, aspirin is not primarily metabolized by CYP2C9.37–40 Thus, it is unlikely that this association reflects genotype-specific differences in tolerance to aspirin therapy. Nonetheless, because low-dose aspirin therapy was included as a stratification variable, we were able to carefully adjust for use of low-dose aspirin in all of our multivariate models to minimize any potential confounding.
Our study has specific clinical implications. Although celecoxib effectively prevents colorectal neoplasia, the associated cardiovascular risk does not permit routine use of the drug for population-based chemoprevention. Despite this, efforts to use genetic information to personalize and optimize chemoprevention have been identified as a high priority and may be warranted for specific patients.41 For example, high dose celecoxib at 400 mg twice daily is currently approved by the Food and Drug Administration for adjunctive treatment of patients with familial adenomatous polyposis. Celecoxib treatment also has been proposed for specific individuals who are at high risk for colorectal cancer, low risk for cardiovascular events, and unable to tolerate routine endoscopic surveillance. Finally, celecoxib treatment is continuing in studies examining its potential role in multi-agent chemoprevention or as an adjunctive treatment in patients with established cancers.42, 43 Our data suggest that for the vast majority of individuals without CYP2C9*3 genotypes, high-dose celecoxib does not confer any additional chemopreventive benefit. Thus, consideration of celecoxib for any chemopreventive strategy should be based on the potential risks and benefits associated with low-dose.
Finally, our study provides proof-of-principle that genetic determinants can influence an individual’s pharmacokinetic response to celecoxib with a significant, clinically apparent impact on outcome.41 Because celecoxib is widely used for other indications, such as treatment of arthritis,44 determining the influence of CYP2C9 genotype on responsiveness to therapy as well as susceptibility to toxicity within other patient populations remains critically important.
In summary, this study observed a pharmacogenetic association between CYP2C9*3 variant genotype and risk of adenoma according to celecoxib dosing within a large, randomized, placebo-controlled trial. Although the number of patients with variant genotypes was relatively small, limiting the statistical power, there was also a potential relationship between CYP2C9 variant genotypes and risk of cardiovascular and thrombotic events according to celecoxib exposure. These data support the potential importance of genetic variability in determining susceptibility to the benefits and hazards of celecoxib. Further research is needed to examine the routine use of genetic information in tailoring treatment with celecoxib across a range of doses, indications for use, and patient populations.
Supplementary Material
Acknowledgments
Portions of this data were previously presented in abstract form at Digestive Disease Week in San Diego, CA on May 17–22, 2008 and the American Association for Cancer Research Molecular Diagnostics in Cancer Therapeutic Development meeting in Philadelphia, PA on September 22–25, 2008. The authors would like to acknowledge Ms. Patrice Soule and Mr. Hardeep Ranu of the Dana-Farber Harvard Cancer Center High Throughput Polymorphism Detection Core for technical assistance.
Grant Support: Dr. Chan is a recipient of the Damon Runyon Cancer Research Foundation Clinical Investigator Award and a career development award from the National Cancer Institute (CA107412). Dr. Zauber and Dr. Bertagnolli are the recipients of funding from the National Cancer Institute (N01-CN95015). The APC trial was sponsored by the National Cancer Institute and Pfizer Inc through a clinical trials agreement. Representatives of the NCI, Pfizer Inc, and affiliated academic institutions participated in the analysis and interpretation of the data.
NIH Grants: CA107412
CA127003
CA131504
CA137178
NO1-CN95015
Footnotes
Financial Disclosures: Dr. Zauber and Dr. Bertagnolli are the recipients of research funding from Pfizer Inc. Dr. Breazna, Dr. Rosenstein, and Dr. Eagle are employees of and shareholders in Pfizer Inc. Dr. Hawk has served as a consultant for Pozen Pharmaceutical Development Company. Dr. Chan, Ms. Hsu, and Dr. Hunter have no conflicts of interest. The statistical analysis of the entire data sets pertaining to efficacy and safety has been independently confirmed by Dr. Zauber, who is not employed by any corporate entity. The corresponding author had full access to all of the data and takes full responsibility for the veracity of the data and analysis.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Use of colorectal cancer tests--United States 2002, 2004, and 2006. MMWR Morb Mortal Wkly Rep. 2008;57:253–8. [PubMed] [Google Scholar]
- 2.Bertagnolli MM, Eagle CJ, Zauber AG, Redston M, Solomon SD, Kim K, Tang J, Rosenstein RB, Wittes J, Corle D, Hess TM, Woloj GM, Boisserie F, Anderson WF, Viner JL, Bagheri D, Burn J, Chung DC, Dewar T, Foley TR, Hoffman N, Macrae F, Pruitt RE, Saltzman JR, Salzberg B, Sylwestrowicz T, Gordon GB, Hawk ET. Celecoxib for the prevention of sporadic colorectal adenomas. N Engl J Med. 2006;355:873–84. doi: 10.1056/NEJMoa061355. [DOI] [PubMed] [Google Scholar]
- 3.Arber N, Eagle CJ, Spicak J, Racz I, Dite P, Hajer J, Zavoral M, Lechuga MJ, Gerletti P, Tang J, Rosenstein RB, Macdonald K, Bhadra P, Fowler R, Wittes J, Zauber AG, Solomon SD, Levin B. Celecoxib for the prevention of colorectal adenomatous polyps. N Engl J Med. 2006;355:885–95. doi: 10.1056/NEJMoa061652. [DOI] [PubMed] [Google Scholar]
- 4.Solomon SD, McMurray JJ, Pfeffer MA, Wittes J, Fowler R, Finn P, Anderson WF, Zauber A, Hawk E, Bertagnolli M. Cardiovascular Risk Associated with Celecoxib in a Clinical Trial for Colorectal Adenoma Prevention. N Engl J Med. 2005;352:1071–80. doi: 10.1056/NEJMoa050405. [DOI] [PubMed] [Google Scholar]
- 5.Lee CR, Goldstein JA, Pieper JA. Cytochrome P450 2C9 polymorphisms: a comprehensive review of the in-vitro and human data. Pharmacogenetics. 2002;12:251–63. doi: 10.1097/00008571-200204000-00010. [DOI] [PubMed] [Google Scholar]
- 6.Miners JO, Birkett DJ. Cytochrome P4502C9: an enzyme of major importance in human drug metabolism. Br J Clin Pharmacol. 1998;45:525–38. doi: 10.1046/j.1365-2125.1998.00721.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takanashi K, Tainaka H, Kobayashi K, Yasumori T, Hosakawa M, Chiba K. CYP2C9 Ile359 and Leu359 variants: enzyme kinetic study with seven substrates. Pharmacogenetics. 2000;10:95–104. doi: 10.1097/00008571-200003000-00001. [DOI] [PubMed] [Google Scholar]
- 8.Tang C, Shou M, Rushmore TH, Mei Q, Sandhu P, Woolf EJ, Rose MJ, Gelmann A, Greenberg HE, De Lepeleire I, Van Hecken A, De Schepper PJ, Ebel DL, Schwartz JI, Rodrigues AD. In-vitro metabolism of celecoxib, a cyclooxygenase-2 inhibitor, by allelic variant forms of human liver microsomal cytochrome P450 2C9: correlation with CYP2C9 genotype and in-vivo pharmacokinetics. Pharmacogenetics. 2001;11:223–35. doi: 10.1097/00008571-200104000-00006. [DOI] [PubMed] [Google Scholar]
- 9.Sandberg M, Yasar U, Stromberg P, Hoog JO, Eliasson E. Oxidation of celecoxib by polymorphic cytochrome P450 2C9 and alcohol dehydrogenase. Br J Clin Pharmacol. 2002;54:423–9. doi: 10.1046/j.1365-2125.2002.01660.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scordo MG, Pengo V, Spina E, Dahl ML, Gusella M, Padrini R. Influence of CYP2C9 and CYP2C19 genetic polymorphisms on warfarin maintenance dose and metabolic clearance. Clin Pharmacol Ther. 2002;72:702–10. doi: 10.1067/mcp.2002.129321. [DOI] [PubMed] [Google Scholar]
- 11.Higashi MK, Veenstra DL, Kondo LM, Wittkowsky AK, Srinouanprachanh SL, Farin FM, Rettie AE. Association between CYP2C9 genetic variants and anticoagulation-related outcomes during warfarin therapy. JAMA. 2002;287:1690–8. doi: 10.1001/jama.287.13.1690. [DOI] [PubMed] [Google Scholar]
- 12.Kirchheiner J, Stormer E, Meisel C, Steinbach N, Roots I, Brockmoller J. Influence of CYP2C9 genetic polymorphisms on pharmacokinetics of celecoxib and its metabolites. Pharmacogenetics. 2003;13:473–80. doi: 10.1097/00008571-200308000-00005. [DOI] [PubMed] [Google Scholar]
- 13.Kirchheiner J, Brockmoller J. Clinical consequences of cytochrome P450 2C9 polymorphisms. Clin Pharmacol Ther. 2005;77:1–16. doi: 10.1016/j.clpt.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 14.Crespi CL, Miller VP. The R144C change in the CYP2C9*2 allele alters interaction of the cytochrome P450 with NADPH:cytochrome P450 oxidoreductase. Pharmacogenetics. 1997;7:203–10. doi: 10.1097/00008571-199706000-00005. [DOI] [PubMed] [Google Scholar]
- 15.Fries S, Grosser T, Price TS, Lawson JA, Kapoor S, DeMarco S, Pletcher MT, Wiltshire T, FitzGerald GA. Marked interindividual variability in the response to selective inhibitors of cyclooxygenase-2. Gastroenterology. 2006;130:55–64. doi: 10.1053/j.gastro.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 16.Chan AT, Tranah GJ, Giovannucci EL, Hunter DJ, Fuchs CS. A prospective study of genetic polymorphisms in the cytochrome P450 2C9 enzyme and the risk of distal colorectal adenoma. Clin Gastro Hepatol. 2004;2:704–12. doi: 10.1016/s1542-3565(04)00294-0. [DOI] [PubMed] [Google Scholar]
- 17.Pilotto A, Seripa D, Franceschi M, Scarcelli C, Colaizzo D, Grandone E, Niro V, Andriulli A, Leandro G, Di Mario F, Dallapiccola B. Genetic susceptibility to nonsteroidal anti-inflammatory drug-related gastroduodenal bleeding: role of cytochrome P450 2C9 polymorphisms. Gastroenterology. 2007;133:465–71. doi: 10.1053/j.gastro.2007.05.025. [DOI] [PubMed] [Google Scholar]
- 18.Kalbfleisch JD, Prentice RL. The statistical analysis of failure time data. J. Wiley; 2002. [Google Scholar]
- 19.Delong DM, Guirguis GH, So YC. Efficient computation of subset selection probabilities with application to Cox regression. Biometrika %R. 1994;81:607–611. doi: 10.1093/biomet/81.3.607. [DOI] [Google Scholar]
- 20.Dickmann LJ, Rettie AE, Kneller MB, Kim RB, Wood AJ, Stein CM, Wilkinson GR, Schwarz UI. Identification and functional characterization of a new CYP2C9 variant (CYP2C9*5) expressed among African Americans. Mol Pharmacol. 2001;60:382–7. doi: 10.1124/mol.60.2.382. [DOI] [PubMed] [Google Scholar]
- 21.Aithal GP, Day CP, Kesteven PJ, Daly AK. Association of polymorphisms in the cytochrome P450 CYP2C9 with warfarin dose requirement and risk of bleeding complications. Lancet. 1999;353:717–9. doi: 10.1016/S0140-6736(98)04474-2. [DOI] [PubMed] [Google Scholar]
- 22.Malhi H, Atac B, Daly AK, Gupta S. Warfarin and celecoxib interaction in the setting of cytochrome P450 (CYP2C9) polymorphism with bleeding complication. Postgrad Med J. 2004;80:107–9. doi: 10.1136/pmj.2003.006478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Visser LE, van Schaik RH, van Vliet M, Trienekens PH, De Smet PA, Vulto AG, Hofman A, van Duijn CM, Stricker BH. The risk of bleeding complications in patients with cytochrome P450 CYP2C9*2 or CYP2C9*3 alleles on acenocoumarol or phenprocoumon. Thromb Haemost. 2004;92:61–6. doi: 10.1160/TH03-12-0741. [DOI] [PubMed] [Google Scholar]
- 24.Samowitz WS, Wolff RK, Curtin K, Sweeney C, Ma KN, Andersen K, Levin TR, Slattery ML. Interactions between CYP2C9 and UGT1A6 polymorphisms and nonsteroidal anti-inflammatory drugs in colorectal cancer prevention. Clin Gastroenterol Hepatol. 2006;4:894–901. doi: 10.1016/j.cgh.2006.04.021. [DOI] [PubMed] [Google Scholar]
- 25.Yun CH, Shimada T, Guengerich FP. Roles of human liver cytochrome P4502C and 3A enzymes in the 3-hydroxylation of benzo(a)pyrene. Cancer Res. 1992;52:1868–74. [PubMed] [Google Scholar]
- 26.Bauer E, Guo Z, Ueng YF, Bell LC, Zeldin D, Guengerich FP. Oxidation of benzo[a]pyrene by recombinant human cytochrome P450 enzymes. Chem Res Toxicol. 1995;8:136–42. doi: 10.1021/tx00043a018. [DOI] [PubMed] [Google Scholar]
- 27.Michaelis UR, Falck JR, Schmidt R, Busse R, Fleming I. Cytochrome P4502C9-derived epoxyeicosatrienoic acids induce the expression of cyclooxygenase-2 in endothelial cells. Arterioscler Thromb Vasc Biol. 2005;25:321–6. doi: 10.1161/01.ATV.0000151648.58516.eb. [DOI] [PubMed] [Google Scholar]
- 28.Yasar U, Bennet AM, Eliasson E, Lundgren S, Wiman B, De Faire U, Rane A. Allelic variants of cytochromes P450 2C modify the risk for acute myocardial infarction. Pharmacogenetics. 2003;13:715–20. doi: 10.1097/00008571-200312000-00002. [DOI] [PubMed] [Google Scholar]
- 29.Dai D, Zeldin DC, Blaisdell JA, Chanas B, Coulter SJ, Ghanayem BI, Goldstein JA. Polymorphisms in human CYP2C8 decrease metabolism of the anticancer drug paclitaxel and arachidonic acid. Pharmacogenetics. 2001;11:597–607. doi: 10.1097/00008571-200110000-00006. [DOI] [PubMed] [Google Scholar]
- 30.Cardiovascular and cerebrovascular events in the randomized, controlled Alzheimer’s Disease. Anti-Inflammatory Prevention Trial (ADAPT) PLoS Clin Trials. 2006;1:e33. doi: 10.1371/journal.pctr.0010033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Feng GS, Ma JL, Wong BC, Zhang L, Liu WD, Pan KF, Shen L, Zhang XD, Li J, Xia HH, Li JY, Lam SK, You WC. Celecoxib-related gastroduodenal ulcer and cardiovascular events in a randomized trial for gastric cancer prevention. World J Gastroenterol. 2008;14:4535–9. doi: 10.3748/wjg.14.4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Solomon SD, Wittes J, Finn PV, Fowler R, Viner J, Bertagnolli MM, Arber N, Levin B, Meinert CL, Martin B, Pater JL, Goss PE, Lance P, Obara S, Chew EY, Kim J, Arndt G, Hawk E. Cardiovascular risk of celecoxib in 6 randomized placebo-controlled trials: the cross trial safety analysis. Circulation. 2008;117:2104–13. doi: 10.1161/CIRCULATIONAHA.108.764530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bigler J, Whitton J, Lampe JW, Fosdick L, Bostick RM, Potter JD. CYP2C9 and UGT1A6 genotypes modulate the protective effect of aspirin on colon adenoma risk. Cancer Res. 2001;61:3566–9. [PubMed] [Google Scholar]
- 34.Lundblad MS, Ohlsson S, Johansson P, Lafolie P, Eliasson E. Accumulation of celecoxib with a 7-fold higher drug exposure in individuals homozygous for CYP2C9*3. Clin Pharmacol Ther. 2006;79:287–8. doi: 10.1016/j.clpt.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 35.Kramer MA, Rettie AE, Rieder MJ, Cabacungan ET, Hines RN. Novel CYP2C9 promoter variants and assessment of their impact on gene expression. Mol Pharmacol. 2008;73:1751–60. doi: 10.1124/mol.107.044149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.DeLozier TC, Lee SC, Coulter SJ, Goh BC, Goldstein JA. Functional characterization of novel allelic variants of CYP2C9 recently discovered in southeast Asians. J Pharmacol Exp Ther. 2005;315:1085–90. doi: 10.1124/jpet.105.091181. [DOI] [PubMed] [Google Scholar]
- 37.Hutt AJ, Caldwell J, Smith RL. The metabolism of aspirin in man: a population study. Xenobiotica. 1986;16:239–49. doi: 10.3109/00498258609043527. [DOI] [PubMed] [Google Scholar]
- 38.Leemann TD, Transon C, Bonnabry P, Dayer P. A major role for cytochrome P450TB (CYP2C subfamily) in the actions of non-steroidal antiinflammatory drugs. Drugs Exp Clin Res. 1993;19:189–95. [PubMed] [Google Scholar]
- 39.Chen XP, Tan ZR, Huang SL, Huang Z, Ou-Yang DS, Zhou HH. Isozyme-specific induction of low-dose aspirin on cytochrome P450 in healthy subjects. Clin Pharmacol Ther. 2003;73:264–71. doi: 10.1067/mcp.2003.14. [DOI] [PubMed] [Google Scholar]
- 40.Takanohashi T, Koizumi T, Mihara R, Okudaira K. Prediction of the metabolic interaction of nateglinide with other drugs based on in vitro studies. Drug Metab Pharmacokinet. 2007;22:409–18. doi: 10.2133/dmpk.22.409. [DOI] [PubMed] [Google Scholar]
- 41.Lippman SM, Lee JJ. Reducing the “risk” of chemoprevention: defining and targeting high risk--2005 AACR Cancer Research and Prevention Foundation Award Lecture. Cancer Res. 2006;66:2893–903. doi: 10.1158/0008-5472.CAN-05-4573. [DOI] [PubMed] [Google Scholar]
- 42.Ulrich CM, Bigler J, Potter JD. Non-steroidal anti-inflammatory drugs for cancer prevention: promise, perils and pharmacogenetics. Nat Rev Cancer. 2006;6:130–40. doi: 10.1038/nrc1801. [DOI] [PubMed] [Google Scholar]
- 43.Arber N. Cyclooxygenase-2 inhibitors in colorectal cancer prevention: point. Cancer Epidemiol Biomarkers Prev. 2008;17:1852–7. doi: 10.1158/1055-9965.EPI-08-0167. [DOI] [PubMed] [Google Scholar]
- 44.Dai C, Stafford RS, Alexander GC. National trends in cyclooxygenase-2 inhibitor use since market release: nonselective diffusion of a selectively cost-effective innovation. Arch Intern Med. 2005;165:171–7. doi: 10.1001/archinte.165.2.171. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.