Abstract
In comparative proteomic studies, it is important to know the variability associated with sample preparation. In this study we report the strategy of using SILAC (stable isotope labeling by amino acids in cell culture) to evaluate the effect of the variation in sample preparation for quantitative proteomics. Variability can be measured when equal amounts of light and heavy SILAC samples undergo the same sample preparation procedures in parallel, and the two samples are mixed for relative protein quantitation by mass spectrometry. The high quantitative accuracy of SILAC allows for characterization of small variations. First, the reproducibility of immunoprecipitation (IP) and in-gel digestion was evaluated, and the impact of replicate number on quantitative accuracy was characterized. Second, we evaluated the overall variation in a comparative workflow involving three sequential sample preparation steps: IP, SDS-PAGE fractionation, and in-gel digestion. The evaluation of individual sample preparation steps was very valuable for experimental design: the optimal number of replicates for each step could be readily determined and the overall variation of the workflow could be predicted from the variation of the individual steps involved. By using informed experimental design, we demonstrated that the error associated with multiple steps of sample preparation in a comparative experiment can be limited to a reasonably low level.
Keywords: quantitation, variation, SILAC, mass spectrometry, proteomics, sample preparation
Introduction
In quantitative proteomics studies, it is essential to understand the effect of the experimental variation. Errors in quantitation are introduced during the various sample preparation steps and in the measurement of analyte signals. While the error from the later source is usually determined by the instrument used and is often relatively consistent, the error caused by sample preparation is highly variable and strongly affected by the experimental design. Depending on the quantitative methods used, sample preparation can be the major source of error in quantitation.
There are many different quantitative strategies being used in proteomics, and in different strategies sample preparation can have different impacts on quantitative accuracy. For example, for approaches based on stable isotope labeling, the “light” and “heavy” samples can be mixed after differential isotope labeling so that subsequent sample handling will introduce minimal error into quantitation1, 2. In contrast, any sample preparation step done in parallel before sample mixing can introduce considerable error. Stable isotopes can be incorporated into samples at different stages of the workflow. For metabolic stable isotope labeling1, 3, 4, proteins are labeled during cell culture, and heavy and light lysates can be mixed before further sample manipulation. Therefore it allows for extensive sample preparation without compromising quantitation2. In contrast, for chemical protein labeling and peptide labeling methods such as ICAT5 and iTRAQ6, the light and heavy samples can only be mixed at later stages of the workflow, which limits the use of some sample preparation techniques before labeling. As an extreme scenario, in label-free methods samples are never mixed and all sample preparation steps are performed in parallel7–12. Each of these different quantitative approaches has its own strengths and weaknesses and plays a complementary role in proteomics. For example, although metabolic labeling is the best method in terms of quantitative accuracy, it is generally not applicable to samples from animals. In that case, chemical labeling or even label-free approaches become the methods of choice and the variation associated with parallel sample preparation becomes a potential problem. When many and/or complicated sample preparation steps are carried out in parallel, the error introduced can be enormous. Therefore it is important to evaluate this type of error so that the overall variation in an experiment can be estimated.
In addition, evaluation of the error in sample preparation will allow informed experimental design to achieve optimal results for both protein identification and quantitation. As discussed above, chemical labeling and label-free approaches limit the use of sample preparation due to the associated errors. However, sample preparation techniques such as purification, enrichment and fractionation can effectively improve protein identification by reducing sample complexity and increasing sample concentration. In many studies, isolation of sub-proteomes is indispensable (for example, to identify interacting and/or posttranslationally modified proteins by immunoprecipitation). Therefore it is desirable to design the experimental workflow in a way that makes the optimal compromise between protein identification and quantitation. By evaluating the reproducibility of different sample preparation steps, one can make informed decisions about what procedures can be used for a given expectation of quantitative accuracy. Moreover, because random error associated with sample preparation can be reduced by employing multiple replicates, the optimal number of replicates for each sample preparation step can be determined based on error evaluation to achieve the most efficient experimental design.
Previous studies have used two-dimensional electrophoresis (2DE)13, 14 and MALDI-TOF MS profiling for evaluation of sample preparation reproducibility15–17. As a quantitative method, 2DE usually has relatively large variation between runs, which makes it difficult to evaluate small variations in sample preparation. The MS profiling approach by MALDI-TOF can obtain fairly good quantitative accuracy with careful tuning and calibration of the instrument15. However analysis of highly complex samples is often limited by the peak capacity of MALDI-TOF. In this paper we chose to use stable isotope labeling with amino acids in cell culture (SILAC)1 combined with LC-MS/MS as the tool to evaluate the reproducibility of sample preparation because it is the most accurate method for large scale relative protein quantitation18. In addition, the use of LC-MS/MS allows for high throughput analysis of large numbers of proteins for error evaluation. Recently we have characterized the reproducibility of SDS-PAGE protein fractionation and its implication on comparative quantitation.19 In this study the reproducibility of two commonly used sample preparation techniques in comparative proteomics were investigated: immunoprecipitation (IP) and in-gel digestion. We then demonstrated how the error evaluation can help in designing a comparative experiment involving multi-step sample preparation.
Materials and Methods
Cell culture and metabolic labeling
Two populations of NG108 cells (mouse neuroblastoma and rat glioma hybrid) were maintained in Lys- and Arg-depleted Dulbecco’s modified Eagle’s medium (Specialty Media) supplemented with 10% dialyzed fetal bovine serum (Invitrogen), hypoxanthineaminopterin-thymidine (Sigma), 100 units/ml penicillin/streptomycin (Invitrogen) and either normal or 13C6 Lys and 13C6 Arg (Cambridge Isotope Labs) respectively. Cells were grown for at least six divisions to allow full incorporation of labeling amino acids.
Sodium pervanadate treatment
After metabolic labeling, the cells were incubated with 1mM pervanadate at 37 °C for 45 min. The sodium pervanadate solution was prepared by mixing 100 mM of Na3VO4 with an equal amount of 100 mM H2O2. The 50 mM sodium pervanadate solution was used within 5 min to minimize decomposition of the vanadate-hydrogen peroxide complex. The cells were lysed in buffer containing 1% Triton X-100, 150 mM NaCl, 20 mM Tris, pH 8, 0.2 mM EDTA, 2 mM Na3VO4, 2 mM NaF, and protease inhibitors (Complete tablet; Roche Applied Science). Lysates were clarified by centrifugation at 14,000 × g for 20 min.
IP
For each IP experiment, 0.5 ml clarified lysate was incubated with 15 μl agarose-conjugated anti-phosphotyrosine antibody PY-99 (Santa Cruz Biotechnology, Santa Cruz, CA) at 4 °C for 3 h. After incubation, the beads were washed four times with lysis buffer. Precipitated proteins were then eluted twice with 20 μl of a buffer containing 0.2% TFA and 0.5% SDS. The eluate was neutralized using 1M NH4HCO3.
Gel electrophoresis and staining
Samples were separated on 8.6 × 6.8 cm precast 7.5% Tris-HCl gels (Bio-Rad). In some cases, a DNA ladder (1kb plus, Invitrogen) was mixed with protein samples before sample loading. For DNA staining with indoine blue (IB) (Sigma), gels were fixed in 7% acetic acid/40% ethanol for 20 min before they were stained with 0.025% IB in 7% acetic acid/40% ethanol for 25 min. Finally gels were washed with 7% acetic acid/40% ethanol for 5 min. For CBB staining, gels were stained with 0.1% CBB-R250 in 7% acetic acid/40% ethanol for 30 min and destained with 7% acetic acid/40% ethanol until background was clear. While IB is considered to be relatively safe (Sigma-Aldrich, Materials Safety Data Sheets), its toxic effects have not been studied as thoroughly as fluorescent dyes, such as ethidium bromide. Because of its ability to associate strongly with DNA, IB may have a potential for mutagenesis. Standard precautions should be exercised when using IB.
In-gel digestion
Using a modified protocol of Shevchenko et al.20, excised gel bands were cut into small pieces and washed in 25 mM NH4HCO3, 50% acetonitrile; dehydrated with acetonitrile; and dried. Then the gel pieces were rehydrated with 12.5 ng/μl trypsin solution (in 25 mM NH4HCO3) and incubated overnight at 37 °C. Peptides were extracted twice with 5% formic acid/50% acetonitrile followed by a final extraction with acetonitrile. Samples were dried with vacuum centrifugation before further preparation or analysis.
LC-MS/MS
For all LC-MS/MS analysis, an LTQ-Orbitrap hybrid mass spectrometer (Thermo Fisher Scientific) equipped with a nano-ESI source (Jamie Hill Instrument Services) was used. A Nano- Acquity UPLC system (Waters) equipped with a 100-μm × 15-cm reverse phase column (Symmetry C18, Waters) was coupled to the ion trap instrument via a 10-μm-inner diameter PicoTipTM emitter (New Objective). Samples were loaded onto a trap column (180-μm × 2-cm Symmetry C18, Waters) with 2% acetonitrile in 0.1% formic acid for 4 min at 5 μl/min. After sample loading, the flow rate was reduced to 0.4 μl/min and directed through the analytical column, and peptides were eluted by a gradient of 6–40% acetonitrile in 0.1% formic acid over 120 min. Mass spectra were acquired in data-dependent mode with one 60,000 resolution MS survey scan by the Orbitrap and up to five concurrent MS/MS scans in the LTQ for the most intense five peaks selected from each survey scan (or more precisely, the MS/MS precursor peaks are selected from a preliminary 15,000 resolution spectrum taken at the beginning of each 60,000 resolution survey scan). Automatic gain control was set to 500,000 for Orbitrap survey scans and 10,000 for LTQ MS/MS scans. Survey scans were acquired in profile mode and MS/MS scans were acquired in centroid mode. Mascot generic format files were generated from the raw data using DTASuperCharge (version 1.01) and Bioworks (version 3.2, Thermo Fisher Scientific) for database searching.
Database searching
Mascot software (version 2.1.0, Matrix Science, London, UK) was used for database searching. An IPI database containing mouse and rat protein sequences (downloaded November 17, 2006) was used. Peptide mass tolerance was 10 ppm, fragment mass tolerance was 0.8 Da, trypsin specificity was applied with a maximum of one missed cleavage, and variable modifications were 13C6 Lys and 13C6 Arg. To control the false positive rate for protein identification, a decoy database was created by reversing the protein sequences of the original database. Based on the decoy database searching, three filters for protein identification were applied: 1) Peptide score threshold was 20. 2) Protein score threshold was 60. 3) Each protein was identified based on at least two peptides. After applying these filters, no false positive protein hits were found in the reversed database search.
Protein quantitation
SILAC ratios were determined using the open source software MSQuant (version 1.4.0a16) developed by Matthias Mann, Peter Mortensen and colleagues at the University of Southern Denmark. Peptide ratios from automated MSQuant analysis were subjected to manual inspection.
Results and Discussion
The strategy for error evaluation using SILAC
A schematic view of the strategy we used to evaluate sample preparation reproducibility is shown in Figure 1. The light and heavy SILAC samples undergo the same sample preparation procedures in parallel. Then the two samples are mixed for subsequent manipulation and the combined sample is analyzed using LC-MS/MS. Then proteins are quantified and statistical analysis is performed to evaluate the variation based on protein ratios. In this design, the light and heavy samples are mixed immediately after the procedure of interest to avoid introduction of error from subsequent manipulation so that it allows focusing on one specific procedure at a time. Alternatively, the light and heavy samples can be mixed after a sequence of procedures to evaluate the variation in the entire sample preparation workflow.
Figure 1.
Strategy to evaluate reproducibility of sample preparation using SILAC.
The error associated with sample preparation is usually random in nature and can be reduced by increasing the number of replicates (N). Therefore for each sample preparation procedure studied, we tried different numbers of replicates (N=1, 3, and 6) to investigate their influence on quantitation and to determine the optimal number of replicates for each step in sample preparation.
Evaluation of the reproducibility of IP
IP is a widely used technique for highly efficient enrichment of target proteins. In comparative proteomics numerous studies have used IP to pull down protein complexes with the goal of identifying components of functional protein complexes or interacting partners of target proteins. However, the reproducibility of IP has rarely been quantified.
We chose to investigate the reproducibility of anti-phosphotyrosine (pY) IP. As a tool for global isolation of tyrosine phosphorylated proteins, pY IP has been extremely useful in comparative studies for signaling pathways, especially those involving receptor tyrosine kinases (RTKs) 21–25. Because pY IP can isolate a large number of proteins from cell lysates, it is possible to identify a large number of proteins, which facilitates statistical analysis. To further improve the number of identified proteins, cells were treated with sodium pervanadate before lysis. Sodium pervanadate is a protein phosphatase inhibitor that is known to increase the level of tyrosine phosphorylation within the cell 26. Based on anti-pY Western blotting analysis on lysates from sodium pervanadate treated and control NG108 cells, a dramatic increase in tyrosine phosphorylation was observed after sodium pervanadate treatment of the cells (data not shown).
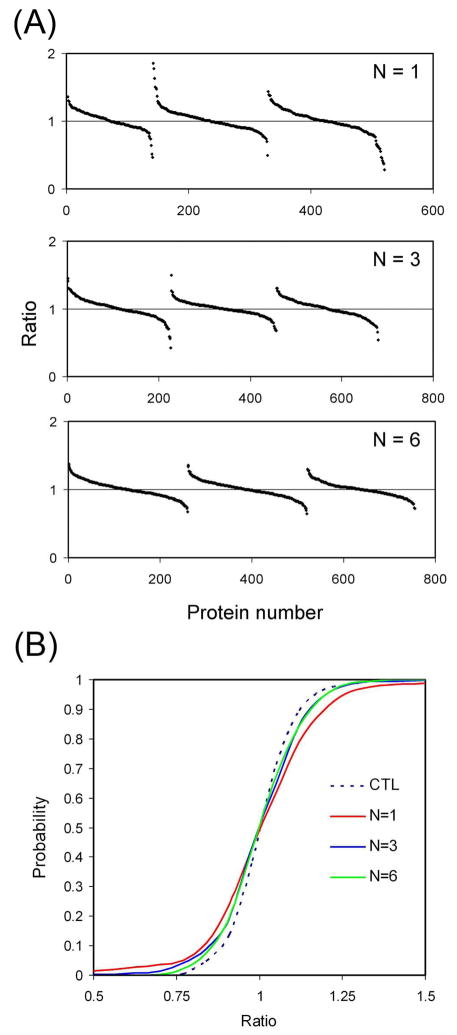
In this experiment, both the isotope labeled and unlabeled cells were treated with sodium pervanadate before lysis, and all labeled lysates were pooled, and all unlabeled lysates were pooled before IP. After the IP, precipitated proteins were eluted and the light and heavy eluates were combined, separated by SDS-PAGE into three fractions, digested in-gel, and analyzed by LC-MS/MS. Different number of replicate IPs (N = 1, 3 and 6) were carried out. For N=3 or N=6, replicate IPs were pooled before SDS-PAGE. The same amount of IPed proteins were loaded onto SDS-PAGE (i.e, one third and one sixth of the IP from the N=3 and N=6 pools were used, respectively). For each replicate number, three independent IP experiments including SDS-PAGE fractionation, in-gel digestion and MS analysis were performed. All identified proteins were quantified by taking the average ratios of their peptides. The peptide ratios were calculated from the ratios of the peak intensities of the heavy and light peptides (Figure 2A and Supplementary Table, which also includes the protein identifications, mascot scores, number of peptides identified per protein, and total ion intensity for all peptides from both heavy and light versions of each protein). It can be observed from Figure 2A that while most proteins showed ratios close to 1, some proteins from the N=1 experiment had considerably higher or lower ratios. It can also be observed that with increasing replicates of IP, protein ratio distributions become more compact, suggesting that the error in quantitation can be reduced by increasing the number of replicates.
Figure 2.
Evaluation of the variation in IP. (A) SILAC ratios of proteins with different numbers of replicates for IP. Proteins from the pull downs were analyzed by LC-MS/MS and quantified. The sorted protein ratios (heavy/light) were plotted for each experiment. IPs were performed one, three, or six times. For each number of replicates (N), three independent experiments were performed, which are represented by the three curves in each panel. (B) Cumulative probability curves for protein quantitation after IP. Quantified proteins from each replicate group (N=1, 3 and 6) were used plot the curves. For the control sample, a 1:1 mixture of heavy and light lysates was used for pY IP.
Cumulative probability plots were generated based on the SILAC ratios to illustrate the quantitative accuracy of IP (Figure 2B). To establish a control for quantitation, the SILAC lysates were mixed (1:1 ratio) before pY IP. Then the IPed proteins were fractionated by SDS-PAGE, digested in-gel, and analyzed by LC-MS/MS. As shown in Figure 2B, all curves from different replicate groups displayed some degree of deviation from the control curve. This reflects the variation caused by performing IP experiments in parallel. When N increased, the curves became closer to the control, indicating increased N can decrease the variability in IP. Note that the curves corresponding to N=3 and 6 were almost identical and both were very close to the control curve, suggesting 3 replicates of IP were sufficient to achieve optimal quantitative accuracy.
Evaluation of the reproducibility of in-gel digestion
Enzymatic protein digestion is probably the most widely used sample preparation technique in proteomic studies. Compared to in-solution digestion, in-gel digestion can be more complicated as it usually involves dicing of gel slices, extensive washing/destaining, and peptide extraction after digestion. These multiple steps may increase the variability and render in-gel digestion less reproducible than in-solution digestion.
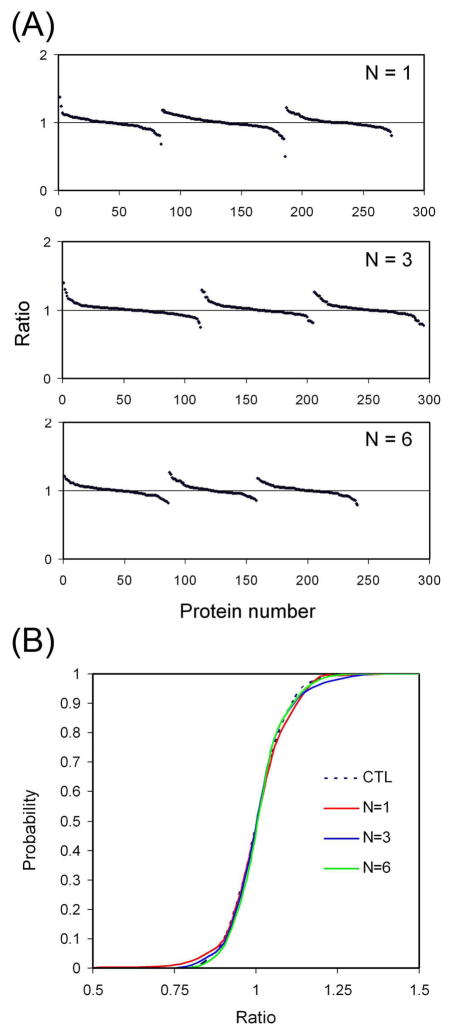
As we began to use SILAC to study the reproducibility of in-gel digestion, a major difficulty in this experiment was to obtain two identical gel pieces that contain exactly the same amount of labeled/unlabeled protein because SDS-PAGE separation of protein and gel cutting introduce additional error into the system. To circumvent this problem, two SDS poly-acrylamide gels were made using light and heavy SILAC lysates respectively. The gels (7.5%) were casted by mixing 30% acrylamide/0.8% bisacrylamide with the lysate (protein concentration: about 0.5 mg/ml) at a 1:3 ratio (v:v) before the addition of ammonium persulfate and TEMED. Gel disks of equal sizes were excised from the gel by pressing the open end of a 1 ml pipette tip (I.D. 7.5 mm, Thermo Fisher Scientific) against the gel. After the digestion, the light and heavy peptide extracts were combined for LC-MS/MS analysis. In-gel digestions were performed one, three, or six times (N = 1, 3 and 6). For N=3 or N=6, replicate digests were pooled before LC-MS/MS analysis. The same amount of tryptic peptides were analyzed in each run (i.e, one third and one sixth of the peptides from the N=3 and N=6 pools were analyzed, respectively). For each replicate number, three independent experiments were performed. The SILAC ratios of the identified proteins are shown in Figure 3A and Supplementary Table.
Figure 3.
Evaluation of the variation in in-gel digestion. (A) SILAC ratios of proteins with different numbers of replicates for in-gel digestion. Protein digests were analyzed by LC-MS/MS and quantified. The sorted protein ratios (heavy/light) were plotted for each experiment. Digestions were performed one, three, or six times. For each number of replicates (N), three independent experiments were performed, which are represented by the three curves in each panel. (B) Cumulative probability curves for protein quantitation after in-gel digestion. Quantified proteins from each replicate group (N=1, 3 and 6) were used to plot the curves. For the control sample, a 1:1 mixture of heavy and light lysates was used.
It can be observed from Figure 3A that all the protein ratios were fairly close to 1. No significant difference can be observed among the results from different replicate groups. Cumulative probability plots were generated to illustrate the quantitative accuracy of in-gel digestion (Figure 3B). To establish a control for quantitation, the SILAC samples labeled with the light and heavy isotopes, respectively, were mixed (1:1 ratio), fractionated by SDS-PAGE and analyzed by LC-MS/MS. The identified proteins were quantified and used as the control. As shown in Figure 3B, all curves from different replicate groups are very close to the control curve. This suggests that our in-gel digestions were highly reproducible. One replicate was sufficient to achieve good quantitative accuracy and increasing replicate number did not significantly improve the quantitative accuracy.
Evaluation of the variation in a multi-step workflow
In this study we chose to investigate a workflow involving three sequential sample preparation steps: pY IP, SDS-PAGE fractionation, and in-gel digestion. In the first step of this workflow, cell lysates from two different conditions are used for pY IP to isolate tyrosine phosphorylated proteins. The IPed proteins are then fractionated by SDS-PAGE and digested in-gel for LC-MS/MS analysis. This workflow has been commonly employed in the study of RTK signaling pathways using SILAC. 21–25 For quantitative experiments, it is generally believed that this workflow can only be employed with SILAC and not other quantitative approaches such as chemical labeling or label-free methods due to concerns about errors associated with the multiple sample preparation steps. In this study, we characterized the errors in relative quantitation inherent to the individual steps to help in experimental design. First we designed the experiment based on error evaluation of each step involved (IP, SDS-PAGE fractionation and in-gel digestion). The optimal replicate number for each step was chosen and the overall quantitative accuracy was predicted. Then the predictions were verified by performing the actual experiment using SILAC.
(1) Experimental design
For a workflow with multiple steps of sample preparation, evaluation of error in individual steps allows prediction of the overall quantitative accuracy. The workflow we studied consists of four major error contributing steps: IP, protein fractionation by SDS-PAGE, in-gel digestion and MSQuant quantification. The variability in effects of pervanadate treatment between the labeled and unlabeled cells was very small as indicated by the fact that the cumulative probability curve of the IP control (Figure 2B) is almost identical to that of the in-gel digestion control (Figure 3B). Therefore pervanadate treatment was not considered in error estimations for the workflow. Because each of the error contributing steps is independent and the error associated with these steps is random, according to the principle of error propagation, the relative standard deviation (RSD) of the workflow is:
| (Equation 1) |
In Equation 1, S is the RSD of the entire workflow; SI, SF, SD, and SQ are the RSDs of IP, fractionation, digestion, and SILAC ratio measurement by mass spectrometry/MSQuant. For calculation of SQ, the protein ratios from the control experiment for in-gel digestion were used (Supporting Information Excel file). For calculation of SI, SF and SD, the following equation was used:
| (Equation 2) |
In Equation 2, Sx stands for SI, SF or SD; Sx_EXP stands for the measured RSD based on protein ratios from the experiment for each sample preparation step; and Sx_CTL stands for the measured RSD for the corresponding control experiment. Based on the data we obtained in this study for IP and in-gel digestion, and the previous study on the variation in SDS-PAGE fractionation,19 the deviation for each step can be calculated and the overall deviation can be easily predicted. Furthermore, the deviations can be calculated for different replicate numbers (N) for each step so that the optimal N value can be determined.
Table 1 shows the calculated RSDs of each sample preparation step and the predicted overall RSDs of the entire workflow with different N values. The RSD for SILAC quantitation (SQ) was calculated to be 0.080. According to the prediction, the major sources of error in the workflow would be IP and SDS-PAGE fractionation. For testing, we chose to use N=3 for IP, N=4 for SDS-PAGE fractionation and N=1 for in-gel digestion. Under these conditions, the predicted overall RSD was 0.139 (the RSD of fractionation when N=4 was calculated to be 0.053 based on the RSDs of N=1, 3 and 6 using non-linear interpolation).
Table 1.
Prediction of relative standard deviation for a multiple-step workflow based on measured errors contributed by each step.
| N | SI | SF | SD | SQ | S |
|---|---|---|---|---|---|
| 1 | 0.152 | 0.197 | 0.046 | 0.080 | 0.265 |
| 3 | 0.089 | 0.067 | 0.037 | 0.080 | 0.142 |
| 6 | 0.072 | 0.037 | 0.000 | 0.080 | 0.114 |
N: replicate number; SI, SF, SD, and SQ: the RSDs contributed by IP, fractionation, digestion, and SILAC ratio measurement by mass spectrometry/MSQuant respectively; S: the predicted RSD for the entire workflow using N replicates of IP, fractionation and digestion.
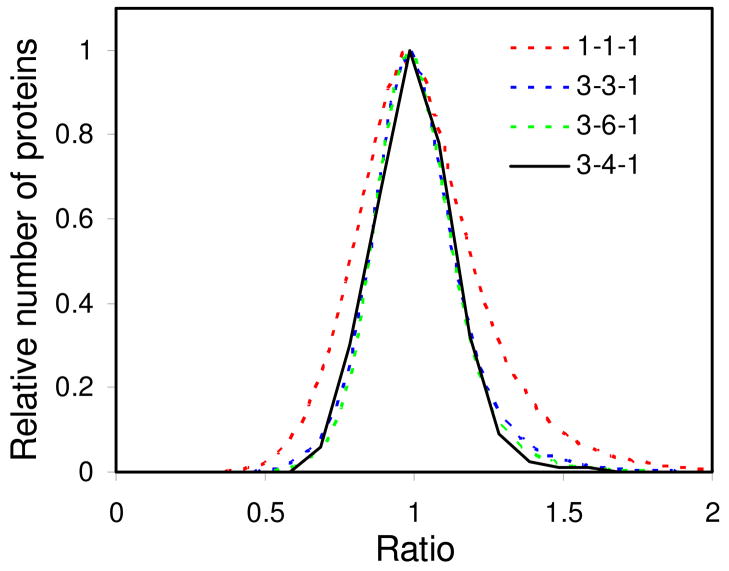
In addition to RSD prediction, the labeled/unlabeled protein ratio distribution for the workflow was simulated based on the measured variation from each sample preparation step. To do this, it was assumed that the contribution to the variation in peak intensity from each experimental step was distributed as the sum of two Gaussian distributions. Distributions of ratios were generated by taking the ratios of 400,000 pairs of random intensity values distributed according to the sum of the two Gaussian distributions. The widths and relative heights of the two Gaussian distributions were determined by fitting the simulated and measured ratio distributions. First, the intensity distribution was determined for the control by fitting the control ratio distribution (data not shown). Second, the intensity distributions of for the IP, SDS-PAGE, and digestion steps were determined (data not shown). Third, the ratio distribution was simulated for the entire experiment with different numbers of replicates (Figure 4). This represents a more comprehensive prediction of the outcome of the quantitative experiment.
Figure 4.
Simulation of protein ratio distribution for a workflow involving multiple sample preparation steps (IP, fractionation and digestion) based on error evaluation of individual steps. The predicted distributions using different replicate numbers for each step are shown with dotted lines. The legend indicates the replicate numbers for IP, fractionation and digestion respectively. The measured distribution (using 3 replicates for IP, 4 replicates for fractionation and 1 replicate for digestion) is shown by a solid line.
(2) Verification by SILAC
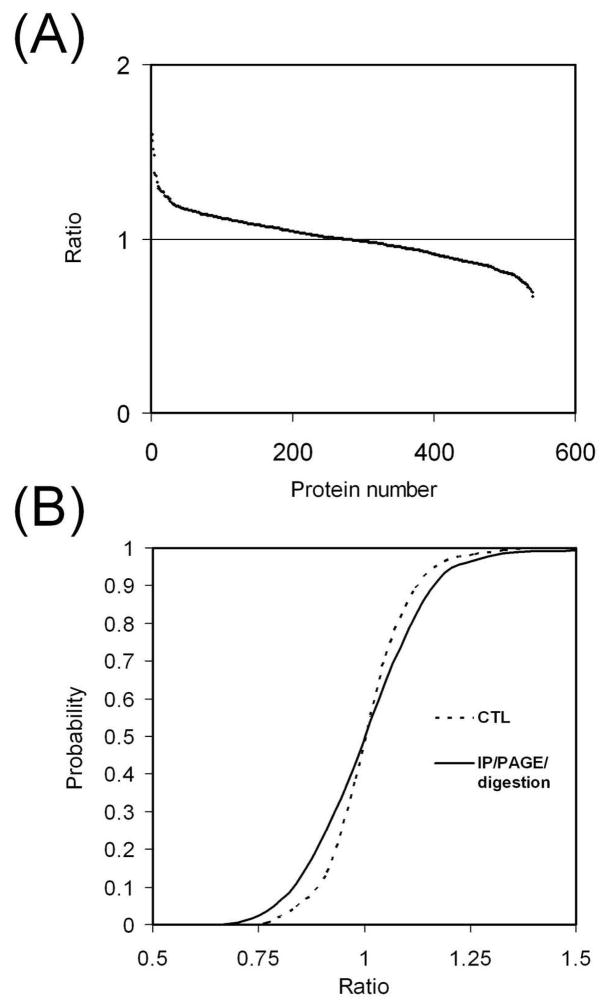
To verify the error prediction for the workflow, we carried out a SILAC experiment to quantify the overall variation in sample preparation. In this experiment, the heavy isotope labeled and unlabeled cells were treated with sodium pervanadate before lysis. Each cell lysate was divided into three aliquots for pY IP. The IPed proteins were eluted and the three eluates were pooled, then divided into four aliquots, mixed with a DNA ladder and separated by SDS-PAGE.19 After electrophoresis each gel lane was cut into eight fractions guided by the DNA markers. For each MW fraction, the four gel slices were pooled for in-gel digestion. After digestion, the light and heavy digests from each MW fraction were combined for LC-MS/MS analysis.
The addition of the DNA ladder produced nicely distributed markers to allow easy but precise gel cutting for fractionation (data not shown)19. The gel cutting would have been much more difficult without the DNA markers because the amount of protein was very small and unable to produce enough number of clear bands to guide gel cutting (data not shown). The identified proteins were quantified and their SILAC ratios are shown in Figure 5A and Supporting Information Excel file. It can be observed from Figure 5A that the ratios were fairly close to 1, with the maximum value of 1.60 and minimum value of 0.67. The RSD of protein ratios was 0.136, very close to the predicted value 0.139.. The measured distribution of the protein ratios (Figure 4) is very close to the simulated distribution curves using N=3 for IP, N= 3 or 6 for fractionation, and N=1 for digestion, indicating our error estimation was accurate. Taken together these results suggest the variation in a multi-step sample preparation can be readily predicted by evaluation of individual steps involved, which would greatly facilitate experimental design for comparative analysis.
Figure 5.
Quantitation of proteins after multi-step sample preparation. The light and heavy samples underwent pY IP, SDS-PAGE fractionation and in-gel digestion separately. The light and heavy digests were combined for LC-MS/MS analysis. For each protein, the ratio was calculated by averaging ratios of its identified peptides. (A) SILAC ratios of all identified proteins. (B) Cumulative probability curve. The control curve for the IP experiment (in Figure 2B) was used as the control for comparison.
The cumulative probability plot was generated (Figure 5B) for the SILAC quantitation. The control curve from the IP experiment (Figure 2B) was used as the control. Figure 5 suggests that although the parallel sample handling does cause variation, with properly experimental design, this variation can be controlled at a low level. After characterization of variation in sample preparation, the probability that the error is greater than a specific value can be easily read out from the cumulative curves (Table 2, based on curves from Figure 2B, 3B and 5B). This information is useful in predicting the cutoff to define significant changes in a comparative analysis and the confidence of analysis.
Table 2.
Error expectations of different sample preparation procedures with different replicate numbers
| Procedure | N | e<25% | e<50% |
|---|---|---|---|
| IP | control | 0.964 | 1.000 |
| 1 | 0.878 | 0.958 | |
| 3 | 0.919 | 0.990 | |
| 6 | 0.938 | 0.998 | |
|
| |||
| in-gel digestion | control | 0.991 | 1.000 |
| 1 | 0.971 | 0.995 | |
| 3 | 0.970 | 1.000 | |
| 6 | 0.992 | 1.000 | |
|
| |||
| IP/fractionation/digestion | 3/4/1 | 0.901 | 0.992 |
N: replicate number; Values in the table represent the probability that the measured ratio error is less than 25% and 50% of the actual ratio.
Another implication of this result is that instead of SILAC, alternative quantitative methods can be used such as chemical labeling, either at the protein or peptide level, or even label-free approaches for this workflow, with predictable compromises in quantitation accuracy. For chemical protein labeling and peptide labeling, the expected quantitative results would follow the cumulative probability curves in Figure 2B and Figure 5B respectively, assuming the labeling techniques used have the same quantitative accuracy for LC-MS analysis as SILAC.
Conclusions
We have demonstrated that SILAC is a powerful tool for evaluating the variation in sample preparation for quantitative proteomics. The degree of error associated with each sample handing procedure and also the optimal number of replicates can be readily determined. This information proved to be valuable for design of comparative experiments. It allows us to evaluate the feasibility of the experiment through error prediction and design the optimal workflow by determining what procedures can be used and how many replicates should be employed for each procedure. We have shown that by applying careful experimental design, fairly complicated sample preparation can be carried out in parallel with only slightly compromised quantitation. This will facilitate our comparative experiments both by broadening the choice of quantitative approaches and by allowing more sophisticated sample preparation.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grants P30 NS050276, S10 RR 017990-01 and NCI Core Grant 2P30 CA 016087 (to T. A. N.).
Abbreviations
- IP
immunoprecipitation
- pY
phosphotyrosine
- 2DE
two-dimensional electrophoresis
- IB
indoine blue
- SILAC
stable isotope labeling with amino acids in cell culture
- RTK
receptor tyrosine kinase
- RSD
relative standard deviation
Footnotes
Supporting Information Available: We provide a detailed list of SILAC ratios for all the proteins identified in all the LC-MS/MS experiments in this study, which also includes the protein identifications, Mascot scores, number of peptides identified per protein, and total ion intensity for all peptides from both heavy and light versions of each protein in an Excel file. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1 (5):376–86. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 2.Ong SE, Mann M. Mass spectrometry-based proteomics turns quantitative. Nat Chem Biol. 2005;1 (5):252–62. doi: 10.1038/nchembio736. [DOI] [PubMed] [Google Scholar]
- 3.Oda Y, Huang K, Cross FR, Cowburn D, Chait BT. Accurate quantitation of protein expression and site-specific phosphorylation. Proc Natl Acad Sci U S A. 1999;96 (12):6591–6. doi: 10.1073/pnas.96.12.6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhu H, Pan S, Gu S, Bradbury EM, Chen X. Amino acid residue specific stable isotope labeling for quantitative proteomics. Rapid Commun Mass Spectrom. 2002;16 (22):2115–23. doi: 10.1002/rcm.831. [DOI] [PubMed] [Google Scholar]
- 5.Gygi SP, Rist B, Gerber SA, Turecek F, Gelb MH, Aebersold R. Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat Biotechnol. 1999;17 (10):994–9. doi: 10.1038/13690. [DOI] [PubMed] [Google Scholar]
- 6.Ross PL, Huang YN, Marchese JN, Williamson B, Parker K, Hattan S, Khainovski N, Pillai S, Dey S, Daniels S, Purkayastha S, Juhasz P, Martin S, Bartlet-Jones M, He F, Jacobson A, Pappin DJ. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol Cell Proteomics. 2004;3 (12):1154–69. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- 7.Higgs RE, Knierman MD, Gelfanova V, Butler JP, Hale JE. Comprehensive label-free method for the relative quantification of proteins from biological samples. J Proteome Res. 2005;4 (4):1442–50. doi: 10.1021/pr050109b. [DOI] [PubMed] [Google Scholar]
- 8.Old WM, Meyer-Arendt K, Aveline-Wolf L, Pierce KG, Mendoza A, Sevinsky JR, Resing KA, Ahn NG. Comparison of label-free methods for quantifying human proteins by shotgun proteomics. Mol Cell Proteomics. 2005;4 (10):1487–502. doi: 10.1074/mcp.M500084-MCP200. [DOI] [PubMed] [Google Scholar]
- 9.Ono M, Shitashige M, Honda K, Isobe T, Kuwabara H, Matsuzuki H, Hirohashi S, Yamada T. Label-free quantitative proteomics using large peptide data sets generated by nanoflow liquid chromatography and mass spectrometry. Mol Cell Proteomics. 2006;5 (7):1338–47. doi: 10.1074/mcp.T500039-MCP200. [DOI] [PubMed] [Google Scholar]
- 10.Wang G, Wu WW, Zeng W, Chou CL, Shen RF. Label-free protein quantification using LC-coupled ion trap or FT mass spectrometry: Reproducibility, linearity, and application with complex proteomes. J Proteome Res. 2006;5 (5):1214–23. doi: 10.1021/pr050406g. [DOI] [PubMed] [Google Scholar]
- 11.Wang W, Zhou H, Lin H, Roy S, Shaler TA, Hill LR, Norton S, Kumar P, Anderle M, Becker CH. Quantification of proteins and metabolites by mass spectrometry without isotopic labeling or spiked standards. Anal Chem. 2003;75 (18):4818–26. doi: 10.1021/ac026468x. [DOI] [PubMed] [Google Scholar]
- 12.Wiener MC, Sachs JR, Deyanova EG, Yates NA. Differential mass spectrometry: a label-free LC-MS method for finding significant differences in complex peptide and protein mixtures. Anal Chem. 2004;76 (20):6085–96. doi: 10.1021/ac0493875. [DOI] [PubMed] [Google Scholar]
- 13.Corzett TH, Fodor IK, Choi MW, Walsworth VL, Chromy BA, Turteltaub KW, McCutchen-Maloney SL. Statistical analysis of the experimental variation in the proteomic characterization of human plasma by two-dimensional difference gel electrophoresis. J Proteome Res. 2006;5 (10):2611–9. doi: 10.1021/pr060100p. [DOI] [PubMed] [Google Scholar]
- 14.Thongboonkerd V, Chutipongtanate S, Kanlaya R. Systematic evaluation of sample preparation methods for gel-based human urinary proteomics: quantity, quality, and variability. J Proteome Res. 2006;5 (1):183–91. doi: 10.1021/pr0502525. [DOI] [PubMed] [Google Scholar]
- 15.Villanueva J, Philip J, Chaparro CA, Li Y, Toledo-Crow R, DeNoyer L, Fleisher M, Robbins RJ, Tempst P. Correcting common errors in identifying cancer-specific serum peptide signatures. J Proteome Res. 2005;4 (4):1060–72. doi: 10.1021/pr050034b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.West-Nielsen M, Hogdall EV, Marchiori E, Hogdall CK, Schou C, Heegaard NH. Sample handling for mass spectrometric proteomic investigations of human sera. Anal Chem. 2005;77 (16):5114–23. doi: 10.1021/ac050253g. [DOI] [PubMed] [Google Scholar]
- 17.West-Norager M, Kelstrup CD, Schou C, Hogdall EV, Hogdall CK, Heegaard NH. Unravelling in vitro variables of major importance for the outcome of mass spectrometry-based serum proteomics. J Chromatogr B Analyt Technol Biomed Life Sci. 2007;847 (1):30–7. doi: 10.1016/j.jchromb.2006.09.048. [DOI] [PubMed] [Google Scholar]
- 18.Ong SE, Kratchmarova I, Mann M. Properties of 13C-substituted arginine in stable isotope labeling by amino acids in cell culture (SILAC) J Proteome Res. 2003;2 (2):173–81. doi: 10.1021/pr0255708. [DOI] [PubMed] [Google Scholar]
- 19.Zhang G, Fenyo D, Neubert TA. Use of DNA Ladders for Reproducible Protein Fractionation by Sodium Dodecyl Sulfate-Polyacrylamide Gel Electrophoresis (SDS-PAGE) for Quantitative Proteomics. J Proteome Res. 2008;7 (2):678–86. doi: 10.1021/pr700601y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem. 1996;68 (5):850–8. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 21.Blagoev B, Mann M. Quantitative proteomics to study mitogen-activated protein kinases. Methods. 2006;40 (3):243–50. doi: 10.1016/j.ymeth.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 22.Blagoev B, Ong SE, Kratchmarova I, Mann M. Temporal analysis of phosphotyrosine-dependent signaling networks by quantitative proteomics. Nat Biotechnol. 2004;22 (9):1139–45. doi: 10.1038/nbt1005. [DOI] [PubMed] [Google Scholar]
- 23.Hinsby AM, Olsen JV, Mann M. Tyrosine phosphoproteomics of fibroblast growth factor signaling: a role for insulin receptor substrate-4. J Biol Chem. 2004;279 (45):46438–47. doi: 10.1074/jbc.M404537200. [DOI] [PubMed] [Google Scholar]
- 24.Kratchmarova I, Blagoev B, Haack-Sorensen M, Kassem M, Mann M. Mechanism of divergent growth factor effects in mesenchymal stem cell differentiation. Science. 2005;308 (5727):1472–7. doi: 10.1126/science.1107627. [DOI] [PubMed] [Google Scholar]
- 25.Zhang G, Spellman DS, Skolnik EY, Neubert TA. Quantitative phosphotyrosine proteomics of EphB2 signaling by stable isotope labeling with amino acids in cell culture (SILAC) J Proteome Res. 2006;5 (3):581–8. doi: 10.1021/pr050362b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huyer G, Liu S, Kelly J, Moffat J, Payette P, Kennedy B, Tsaprailis G, Gresser MJ, Ramachandran C. Mechanism of inhibition of protein-tyrosine phosphatases by vanadate and pervanadate. J Biol Chem. 1997;272 (2):843–51. doi: 10.1074/jbc.272.2.843. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.