Abstract
Human ductal saliva contributes over a thousand unique proteins to whole saliva. The mechanism by which most of these proteins are secreted by salivary glands remains to be determined. The present study used a mass spectrometry-based, shotgun proteomics approach to explore the possibility that many of the proteins found in saliva are derived from exosomes, membrane-bound vesicles of endosomal origin within multivesicular endosomes. Using MudPIT (multidimensional protein identification technology) mass spectrometry, we catalogued 491 proteins in the exosome fraction of human parotid saliva. Many of these proteins were previously observed in ductal saliva from parotid glands (265 proteins). Furthermore, 72 of the proteins in parotid exosomes overlap with those previously identified as urinary exosome proteins, proteins which are also frequently associated with exosomes from other tissues and cell types. Gene Ontology (GO) and KEGG pathway analyses found that cytosolic proteins comprise the largest category of proteins in parotid exosomes (43%), involved in such processes as phosphatidylinositol signaling system, calcium signaling pathway, inositol metabolism, protein export, and signal transduction among others; whereas the integral plasma membrane proteins and associated/peripheral plasma membrane proteins (26%) were associated with extracellular matrix-receptor interaction, epithelial cell signaling, T-cell and B-cell receptor signaling, cytokine receptor interaction, and antigen processing and presentation among other biological functions. In addition, exosomal proteins were linked to specific diseases (e.g. neurodegenerative disorders, prion disease, cancers, type I and II diabetes). Consequently, parotid glands secrete exosomes that reflect the metabolic and functional status of the gland and may also carry informative protein markers useful in the diagnosis and treatment of systemic diseases.
Keywords: Exosomes, parotid saliva, acinar cells, MudPIT, protein markers
Introduction
Salivary glands secrete proteins with numerous functions; e.g. digestion of starches and lipids, anti-viral and anti-bacterial activities, lubrication, and tooth mineralization 1–5. These varied and numerous tasks suggest that saliva contains a large number of unique proteins. Indeed, high-throughput, proteomic analysis of saliva revealed a surprisingly diverse collection of more than a thousand different proteins 2, 6, 7. A relatively few number of abundant proteins packaged primarily in large secretory granules make up more than 90% of the total protein content (about 40 major proteins, e.g. amylase, mucin, histatin, statherin, cystatin, and the acidic, basic, and glycosylated proline-rich proteins) 3, 5. The origins for the majority of the less abundant proteins are unknown; however, the many different classes of proteins found in saliva suggest that multiple secretion pathways are involved.
The apical portions of the cells of a salivary gland acinus, also known as the secretory endpiece, contain densely packed, large secretory granules 8, 9. This secretion pathway, termed the major-regulated pathway, is primarily regulated by β-adrenergic stimulation which increases the intracellular cAMP content leading to activation of protein kinase A 8–10. Secretion of the abundant salivary proteins, and consequently most of the total protein found in saliva, is mediated by exocytosis of the granules in the major-regulated pathway. Although generally found in low abundance, approximately one third of the different proteins found in saliva are also common to blood serum 2 suggesting that the acinar secretory apparatus may be more “leaky” than previously appreciated 11, 12, and/or that salivary gland cells possess specialized receptors that facilitate transcellular movement of many proteins, as is well-known for IgA 13, 14. In addition to the major-regulated pathway, two other membrane-bound pathways have been described, the constitutive-like and the minor-regulated pathways. These latter two pathways secrete relatively small amounts of the total secretory protein compared to the major regulated pathway 8, 15. As the name implies, the constitutive-like pathway secretes continuously and does not appear to be subject to acute regulation 8, 15. In contrast, the minor regulated pathway is activated by either low-level cholinergic or β-adrenergic stimulation, considerably lower than that needed to stimulate the major regulated pathway. Together, the constitutive-like and the minor-regulated pathways are most responsible for the resting salivary secretion of proteins 16 associated with basal parasympathetic stimulation 17. The major-regulated pathway on the other hand, provides most of the salivary proteins during eating 16.
In addition to the above secretion pathways, it appears that most cell types also secrete two varieties of membrane-bound vesicles, exosomes and apoptotic blebs 18, 19. Exosomes were first described as exfoliated membrane vesicles released by different cell lines 20. Morphologic analysis showed that these exosomes have a diameter of 30–100 nm with a rounded shape 19, 21–24. Exosomes are created by the inward budding of the limiting membrane of endosomes; a process that produces intraluminal vesicles (ILVs) 23, 25–30, traps cytosolic material 25, 28, 31–34 and generates multivesicular endosomes (MVEs) 19, 23, 26, 31. Exosomes accumulate within MVEs (also known as multivesicular bodies) and are constitutively released upon fusion of the MVE membrane with the plasma membrane 18, 19, 23, 26–30, 32–35. Fusion of the MVE with the plasma membrane releases the intact membrane-bound exosomes, a distinct exocytosis mechanism. In contrast to exosomes, apoptotic blebs are heterogeneous vesicles which bud directly from the plasma membrane after apoptotic cell death 19, 36. These vesicles contain mainly nuclear and endoplasmic reticulum proteins that are not observed in exosomes 19, 35.
Exosomes are released by a variety of specialized cells including reticulocytes, B-lymphocytes, dendritic cells, T-lymphocytes, mast cells, platelets, intestinal epithelial cells, neuroglial cells, and tumor cells 18, 19, 21, 22, 24, 27, 37–40. In addition, exosomes are present in different physiological fluids such as urine 25, 28, 33, 41, blood 31, pleural effusions 21 and malignant ascites 42.
Recent advances in mass spectrometry 43 have made the field of clinical proteomics increasingly important in the identification of biomarkers for early disease diagnosis and in the development of new treatments 22, 25, 28, 41, 43. Proteomic analysis has demonstrated that certain proteins are common to all exosomes, regardless of their cell origin, although others are cell-type specific, making them potentially attractive biomarker targets 18, 19, 21–23, 25, 27, 31, 34, 35, 37, 39, 44. Human saliva is an ideal biological fluid for this purpose because its collection is non-invasive, low-cost, safe and simple; and, it contains proteins useful for disease detection 45–47. The association between salivary proteins and diseases such as periodontal disease, HIV-immunodeficiency and autoimmune diseases has been demonstrated 48–52. To date, exosome analysis of salivary gland cells and saliva has been restricted to non-neoplastic human salivary gland cell lines 18 and whole saliva 53. Therefore, the aim of the present study was an initial step to identify proteins secreted as human parotid gland exosomes in vivo in order to gain insight into their biological functions related to health and disease processes.
Materials and Methods
Chemicals were purchased from Sigma-Aldrich (St. Louis, MO), or as indicated in the text, with the following exceptions: Tris base (Promega Co., Madison, WI), ε-Amino-n-caproic acid (Calbiochem/EMD Biosciences Inc., LaJolla, CA), ethylene diamine tetraacetic acid (EDTA; J.T. Baker, Phillipsburg, NJ), leupeptin (Bachem Inc., Torrance, CA), and trypsin and lysine C (Roche Applied Science, Indianapolis, IN). The protocol for the collection of human saliva was approved by the University of Rochester Institutional Review Board.
Saliva collection
To avoid the contamination inherent to the collection of whole saliva (a combination of saliva, bacteria and their metabolic products, mucosal cell sloughing, nasal discharge, gingival crevicular fluid and food debris), ductal parotid saliva was obtained from a single, healthy, non-medicated and non-smoking Caucasian male donor (48 years old). Saliva collection was performed under standard conditions, at the University of Rochester, Center for Oral Biology after overnight fasting 2. Briefly, the subject was asked not to eat, drink, or perform any oral hygiene measures at least one hour before salivary collection. Saliva samples were collected between 8:00 a.m. and 10:00 a.m. under stimulation conditions (0.4% citric acid) on ice using a Lashley collector device 54. During salivary collection, a 1/20 volume of protease inhibitor cocktail (0.1 M Tris-HCl (pH7.4,) 0.1 M epsilon amino caproic acid, 0.05 M sodium EDTA, 25 mg/L of pepstatin A, 0.025 M benzamidine-HCl, 0.25 mg/L of leupeptin and 0.5 M phenylmethylsulfonyl fluoride) and a 1/20 volume of 10x PBS were added simultaneously to ensure the preservation of the exosomal proteins and to minimize any potential osmotic pressure differences on cells and vesicles in the hypotonic ductal saliva.
Isolation of parotid exosomes
Isolation of exosomes was accomplished following the protocol of Pisitkun et al 28 with the following minor modifications. Parotid saliva (~30 ml) was centrifuged at 300g for 10 minutes at 4 °C using an SH3000 rotor (Sorvall RC-5C centrifuge; DuPont; Newtown, CT) to remove whole cells and debris. The clear supernatant was centrifuged again at 10,000g at 4 °C for 30 minutes using an SS34 rotor (Sorvall; Newtown, CT) to remove large membrane fragments. The resultant supernatant was dispensed in 5 different SW 50.5 centrifuge tubes and centrifuged at 200,000g for 60 minutes on a Beckman Optima™ XL-100K ultracentrifuge (Beckman Coulter Inc.; Fullerton, CA) to obtain the low-density, exosome-rich pellet. Total exosomal protein was calculated at ~80 μg in the 30 ml of collected saliva (BCA Protein Assay Kit, Pierce; Rockford, IL).
Sample preparation for proteomic analysis
Protein pellets were re-suspended in digestion buffer (8 M urea, 100 mM Tris-HCl; pH 8.5). The mixture was brought to 5 mM Tris[2-carboxyethyl] phosphine (TCEP) and incubated at room temperature for 15 min. Iodoacetamide was then added to a final concentration of 10 mM and the resultant mixture was incubated at room temperature for 20 min in the dark, when Lys-C was added at 1:50 (enzyme to substrate ratio) followed by incubation at 37 °C for 4 hr. The digest was then diluted to achieve a final concentration of 2 M urea with 100 mM Tris-HCl; pH 8.5 and trypsin digestion (1:50 protease to protein ratio) was carried out at 37 °C overnight. Protein digestion was stopped by the addition of formic acid at 4% final concentration. Digested samples were stored at −80 °C without further treatment until mass spectrometry analysis.
Multidimensional chromatography coupled tandem mass spectrometry
Digested peptide mixtures were pressure-loaded onto a fused silica capillary column packed with a 3 cm 5-μm Partisphere strong cation exchanger (SCX, Whatman, Clifton, NJ) and a 3 cm 5-μm Aqua C18 material (RP, Phenomenex, Ventura, CA), with a 2 μm filtered union (UpChurch Scientific, Oak Harbor, WA) attached to the SCX end. The column was washed with buffer containing 95% water, 5% acetonitrile, and 0.1% formic acid. After desalting, a 100-μm i.d. capillary with a 5-μm pulled tip packed with 10 cm 3-μm Aqua C18 material was attached to the filter union, and the entire split-column was placed inline with an Agilent 1100 quaternary HPLC (Agilent, Palo Alto, CA) and analyzed using a modified 6-step separation described previously 55. Three buffer solutions were used: 5% acetonitrile/0.1% formic acid (buffer A); 80% acetonitrile/0.1% formic acid (buffer B), and 500 mM ammonium acetate/5% acetonitrile/0.1% formic acid (buffer C). The first step consisted of a 50 min gradient from 0 to 100% buffer B. Steps 2–6 had the following profile: 3 min of 100% buffer A, 5 min of X% buffer C, a 10 min gradient from 0 to 10% buffer B, and a 70 min gradient from 10 to 45% buffer B. The 5 min buffer C percentages (X) were 10, 30, 55, 70 and 100%, respectively, for the 2–6 step analysis. As peptides were eluted from the microcapillary column, they were electrosprayed directly into an LTQ linear ion trap mass spectrometer (Thermo Fisher Scientific, San Jose, CA) with the application of a distal 2.5 kV spray voltage. A cycle of one full-scan mass spectrum (400–1400 m/z) followed by 6 data-dependent tandem mass (MS/MS) spectra at a 35% normalized collision energy was repeated continuously throughout each step of the multidimensional separation. Application of mass spectrometer scan functions and HPLC solvent gradients was controlled by the Xcalibur data system (Thermo Fisher Scientific, San Jose, CA).
Interpretation of tandem mass spectra data sets
SEQUEST 56 was used for MS/MS database search. The validity of peptide/spectrum matches was assessed using SEQUEST-defined parameters, the cross-correlation score (XCorr) and normalized difference in cross-correlation scores (ΔCn). Distribution of XCorr and (ΔCn) values for direct and decoy database hits was obtained, and the two subsets were separated by quadratic discriminant analysis. Full separation of the direct and decoy subsets is not generally possible. Therefore, the discriminant score was set such that a false-positive rate of 5% at the protein level was determined based on the number of accepted decoy proteins. This procedure was independently done on data subsets for charge states +1, +2, and +3. DTASelect 57 was used to select and sort peptide/spectrum matches passing this set of criteria. The human protein database used was the EBI International Protein Index (IPI) protein database version 3.24 (December 1, 2006). The reverse protein sequences of the IPI database were used as the decoy database. Proteins were considered detected if they were identified by at least two peptides. All the parotid exosome proteins that were inferred by the same set of peptides (equivalent protein identifications) were clustered by using the DTASelect program 57. DTASelect assembles identified peptides into proteins and protein groups by using a parsimony principle in which the minimum set of proteins accounts for all the observed peptides.
Data centralization and analysis
A human parotid exosome database was created at The Scripps Research Institute to serve as the central repository for the parotid exosome proteome. Access online to this repository is provided for the research community at http://fields.scripps.edu/public/project/saliva_exosome/. Protein function annotation using the Gene Ontology (GO) was carried out using the GoMiner software available at: http://discover.nci.nih.gov/gominer/. To reveal which pathways are significantly represented by the identified proteins, pathway analysis was performed using KEGG (Kyoto Encyclopedia of Genes and Genomes) database available at: http://www.genome.jp/kegg/pathway.html.
Transmembrane domain prediction
The prediction of transmembrane helices of the identified parotid exosome proteins was carried out using the TMHMM 2.0 web-based program available at http://www.cbs.dtu.dk/services/TMHMM-2.0/ 58.
Comparison of the human parotid exosomes to the human salivary proteome and urinary exosomes
The database corresponding to the human parotid salivary proteome (914 proteins) was downloaded from the central repository hosted at the http://www.hspp.ucla.edu, while the urinary exosome database (304 proteins) was downloaded from the National Heart, Lung and Blood Institute http://dir.nhlbi.nih.gov/papers/lkem/exosome/ 28. This urinary exosome protein database was mapped to the IPI v.3.24 for comparison purposes with the human parotid salivary and exosome proteomes.
Immunoelectron Microscopy
Parotid saliva exosomes mixed 1:1 with 4% paraformaldehyde, were diluted with 10 mM Tris-maleate buffer, pH 7.4, applied to a Formvar-carbon coated specimen grid, and stained with 2% ammonium molybdate or 2% uranyl acetate. The preparations were examined and photographed in a Philips CM10 transmission electron microscope (TEM; Philips, Eindhoven, The Netherlands) at 80 kV. Developed negatives were scanned at 1200 pixels/inch on an Epson Perfection V750 Pro scanner (Epson, Long Beach, CA), and levels and contrast were adjusted in Adobe Photoshop CS2 (version 9.0.2). Exosome diameters were measured using the public domain software, ImageJ (version 1.38x), and a histogram constructed in Microsoft Excel 2004 (version 11.3.7).
For immunogold labeling, exosomes were applied to Formvar-carbon coated nickel grids, treated with 1% bovine serum albumin/5% normal goat serum to block non-specific binding, and incubated with anti-TSG101 diluted 1:20 in blocking solution. The grids were then rinsed with phosphate buffered saline (PBS) and incubated with 10 nm gold-labeled goat anti-mouse IgG (Amersham/GE Healthcare Life Sciences, Piscataway, NJ). After rinsing with PBS and distilled water, the grids were negatively stained with uranyl acetate and examined in the TEM.
Gel electrophoresis and Western blot
Ten μg of parotid exosome protein was heated at 70 °C for 10 minutes prior to separation in NuPAGE® Novex 4–12% Bis-Tris polyacrylamide gradient gels or in a NativePAGE™ Novex® 4–16 % Bis-Tris gel (Invitrogen, Carlsbad, CA). Protein was transferred onto polyvinylidene (PVDF) membranes (Invitrogen, Carlsbad, CA) for 1 hr using 1x NuPAGE transfer buffer. Membranes were blocked overnight at 4 °C with 5% non-fat dry milk in 25 mM Tris-HCl pH 7.5, 150 mM NaCl (TBS) and then incubated with rabbit polyclonal antibody raised against the mouse/rat AQP5 C-terminal peptide sequence (LL639, residues 243–265) 59, mouse polyclonal antibody against the exosome-specific protein AIP1/ALIX (residues 375–580) (BD Transduction Laboratories; BD Biosciences Pharmigen Biotechnology Inc.; San Diego, CA), mouse monoclonal [1D6] against the exosomal protein marker TAPA1/CD81, or mouse monoclonal [MEM-259] against the exosomal protein marker CD63 (Abcam Inc., Cambridge, MA); 1:1,000; 1:250; 1:1,000; 1:800 dilutions, respectively in 2.5% non-fat dry milk solution at 4 °C overnight. After washing with TBS containing 0.1% Tween-20 (TBST), the membranes were incubated with horseradish peroxidase-conjugated goat anti-rabbit IgG secondary antibody (Pierce) at a dilution of 1:2,500 or with goat anti-mouse IgG H&L (HRP) antibody (Bio-Rad; Hercules, CA) at a dilution of 1:3,000 in TBST/2.5% non-fat dry milk for 1 hr at room temperature. Labeled proteins were visualized using enhanced chemiluminescence (ECL detection kit, GE/Amersham Biosciences; Piscataway, NJ).
Results
Proteomic analysis of parotid exosomes
Multidimensional protein identification technology (MudPIT) was used to identify the protein composition of parotid exosomes based on its robust sensitivity for analyzing complex protein mixtures 60, 61. Database searches of the MS/MS data by SEQUEST 56 followed by DTASelect analysis 57 identified a total of 3044 peptides which matched to 875 redundant proteins present in parotid exosomes. Clustering analysis of equivalent protein identifications by DTASelect yielded a total of 491 unique parotid exosome clusters with a minimum of two peptides for each identified protein (Supplemental table #1).
Protein annotation analysis
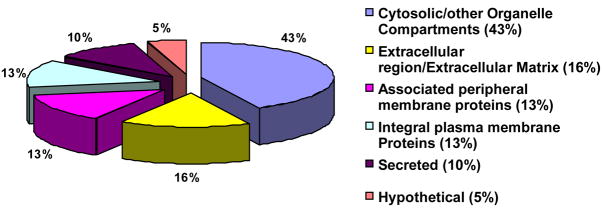
Parotid exosome proteins were annotated according to the UniProtKB/Swiss-Prot http://www.expasy.ch/sprot/sprot_details.html, European Bioinformatics Institute (EBI) http://www.ebi.ac.uk/ or UniProtKB/TrEMBL http://www.ebi.ac.uk/trembl/ knowledgebases. Results demonstrated that of the 491 proteins detected in parotid exosomes 43% were of cytosolic origin, followed by integral plasma membrane proteins and associated/peripheral plasma membrane proteins (26%; 13% in each category), extracellular matrix/extracellular region proteins (16%), secreted (10%) and hypothetical proteins (5%) [Fig #1].
Figure 1. Annotation of identified parotid exosome proteins by subcellular location.
Protein classification was obtained by using UniProtKB/Swiss-Prot, EBI and UniProtKB/TrEMBL databases. The majority of exosomal proteins were allocated to the cytosolic compartment, whereas the smallest number was identified as hypothetical.
As expected for membrane-bound compartments, parotid exosomes contained 110 integral membrane proteins (IMP) and 65 peripheral membrane proteins associated with the membrane of the multivesicular endosomes, while a large number of cytosolic proteins were engulfed in the intraluminal vesicles lumen during exosome formation 28, 34. In regards to the IMPs present in parotid exosomes, these can be further sub-divided into two groups according to their subcellular localization: a) plasma membrane proteins (58%) and b) membrane proteins expressed in organelles (42%). Integral plasma membrane proteins were annotated according to their function: pumps (2%), channel proteins (5%), solute carriers (14%), antigen presenting/defense (5%), enzyme regulator/catalytic activities (14%), calcium binding/ion transport (2%), unknown function (6%), receptor activity (22%), cell differentiation (6%), signaling pathway/cytokines (13%), and cell adhesion/cell motility (11%) [Fig #2A]. Meanwhile, analysis of the peripheral membrane proteins showed that 26% of these proteins are involved in signaling pathways, 17% are implicated in catalytic activities, 15% are engaged in assembly/self association, 14% are linked to endocytosis/exocytosis/vesicle trafficking, 9% are involved in calcium binding, 6% are associated with antigen presenting/defense, 6% are related with cell adhesion, 3% are linked to cell motility, 2% are involved in ion binding/transport and the remaining 2% are implicated in actin binding [Fig #2B].
Figure 2. Protein annotation using UniProtKB/Swiss-Prot, EBI and UniProtKB/TrEMBL databases.
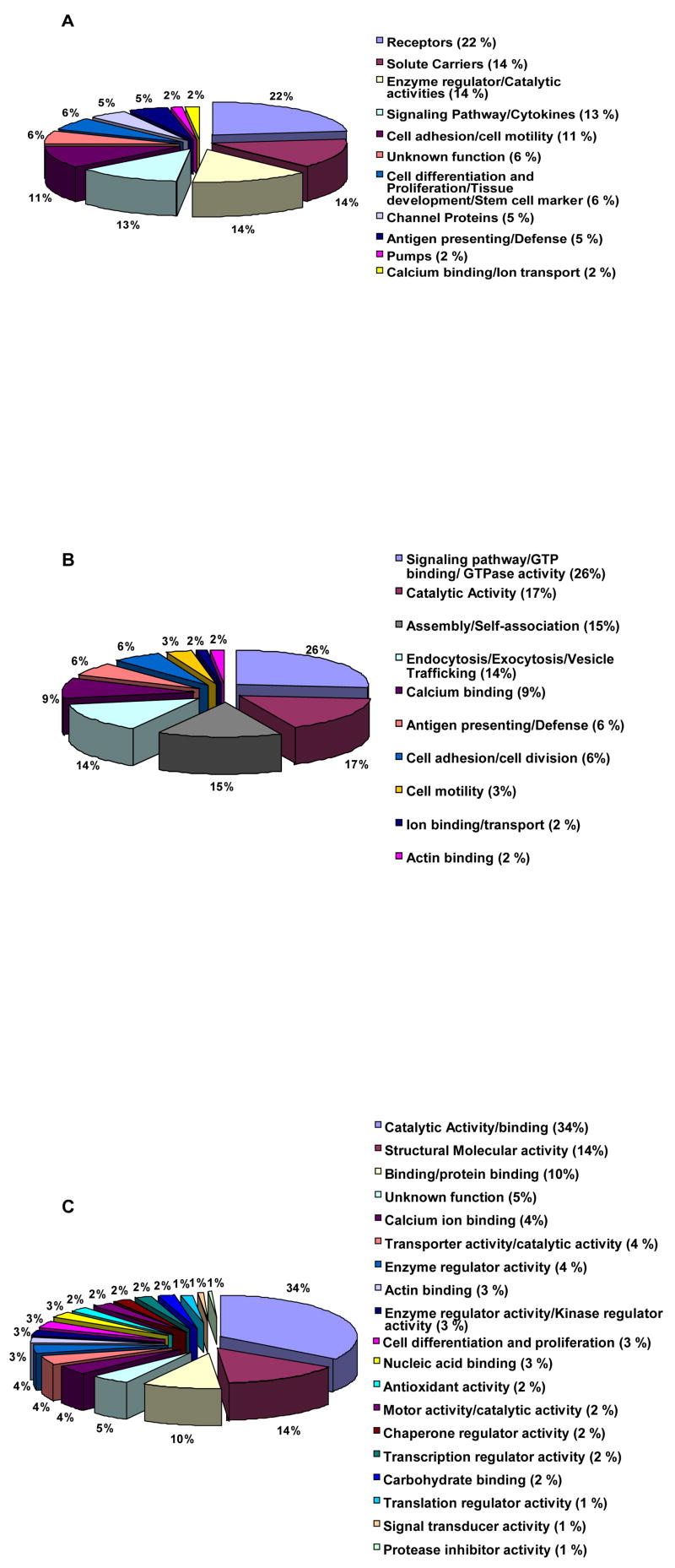
A. Annotation of identified parotid exosome integral plasma membrane proteins by function. Twenty-two % of the proteins identified were receptors and the minority (2%) was represented by pumps and/or calcium binding/ion transport proteins. B. Annotation of identified parotid exosome peripheral membrane proteins by function. The bulk of parotid exosomal proteins was associated with proteins involved in signaling pathways, while lesser amounts of proteins were linked to ion binding/transport and actin binding activities. C. Annotation of identified parotid exosome cytosolic proteins by function. Thirty-four % of the cytosolic proteins were associated with catalytic activities whilst only 0.5% with protease inhibitor activity.
The 210 cytosolic proteins found in parotid exosomes were classified as proteins with: catalytic/binding activity (34%), structural molecular activity (14%), unknown function (5%), chaperone regulator activity (2%), enzyme regulator activity (4%), binding/protein binding activity (10%), calcium ion binding activity (4%), transcription regulator activity (2%), enzyme regulator activity/kinase regulator activity (3%), antioxidant activity (2%), transporter activity/catalytic activity (4%), cell differentiation (3%), motor activity (2%), nucleic acid binding (3%), signal transducer activity (1%), carbohydrate binding (2%), translation regulator activity (1%), protease inhibitor activity (0.5%) and actin binding (3%) [Fig #2C]. Extracellular matrix/extracellular region proteins and secreted molecules (129 proteins in total) were also found to be associated with parotid exosomes. Some of these proteins have been identified by mass spectrometry in the exosomes of other tissues 21, 27, 28. Annotation of the extracellular matrix/extracellular region proteins and secreted molecules showed that they are involved in different functions such as catalytic activity (12%), structural molecule activity (13%), lipid binding (4%), immune activity (16%), cytokine activity (3%), antigen processing and presentation (2%). Complete and descriptive information regarding the annotation of parotid exosome proteins by gene, function and prediction of transmembrane helices is provided in [Supplemental Table #2].
Proteins involved in exosome biogenesis
MudPIT analysis identified numerous proteins in parotid exosomes that are thought to be involved in exosome formation and exocytosis into the external milieu. Proteins such as PDCD6IP protein (Alix), ubiquitin activating enzyme E1 (UBA1), vacuolar protein sorting-associated protein 28 (VPS28) and vacuolar protein sorting-associating protein 4A (VPS4A) are involved in the endosomal sorting complex required for transport (ESCRT). Chaperone proteins like heat shock 70 kDa protein 1 (HSPA1A) and heat shock protein 90 (HSP90B1) participate in membrane translocation, processing and transport of secreted proteins. Annexins (ANXA4), clathrin heavy chain 1 (CLTC), and Rab proteins (RaB5C) are engaged in transport and fusion. In addition, during the formation of the ILVs major components of the cytosolic compartment are trapped in parotid exosomes such as enzymes (dipeptidyl-peptidase 1, glucose-6 phosphate isomerase, pyruvate kinase 3 isoform 1), elongation factors (elongation factor 1-alpha 1, elongation factor 2), adaptors and signaling proteins (14-3-3 β, ε, γ, η, δ, ζ proteins), actins and tubulins. These proteins are common exosomal components, having been observed in all exosomes regardless of their cell origin 27, 34.
Numerous examples of cell type specific proteins have been identified 28, 31, 34, 38, 40. Data obtained from mass spectrometry analysis demonstrated the presence of unique parotid exosomal proteins involved in processes such as: a) exosome biogenesis, endocytosis, exocytosis, vesicular trafficking and fusion (RaB27B, Rab22A, TWF2, SLC9A3R1, TINAGL1, MYO1B, VTA1, S100A6), b) proteins associated with the salivary secretion mechanism (Aqp5, ATPase; 62), c) proteins related to epithelial-specific markers (cytokeratins; 18), d) antigen presenting molecules (HLA-DRA, HLA-B, HLA-A) among others. These proteins may reflect the unique characteristics of the exosomes secreted by parotid cells.
Gene Ontology (GO) analysis
In order to obtain a functional overview of parotid exosome proteins, we further analyzed the parotid exosome proteome using the GoMiner program by searching against gene ontology databases (version January, 2008). The results showed that 336 proteins mapped to specific biological processes, 375 were associated with diverse cellular components and 377 proteins had other molecular functions [Supplemental tables #3, #4, and #5 respectively]. In regards to “biological process”, proteins were classified according to different categories. The highest number mapped to proteins involved in cellular process (261 proteins), followed by proteins associated with metabolic process (171 proteins), localization (102 proteins), developmental process (101 proteins), multicellular organismal process (95 proteins), biological regulation (93 proteins), response to stimulus (88 proteins), establishment of localization (82 proteins), immune system process (40 proteins), biological adhesion (36 proteins), gene expression (18 proteins), and 46 proteins associated with other processes (e.g. reproduction, multi-organism process, locomotion, growth, maintenance of localization, pigmentation, viral reproduction and cell killing) [Fig #3A]. With regard to “cellular component”, the majority of the proteins were assigned to the cell and cell part (306 proteins in each category), while 176 proteins mapped to organelle, 120 proteins to the extracellular region, 108 proteins to organelle part, 54 proteins to macromolecular complex, 48 to extracellular region part, 24 to extracellular matrix, 21 to membrane-enclosed lumen, 10 to extracellular matrix part, and 7 to the envelope [Fig #3B]. Based on the “molecular function”, the highest distribution was associated with binding (311 proteins) and catalytic activity (151 proteins). The remaining proteins were linked to different activities such as structural molecule activity (53 proteins), molecular transducer activity (40 proteins), transporter activity (32 proteins), enzyme regulator activity (23 proteins), antioxidant activity (8 proteins), motor activity and transcription regulator activity (5 proteins each), translation regulator activity (3 proteins) and chaperone regulator activity and auxiliary transport protein activity (1 protein each) [Fig #3C].
Figure 3. Gene Ontology annotation of parotid exosome proteins.
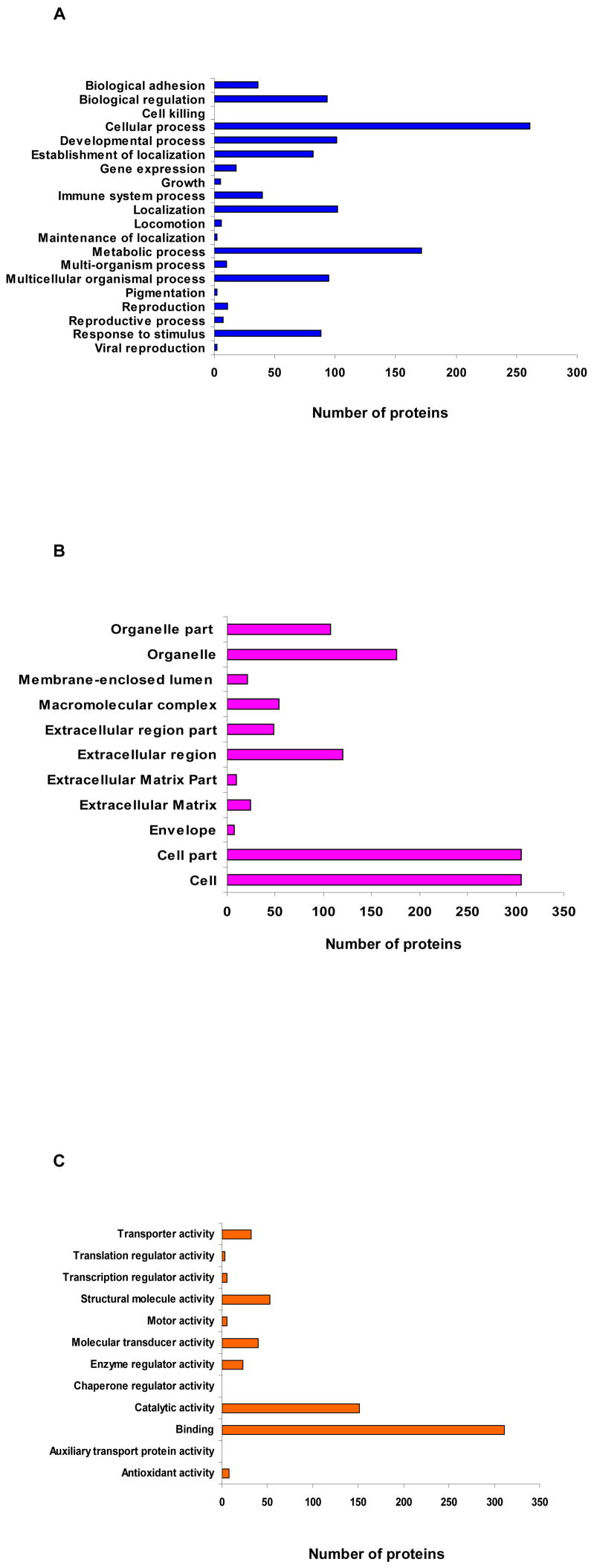
A. GO annotation by biological process. Allocation of parotid exosome proteins by biological process showed that the greatest number of these proteins (261) was allocated to cellular process and the smallest (1) to cell killing. B. GO annotation by cellular component. Allocation of parotid exosome proteins by cell component demonstrated that the majority belonged to the cell part and cell (306 each), while the minority resided in the envelope (7). C. GO annotation by molecular function. Allocation of parotid exosome proteins by molecular function confirmed that the highest number of proteins (311) was associated with binding activities and the lowest (1 each) with auxiliary transport protein activity and chaperone regulator activity.
Analysis of the hypothetical proteins
A total of 23 hypothetical proteins were identified, representing 5% of the total human parotid exosome proteome. Annotation on UniProtKB/Swiss-Prot, EBI and UniProtKB/TrEMBL databases suggests that 5 proteins (1%) carry out different functions such as immune activity, glycolysis, carbohydrate binding, protease enzyme activity and assembly (1 protein per each category). The functions of the remaining 18 proteins (4%) are unknown.
The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis
KEGG pathway analysis (version September, 2007) was performed on the identified parotid exosome proteins to evaluate which pathways were significantly represented. One hundred and ten different pathways were linked to the parotid exosomes. Proteins were involved in processes such as cell junction (30 proteins), N-glycan biosynthesis (11 proteins), glycan structures-biosynthesis (18 proteins), cholera-infection (9 proteins), pathogenic Escherichia coli infection (10 proteins), epithelial cell signaling in Helicobacter pylori infection (11 proteins), ECM-receptor interaction (12 proteins), glycolysis/gluconeogenesis (9 proteins), glycosphingolipid biosynthesis-Neo-Lactose series (5 proteins) among others (p ≤ 0.001). This analysis implicates exosomes in the regulation of numerous diverse biological functions such as cell-cell interactions/communication 63, 64, invasion and migration 65, transference of luminal antigenic information 40, and transmission of pathogens 66. It is interesting to note that some parotid exosome proteins were implicated in pathways associated with systemic diseases, for instance, proteins involved in Alzheimer’s disease, neurodegenerative disorders, Parkinson’s disease, cholera infection, Huntington’s disease, long-term depression, Prion disease, Dentatorubropallidoluysian atrophy (DRPLA), glioma, type I and II diabetes, and different types of carcinomas (renal cell carcinoma, pancreatic carcinoma, prostate cancer, small cell lung cancer, bladder cancer, melanoma) making exosomes an attractive source for exploring biomarkers (Supplemental table #6). Although not statistically significant (p>0.05), parotid exosomes also contained proteins involved in immunological pathways such as Toll-like receptor signaling pathway, T-cell receptor signaling pathway, B-cell receptor signaling pathway, cytokine-cytokine receptor interaction, natural killer cell mediated-cytotoxicity consistent with a role for exosomes in the regulation of immune response 67, 68.
Comparison of human parotid exosome with human parotid saliva proteomes
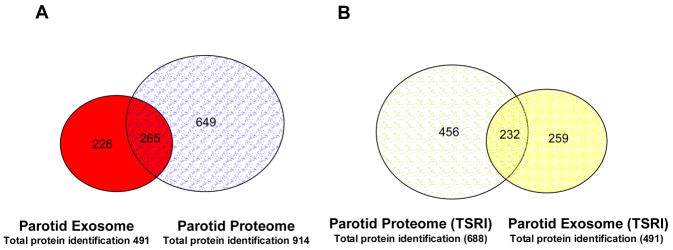
To gain a better understanding of the relationship of parotid exosomal proteins to those proteins detected in parotid ductal saliva, we compared the parotid exosome proteome with the 914 proteins from the parotid salivary proteome 2. Not surprisingly, the results demonstrated that 265 proteins found in the parotid exosome were also detected in the parotid salivary proteome [Supplemental table #7 and Fig #4A]. The vast majority of the 265 exosomal proteins detected in parotid saliva were expressed in low or medium abundance in the parotid proteome, suggesting that the origin of many of these proteins in ductal saliva may be exosomal. Consistent with this proposal, these overlapping proteins were generally the most abundant proteins detected in the parotid exosomal proteome. Moreover, the majority of the proteins found in low abundance in parotid exosomes were not detected in parotid saliva, indicating that the exosome isolation procedure had enriched for proteins not detectable in parotid ductal saliva. Importantly, the considerable overlap between the parotid ductal saliva proteome and the parotid exosomal proteome confirms the robust and sensitive nature of MudPIT analysis, and furthermore, suggests that it may not be necessary to perform the labor intensive process of isolating exosomes to monitor the expression of many of the clinically relevant biomarkers secreted as exosomes. To eliminate subject variation, we also compared data generated by The Scripps Research Institute (TSRI) from parotid saliva collected from the same donor as used in the parotid exosome experiments. Results showed that 232 proteins (25%) were common to both proteomes (supplemental table #8) while 456 were unique for the salivary proteome and 259 to parotid exosomes [Fig #4B]. Parotid ductal saliva from the same donor used in this study contained 232 overlapping exosomal proteins, similar to the overlapping number of exosomal proteins (265) found in a population of subjects 2.
Figure 4. Comparisons between parotid exosome and parotid salivary proteomes.
A. Venn diagram showing the overlap of proteins between the parotid exosome and the parotid salivary proteomes. Twenty-three % of the proteins were found in both proteomes, whereas 20% were unique parotid exosome proteins and 57% were unique to the parotid saliva. B. Venn diagram showing the overlap of proteins between the parotid exosome and the parotid salivary proteomes from the same donor. Twenty-five % proteins were found in both proteomes, whereas 27% were unique parotid exosome proteins and 48% were unique to the parotid saliva.
Moreover, since different cell-type specific exosomal proteomes have both unique and common proteins 21, 22, 27, 28, 31, 69, we compared the parotid exosome proteome with the parotid saliva proteome 2 and the urinary proteome 28. Only 46 proteins which have been associated with exosomes were present in the three different proteomes 18, 19, 22, 24, 27, 28, 30, 31, 33–36, 38, 39, 44 [Supplemental table #9 and supplemental Fig #1]. Additionally, MudPIT analysis was run on urinary exosomes obtained from the same saliva donor used in the present study (data not shown). We detected 146 proteins in the urinary exosome that overlapped with those previously reported 28 confirming the reproducibility of the exosome fractionation procedure.
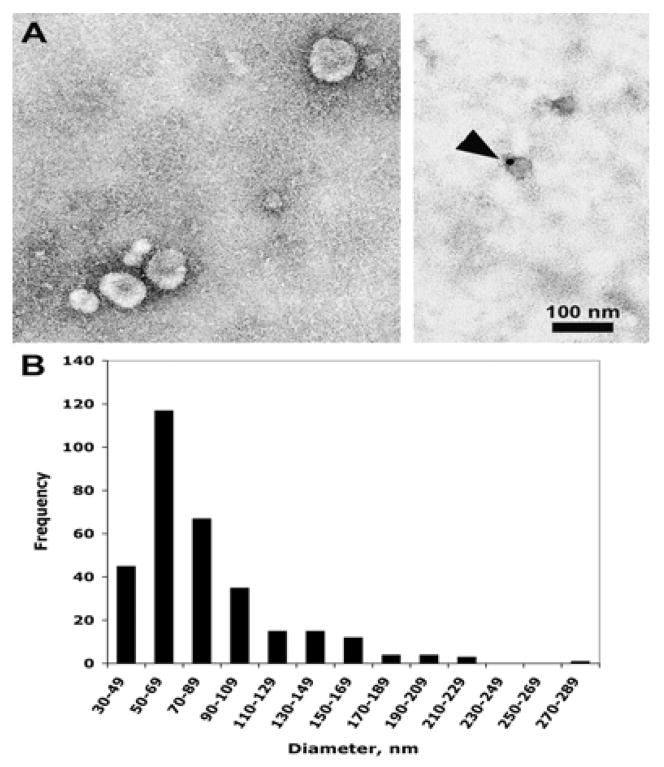
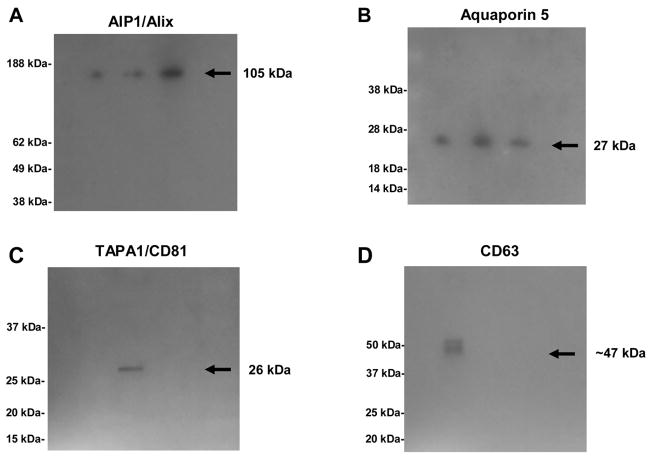
To further validate our MudPIT data, immunoelectron microscopy and western blotting analyses were performed on parotid exosome samples. The electron microscopy results depicted in the left panel of Fig #5A show the negative stained image of uniformly rounded exosomes with a mean size of 81.58 nm (SE=2.17 nm) and a median size of 69.45 nm, as summarized in the corresponding histogram (Fig #5B). These results are in agreement with previous reports 22, 28, 31, 34, 53. Additionally, immunogold labeling with anti-TSG101 antibody revealed the presence of the tumor-susceptibility gene 101 protein in parotid exosomes [Fig #5A; right panel]. The role of this protein in the transport of membrane proteins in the endocytic pathway is well established 24, 27, 29, 33–36, implicating this protein in the sorting of exosomal proteins during parotid exosome formation as well. In addition, western blots performed on parotid exosome samples (n=3 different preparations) confirmed the presence of representative proteins identified by MudPIT. Apical plasma membrane proteins are sequestered during exosome formation 28, 33, 34. Therefore, as expected for salivary gland exosomes, Fig #6B shows a band of ~ 27 kDa that corresponds to the estimated molecular mass of Aquaporin 5 (AQP5), a relatively abundant water channel present in the apical plasma membrane of salivary gland acinar cells 70. Fig #6A shows that the mouse polyclonal antibody against the exosome-specific AIP1/Alix recognized a band of the predicted size for this protein at ~ 105 kDa. AIP1/Alix forms part of the ESCRT endosomal sorting complex necessary for ILV (pro-exosomes) formation 33. Note that the band intensities for AIP1/Alix and AQP5 were comparable in the three individual exosomal preparations. Further verification of the endosomal origin of parotid exosomes was achieved by immonodetection of TAPA1/CD81 and CD63 exosomal markers [Figs #6C and 6D, respectively]. These proteins belong to the tetraspanin family and are highly enriched in exosomes where they are involved in the formation of membrane microdomains and large protein networks associated with the endocytic pathway 27, 29, 31, 44.
Figure 5. Immunoelectron microscopy of parotid exosomes.
A. Electron microscopy of parotid exosomes. Left panel, ammonium molybdate negative stain; right panel, immunogold labeling with anti-TSG101 (50 μg/ml). The arrowhead indicates a labeled exosome. B. Frequency distribution of parotid exosomes by diameter. A total of 318 exosomes were measured.
Figure 6. Western blot analyses of parotid exosome proteins isolated from human parotid saliva.
Panel A: AIP1/Alix parotid exosome protein (n=3) was detected using an anti AIP1/Alix polyclonal antibody from Abcam (Cambridge, MA). The expected molecular weight of this protein was around ~ 105 kDa. Panel B: Aquaporin-5 parotid exosomal protein (n=3) was detected with a polyclonal antibody kindly provided by Dr. Anil Menon (Department of Molecular Genetics, Biochemistry and Microbiology; University of Cincinnati, OH). This antibody recognized a protein of ~27 kDa. Panel C: TAPA1/CD81 and Panel D: CD63 (non-reducing condition) parotid-exosome proteins involved in biogenesis process were detected using monoclonal antibodies from Abcam (Cambridge, MA). The corresponding molecular weights of these proteins were ~ 26 kDa and ~ 47 kDa respectively.
Discussion
In the past two decades, major advances in mass spectrometric ionization techniques have made possible the identification of macromolecules and protein complexes in health and disease processes, revolutionizing the biomarker research field 43. To achieve sensitive and accurate detection of biomarkers in biological fluids, multiple fractionation steps have been proposed to reduce sample complexity 43, 71–75. One strategy that has been utilized with success is MudPIT 55, a shotgun proteomic approach to identify with great sensitivity and accuracy medium and low abundant proteins from complex protein mixtures by using a combination of strong cation exchange chromatography with reverse phase separations 61. Our data showed an improved mass accuracy, resolution and sensitivity with MudPIT technology for identifying parotid exosome proteins with great coverage. Furthermore, these data provide for the first time a comprehensive analysis of the secretion of exosomes in parotid ductal saliva. Exosomes represent an important pathway by which salivary gland cells secrete proteins in saliva, both in a constitutive 18, 19, 35 and regulated manner 76. Of the total 914 proteins identified in the parotid salivary proteome 2, 265 (23% overlapping) were common to the parotid exosomal proteome. The presence of these proteins in exosomes likely explains the existence of cellular proteins like plasma membrane (polymeric-immunoglobulin receptor precursor, 4F2 cell-surface antigen heavy chain), and cytosolic proteins (Ras GTPase-activating-like protein IQGAP1, 14-3-3 protein zeta/delta, clathrin heavy chain 1, peroxiredoxin-2, syntenin-1 and syntenin-3, annexin-A1, glucose-6-phosphate isomerase, actins, tubulins, keratins etc.) in ductal saliva. For example, the exosomal proteins found were of integral plasma membrane origin (16 proteins; 2%), from peripheral plasma membrane (27 proteins; 3%), and 118 cytoplasmic proteins (13%). Additionally, using immunoelectron microscopy and immunoblotting techniques the origin of five proteins (TSG101, Aquaporin-5, Alix, CD81 and CD63) identified by MudPIT were confirmed as proteins associated with endomembrane vesicles. Morphological analyses performed on exosomes derived either from cell lines 18, 19, 37 or body fluids such as urine 25, 28, blood 31, malignant pleural effusions 21 and amniotic fluid 25, as well as our electron microscopy findings reveal that mammalian exosomes have common characteristics such as structure (membranes with lipid bilayer), shape (round-shaped), size (30–100 nm), and density 19, 31, 35.
Many common exosomal proteins found in other tissues were identified in parotid exosomes like heat shock proteins (HSP90 and HSP70), cytoskeletal components (actins, cofilin-1, tubulins), proteins implicated in translation (elongation factor 1α1, elongation initiation factor-4A), proteins involved in signal transduction (14-3-3, syntenin, G- proteins), proteins associated with intracellular membrane fusion and transport (annexins, small GTPases, ADP-ribosylation factor 1 and Rab family members), enzymes implicated in different metabolic processes 19, 27, 35 as well as membrane bound proteins that belong to the tetraspanin family (C9, CD82, CD81) 19, 29, 31, 34, 44. In addition, we found that parotid exosomes lacked endoplasmic reticulum or nuclear resident proteins, distinguishing them from apoptotic bodies or shed membranes 19, 22, 31. However, some of the identified proteins were associated with pro- apoptotic or anti-apoptotic activities such as 14-3-3, galectin-3, galectin-7, peroxiredoxin-2, and thioredoxin domain containing protein 5 precursor, suggesting their possible cell-protector role.
Cumulative evidence suggests a role of the ESCRT family in protein sorting associated with MVE generation 24, 28, 29, 34, 35. These multimeric protein complexes direct specific protein-protein and protein-lipid interactions causing a deformity in the endosomal-limiting membrane, and thus promoting the inward budding of the ILVs 76, 77. In the present study, we also identified proteins implicated in the endosomal sorting complex required for transport, e.g. ESCRT-I (VPS28) and ESCRT-III (VPS4A), in addition to VTA1 (trafficking of multivesicular bodies) and Alix (necessary for the targeting of endosomes) 28, 33–35, 76. Moreover, proteins consistent with an alternative pathway for sorting cargo in the MVEs completely independent of the ESCRT machinery 30 were also detected such as UDP-glucose ceramide glucosyltransferase-like 1 isoform 1, UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase 1 membrane-bound form, Phosphatidylethanolamine-binding protein 1. This latter mechanism depends on raft-based microdomains in which the cone-shaped lipid ceramide induces a spontaneous negative curvature on the MVE membrane by creating an area difference between the membrane leaflets and merging small microdomains into larger domains, thus promoting domain-induced budding. During this process, interaction with tetraspanins and GPI-anchored proteins is necessary for ILV formation 24, 26, 63. Little is known about the exosome biogenesis pathway in epithelial cells; however the detection of proteins associated with these two mechanisms of exosome formation in parotid exosomes suggests that cell-type- and cargo-specific differences may be present in parotid gland cells 76.
Besides the proteins implicated in the process of exosome formation, we detected proteins involved in many other biological functions including proteins exposed on the exosomal surface involved in cell-adhesion (Von Willebrand factor A domain-related protein isoform 1, MFGE8, LAMB2, LAMC1), suggesting another cell-communication mechanism for exosome targeting to effector molecules, cell fusion and exchange of antigenic information 18, 19. Parotid exosomes also exhibited unique molecular signatures. These proteins included: Aquaporin 5, an apical plasma membrane channel involved in water regulation during salivary secretion 59; different solute carrier molecules involved in transporter activities during salivation (SLC12A2, SLC9A1) 62, 78, 79; CD59 and Decay-accelerating factor splicing variant 4, co-stimulatory molecules of the immune system 80, 81; cytokeratins, specific epithelial markers 18; cytokines (TNFSF10, TNFSF13), implicated in tumor growth regulation and in mediation of immunological process 23; and MUC1 and CD44, involved in cell-adhesion and cell-cell interactions 82. These findings are in agreement with previous reports where exosomal proteins vary according to their cell origin 19, 22, 24, 37, 69. Furthermore, MudPIT analysis revealed the presence of both exclusive and common salivary gland protein markers indicating the possible origin of parotid exosomes from ductal (SLC5A583, Duox2 84, galectin 385), acinar (AQP5 59, TMEM16A 86) or both cell types (desmoglein 287, DPP IV88).
Given that parotid exosomes contain numerous cytokines and antigen presenting proteins, it may be worthwhile to explore their potential role in autoimmune diseases such as Sjögren’s syndrome. To date, one study conducted on human non-neoplastic salivary gland epithelial cells (SGECs) by Kapsogeorgou et al 18 has demonstrated that salivary gland epithelial cell exosomes contain autoantigenic Ro/SSA, La/SSB and Sm RNPs (ribonucleoproteins), proteins associated with Sjögren’s syndrome. Exosomes are known to mediate antigen presentation either through their surface receptors (major histocompatibility complex, antigenic-peptide complexes, and co-stimulatory molecules) or by antigenic transfer to antigen presenting cells (APCs) 29, 68. Thus, it may prove important to define the immunological role of exosomes in Sjögren’s syndrome disease and progression.
Some extracellular component proteins were detected at low levels in the parotid exosomal preparation such as immunoglobulins (IgGs; 21, 28), complement proteins, cystatin D, carbonic anhydrase 6 and alpha-amylase 28. These extracellular secreted proteins are highly expressed in human parotid saliva, suggesting that incorporation of these proteins may have occurred during exosome formation. Alternatively, some minor contamination may have occurred during the isolation procedure of the exosomes. Taking into consideration the high sensitivity of MudPIT analysis and the high abundance of these salivary proteins, it is not that surprising to detect them. Furthermore, Bard et al 21 also found several IgGs and complement proteins in human malignant pleural effusions suggesting the presence of both cellular and humoral immunity and the possible trapping of these proteins during the ultracentrifugation step. Nevertheless, we can not discard the possibility of a cell-cell interaction taking place between the exosome plasma membrane antigen presenting-defense and mediators of immunological process molecules with the immunoglobulins per se or the possible generation of these proteins (IgGs and complement proteins) from locally stimulated B-lymphocytes 89, consistent with the biological role of exosomes in regulation of the immune response 35, 44, 67, 68.
New proteomic methods have revealed the protein complexity of exosomal vesicles, including cell surface proteins, cytosolic proteins as well as the intracellular machinery that is responsible for exosome formation and extracellular release 24, 28, 30, 33, 34, 69. These previous studies have demonstrated that endomembrane vesicles are secreted in the urine, blood, plasma, amniotic fluid and malignant pleural effusions. Here we show that these vesicles are also secreted in parotid saliva. This process may be regulated by an increase in the intracellular calcium concentration 90 which stimulates exosome release in epithelial cells 76. Numerous proteins participate during exosomal secretion such as dynein and kinesin which mediate the movement of endosomes 91, RHO-A, different RAB proteins, GTPases and syntaxin proteins (syntaxin-binding protein 2) 40 which interact at the apical membrane site of parotid acinar cells 8, 76 to promote exocytosis through the V0 sectors of the V-ATPase (ATP6V0A4) by forming a proteolipid pore during exocytic fusion of the MVEs with the plasma membrane 76. Furthermore, Valadi et al 92 demonstrated that in addition to proteins, exosomes from mouse mast cell line (MC/9), human mast cell line (HMC-1) as well as bone marrow-derived mouse mast cells (BMMC) contain a variety of mRNA and microRNA molecules. These results suggest that exosomes may be involved in a novel mechanism of cell-cell interaction and communication in mammalian cells 63, 64. This process may be important in neurodegenerative diseases (Prion diseases, Alzheimer’s disease) and HIV-transmissible disease since the severity of these diseases is related to cell-to-cell uptake mechanism 24, 35, 93. Of possible significance is the KEGG analysis finding that parotid exosome proteins were associated with different disease conditions (e.g. neurodegenerative diseases, cancers).
In summary, MudPIT technology generated an in-depth analysis of the parotid exosome proteome by achieving efficient on-line peptide separation, high mass accuracy and analytical sensitivity. Our proteomic analysis provides a first step in analyzing a large set of parotid exosome proteins that may be important in future studies. The protein composition of the parotid exosome revealed important insights into the generation and physiology of these membrane vesicles. Future studies are needed to elucidate the biological functions of exosomes in healthy and disease subjects. Variables such as gender, age and ethnicity will need to be taken into account when exploring the use of exosomal proteins as biomarkers for diagnostic and disease progression purposes.
Supplementary Material
Supplemental Figure 1. Venn diagram showing the overlap of proteins among the urinary exosome, the parotid exosome and the parotid salivary proteomes. Three % of the proteins were common to the three different proteomes. The remaining protein identifications (urinary exosome: 16%; parotid exosome: 27% and parotid saliva 54%) were unique to the cell/fluid of origin.
Supplemental table #1: Parotid exosome cluster composition with a minimum of two peptides identified.
Supplemental table #2: Annotation of parotid exosome proteins.
Supplemental table #3: Gene ontology annotation of parotid exosome proteins by biological process.
Supplemental table #4: Gene ontology annotation of parotid exosome proteins by cellular component.
Supplemental table #5: Gene ontology annotation of parotid exosome proteins by molecular function.
Supplemental table #6: KEGG annotation of parotid exosome proteins.
Supplemental table #7: Protein evidence corresponding to common proteins present between the parotid exosome proteome and the parotid saliva proteome.
Supplemental table #8: Protein evidence corresponding to common proteins present between the parotid salivary proteome and the exosome proteome from the same donor.
Supplemental table #9: Protein evidence corresponding to common proteins present among the three different proteomes (urinary exosome, parotid exosome and salivary exosome).
Acknowledgments
This work was supported in part by NIH grants UO1 DE016267 (JRY), P41 RR011823 (JRY), RO1 DE09692 (JEM) and RO1 DE08921 (JEM). MG-B was supported in part by T32 DE07202. BL is supported by a CFFT computational fellowship BALCH05X5.
Footnotes
Supporting information:
Detailed information summarizing parotid exosome cluster composition, annotation of parotid exosomes, protein evidence corresponding to common proteins, as well as gene ontology and KEGG annotations of the parotid exosomal proteins are available free at http://pubs.acs.org.
References
- 1.Bennick A. J Oral Biosci. 2007;49:24–26. [Google Scholar]
- 2.Denny P, Hagen FK, Hardt M, Liao L, Yan W, Arellanno M, Bassilian S, Bedi GS, Boontheung P, Cociorva D, Delahunty CM, Denny T, Dunsmore J, Faull KF, Gilligan J, Gonzalez-Begne M, Halgand F, Hall SC, Han X, Henson B, Hewel J, Hu S, Jeffrey S, Jiang J, Loo JA, Ogorzalek Loo RR, Malamud D, Melvin JE, Miroshnychenko O, Navazesh M, Niles R, Park SK, Prakobphol A, Ramachandran P, Richert M, Robinson S, Sondej M, Souda P, Sullivan MA, Takashima J, Than S, Wang J, Whitelegge JP, Witkowska HE, Wolinsky L, Xie Y, Xu T, Yu W, Ytterberg J, Wong DT, Yates JR, 3rd, Fisher SJ. J Proteome Res. 2008;7:1994–2006. doi: 10.1021/pr700764j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oppenheim FG, Salih E, Siqueira WL, Zhang W, Helmerhorst EJ. Ann N Y Acad Sci. 2007;1098:22–50. doi: 10.1196/annals.1384.030. [DOI] [PubMed] [Google Scholar]
- 4.Tabak LA. Pediatr Dent. 2006;28:110–117. discussion 192–118. [PubMed] [Google Scholar]
- 5.Vitorino R, Lobo MJ, Ferrer-Correira AJ, Dubin JR, Tomer KB, Domingues PM, Amado FM. Proteomics. 2004;4:1109–1115. doi: 10.1002/pmic.200300638. [DOI] [PubMed] [Google Scholar]
- 6.Siqueira WL, Salih E, Wan DL, Helmerhorst EJ, Oppenheim FG. J Dent Res. 2008;87:445–450. doi: 10.1177/154405910808700508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo T, Rudnick PA, Wang W, Lee CS, Devoe DL, Balgley BM. J Proteome Res. 2006;5:1469–1478. doi: 10.1021/pr060065m. [DOI] [PubMed] [Google Scholar]
- 8.Castle AM, Huang AY, Castle JD. J Cell Sci. 2002;115:2963–2973. doi: 10.1242/jcs.115.14.2963. [DOI] [PubMed] [Google Scholar]
- 9.Turner RJ, Sugiya H. Oral Dis. 2002;8:3–11. doi: 10.1034/j.1601-0825.2002.10815.x. [DOI] [PubMed] [Google Scholar]
- 10.Ishikawa Y, Cho G, Yuan Z, Skowronski MT, Pan Y, Ishida H. J Pharmacol Sci. 2006;100:495–512. doi: 10.1254/jphs.crj06007x. [DOI] [PubMed] [Google Scholar]
- 11.Mazariegos MR, Tice LW, Hand AR. J Cell Biol. 1984;98:1865–1877. doi: 10.1083/jcb.98.5.1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oliver C, Hand AR. J Cell Biol. 1978;76:207–229. doi: 10.1083/jcb.76.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brandtzaeg P. Ann N Y Acad Sci. 2007;1098:288–311. doi: 10.1196/annals.1384.012. [DOI] [PubMed] [Google Scholar]
- 14.Rojas R, Apodaca G. Nat Rev Mol Cell Biol. 2002;3:944–955. doi: 10.1038/nrm972. [DOI] [PubMed] [Google Scholar]
- 15.Castle JD. Ann N Y Acad Sci. 1998;842:115–124. doi: 10.1111/j.1749-6632.1998.tb09639.x. [DOI] [PubMed] [Google Scholar]
- 16.Huang AY, Castle Anna M, Hinton Barry T, David Castle J. J Biol Chem. 2001;276:22296–22306. doi: 10.1074/jbc.M100211200. [DOI] [PubMed] [Google Scholar]
- 17.Emmelin N. J Dent Res. 1987;66:509–517. doi: 10.1177/00220345870660022101. [DOI] [PubMed] [Google Scholar]
- 18.Kapsogeorgou EK, Abu-Helu RF, Moutsopoulos HM, Manoussakis MN. Arthritis Rheum. 2005;52:1517–1521. doi: 10.1002/art.21005. [DOI] [PubMed] [Google Scholar]
- 19.Thery C, Boussac M, Veron P, Ricciardi-Castagnoli P, Raposo G, Garin J, Amigorena S. J Immunol. 2001;166:7309–7318. doi: 10.4049/jimmunol.166.12.7309. [DOI] [PubMed] [Google Scholar]
- 20.Trams EG, Lauter CJ, Salem N, Jr, Heine U. Biochim Biophys Acta. 1981;645:63–70. doi: 10.1016/0005-2736(81)90512-5. [DOI] [PubMed] [Google Scholar]
- 21.Bard MP, Hegmans JP, Hemmes A, Luider TM, Willemsen R, Severijnen LA, van Meerbeeck JP, Burgers SA, Hoogsteden HC, Lambrecht BN. Am J Respir Cell Mol Biol. 2004;31:114–121. doi: 10.1165/rcmb.2003-0238OC. [DOI] [PubMed] [Google Scholar]
- 22.Mears R, Craven RA, Hanrahan S, Totty N, Upton C, Young SL, Patel P, Selby PJ, Banks RE. Proteomics. 2004;4:4019–4031. doi: 10.1002/pmic.200400876. [DOI] [PubMed] [Google Scholar]
- 23.O’Neill HC, Quah BJ. Sci Signal. 2008;1:pe8. doi: 10.1126/stke.16pe8. [DOI] [PubMed] [Google Scholar]
- 24.Vella LJ, Sharples RA, Nisbet RM, Cappai R, Hill AF. Eur Biophys J. 2008;37:323–332. doi: 10.1007/s00249-007-0246-z. [DOI] [PubMed] [Google Scholar]
- 25.Keller S, Rupp C, Stoeck A, Runz S, Fogel M, Lugert S, Hager HD, Abdel-Bakky MS, Gutwein P, Altevogt P. Kidney Int. 2007;72:1095–1102. doi: 10.1038/sj.ki.5002486. [DOI] [PubMed] [Google Scholar]
- 26.Marsh M, van Meer G. Science. 2008;319:1191–1192. doi: 10.1126/science.1155750. [DOI] [PubMed] [Google Scholar]
- 27.Olver C, Vidal M. Subcell Biochem. 2007;43:99–131. doi: 10.1007/978-1-4020-5943-8_7. [DOI] [PubMed] [Google Scholar]
- 28.Pisitkun T, Shen RF, Knepper MA. Proc Natl Acad Sci U S A. 2004;101:13368–13373. doi: 10.1073/pnas.0403453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thery C, Zitvogel L, Amigorena S. Nat Rev Immunol. 2002;2:569–579. doi: 10.1038/nri855. [DOI] [PubMed] [Google Scholar]
- 30.Trajkovic K, Hsu C, Chiantia S, Rajendran L, Wenzel D, Wieland F, Schwille P, Brugger B, Simons M. Science. 2008;319:1244–1247. doi: 10.1126/science.1153124. [DOI] [PubMed] [Google Scholar]
- 31.Caby MP, Lankar D, Vincendeau-Scherrer C, Raposo G, Bonnerot C. Int Immunol. 2005;17:879–887. doi: 10.1093/intimm/dxh267. [DOI] [PubMed] [Google Scholar]
- 32.Hoorn EJ, Pisitkun T, Zietse R, Gross P, Frokiaer J, Wang NS, Gonzales PA, Star RA, Knepper MA. Nephrology (Carlton) 2005;10:283–290. doi: 10.1111/j.1440-1797.2005.00387.x. [DOI] [PubMed] [Google Scholar]
- 33.Knepper MA, Pisitkun T. Kidney Int. 2007;72:1043–1045. doi: 10.1038/sj.ki.5002510. [DOI] [PubMed] [Google Scholar]
- 34.van Niel GP-CI, Simoes S, Raposo G. J Biochem (Tokyo) 2006;140:13–21. doi: 10.1093/jb/mvj128. [DOI] [PubMed] [Google Scholar]
- 35.Keller S, Sanderson MP, Stoeck A, Altevogt P. Immunol Lett. 2006;107:102–108. doi: 10.1016/j.imlet.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 36.Chertova E, Chertov O, Coren LV, Roser JD, Trubey CM, Bess JW, Jr, Sowder RC, 2nd, Barsov E, Hood BL, Fisher RJ, Nagashima K, Conrads TP, Veenstra TD, Lifson JD, Ott DE. J Virol. 2006;80:9039–9052. doi: 10.1128/JVI.01013-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blanchard N, Lankar D, Faure F, Regnault A, Dumont C, Raposo G, Hivroz C. J Immunol. 2002;168:3235–3241. doi: 10.4049/jimmunol.168.7.3235. [DOI] [PubMed] [Google Scholar]
- 38.Fevrier B, Vilette D, Archer F, Loew D, Faigle W, Vidal M, Laude H, Raposo G. Proc Natl Acad Sci U S A. 2004;101:9683–9688. doi: 10.1073/pnas.0308413101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hegmans JP, Bard MP, Hemmes A, Luider TM, Kleijmeer MJ, Prins JB, Zitvogel L, Burgers SA, Hoogsteden HC, Lambrecht BN. Am J Pathol. 2004;164:1807–1815. doi: 10.1016/S0002-9440(10)63739-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Niel G, Raposo G, Candalh C, Boussac M, Hershberg R, Cerf-Bensussan N, Heyman M. Gastroenterology. 2001;121:337–349. doi: 10.1053/gast.2001.26263. [DOI] [PubMed] [Google Scholar]
- 41.Pisitkun T, Johnstone R, Knepper MA. Mol Cell Proteomics. 2006;5:1760–1771. doi: 10.1074/mcp.R600004-MCP200. [DOI] [PubMed] [Google Scholar]
- 42.Gutwein P, Stoeck A, Riedle S, Gast D, Runz S, Condon TP, Marme A, Phong MC, Linderkamp O, Skorokhod A, Altevogt P. Clin Cancer Res. 2005;11:2492–2501. doi: 10.1158/1078-0432.CCR-04-1688. [DOI] [PubMed] [Google Scholar]
- 43.Lu MFKF, Whitelegge JP, He J, Shen D, Saxton RE, Chang HR. Biomarker Insights. 2007;2:347–360. [PMC free article] [PubMed] [Google Scholar]
- 44.van Niel G, Heyman M. Am J Physiol Gastrointest Liver Physiol. 2002;283:G251–255. doi: 10.1152/ajpgi.00102.2002. [DOI] [PubMed] [Google Scholar]
- 45.Hu S, Loo JA, Wong DT. Ann N Y Acad Sci. 2007;1098:323–329. doi: 10.1196/annals.1384.015. [DOI] [PubMed] [Google Scholar]
- 46.Hu S, Loo JA, Wong DT. Expert Rev Proteomics. 2007;4:531–538. doi: 10.1586/14789450.4.4.531. [DOI] [PubMed] [Google Scholar]
- 47.Wong DT. J Am Dent Assoc. 2006;137:313–321. doi: 10.14219/jada.archive.2006.0180. [DOI] [PubMed] [Google Scholar]
- 48.Hu S, Wang J, Meijer J, Ieong S, Xie Y, Yu T, Zhou H, Henry S, Vissink A, Pijpe J, Kallenberg C, Elashoff D, Loo JA, Wong DT. Arthritis Rheum. 2007;56:3588–3600. doi: 10.1002/art.22954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kalk WW, Vissink A, Stegenga B, Bootsma H, Nieuw Amerongen AV, Kallenberg CG. Ann Rheum Dis. 2002;61:137–144. doi: 10.1136/ard.61.2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kaufman E, Lamster IB. J Clin Periodontol. 2000;27:453–465. doi: 10.1034/j.1600-051x.2000.027007453.x. [DOI] [PubMed] [Google Scholar]
- 51.Malamud D. Am J Med. 1997;102:9–14. doi: 10.1016/s0002-9343(97)00032-6. [DOI] [PubMed] [Google Scholar]
- 52.Pijpe J, Kalk WW, Bootsma H, Spijkervet FK, Kallenberg CG, Vissink A. Ann Rheum Dis. 2007;66:107–112. doi: 10.1136/ard.2006.052647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ogawa Y, Kanai-Azuma M, Akimoto Y, Kawakami H, Yanoshita R. Biol Pharm Bull. 2008;31:1059–1062. doi: 10.1248/bpb.31.1059. [DOI] [PubMed] [Google Scholar]
- 54.Lashley KS. J Exp Psychol. 1916;1:461–493. [Google Scholar]
- 55.Washburn MP, Wolters D, Yates JR., 3rd Nat Biotechnol. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 56.Eng J, McCormack A, Yates JR., 3rd J Am Soc Mass Spectrom. 1994;5 doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 57.Tabb DL, McDonald WH, Yates JR., 3rd J Proteome Res. 2002;1:21–26. doi: 10.1021/pr015504q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 59.Krane CM, Melvin JE, Nguyen HV, Richardson L, Towne JE, Doetschman T, Menon AG. J Biol Chem. 2001;276:23413–23420. doi: 10.1074/jbc.M008760200. [DOI] [PubMed] [Google Scholar]
- 60.Washburn MP. Brief Funct Genomic Proteomic. 2004;3:280–286. doi: 10.1093/bfgp/3.3.280. [DOI] [PubMed] [Google Scholar]
- 61.Yates JR., 3rd Annu Rev Biophys Biomol Struct. 2004;33:297–316. doi: 10.1146/annurev.biophys.33.111502.082538. [DOI] [PubMed] [Google Scholar]
- 62.Melvin JE, Yule D, Shuttleworth T, Begenisich T. Annu Rev Physiol. 2005;67:445–469. doi: 10.1146/annurev.physiol.67.041703.084745. [DOI] [PubMed] [Google Scholar]
- 63.de Gassart AGC, Hoekstra D, Vidal M. Traffic. 2004;5:896–903. doi: 10.1111/j.1600-0854.2004.00223.x. [DOI] [PubMed] [Google Scholar]
- 64.Fevrier B, Raposo G. Curr Opin Cell Biol. 2004;16:415–421. doi: 10.1016/j.ceb.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 65.Ginestra ALPM, Saladino F, Cassarà D, Nagase H, Vittorelli ML. Anticancer Res. 1998;18:3433–3437. [PubMed] [Google Scholar]
- 66.Fevrier B, Vilette D, Laude H, Raposo G. Traffic. 2005;6:10–17. doi: 10.1111/j.1600-0854.2004.00247.x. [DOI] [PubMed] [Google Scholar]
- 67.Kapsogeorgou EKMH, Manoussakis MN. J Immunol. 2001;1:3107–3113. doi: 10.4049/jimmunol.166.5.3107. [DOI] [PubMed] [Google Scholar]
- 68.Thery C, Duban L, Segura E, Veron P, Lantz O, Amigorena S. Nat Immunol. 2002;3:1156–1162. doi: 10.1038/ni854. [DOI] [PubMed] [Google Scholar]
- 69.Simpson RJ, Jensen SS, Lim JW. Proteomics. 2008;8:4083–4099. doi: 10.1002/pmic.200800109. [DOI] [PubMed] [Google Scholar]
- 70.Konttinen YT, Tensing EK, Laine M, Porola P, Tornwall J, Hukkanen M. J Rheumatol. 2005;32:1071–1075. [PubMed] [Google Scholar]
- 71.Hu S, Xie Y, Ramachandran P, Ogorzalek Loo RR, Li Y, Loo JA, Wong DT. Proteomics. 2005;5:1714–1728. doi: 10.1002/pmic.200401037. [DOI] [PubMed] [Google Scholar]
- 72.Smith RD. Biotechniques. 2006;41:147–148. doi: 10.2144/000112217. [DOI] [PubMed] [Google Scholar]
- 73.Tao WAWB, O’Brien R, Eng JK, Li XJ, Bodenmiller B, Watts JD, Hood L, Aebersold R. Nat Methods. 2005;2:591–598. doi: 10.1038/nmeth776. [DOI] [PubMed] [Google Scholar]
- 74.Wang H, Hanash S. Mass Spectrom Rev. 2005;24:413–426. doi: 10.1002/mas.20018. [DOI] [PubMed] [Google Scholar]
- 75.Zhang HYEC, Li XJ, Mallick P, Kelly-Spratt KS, Masselon CD, Camp DG, II, Smith RD, Kemp CJ, Aebersold R. Mol Cell Proteomics. 2005;4:144–155. doi: 10.1074/mcp.M400090-MCP200. [DOI] [PubMed] [Google Scholar]
- 76.Lakkaraju A, Rodriguez-Boulan E. Trends Cell Biol. 2008;18:199–209. doi: 10.1016/j.tcb.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Williams RL, Urbe S. Nat Rev Mol Cell Biol. 2007;8:355–368. doi: 10.1038/nrm2162. [DOI] [PubMed] [Google Scholar]
- 78.Evans RL, Park K, Turner RJ, Watson GE, Nguyen HV, Dennett MR, Hand AR, Flagella M, Shull GE, Melvin JE. J Biol Chem. 2000;275:26720–26726. doi: 10.1074/jbc.M003753200. [DOI] [PubMed] [Google Scholar]
- 79.Park K, Evans RL, Melvin JE. J Korean Med Sci. 2000;15(Suppl):S5–6. doi: 10.3346/jkms.2000.15.S.S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kimberley FC, Sivasankar B, Paul Morgan B. Mol Immunol. 2007;44:73–81. doi: 10.1016/j.molimm.2006.06.019. [DOI] [PubMed] [Google Scholar]
- 81.Miwa T, Maldonado MA, Zhou L, Yamada K, Gilkeson GS, Eisenberg RA, Song WC. Am J Pathol. 2007;170:1258–1266. doi: 10.2353/ajpath.2007.060601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Goodison S, Urquidi V, Tarin D. Mol Pathol. 1999;52:189–196. doi: 10.1136/mp.52.4.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Liu B, Herve J, Bioulac-Sage P, Valogne Y, Roux J, Yilmaz F, Boisgard R, Guettier C, Cales P, Tavitian B, Samuel D, Clerc J, Brechot C, Faivre J. Gastroenterology. 2007;132:1495–1503. doi: 10.1053/j.gastro.2007.01.044. [DOI] [PubMed] [Google Scholar]
- 84.Geiszt M, Witta J, Baffi J, Lekstrom K, Leto TL. FASEB J. 2003;17:1502–1504. doi: 10.1096/fj.02-1104fje. [DOI] [PubMed] [Google Scholar]
- 85.Xu XC, Sola Gallego JJ, Lotan R, El-Naggar AK. Int J Oncol. 2000;17:271–276. doi: 10.3892/ijo.17.2.271. [DOI] [PubMed] [Google Scholar]
- 86.Yang YD, Cho H, Koo JY, Tak MH, Cho Y, Shim WS, Park SP, Lee J, Lee B, Kim BM, Raouf R, Shin YK, Oh U. Nature. 2008;455:1210–1215. doi: 10.1038/nature07313. [DOI] [PubMed] [Google Scholar]
- 87.Andreadis D, Epivatianos A, Poulopoulos A, Nomikos A, Christidis K, Papazoglou G, Antoniades D, Barbatis C. Oral Oncol. 2005;41:799–805. doi: 10.1016/j.oraloncology.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 88.Sahara N, Suzuki K. Cell Tissue Res. 1984;235:427–432. doi: 10.1007/BF00217869. [DOI] [PubMed] [Google Scholar]
- 89.Telvi L, Jaubert F, Eyquem A, Andreux JP, Labrousse F, Chretien J. Thorax. 1979;34:389–392. doi: 10.1136/thx.34.3.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Savina A, Furlan M, Vidal M, Colombo MI. J Biol Chem. 2003;278:20083–20090. doi: 10.1074/jbc.M301642200. [DOI] [PubMed] [Google Scholar]
- 91.Lebrand C, Corti M, Goodson H, Cosson P, Cavalli V, Mayran N, Faure J, Gruenberg J. EMBO J. 2002;21:1289–1300. doi: 10.1093/emboj/21.6.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 93.Vella LJSR, Lawson VA, Masters CL, Cappai R, Hill AF. J Pathol. 2007;211:582–590. doi: 10.1002/path.2145. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Venn diagram showing the overlap of proteins among the urinary exosome, the parotid exosome and the parotid salivary proteomes. Three % of the proteins were common to the three different proteomes. The remaining protein identifications (urinary exosome: 16%; parotid exosome: 27% and parotid saliva 54%) were unique to the cell/fluid of origin.
Supplemental table #1: Parotid exosome cluster composition with a minimum of two peptides identified.
Supplemental table #2: Annotation of parotid exosome proteins.
Supplemental table #3: Gene ontology annotation of parotid exosome proteins by biological process.
Supplemental table #4: Gene ontology annotation of parotid exosome proteins by cellular component.
Supplemental table #5: Gene ontology annotation of parotid exosome proteins by molecular function.
Supplemental table #6: KEGG annotation of parotid exosome proteins.
Supplemental table #7: Protein evidence corresponding to common proteins present between the parotid exosome proteome and the parotid saliva proteome.
Supplemental table #8: Protein evidence corresponding to common proteins present between the parotid salivary proteome and the exosome proteome from the same donor.
Supplemental table #9: Protein evidence corresponding to common proteins present among the three different proteomes (urinary exosome, parotid exosome and salivary exosome).