Abstract
A combined approach using mass spectrometry, a novel neuron affinity capture technique, and Drosophila melanogaster genetic manipulation has been developed to characterize the expression and localization of neuropeptides in the adult D. melanogaster brain. In extract from the whole adult brain, 42 neuropeptides from 18 peptide families were sequenced. Neuropeptide profiling also was performed on targeted populations of cells which were enriched with immunoaffinity purification using a genetically expressed marker.
Keywords: neuropeptide, Drosophila, mass spectrometry, MALDI-TOF, biogenic amine, dopamine, serotonin
Introduction
In many invertebrates and vertebrates, neuropeptides serve important neurohormonal and neuromodulatory roles in a diverse array of behaviors including locomotion,1-3 feeding,4-7 aggression,8-13 learning and memory,14,15 reproduction16,17 and circadian rhythms.18 Numerous peptides have been identified in Drosophila melanogaster using database mining or peptidomic approaches in the larval nervous system.19,20 However, since many behavioral studies are performed in the adult animal, it is important to further refine characterization of neuropeptides both with regard to the expressed amino acid sequence in the mature adult nervous system and to localization within the brain. To address these issues, we characterized neuropeptide expression and function in the adult D. melanogaster brain by (1) sequencing 42 neuropeptides extracted from adult brain tissue and (2) identifying the subsets of peptides found in two well-defined neural circuits using a novel method of neuronal tissue enrichment.
Previous studies of adult D. melanogaster peptide expression were performed using matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectral-profiling of individually dissected cells from the adult brain.21,22 In some cases, the neurons of interest were genetically labeled by green fluorescent protein (GFP) facilitating dissection and direct peptide profiling by MALDI-TOF mass spectrometry (MS). This approach is built upon in this study by using the D. melanogaster GAL4-UAS gene targeting system23 to label and selectively enrich for population of cells in the fruit fly brain. The dimmed-GAL4 line, which expresses in a large subset of peptidergic neurons in fly brains,24 and the dopa decarboxylase-GAL4 line, which expresses in dopamine- and serotonin-containing neurons,25 were used to selectively label each respective subset of targeted cells with GFP and mouse-CD8 (mCD8), a cell-surface antigen. This targeted manipulation of selected cell populations enabled immunoaffinity purification and subsequent peptide-profiling by MALDI-TOF MS. These results demonstrate that it is possible to use genetically expressed cell-surface antigens to facilitate sample-preparation based on cellular identity.
Experimental Procedures
Flies
Flies were cultured at 25 °C, except where noted otherwise, on a standard cornmeal-yeast-agar medium. The following D. melanogaster stocks were used: dimm (c929)-GAL4 from P. Taghert (Washington University, St. Louis, MO); Ddc-GAL4 from J. Hirsch (University of Virginia, Charlottesville, VA); NPF-GAL4 from P. Shen (University of Georgia, Athens, GA); and UAS-mCD8-GFP from the Bloomington Stock Center (Bloomington, IN).
Preparation of Extracts from Whole Brain
A total of 200 brains from adult male and female Canton S flies were dissected, pooled and frozen at -80 °C. The tissue was homogenized in 200 μL of ice-cold acidified methanol (90% methanol, 1% glacial acetic acid) using a polypropylene-coated pestle (Kontes, Vineland, NJ). Following an overnight incubation at -20 °C, the extract was centrifuged at 16 100 RCF for 10 min and the supernatant removed. After evaporation by vacuum centrifugation to near dryness, 20 μL of 10% acetonitrile with 0.1% formic acid was added to the sample.
GAL4-UAS Mediated Cell Enrichment
A total of 100 brains were dissected from adult flies of each of the following genotypes: w; dimm (c929)-GAL4, UAS-mCD8-GFP or w; UAS-mCD8-GFP; Ddc-GAL4. The dissected tissue was placed in ice-cold dissection saline composed of the following (in mM): 126 NaCl, 5.4 KCl, 0.17 NaH2PO4, 0.22 KH2PO4, 33.3 glucose, 43.8 sucrose, and 9.9 HEPES, pH 7.4.26 Once the dissections were completed, the saline was gently removed and replaced with dissecting saline containing 0.4 mg/mL dispase (Roche Diagnostics, Indianapolis, IN) and 0.1 mg/mL collagenase I (Worthington Biochemical Corp., Lakewood, NJ) for 1 h at room temperature.27 Following two washes in phosphate buffered saline (PBS), pH 7.4 (Gibco, Carlsbad, CA) enriched with 0.5% bovine serum albumin and 2 mM EDTA, cells were suspended in 80 μL of the enriched PBS solution by gentle trituration through a narrow fire-polished Pasteur pipet. Twenty microliters of monoclonal anti-mCD8 antibody conjugated to paramagnetic beads (Miltenyi Biotec, Inc., Auburn, CA) was added to the sample and the mixture was incubated at 4 °C for 20 min. Labeled cells were enriched with two consecutive purification columns according to the manufacturer’s instructions. Following elution from the column, the cells were centrifuged at 0.8 RCF for 3 min and the supernatant was removed and replaced with 40 μL of acidified methanol. The mixture was sonicated briefly and incubated overnight at 4 °C. The acidified methanol was evaporated by vacuum centrifugation and the sample was solubilized in 0.1% trifluoroacetic acid (TFA), 10% acetonitrile (ACN). Peptide extracts were desalted using C18-packed pipet tips (ZipTips, Millipore, Billerica, MA) according to the manufacturer’s instructions.
N-Terminal Amino Group Acetylation
Acidified methanol was removed from the enriched cell preparations using vacuum centrifugation as described above and the extract was dissolved in 4 μL of 50 mM ammonium bicarbonate (pH 8.5). An equivalent volume of acetylating solution (3:1 methanol/acetic anhydride) was added and the samples were incubated at room temperature for 30 min followed by removal of the reagents by vacuum centrifugation.
Fluorescent Assisted Cell Sorting (FACS) Analysis
Flow cytometric analysis of live GFP-expressing cells from D. melanogaster brains was performed on a Becton Dickinson FACSAria instrument (San Jose, CA) equipped with 3 lasers and DiVa software. The cells were isolated and collected using the immunoaffinity column as described above and chilled on ice prior to sorting. All flow cytometry analysis was conducted by using excitation at 488 and 633 nm; GFP fluorescence was detected with a 530/30 nm bandpass filter. For each sample, 10 000–20 000 cells were gated using forward light scatter. For reference, GFP labeled cells from the experimental dimm (c929)-GAL4 and Ddc-GAL4 lines were compared with suspensions from fly brains not expressing GFP (yw; UAS-mCD8-GFP) in order to establish the background autofluoresence and detection sensitivity. The cell samples were stained with the viability dye TOPRO-3 (Molecular Probes, Eugene, OR) or a nuclear stain DRAQ5 (Alexis Biochemicals, San Diego, CA) to define the gate for damaged cells. The FlowJo program (Tree-star, Ashland, OR) was used for off-line anaylsis.
Imagestream 100 Acquisition and Analysis
A total of 10 000–20 000 event image files were acquired at rates up to 200 events/s using a multispectral Imagestream 100 imaging cytometer (Amnis, Seattle, WA) equipped with a 488 nm cw laser. Single color controls were used to calculate a spectral crosstalk matrix, and the compensated files were analyzed using image-based statistical algorithms available from the IDEAS statistical analysis software package (Amnis, Seattle, WA). To separate cells from debris and clumps, individual, nondoublet cells were selected by comparing the bright-field aspect ratio to the bright-field area. Single cells were rounder (aspect ratio, ∼1) and smaller than clumps of cells. Identification of cells also was based on selection of distinct cell populations above background fluorescence and confirmed visually by the appearance of cytoplasmic and circumferential fluorescence signals, respectively.
Immunocytochemistry and Brain Imaging
Brains from dimm (c929)-GAL4/UAS-mCD8-GFP transgenic animals were dissected from adult male and female flies in PBS, pH 7.4, fixed at 4 °C for 30 min, rinsed with several washes of PBS containing 0.1% Triton-X 100 (PBT), and then blocked in PBT with 5% normal goat serum at room temperature for 1 h. Subsequently, the tissues were incubated overnight at 4 °C with anti-short Neuropeptide F (sNPF) polyclonal antibody (1:500 in blocking medium; gift from Dr. Ping Shen, University of Georgia, GA) and anti-NC82 monoclonal antibody, a neuropil marker (1:100 in blocking medium; gift from Dr. Alois Haufbauer, University of Regensburg, Germany). The following day, tissues were rinsed several times with PBT and incubated with goat-anti-rabbit secondary antibody conjugated with Alexa 594 (1:200) and goat-anti-mouse secondary antibody conjugated with Cy5 (1:200) in blocking medium for 2 h at room temperature. Following several rinses with PBT, tissues were mounted on glass slides using fluorescent mounting medium (Vectashield; Vector Laboratories, Burlingame, CA). For colocalization of sNPF and serotonin, brains from flies with the NPF-GAL4:UAS-mCD8-GFP genotype were prepared as above and incubated with a rabbit polyclonal antibody to serotonin (1:1000 in blocking medium; Sigma-Aldrich; St. Louis, MO). Confocal images were taken using a Zeiss LSM META 510 confocal microscope (Thornwood, NY), processed with LSM 510 image examiner and an Olympus BX61W1 FluoView confocal microscope (Center Valley, PA), and processed with Fluoview 1.7A and ImageJ software (available at http://rsb.info.nih.gov/ij; developed by Wayne Rasband, National Institutes of Health, Bethesda, MD).
Mass Spectrometry
In the course of this study, mass measurements were obtained using several different instrument configurations depending on the analytical question, sample complexity, and availability of the instruments. Highly concentrated, complex mixtures were analyzed using liquid chromatography (LC) in conjunction with on-line electrospray ionization (ESI-) or off-line MALDI-Fourier Transform (FT) mass spectrometry. Less complex mixtures requiring high detection sensitivity were analyzed using MALDI-TOF, MALDI-FT, and/or MALDI-TOF/TOF MS.
Capillary LC-ESI-Tandem MS
Capillary liquid chromatography coupled with electrospray ionization quadrupole time-of-flight tandem mass spectrometry (CapLC-ESI-Q-TOF MS/MS) experiments were conducted using a Waters CapLC coupled to a hybrid Micromass Q-TOF system (Waters Corp., Milford, MA). The CapLC contains three pumps, A and B for gradient formation and C for sample injection and delivery, a stream selector (Valco Instruments, Houston, TX), and a Waters autosampler. Solvent A was 5% acetonitrile in 0.1% formic acid and solvent B was 95% acetonitrile in 0.1% formic acid. A micro-T with a 1-m length of capillary tubing (24 μm i.d., Polymicro Technologies, Phoenix, AZ) was used to split the gradient before the capillary column to produce a flow rate of 200 nL/min. Chromatographic separation was performed using a precolumn (300 μm i.d.,1 mm length, C18 PepMap 300, 5 μm, 300 Å, LC-Packings) acting as a reverse phase support to trap and concentrate the peptides and to desalt. An aliquot of 1.4 μL of the sample corresponding to 14 of the 200 brains was loaded onto the precolumn equilibrated with an isocratic flow at 20 μL/min of 5% acetonitrile/0.1% formic acid. After 3 min, the valve position of the stream selector was switched, placing the precolumn online with the analytical capillary column (75 μm i.d., 150 mm length, packed with 3 μm of C18 particles, Dionex Corp., Sunnyvale, CA). Separation was conducted using a linear gradient from 5% B to 70% B in 65 min, then to 100% B in 10 min. The gradient was kept at 100% B for 15 min, followed by a linear gradient to 5% B in 10 min. The column was equilibrated at 5% B for 15 min before starting the next run.
Mass spectra were recorded in positive ion mode. The capillary voltage was set at 3.8 kV, the cone voltage at 40 V, and the source block temperature at 90 °C. A data-dependent acquisition method was employed for the MS survey scan and the selection of precursor ions and subsequent MS/MS of the selected parent ions. The MS/MS peak detection window was set at 3 Da. The collision energy was set according to the ion charge state and the mass-to-charge ratio (m/z) and varied from 16 to 55 eV. The MS scan range was set from m/z 300–2000 and the MS/MS scan from m/z 50–2000. A lock-mass was utilized to correct the mass shift during the sample run. The baffle switched between the analyte and reference position at a frequency of 10 s to sequentially sample the ions from the analyte and reference solution (1 pmol/μL proglufibrinopeptide in 50:50 acetonitrile and water with 0.1% formic acid) to the mass spectrometer.
Off-line HPLC with MALDI-FTMS Analysis
The portion of the extract from 200 adult D. melanogaster brains not used for CapLC-tandem MS experiments was fractionated on a 1.0 mm i.d. Vdac C18 column using a Dnamax HPLC sstem (Rainin, Palo Alto, CA). Solvent A was 0.1% formic acid and solvent B was 0.1% formic acid in acetonitrile. The gradient used was from 5% B to 20% B over 8 min; 20% B to 50% B over 90 min; and 50% B to 95% B over 22 min at a flow rate of 0.05 mL/ min. A total of 26 fractions were collected at 5 min intervals. The LC fractions were concentrated to drness in a SpeedVac and reconstituted in 15 μL of H2O for MALDI FTMS. One microliter of saturated 2,5-dihdroxbenzoic acid matrix (Sigma-Aldrich, St. Louis, MO) and 1 μL of each reconstituted fraction were mixed on the facet of a metal target and allowed to crstallize in the air. MALDI FTMS was performed on a 7-T Ion-Spec FTMS sstem (Varian, Inc., Palo Alto, CA) equipped with a 337 nm N2 laser for desorption/ionization. Mass spectra were collected in positive ion mode using an in-cell accumulation (ICA) method whereby multiple ionization/desorption events occur prior to excitation and detection events. The experimental settings used were similar to those described in a previous paper.28 The mass accuracy of the instrument was approximately 50 ppm. Some of the data were processed using BUDA (available at http://www.bumc.bu.edu/ftms/buda; developed by Peter O’Connor, Boston University, MA). Substance P (Sigma-Aldrich, St. Louis, MO) and vasopressin (Sigma-Aldrich, St. Louis, MO) were spotted on a separate facet of the target and used for internal calibration as previously described.29
MALDI-TOF MS Analysis of Extract from Immunoaffinity-Purified Tissue
A 0.5 μL aliquot of the eluant was mixed with 0.5 μL of α-cano-4-hdroxycinnamic acid (saturated solution in 70% MeOH). The MALDI mass spectra were obtained by using a Voyager-DE Pro mass spectrometer (Applied Biosystems, Foster City, CA) equipped with a 337 nm N2 laser. Positive-ion mass spectra (m/z 500–5000) were acquired in both linear and reflectron modes. In reflectron mode, the acceleration voltage was set at 20 kV. At least 50–100 laser shots were applied, depending on signal intensity. The mass accuracy of the instrument was about 500 ppm. Bradkinin (Sigma-Aldrich, St. Louis, MO) and bovine insulin (Sigma-Aldrich, St. Louis, MO) were used for mass calibration.
MALDI-TOF/TOF MS Analysis
MALDI-TOF/TOF spectra were acquired in the LIFT mode on an Ultraflex II TOF/TOF mass spectrometer (Bruker Daltonics, Inc., Billerica, MA) equipped with a Smartbeam diode-pumped solid state laser with tunable frequency 1–200 Hz. For TOF/TOF Analysis, the acceleration voltage in the source was set at 8.0 kV, while the reflectron voltage was set at 29.5 kV. Mass spectra were acquired in positive ion mode with 200–400 laser shots. Accuracy in reflectron mode was estimated to be better than 200 ppm with external calibration. Samples were prepared by mixing 0.5 μL of the extract from immunoaffinity-purified tissue with 0.5 μL of α-cyano-4-hydroxycinnamic acid (saturated solution in 70% MeOH). Masses were annotated using the FlexAnalysis software package (Bruker Daltonics, Inc., Billerica, MA). Sequences were checked for homology to known peptides by a database search using BioTools software (Bruker Daltonics, Inc., Billerica, MA) and Mascot search engine (http://www. matrixscience.com).
Peptide Identification
Four different approaches were used for peptide identification. (1) The raw data file was transformed into peak list (PKL) files by ProteinLynx software (version 2.1, Micromass, Waters Corp., Milford, MA) and the PKL file was submitted to Mascott search (http://www.matrixscience.com). Fruit fl taxonomy restriction was used and C-amidation, tyrosine-sulfation, N-pyroglutamic acid modification, and methionine-oxidation were set as variable modifications. (2) The MS/MS de novo sequencing was performed with a combination of manual sequencing and automatic sequencing by PepSeq software (Waters Corp., Milford, MA). (3) The fragmentation pattern of an ion with an experimental mass corresponding to the calculated mass of a known or predicted D. melanogaster neuropeptide was compared with the theoretical fragmentation spectrum of the peptide obtained from ProteinProspector (http://prospector.ucsf.edu) to check the match. (4) The fragmentation spectrum was first analyzed manually and the deduced amino acid sequence used in a BLAST search against the Flybase database (http://www.flybase.org) to check if the sequence belonged to a D. melanogaster gene-encoded fragment.
For peptide identifications based on FTMS or MALDI-TOF measurements, assignments were made if the difference between the measured and theoretical monoisotopic values was within the mass accuracy range for each of the instruments (i.e., 50 ppm for FTMS and 500 ppm for MALDI-TOF MS). For certain peptides, supporting sequence information was provided by MS/MS analysis and detection of expected fragments (see MALDI-TOF/TOF section). Chemical derivatization using acetylation of primary amino groups (found on unblocked N-termini and on Lys residues) also assisted peptide identification.
Results and Discussion
Three main approaches have been used in previous studies to characterize peptide expression in the fruit fly: (1) bioinformatic analysis of the D. melanogaster genome;30 (2) LC/MS/MS Analysis of extracts prepared from larval-stage animals,19,20 and (3) direct MALDI TOF profiling of neurons dissected from subregions of the adult fly brain.21,22 The results of our current study complement the previous findings by identifing novel peptides using MS/MS structural analysis of extracts from adult brain and by using MS-based identification methods on isolated populations of adult neurons. Despite the largely redundant inventory of peptides found when comparing larval to adult tissue, it was important to perform this peptidomic analysis in the adult CNS. Since peptides likely serve many essential roles in behavior in adult animals, it is highly desirable to know the precise sequence of the ligands involved in these behaviors and to understand the secondary modifications undergone by these substances.
Characterization of Neuropeptides from Adult D. melanogaster Brains
When a combination of LC/MS/MS and off-line HPLC followed by MALDI-FTMS analysis was used, 42 neuropeptides encoded by 18 D. melanogaster genes were identified from the extract of adult D. melanogaster brain. For a complete list of encoded genes, neuropeptide families, amino acid sequences, and for a comparison with earlier studies of other investigators, see Table 1 and Supporting Information Table 1. The analysis of the peptide extract with capillary liquid chromatography coupled on-line with ESI-QTOF MS/MS yielded 26 fully sequenced peptides, with m/z values and MS/MS fragmentation patterns supporting the predicted amino acid sequence after processing of the prepropeptide at mono- or dibasic cleavage sites, and in many cases, after C-terminal amidation. Eight precursor molecules or fragments of identified peptides also were sequenced (highlighted in blue, Table 1), while six peptides that were not obvious precursors or fragments of identified peptides and whose gene transcript did not predict flanking basic cleavage sites were identified (highlighted in gray, Table 1). Four of the neuropeptides had not been reported previously in fruit flies (highlighted in yellow, Table 1). The extract also was characterized using HPLC followed by MALDI-FTMS analysis of individual fractions. FTMS offers higher mass accuracy and resolving power and can provide different complementary peptide coverage. Thus, in addition to detecting many of the peptides found with ESI-Q-TOF sequencing, this approach also resulted in finding two additional peptides (one from the Allatostatin B family and the other a Corazonin peptide) that were not observed with the Q-TOF analysis. Examples of peptide identification by either ESI-tandem MS or exact mass MALDI-FTMS are shown in Figure 1. The neuropeptides and their short forms reported in this work are grouped into families and described in what follows.
Table 1.
Summary of Amino Acid Sequences of the Peptides Identified in This Study (Using LC-ESI-QTOF-MS/MS and/or Exact Mass Measurements Employing Off-line HPLC MALDI-FTMS) and Previously Identified Peptides
| Encoding genea |
Peptide Family | Neuropeptide Sequenceb |
Previous work | This work | |||
|---|---|---|---|---|---|---|---|
| Larval Tissuec,d | Adult Tissuee | Adult Tissue | |||||
| 1-D LC/MS | 2-D LC/MS | Direct MALDI MS | ESO-Q-TOF MS | MALDI-FTMS | |||
| CG1171f | Adipokinetic hormone (Akh) | pQL TFSPDWa | + | + | |||
| c.pQL TFSPDWGK.R | + | + | |||||
| CG 13633g Y/F(X)FGL-mide |
Allatostatin A(Ast) | KR.LPVYNFGLa.GKR | + | ||||
| KR.SRPYSFGLa.GKR | + | + | + | ||||
| KR.VERYAFGLa.GRR | + | ||||||
| RR.AYMYTNGGPGM.KR | + | ||||||
| KR.TTRPQPFNFGLa.GRR | + | + | + | + | + | ||
| CG6456h W(X)6Wa |
Allatostatin B (Myoinhibiting peptide) |
KR.AWKSMNVAWa.GKR | + | + | |||
| KR.DQWQKLHGGWa.GKR | + | + | + | ||||
| KR.RQAQGWNKFRGAWa.GKR | + | + | + | ||||
| CG14919i | Allatostain C(Ast C) | L.FAQYRPTSYSA.Y | + | ||||
| A.YLRSPTYGNVNEL.Y | + | ||||||
| CG3302j | Corazonin(Crz) | G.pQTFQYSRGWTNa.GKR | + | + | + | + | |
| CG6440k | Dromyosupressin (Dms) | KR.TDVDHVFLRFa.GKR | + | + | + | + | + |
| KRT.DVDHVFLRFa.GKR | + | ||||||
| KR.TDVDHVFLR.FGKR | + | ||||||
| CG18090l | Drosulfakinin (Dsk) | KR.FDDYGHMRFa.GKR | + | + | + | ||
| KR.FDDYGHmRFa.GKR | + | ||||||
| KR.GGDDQFDDYGHMRFa.GR | + | + | + | ||||
| CG2346m | FMRFamide | KR.AAMDRYa.GR | + | ||||
| R.SDNFMRFa.GR | + | + | + | ||||
| R.PDNFMRFa.GR | + | + | + | ||||
| R.SAPQDFVRSa.GK | + | + | |||||
| GK.MDSNFIRFa.GK | + | + | |||||
| R.TPAEDFMRFa.GR | + | + | + | + | |||
| RR.SVQDNFMHFa.GKR | + | ||||||
| R.SPKQDFMRFa.GR | + | ||||||
| R.DPKQDFMRFa.GR | + | + | + | + | |||
| RD.PKQDFMRFa.GR | + | ||||||
| CG6371n | HUGIN | KK.SVPFKPRLa.GKR | + | + | + | + | |
| CG13480o | Leucokinin (Drm-kinin) | KR.NSVVLGKKQRFHSWGa.GKR | + | + | + | + | |
| CG13968p | Short Neuropeptide Fs (short NPFs) |
RK.PQRLRWa.GR | + | ||||
| R.SPSLRLRFa.GR | + | + | + | + | |||
| R.KPMRLRWa.GR | + | ||||||
| KR.WFGDVNQKPHR | + | + | + | + | |||
| R.SDPDMLNSIVE.KR | + | + | + | + | |||
| R.SDPDmLNSIVE.KR | + | ||||||
| RK.AQRSPSLRLRFa.GR | + | ||||||
| CG3441q | Neuropeptide-like precursor 1 (Nplp 1) |
KR.SVAALAAQGLLNAPK.R | + | + | + | + | + |
| KR.YIGSLARAGGLMTYa.GKR | + | + | + | + | |||
| KR.NLGALKSSPVHGVQQ.KR | + | + | |||||
| KR.NVGTLARDFQLPIPNa.GKR | + | + | + | + | + | ||
| KR.NLGALKSSPVHGVQ.QKR | + | ||||||
| KR.YIGSLARAGGLMT.YGKR | + | ||||||
| CG11051r | Neuropeptide-like precursor 2 (Nplp2) |
L.TKAQGDFNEF.I | + | ||||
| R.EESNPAQEFLTK.A | + | ||||||
| K.AQGDFNEFIEKLK.A | + | ||||||
| R.EESNPAQEFLTKAQGDF.N | + | ||||||
| R.EESNPAQEFLTKAQGDFNEF.I | + | ||||||
| F.LTKAQGDFNEF.I | + | ||||||
| CG6496s | Pigment dispersing factor (PDF) | KR.NSELINSLLSLPKNMNDAa.GK | + | ||||
| CG15520t F(X)PPLamide or PRVamide |
Pyrokinin (Capa) | KK.ASGLVAFPRVa.GR | + | + | + | ||
| RR.GANMGLYAFPRVa.GR | + | + | + | ||||
| KRT.GPSASSGLWFGPRLa.GKR | + | + | + | + | |||
| KR.TGPSASSGLWFGPRLa.GKR | + | + | + | + | |||
| CG33527u | SIFamide | A.AYRKPPFNGSIFa.GKR | + | + | |||
| AA.YRKPPFNGSIFa.GKR | + | + | + | + | |||
| CG14734v F(X)1G(X)2Ramide |
Tachykinin-like peptides | KR.APTGFTGMRa.GKR | + | ||||
| KK.APLAFVGLRa.GK | + | ||||||
| KR.APNGFLGMRa.GKR | + | + | + | ||||
| KR.APTSSFIGMRa.GKK | + | + | |||||
| KR.APVNSFVGMRa.GKK | + | + | + | + | |||
| CG15231w | Immune-induced peptide (IM4) | P.GTVLIQTDNTQYIRTa.G. | + | + | |||
Drosophila computated gene (CG) number.
Amino acid sequences of peptides flanked by the predicted cleavage sites (indicated by •). Fields in yellow indicate previously unidentified peptides. Fields in blue indicate precursors or fragments of identified peptides. Fields in gray indicate peptides not predicted to be flanked by mono- or dibasic cleavage sites according to the encoding gene.
One-dimensional LC/MS on extract from larval nervous system (ref. 20).
Two-dimensional LC/MS on extract from larval nervous system (ref. 19).
Peptide expression characterized by direct MALDI-TOF MS analysis on dissected tissue (refs 21, 22).
Ref 37.
Ref 39.
Ref 40.
Ref 42.
Ref 48.
Ref 51.
Ref 58.
Ref 63.
Ref 20.
Ref 20.
Ref 71.
Ref 76.
Ref 79.
Ref 80. m, oxidized Met residue; q, pyroglutamic acid; X, any amino acid.
Figure 1.
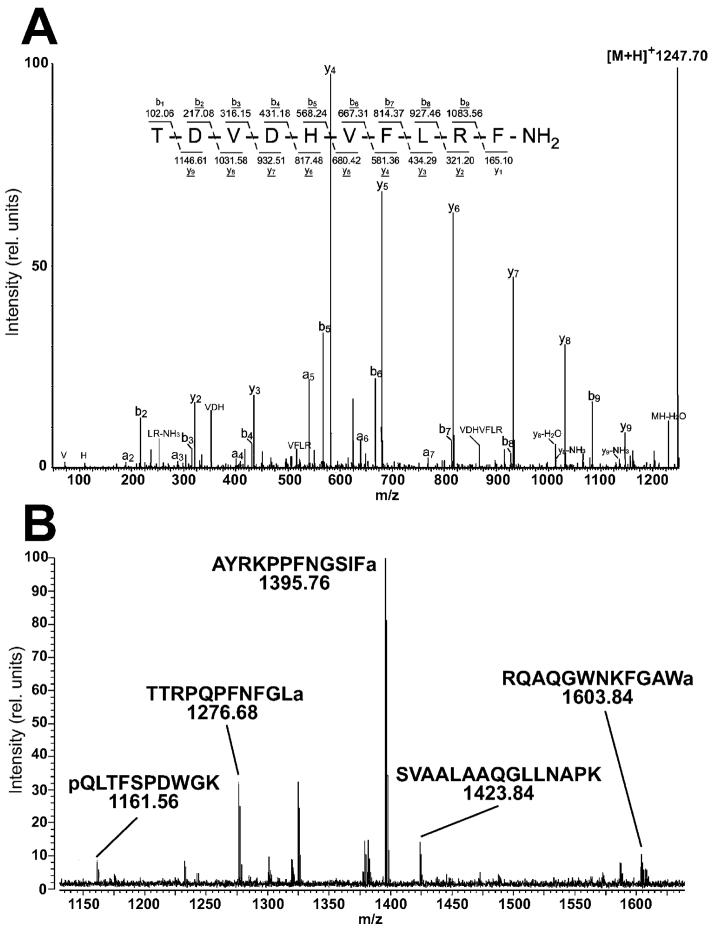
Examples of mass spectra obtained by LC-ESI-QTOF-CID MS and off-line HPLC/MALDI-FTICR MS analysis from extracts of pooled D. melanogaster brain tissue. (A) ESI-Q-TOF MS/MS analysis of the isolated protonated molecular ion with m/z 1247.70 generated almost complete y- and b-ion series and resulted in identification of this molecule as Dromyosuppressin. The fragment ions are labeled according to the nomenclature of Biemann.105 The sequence is consistent with the predicted product encoded by gene CG6440.48 (B) MALDI-FT mass spectrum of one LC fraction obtained by HPLC separation of the extract. The quasimolecular ions [M + H]+ of several neuropeptides or intermediate processing products were present in this LC fraction including the AKH processing intermediate (m/z 1161.56), an Allatostatin A peptide (m/z 1276.68), a member of the SIFamide family (m/z 1395.76), an Allatostatin B peptide (m/z 1603.84), and a member of the Nplp1 family (m/z 1423.84).
Adipokinetic Hormone (AKH, CG1171)
Members of this family function as homeostatic regulators, controlling the release of energy-yielding lipid and carbohydrate substrates during flight.31,32 A member of the AKH family of arthropod neuropeptides (pQLTFSPDWa) has been identified previously in fruit fly extracts using automated Edman degradation and fast atom bombardment (FAB) mass spectrometry.33 The gene, its expression pattern, and the function of the peptide as a cardioaccelerator were subsequently characterized.34 An incompletely processed form of this peptide was identified in the present work, with the N-terminal end still in the proglutamic acid form and the C-terminal residue remaining nonamidated.
Allatostatins (CG13633, CG6456, and CG14919)
The insect Allatostatins are a diverse group of neuropeptides that act on the corpora allata to block the release of juvenile hormone and on the gut to block smooth muscle contraction.35 These peptides also modulate motoneurons and muscle activity in the adult animal. The preprohormones of three types of Allatostatins in D. melanogaster were cloned in earlier studies.36-40 The CG13633 gene encodes a large group of A-type (cockroach-type) Allatostatins with a common C-terminal sequence Y/F(X)FGLamide. We identified two CG13633-encoded peptides (LPVYNFGL-amide and TTRPQPFNFGL-amide), three of which have been found before; LPVYNFGLa has not been reported in earlier publications. CG6456 encodes a B-type (cricket-type) Allatostatin with a shared C-terminal sequence W(X)6Wamide. Of the five putative CG6456 Drosophila encoded peptides, we detected RQAQGWNKFRGAWa with both on-line ESI-Q-TOF and off-line HPLC MALDI-FT mass spectrometry and DQWQKLHGGWa with MALDI-FTMS onl. The peptide RQAQGWNKFRGAWa was not detected by earlier investigators in the larval nervous system, 19,20 raising the possibility that its expression is developmentally regulated. CG14919 encodes a single peptide with the predicted sequence QVRYRQCYFNPISCF (Allatostatin C). Allatostatin C is not amidated at the C-terminus in D. melanogaster, and the overall sequence is unrelated to the A- and B-types. The predicted Allatostatin C was not identified in our studies, but two fragments encoded by the same gene were found and these are not flanked by common basic cleavage sites. Allatostatin C in D. melanogaster differs by a single amino acid residue (F → Y in position 4) from the C-type Allatostatin in Manduca sexta.41
Corazonin (CG3302)
Corazonin is a cardioacceleratory peptide first isolated from the corpora cardiaca of the American cockroach.42 The peptide has been shown to stimulate heart-beat frequency and hyperneural muscle in the cockroach and to influence pigmentation in nymphs and locusts.43 Thus far, no function has been attributed to Corazonin in D. melanogaster. The peptide sequence (pQTFQYSRGWTNa) is blocked at the N-terminus with pyroglutamic acid. The blocking and charge sequestering effect caused by an internal Arg residue leads to a low ionization and fragmentation efficiency of this peptide using LC-ESI-QTOF MS. This may explain the reason Corazonin was detected only in the MALDI-FTMS analysis.
Dromyosuppressin (CG6440)
In the fruit fly, the Dromyosuppressin peptide is expressed in the nervous system in addition to reproductive and gastrointestinal tissue.44 Endogenous application of the peptide results in contractions of the heart45 and foregut.46,47 This study identified the only neuropeptide encoded by the Dromyosuppressin gene, TDVDHVLRFamide.48
Drosulfakinins (DSK, CG18090)
Sulfakinins, showing sequence similarity to vertebrate Gastrin- and CCK-like peptides, have been found in a large number of insect and mammalian species.49,50 These peptides also have been identified as modulators of motor neurons and muscle activity in a number of peripheral organs.43 The gene coding for the fruit fly Sulfakinin peptides has been cloned and sequenced.51 It predicts three putative peptides, DSK-0 (NQKTMSFa), DSK-I (FDDYGHMRFa), and DSK-II (GGDDQFDDYGHMRFa).51 A sulfated Tyr residue is required for the biological activity of Sulfakinins,52 but we isolated DSK-I and DSK-II only in nonsulfated forms. It is possible that the peptide was desulfated during sample preparation or that the sulfated form is not detected in positive ion mode. An oxidized form of DSK-I (at the Met residue) was found in our studies in addition to the unmodified version. This modification may be an artifact from sample preparation, though it should be noted that, with the exception of one of the short Neuropeptide F products, oxidized versions of other Met-containing peptides were not detected.
FMRFamides and FMRFamide-Related Peptides (CG2346)
FMRFamide family peptides have been shown to evoke muscle contractions at neuromuscular junctions in D. melanogaster.53 Immunocytochemical studies demonstrate FMRFamide staining in neurohemal release regions as well, suggesting a hormonal role for these peptides.54 The FMRFamide gene encodes a number of peptides with FMRFamide or related sequences at their C-termini.55,56 We identified four FMRFamide-related peptides in these studies. Of these, the peptide AAMDRYa was not reported in previous papers and the peptide SAPQDFVRSa was not discovered in pooled larval central nervous system extracts. The present studies did not find significant amounts of several of the six FMRFamide-related peptides identified in the earlier larval studies (Table 1) most likely because tissues from the central nervous sstem (CNS) rather than the periphery were analyzed. It is possible that the peptide PKQDFMRFa is a fragment formed from the cleavage of the N-terminal Asp-Pro peptide bond found in DPKQDFMRFa. This cleavage has been shown to be particularly labile in mild acidic conditions.57
HUGIN (CG6371)58
The HUGIN peptide is one of two D. melanogaster Pyrokinin genes. Peptides belonging to this family are widely distributed in insect species and contain C-terminal F(X)PRLamide or PRVamide motifs.59,60 The HUGIN geneencodes two predicted peptides: hug γ (QLQSNGEPAYRVRTPRLa) and Drm-PK-2 (SVPFKPRLGa).61 The Drm-PK-2 peptide was found in our work. The physiological effects of HUGIN gene products include the initiation of feeding and a decrease in the inhibition of feeding from novel sources.
Leucokinin (CG13480)
Leucokinin is likely to function as a homeostatic regulatory by influencing fluid secretion.62,63 Both the doubly- and triply charged ions of D. melanogaster Leucokinin were detected in this work in the pooled adult brain extracts. The product spectrum of these precursor ions corresponds to the sequence of NSVVLGKKQRFHSWG. This peptide also was found in extracts of the CNS of larval fruit flies.20
Short NPF-like Peptides (sNPF, CG13968)
sNPF, the fly homologue of Neuropeptide Y, has been shown to modulate feeding and social behavior in larvae7 and to mediate sensitivit to ethanol sedation64 and aggressive behavior65 in adults. Five sequences encoded by the sNPF gene30,66 were identified in this study. Two of these peptides, PQRLRWa and KPMRLRWa, were not reported in previous papers.
Nplp1 (CG3441)
The Neuropeptide-like precursor 1 peptide family was first identified using a peptidomics analysis of larval nervous tissue.19,20 Four members of the family were sequenced in these studies. We identified 4 of the predicted famil members and 2 possible precursors or fragment molecules. These peptides have no identified function, though their expression in a subset of Drosophila clock neurons suggests a possible role in circadian control.67
Pigment Dispersing Factor (PDF, CG6496)
First discovered in crustaceans,68 PDF in D. melanogaster is expressed by the ventral subset of lateral pacemaker neurons69 important in the regulation of circadian output.70-72 Loss of PDF expression or ablation of the PDF neurons produces abnormal locomotor rhythms.72 A recent study using direct MALDI-TOF MS analysis of tissue showed biochemical expression of the amidated peptide in the adult fly brain.21 The amidated form of PDF was identified in our work using ESI-Q-TOF-CID sequencing.
Pyrokinin
Two D. melanogaster Pyrokinin genes have been identified: Capa (CG15520) and HUGIN (CG6371).58 Peptides belonging to this family are involved in the regulation of a wide variety of physiological activities including fluid secretion,54,62 hindgut contractility, pheromone biosynthesis, cuticular melanization, diapause induction and pupariation.73 From the gene sequence, Capa encodes at least three putative neuropeptides: two are similar in sequence to the cardioacceleratory peptide CAP2b found in M. sexta74 and one is a Pyrokinin-like peptide. In the gene transcript, each encoded peptide is flanked by consensus processing sites and all the peptides have C-terminal Gly residues.75 We found no CAP2b-like peptides, but the full Pyrokinin peptide and a form with one amino acid truncated from the N-terminus were identified with high confidence using both Q-TOF and FTMS.
SIFamide (CG3352776)
The present study identified SIFamide and its N-terminal truncated form. This fragment is a homologue of a 12-amino acid peptide purified from the blowfly that stimulates oviduct contraction in Locusta migratoria.77 In D. melanogaster, the SIFamide peptide is expressed in only four neurons of the pars intercerebralis shown to modulate adult courtship behavior.78
Tachykinin-like Peptides (TKLPs, CG14734)
Found in both vertebrates and invertebrates, a large and diverse number of peptides belong to the TKLP family. In invertebrates, these peptides are identified by a common C-terminal F(X)1G(X)2 Ramide motif. In adult D. melanogaster, TKLPs have been shown to modulate odor perception and locomotor activity.79 Two TKLPs were discovered in this work: APNGFLGMRa, identified with FTMS, and APVNSFVGMRa, sequenced using Q-TOF mass spectrometry.
Other Peptides and Fragments
A Mascot search of the processed raw data identified fragments derived from several other peptide families such as Allatostatin C, Nplp2,20 and Immune-induced peptide (IM4).80 These fragments could be formed by chemical or enzymatic breakdown during preparation of the brain extracts. Regardless of the mechanism of degradation, however, detection of these fragments suggests that the parent peptides should be present in the adult brain. Further studies will be necessary, however, to determine their full-length amino acid sequences.
Compared to studies from extracts of larval tissue,19,20 the set of D. melanogaster peptides found in the adult CNS overlapped but were not identical. In our work, peptides from the FMRFamide, short NPF, and Allatostatin A and C families were sequenced that had not been identified previously. Conversely, certain peptides from the Cap2b FMRFamide, Allatostatin, Adipokinetic hormone, and Tachykinin-like families were found in the earlier studies but not in this work. These differences could result from developmental changes in peptide expression, but also could be accounted for by variation in sample preparation. Notably, a number of peptides predicted by genomic sequences have not been found in any studies, including this one. These could be peptides that are degraded rapidly, present at low concentrations, processed and expressed during a transient time window, expressed in tissues other than the CNS, or simply peptides that are not translated.
Neuron Subtype-Specific Localization of Peptides Using Genetically Mediated Immunoaffinity Purification
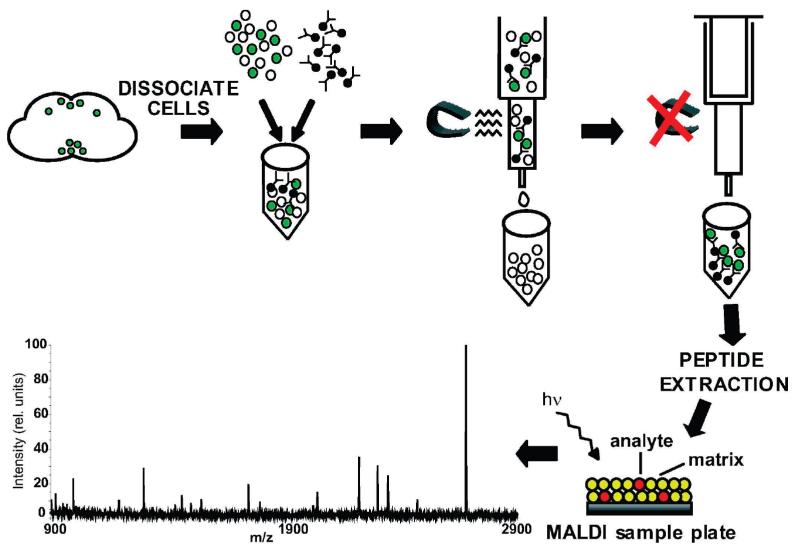
To localize peptides in subtypes of neurons in the adult brain, the GAL4-UAS system was used to express a surface antigen consisting of the extracellular and transmembrane domains of the mouse CD8 protein fused to GFP (mCD8-GFP).81 Two different drivers were used to specify expression of the cell surface antigen on select populations of cells: (1) dimm (c929)-GAL4, a marker of primarily neuropeptidergic cells; and (2) Ddc-GAL4, a marker of dopaminergic and serotonergic cells. Expression of this cell surface antigen on the cell membrane allowed labeled cells to be isolated via immunoaffinity purification and the respective peptide content of each cell population to be assayed by mass spectrometry (Figure 2). This method had been used to isolate ovary border cells in D. melanogaster,82 but to our knowledge, it has not been used previously for neuronal enrichment, particularly in conjunction with mass spectrometr.
Figure 2.
Outline of method used to enrich for labeled populations of neurons in the adult D. melanogaster brain. Adult brains genetically labeled with the surface marker mCD8-GFP using the GAL4-UAS expression system are dissociated into single cell preparations and incubated with paramagnetic beads conjugated to a monoclonal anti-mCD8 antibody. The mixture is placed in a magnetic column allowing antibody-captured cells to be retained in the column. When the magnet is removed, the cells are eluted. Following an acidified methanol extraction, the contents of the immunoaffinity-purified cells are analyzed by MALDI-TOF mass spectrometry.
Purity of Cell Samples Estimated by FACS
Following immunoaffinity enrichment, FACS analysis determined that GFP and the nuclear stain DRAQ5 was expressed in more than 55% of the cells isolated using the dimm (c929)-GAL4 driver and more than 35% of the cells isolated using the Ddc-GAL4 driver (Figure 3). It is estimated that GFP-positive cells represented less than 0.5–1% of the total cell population in both of these lines; therefore, this method resulted in a 30- to 50-fold enrichment of GFP-positive cells. When using a third driver (Tyrosine hdroxylase-GAL4) which is expressed in less than approximately 0.1% of the total number of neurons in the central brain, no significant enrichment was observed. It may be suggested that this is the lower limit of the immunoaffinity technique and that this method is better suited for isolation from tissue in which GFP is expressed at least above 0.5% of the original cell population. Longer incubation times with the immunoaffinity reagents may help, but this comes at the cost of cell viability.
Figure 3.
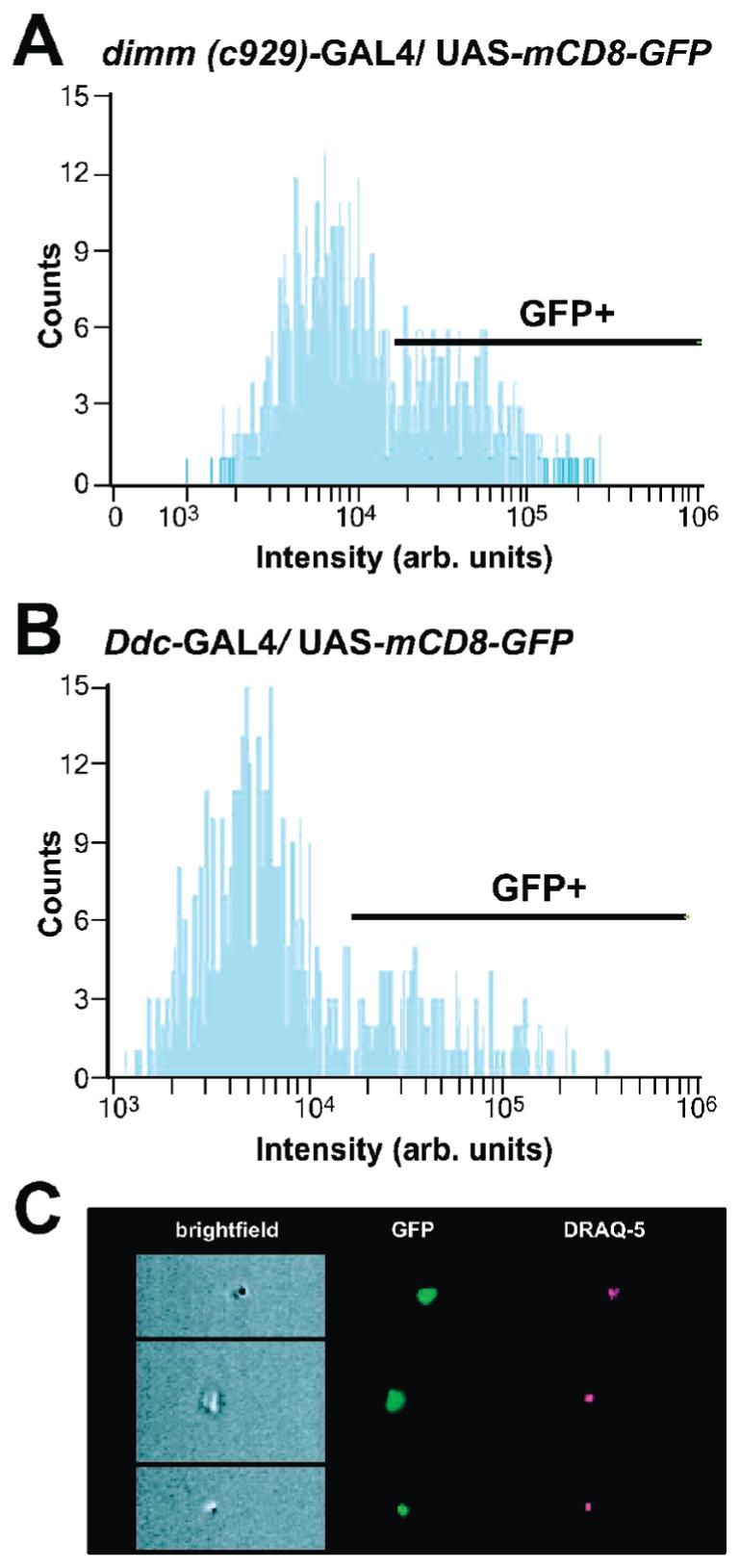
The efficiency of the immunoaffinity enrichment method was assessed by FACS analysis. The histograms demonstrate the counts of DRAQ5-positive cells expressing GFP in cell suspension after immunoaffinity enrichment. With the use of the dimm (c929)-GAL4 driver, 55% of the cells were GFP-positive (A). The line indicates the gate intensity threshold for GFP-positive fluorescence. With the use of the Ddc-GAL4 driver, 35% of the cells were GFP-positive (B). Live cells were distinguished from necrotic cells using an imaging flow cytometer which provided simultaneous analysis of morphological features (brightfield illumination), GFP fluorescence (green), and DRAQ5 staining (red). Representative images of cells isolated with the dimm (c929)-GAL4 driver are shown (C).
Enrichment and MS Characterization of Genetically Labeled Cells
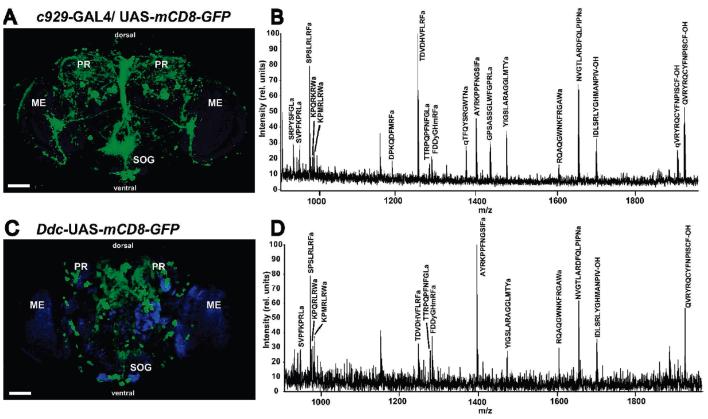
The dimm (c929)-GAL4 driver is expressed in many neuroendocrine and neurosecretory cells24 and was therefore used to ask whether adult neurons would retain their peptide content during the isolation procedure and to determine whether the peptide concentration from the captured cells would be adequate for MS detection. When the dimm (c929)-GAL4 driver line was crossed with UAS-mCD8-GFP flies, progeny of this cross express GFP strongly in the subesophageal ganglion (SOG), in processes around the foramen, and in various neurosecretory cells in the medial superior protocerebrum59 (Figure 4A). Many peptide families including Allatostatin A, B, and C,83 Cap2b,84 Corazonin,85 Dromyosuppressin,44 Drosulfakinin.22,86 Diuretic hormones,83 FMRFamide peptides,87,88 SIFamide peptides,77 PDF,43,89 Pyrokinn,90 HUGIN,91 the short NPFs,7 and Tachykinin-like peptides43 have been shown by immunocytochemistry and enhancer trap analysis to express in the SOG in D. melanogaster and other insects. Mass spectrometric analysis of the peptide extract from cells isolated using this immunoaffinity procedure detected numerous ions with m/z ratios corresponding to known and predicted neuropeptides (Figure 4B). These signals represented a large subset of the peptides that were identified in the adult whole brain extracts. In addition, several signals were seen that could not be assigned to known peptide sequences, indicating the presence of novel peptides, intermediates in the processing of peptides, or fragments derived from the proteolytic breakdown of proteins or peptides. From the dimm (c929)-GAL4 driver, 29 peptides were identified, including members of the Allatostatin family, Corazonin peptides, Dromyosuppressin, Drosulfakinin, FMRFamide, HUGIN, short NPFs, and Nplp1 peptide (Table 2; Supporting Information Table 2). The amino acid sequences of many of these peptides were confirmed by ESI-Q-TOF CID analysis of extract from whole brain, including the novel peptide KPMRLRWamide. It was not possible to perform ESI mass spectral analysis on the immunopurified extracts due to insufficient amount of material. Control experiments in which parent lines were carried through the same extraction and mass spectrometric analysis showed no peaks corresponding to known peptides (data not shown).
Figure 4.
Frontal confocal microscope images of D. melanogaster adult brain tissue genetically labeled with the cell-surface antigen mCD8-GFP (green) and the corresponding mass spectral profile of extract from cell populations enriched using GAL4-UAS-mediated immunoaffinity purification. Counterstaining with the anti-NC82 antibody was used to mark the neuropil regions (blue). The dimm (c929)- (A) or Ddc-GAL4 (C) driver lines targeted, respectively, primarily peptidergic or dopaminergic and serotonergic cells. Dorsal and ventral aspects of the fly brain are indicated. MALDI-TOF MS analysis of extract from neuronal subpopulations enriched using the dimm (c929)-GAL4 driver gave the broadest peptide profile (B), whereas isolation with the Ddc-GAL4 driver (D) resulted in simpler profile. Signals of identified peptide ion species have been labeled with the predicted amino acid sequence. PR, protocerebrum; ME, medulla; SOG, subesophageal ganglion. Scale bar: 50 μm.
Table 2.
Peptides Identified Using the Immunoaffinity Cell-Enrichment/Mass Spectrometry Method with the dimm (c929)-, or Ddc-GAL4 Driver Lines (Indicated by Fields in Gray)
| Calculated mass of [M+H]+ iona |
Peptide Family | Predicted peptide sequenceb |
dimm-GAL4 enriched cells |
Ddc-GAL4 enriched cells |
|---|---|---|---|---|
| 925.49 | Allatostatin A | SRPYSFGLa | 1 | |
| 953.52 | Allatostatin A | VERYAFGLa | ||
| 1276.68 | Allatostatin A | TTRPQPFNFGLa | 1,2 | 1 |
| 1091.55 | Allatostatin B (Myoinhibiting peptide) | AWKSMNVAWa | ||
| 1253.62 | Allatostatin B (Myoinhibiting peptide) | DQWQKLHGGWa | ||
| 1374.66 | Allatostatin B (Myoinhibiting peptide) | EPTWNNLKGMWa | ||
| 1603.84 | Allatostatin B (Myoinhibiting peptide) | RQAQGWNKFRGAWa | 1 | 1 |
| 1906.87 | Allatostatin C | qVRYRQCYFNPISCF-OH | 1 | 1 |
| 1923.90 | Allatostatin C | QVRYRQCYFNPISCF-OH | 1 | 1 |
| 1015.61 | Cap2b | ASGLVAFPRVa | ||
| 1294.67 | Cap2b | GANMGLYAFPRFa | ||
| 1406.78 | Cap2b | WAHLLALQQVLD-OH | ||
| 1076.57 | Corazonin | APVNSFVGMRa | ||
| 1369.63 | Corazonin | qTFQYSRGWNa | 1 | |
| 1247.65 | Dromyosuppressin | TDVDHVFLRFa | 1,2,3 | 1 |
| 854.42 | Drosulfakinin | NQKTMSFa | ||
| 1282.47 | Drosulfakinin | FDDyGHmRFa | 1 | 1 |
| 1738.67 | Drosulfakinin | GGDDQFDDyGHMRF | 1 | |
| 925.44 | FMRFamide | PDNFMRFa | ||
| 1005.51 | FMRFamide | SAPQDFVRSa | ||
| 1154.58 | FMRFamide | SPKQDFMRFa | ||
| 1182.57 | FMRFamide | DPKQDFMRFa | 1 | 1 |
| 942.59 | HUGIN | SVPFKPRLa | 1 | 1 |
| 1698.90 | Lipoprotein | IDLSRLYGHMANPIV-OH | 1,2 | 1 |
| 1423.84 | Nplp 1 | SVAALAAQGLLNAPK-OH | 1 | |
| 1471.77 | Nplp1 | YIGSLARAGGLMTYa | 1,2,3 | 1 |
| 1653.91 | Nplp1 | NVGTLARDFQLPIPNa | 1,2,3 | 1 |
| 1972.02 | NSELINSLLSLPKNMNDa | 2 | ||
| 1430.75 | Pyrokinin/Cap-2b | GPSASSGLWFGPRLa | 1,2 | |
| 1531.80 | Pyrokinin/Cap-2b | TGPSASSGLWFGPRLa | 1 | |
| 974.59 | Short NPF | SPSLRLRFa | 1,3 | 1 |
| 982.61 | Short NPF | KPQRLRWa | 1 | 1 |
| 985.59 | Short NPF | KPMRLRWa | 1 | 1 |
| 1203.62 | Short NPF | WFGDVNQKPI-OH | ||
| 1219.55 | Short NPF | SDPDMLNSIVE-OH | ||
| 1329.79 | Short NPF | AQRSPSLRLRFa | 1 | |
| 1395.75 | SIFamide | AYRKPPFNGSIFa | 1,2,3 | 1 |
| 936.47 | Tachykinin-like peptide | APTGFTGMRa | ||
| 942.59 | Tachykinin-like peptide | APLAFVGLRa | 1 | 1 |
| 961.50 | Tachykinin-like peptide | APNGFLGMRa | 1 | |
| 1065.55 | Tachykinin-like peptide | APTSSFIGMR | 1 | 1 |
| 4589.39 | Tachykinin-like peptide | FIPINNRLSDVLQSLEEERLRDSLLQDFFDRVAGRDGSAVa |
Monoisotopic mass.
Peptide sequences and amidation were predicted based on flanking mono- and dibasic cleavage sites and a C-terminal Gly residue encoded by the peptide gene. 1: Acetylation reaction supported the predicted number of Lys residues or an N-terminal pyroglutamic acid or Proresidue. 2: MALDI-FTMS measurements indicated that the experimental mass of the protonated ions was within 0.1 Da of the calculated mass of the predicted peptide sequence. 3: Amino acid sequence was supported by the pattern of MALDI-TOF/TOF fragmentation. m, oxidized Met residue; q, pyroglutamic acid; y, sulfated Tyr residue.
Additional Methods for Confirmation of Peptide Identification
The MALDI-TOF mass spectral analysis of immunoaffinity-purified cell extract was sufficiently sensitive to detect low levels of material; however, confident peptide assignments could not be made in several cases due to insufficient mass accuracy and resolving power. To address these limitations, three methods were used to verify the identity of the peaks observed in extracts of immunoaffinity-enriched tissue. These techniques utilized (i) high mass accuracy measurement with FTMS, (ii) sequence analysis using MALDI-TOF/TOF mass spectrometry for ions with lower abundance, and (iii) chemical derivatization of primary amine groups by acetic anhydride. Many of the original assignments were supported using FTMS, a method that provides mass accuracy within 50 ppm (Supporting Information Table 3).
For several species of ions, mass measurement alone was not sufficient to distinguish between two possible peptide assignments. In these cases, sequence information was obtained using MALDI-TOF/TOF Analysis. For example, the peptide ion observed at m/z 1247.62 could be attributed to either the myoinhibiting peptide RAWQSLQSSWamide (theoretical m/z 1247.63) or the Dromyosuppressin peptide TDVDHVFLRFamide (m/z 1247.65). Similarly, the ion observed at m/z 942.72 could be identified as the HUGIN peptide SVPFKPRLamide (m/z 942.59) or a Tachykinin-related peptide APLAFVGLRamide (m/z 942.59). For the ion with m/z 1247.62, fragmentation analysis obtained with MALDI-TOF/TOF MS was consistent with the sequence for Dromyosuppressin (Supporting Information Figure 1A). The MS/MS sequence for the ion with m/z 942.72 is consistent with that for Tachykinin-like peptide (Supporting Information Figure 1B).
Acetylation of extract from immunopurified cells provided a third means for sequence analysis. The derivatization targeted primary amine groups, unblocked N-termini, and internal Lys residues. By counting the number of acetyl groups that are added following the reaction (indicated by a mass shift increase of 42.04 Da), this reaction was useful for detecting N-terminal Pro (which will not be acetylated) and differentiating peptide sequence based on the presence or absence of Lys. Mass spectral analysis of the extract following acetylation revealed signals consistent with the expression of the HUGIN peptide and two members of the Allatostatin A and C families. A signal with m/z 1026.6 was detected in the derivatized extract, an observation consistent with the addition of two acetyl groups, likel to the N-terminus and to the primary amine of the Lys group of HUGIN. Successful acetylation of the signal with m/z 925.49 indicated that the N-terminus corresponded to one of the Allatostatin A peptides (SRPYSFGLa) rather than an FMRFamide peptide (PDNFMRFa) (Supporting Information Table 2). For another member of the Allatostatin family, Allatostatin C, examination of the acetylated extract revealed that both the N-terminal blocked (with pyroglutamic acid; m/z 1904.56) and unblocked (with glutamic acid; m/z 1921.58) forms are expressed. Following derivatization, a signal with m/z 1963.84 was detected, indicating a 42.04 mass shift from acetylation of the unblocked peptide. However, the signal corresponding to the N-terminal blocked peptide also was detected in the derivatized extract. An ion with m/z 1946.60 (indicating acetylation) was not observed (Supporting Information Table 2). Both forms of Allatostatin C showed an m/z that was 2 Da less than the calculated monoisotopic mass, suggesting the presence of disulfide bonds between the two Cys residues of the peptide.
Enrichment and MS Characterization of Dopa Decarboxylase-GAL4 Labeled Cells
To begin a survey of the peptides colocalized with dopamine (DA) and/or serotonin (5HT) in neurons, we used the immunoaffinity purification procedure to enrich for cells labeled by mCD8-GFP using the Ddc-GAL4 driver (Figure 4C). In extracts of these cells, ions with m/z corresponding to the calculated mass of the protonated molecular ion of Allatostatin B, both forms of Allatostatin C, Dromyosuppressin, HUGIN, SIFamide, Nplp1, some of the short NPFs, and several of the Tachykinin-like peptides were found (Figure 4D; Table 2; Supporting Information Table 2). In total, 18 peptides from 10 different families were present, representing a subset of the ones found using the dimm (c929)-GAL4 driver.
A comparison of the ions detected using the dimm (c929)-GAL4 and Ddc-GAL4 drivers showed an absence of signals with m/z corresponding to the Cap2b, FMRFamide, Corazonin, Pyrokinin, and several of the Tachykinin-like peptides in tissue enriched using the Ddc-GAL4 driver, suggesting that these peptides are not coexpressed with DA or 5HT. Such findings are consistent with recent studies showing no colocalization of 5HT and PDH (equivalent to the fruit fly PDF) in other insects92 and no colocalization of FMRFamide peptides and 5HT in D. melanogaster.93
Verification of Peptide Expression Using Immunocytochemistry
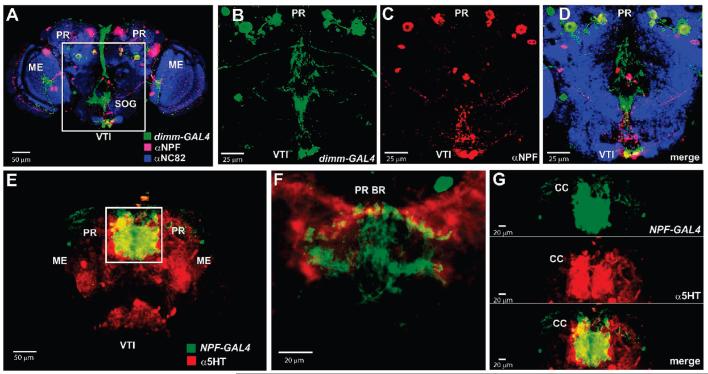
Immunocytochemistry was used in adult brains to confirm that some of the predicted peptides colocalized with the GFP signal driven by the dimm (c929)-GAL4 or Ddc-GAL4 lines. For the short Neuropeptide F family, six paired cells in the protocerebrum and three cells in the SOG showed coexpression of both dimm (c929)-GAL4-driven GFP and the short NPF antibod (Figure 5A-D). Immunoctochemist also confirmed co-expression of Tachykinin and PDF within dimm-labeled neurons (data not shown). Consistent with the results from the Ddc-GAL4 enrichment experiments, immunolocalization showed overlapping 5HT staining and short NPF gene expression in neurons and processes of the central protocerebrum and in the processes of the central complex (Figure 5E-G).
Figure 5.
Frontal images of D. melanogaster adult brain tissue labeled using immunocytochemistry and the GAL4-UAS expression system. Immmunocytochemical staining was performed to confirm the colocalization of short Neuropeptide F (NPF) expression within dimm (c929)-GAL4 circuitry and with the biogenic amine serotonin (5HT). Colocalization of anti-short NPF antibody staining (red) and dimm (c929)-GAL4-driven GFP expression (green) was observed in paired cells in the protocerebrum and in the subesophageal ganglion (yellow, A). A higher magnification view is shown of the framed region in A (B–D). Neuropil regions were labeled with the anti-NC82 antibody (blue). Colocalization of anti-5HT staining (red) with NPF-GAL4-driven GFP expression was observed in the central protocerebrum (yellow, E). Higher magnification images of the framed area showed colocalization in cell bodies and processes in the protocerebral bridge (F) and central complex neuropils (G). These observations were consistent with the results of the immunoaffinity cell-enrichment/MS approach which indicated that short NPFs are expressed within dimm (c929)-GAL4 and Ddc-GAL4-labeled cells. CC, central complex; ME, medulla: PR, protocerebrum; PR BR, protocerebral bridge; SOG, subesophageal ganglion; VTI, ventral tritocerebrum.
Some peptides were expected to be found based on the gene transcript but were not, including Adipokinetic Hormone, Droleucokinin, the nonamidated form of Nplp1 (SVAALAAQGLLNAPK-OH) and two nonamidated peptides from the short NPF family (SDPDMLNSIVE-OH and WFGDVNQKPI-OH). Other members of these peptide families were detected using either the dimm (c929)-GAL4 or Ddc-GAL4 driver. In addition, the corresponding ions for these peptides were detected in extracts from whole brain using ESI-Q-TOF MS. It ma be that these peptides were not detected due to rapid degradation, low levels of expression, or possibly, cell-specific splicing of the encoding gene.94,95 Interestingly, ion signals corresponding to only two members of the FMRFamide family were detected using this method. This is consistent with the localization of dimm (c929)-GAL4 in some but not all of the neurons that show FMRFamide immunostaining.24
The combination of the GAL4-UAS system, immunoaffinity purification, and mass spectrometric analysis used here begins the characterization of the peptide content of defined subpopulations of neurons. Man peptides isolated using the dimm (c929)-GAL4 driver corroborated previous direct tissue profiling studies of the SOG and of neurosecretory cells within the protocerebrum.22 In addition, several novel peptides, whose sequences were confirmed by LC/MS/MS, also were identified. The detection of additional peptides highlights the advantage of affinity isolation of specific populations of cells by showing reduced sample complexity and enhancement in the dynamic range of analysis. Several peptides found in earlier studies were not detected using this method, including SDNFMRFamide, and the FMRFamide-related peptide SAPQDFVRSamide. This could be because the dimm (c929)-GAL4 driver does not label all neurosecretory neurons96 or because some of these peptides are expressed at levels too low to be detected using these protocols. Using GAL4 lines for other neuronal markers of peptidergic identity such as neuropeptide processing enyzmes may provide a more complete inventor of expressed peptides.
One focus of the studies presented here was to search for colocalization of peptides with biogenic amines. Amines and peptides frequently are colocalized in the same neurons and, in some cases, have been shown to modulate downstream effects of each other;97-100 however, colocalization has been less well-studied in the fruit fly. In that regard, it is interesting that the number of peptides found associated with amine-expressing cells was smaller than in the dimm-enriched cell population. Possible explanations for this observation include limits of instrument sensitivity with the small numbers of amine-containing neurons found in fly brains; a reduced efficiency of cell isolation due to small numbers of GFP-positive neurons or lower levels of GFP-expression; and of greatest interest, relatively few peptides are expressed in aminergic neurons. The first two potential explanations represent possible limits of the technique. However, even when pooling together the enriched population from multiple isolation experiments, we found no differences in the peptide profiles using either the dimm (c929)-GAL4 or Ddc-GAL4 driver (data not shown), suggesting that issues of peptide abundance are not a major contributing factor to the limited number of peptides identified. In the future, it would be interesting to compare the peptide content of dopaminergic- and serotonergic-cells with that of tyramine and octopamine-containing cells using available GAL-4 driver lines.101 Both tyramine and octopamine also have important roles in Drosophila physiology and behavior.3,102-104
Conclusions
In the present study, we provide extensive biochemical characterization of the peptides expressed in the adult brain of D. melanogaster and use a novel method for the isolation of subgroups of neurons followed by analysis of their neuropeptide profiles. As with any wide-scale screening method, secondary forms of confirmation of these results will be required, particularly since the purity of the enriched neuron populations remains at approximately 35–60% and could therefore result in a number of false positive or negative results. Nevertheless, this combined genetic labeling/mass spectrometry approach provides a list of candidate peptides likely to be coexpressed with amines in D. melanogaster. In follow-up studies, immunocytochemistry and/or in situ hybridization can be used in targeted ways to bring the resolution of the colocalization of amines and identified peptides to an individual neuron level.
Supplementary Material
Acknowledgment
This work was supported by a National Institute of Mental Health National Research Service Award grant (J.Y.Y.) and research grants from the National Institute of Diabetes and Digestive and Kidney Diseases DK071801 (L.L.), and the National Institute of General Medical Sciences GM074675 and GM067645 (E.A.K.). We wish to acknowledge members of the Kravitz laboratory (Olga Alekseyenko, Sarah Certel, Yick-Bun Chan, Jill Penn, Maria de la Paz Fernandez) for helpful discussions and, in particular, Adelaine Leung for acquiring some of the confocal microscopy images. We would also like to thank Rochelle Witt and Jessica Saulnier for excellent technical advice. Finally, we are grateful to Dr. Klaus Dreisewerd for critical reading of the manuscript, Dr. David Core for use of the confocal microscope, and Drs. Alois Haufbauer, Randy Hewes, Jay Hirsh, Paul Taghert, and Ping Shen for generous gifts of reagents.
Footnotes
Supporting Information Available: Supplementary Figure 1, MALDI TOF/TOF MS analysis of two selected ions from immunoaffinity purified extract; Supplementary Table 1, the theoretical and observed m/z values of monoisotopic species identified in adult D. melanogaster brain extract by ESIQ-TOF tandem MS or accurate mass MALDI-FTMS measurement; Supplementary Table 2, the theoretical and observed monoisotopic m/z values of peptides identified from adult D. melanogaster cell immunoaffinity enrichment method; Supplementary Table 3, the theoretical and observed m/z values of peptides found in the extract from the dimm (c929)-GAL4 line using MALDI FTMS analysis. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Alford S, Schwartz E, di Prisco G. Viana. The pharmacology of vertebrate spinal central pattern generators. Neuroscientist. 2003;9(3):217–28. doi: 10.1177/1073858403009003014. [DOI] [PubMed] [Google Scholar]
- 2.Crisp KM, Mesce KA. Beyond the central pattern generator: amine modulation of decision-making neural pathways descending from the brain of the medicinal leech. J. Exp. Biol. 2006;209(Pt 9):1746–56. doi: 10.1242/jeb.02204. [DOI] [PubMed] [Google Scholar]
- 3.Saraswati S, Fox LE, Soll DR, Wu CF. Tyramine and octopamine have opposite effects on the locomotion of Drosophila larvae. J. Neurobiol. 2004;58(4):425–41. doi: 10.1002/neu.10298. [DOI] [PubMed] [Google Scholar]
- 4.Erickson JC, Clegg KE, Palmiter RD. Sensitivity to leptin and susceptibility to seizures of mice lacking neuropeptide Y. Nature. 1996;381(6581):415–21. doi: 10.1038/381415a0. [DOI] [PubMed] [Google Scholar]
- 5.Kyrkouli SE, Stanley BG, Leibowitz SF. Galanin: stimulation of feeding induced by medial hypothalamic injection of this novel peptide. Eur. J. Pharmacol. 1986;122(1):159–60. doi: 10.1016/0014-2999(86)90175-5. [DOI] [PubMed] [Google Scholar]
- 6.Pelleymounter MA, Cullen MJ, Baker MB, Hecht R, Winters D, Boone T, Collins F. Effects of the obese gene product on body weight regulation in ob/ob mice. Science. 1995;269(5223):540–3. doi: 10.1126/science.7624776. [DOI] [PubMed] [Google Scholar]
- 7.Wu Q, Wen T, Lee G, Park JH, Cai HN, Shen P. Developmental control of foraging and social behavior by the Drosophila neuropeptide Y-like system. Neuron. 2003;39(1):147–61. doi: 10.1016/s0896-6273(03)00396-9. [DOI] [PubMed] [Google Scholar]
- 8.Ferris CF, Melloni RH, Jr., Koppel G, Perry KW, Fuller RW, Delville Y. Vasopressin/serotonin interactions in the anterior hypothalamus control aggressive behavior in golden hamsters. J. Neurosci. 1997;17(11):4331–40. doi: 10.1523/JNEUROSCI.17-11-04331.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferris CF, Potegal M. Vasopressin receptor blockade in the anterior hypothalamus suppresses aggression in hamsters. Physiol. Behav. 1988;44(2):235–9. doi: 10.1016/0031-9384(88)90144-8. [DOI] [PubMed] [Google Scholar]
- 10.Kravitz EA. Serotonin and aggression: insights gained from a lobster model system and speculations on the role of amine neurons in a complex behavior. J. Comp. Physiol., A. 2000;186(3):221–38. doi: 10.1007/s003590050423. [DOI] [PubMed] [Google Scholar]
- 11.Nelson RJ, Trainor BC. Neural mechanisms of aggression. Nat. Rev. Neurosci. 2007;8(7):536–46. doi: 10.1038/nrn2174. [DOI] [PubMed] [Google Scholar]
- 12.Stevenson PA, Hofmann HA, Schoch K, Schildberger K. The fight and flight responses of crickets depleted of biogenic amines. J. Neurobiol. 2000;43(2):107–20. [PubMed] [Google Scholar]
- 13.White SA, Nguyen T, Fernald RD. Social regulation of gonadotropin-releasing hormone. J. Exp. Biol. 2002;205(Pt 17):2567–81. doi: 10.1242/jeb.205.17.2567. [DOI] [PubMed] [Google Scholar]
- 14.Margulies C, Tully T, Dubnau J. Deconstructing memory in Drosophila. Curr. Biol. 2005;15(17):R700–13. doi: 10.1016/j.cub.2005.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McGuire SE, Deshazer M, Davis RL. Thirty years of olfactory learning and memory research in Drosophila melanogaster. Prog. Neurobiol. 2005;76(5):328–47. doi: 10.1016/j.pneurobio.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 16.Aigaki T, Fleischmann I, Chen PS, Kubli E. Ectopic expression of sex peptide alters reproductive behavior of female D. melanogaster. Neuron. 1991;7(4):557–63. doi: 10.1016/0896-6273(91)90368-a. [DOI] [PubMed] [Google Scholar]
- 17.Fox AS, Mead CG, Munyon IL. Sex peptide of Drosophila melanogaster. Science. 1959;129(3361):1489–90. doi: 10.1126/science.129.3361.1489. [DOI] [PubMed] [Google Scholar]
- 18.Hardin PE. From biological clock to biological rhythms. genomebiology. 2000;1(4) doi: 10.1186/gb-2000-1-4-reviews1023. REVIEWS1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baggerman G, Boonen K, Verleyen P, De Loof A, Schoofs L. Peptidomic Analysis of the larval Drosophila melanogaster central nervous system by two-dimensional capillary liquid chromatography quadrupole time-of-flight mass spectrometry. J. Mass Spectrom. 2005;40(2):250–60. doi: 10.1002/jms.744. [DOI] [PubMed] [Google Scholar]
- 20.Baggerman G, Cerstiaens A, De Loof A, Schoofs L. Peptidomics of the larval Drosophila melanogaster central nervous system. J. Biol. Chem. 2002;277(43):40368–74. doi: 10.1074/jbc.M206257200. [DOI] [PubMed] [Google Scholar]
- 21.Neupert S, Johard HA, Nassel DR, Predel R. Single-cell peptidomics of drosophila melanogaster neurons identified by Gal4-driven fluorescence. Anal. Chem. 2007;79(10):3690–4. doi: 10.1021/ac062411p. [DOI] [PubMed] [Google Scholar]
- 22.Predel R, Wegener C, Russell WK, Tichy SE, Russell DH, Nachman RJ. Peptidomics of CNS-associated neurohemal systems of adult Drosophila melanogaster: a mass spectrometric survey of peptides from individual flies. J. Comp. Neurol. 2004;474(3):379–92. doi: 10.1002/cne.20145. [DOI] [PubMed] [Google Scholar]
- 23.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118(2):401–15. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 24.Hewes RS, Park D, Gauthier SA, Schaefer AM, Taghert PH. The bHLH protein Dimmed controls neuroendocrine cell differentiation in Drosophila. Development. 2003;130(9):1771–81. doi: 10.1242/dev.00404. [DOI] [PubMed] [Google Scholar]
- 25.Johnson WA, McCormick CA, Bray SJ, Hirsh J. A neuron-specific enhancer of the Drosophila dopa decarboxylase gene. Genes Dev. 1989;3(5):676–86. doi: 10.1101/gad.3.5.676. [DOI] [PubMed] [Google Scholar]
- 26.Su H, O’Dowd DK. Fast synaptic currents in Drosophila mushroom body Kenyon cells are mediated by alpha-bungarotoxin-sensitive nicotinic acetylcholine receptors and picrotoxin-sensitive GABA receptors. J. Neurosci. 2003;23(27):9246–53. doi: 10.1523/JNEUROSCI.23-27-09246.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wegener C, Hamasaka Y, Nassel DR. Acetylcholine increases intracellular Ca2+ via nicotinic receptors in cultured PDF-containing clock neurons of Drosophila. J. Neurophysiol. 2004;91(2):912–23. doi: 10.1152/jn.00678.2003. [DOI] [PubMed] [Google Scholar]
- 28.Kutz KK, Schmidt JJ, Li L. In situ tissue Analysis of neuropeptides by MALDI FTMS in-cell accumulation. Anal. Chem. 2004;76(19):5630–40. doi: 10.1021/ac049255b. [DOI] [PubMed] [Google Scholar]
- 29.O’Connor PB, Costello CE. Internal calibration on adjacent samples (InCAS) with Fourier transform mass spectrometry. Anal. Chem. 2000;72(24):5881–5. doi: 10.1021/ac000770t. [DOI] [PubMed] [Google Scholar]
- 30.Hewes RS, Taghert PH. Neuropeptides and neuropeptide receptors in the Drosophila melanogaster genome. Genome Res. 2001;11(6):1126–42. doi: 10.1101/gr.169901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gronke S, Muller G, Hirsch J, Fellert S, Andreou A, Haase T, Jackle H, Kuhnlein RP. Dual lipolytic control of body fat storage and mobilization in Drosophila. PLoS Biol. 2007;5(6):e137. doi: 10.1371/journal.pbio.0050137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Van der Horst DJ. Insect adipokinetic hormones: release and integration of flight energy metabolism. Comp. Biochem. Physiol., Part B: Biochem. Mol. Biol. 2003;136(2):217–26. doi: 10.1016/s1096-4959(03)00151-9. [DOI] [PubMed] [Google Scholar]
- 33.Schaffer MH, Noyes BE, Slaughter CA, Thorne GC, Gaskell SJ. The fruitfly Drosophila melanogaster contains a novel charged adipokinetic-hormone-family peptide. Biochem. J. 1990;269(2):315–20. doi: 10.1042/bj2690315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Noyes BE, Katz FN, Schaffer MH. Identification and expression of the Drosophila adipokinetic hormone gene. Mol. Cell. Endocrinol. 1995;109(2):133–41. doi: 10.1016/0303-7207(95)03492-p. [DOI] [PubMed] [Google Scholar]
- 35.Nichols R, Bendena WG, Tobe SS. Myotropic peptides in Drosophila melanogaster and the genes that encode them. J. Neurogenet. 2002;16(1):1–28. [PubMed] [Google Scholar]
- 36.Lenz C, Williamson M, Grimmelikhuijzen CJ. Molecular cloning and genomic organization of a second probable allatostatin receptor from Drosophila melanogaster. Biochem. Biophys. Res. Commun. 2000;273(2):571–7. doi: 10.1006/bbrc.2000.2964. [DOI] [PubMed] [Google Scholar]
- 37.Lenz C, Williamson M, Grimmelikhuijzen CJ. Molecular cloning and genomic organization of an allatostatin preprohormone from Drosophila melanogaster. Biochem. Biophys. Res. Commun. 2000;273(3):1126–31. doi: 10.1006/bbrc.2000.3062. [DOI] [PubMed] [Google Scholar]
- 38.Lenz C, Williamson M, Hansen GN, Grimmelikhuijzen CJ. Identification of four Drosophila allatostatins as the cognate ligands for the Drosophila orphan receptor DAR-2. Biochem. Biophys. Res. Commun. 2001;286(5):1117–22. doi: 10.1006/bbrc.2001.5475. [DOI] [PubMed] [Google Scholar]
- 39.Williamson M, Lenz C, Winther AM, Nassel DR, Grimmelikhuijzen CJ. Molecular cloning, genomic organization, and expression of a B-type (cricket-type) allatostatin preprohormone from Drosophila melanogaster. Biochem. Biophys. Res. Commun. 2001;281(2):544–50. doi: 10.1006/bbrc.2001.4402. [DOI] [PubMed] [Google Scholar]
- 40.Williamson M, Lenz C, Winther AM, Nassel DR, Grimmelikhuijzen CJ. Molecular cloning, genomic organization, and expression of a C-type (Manduca sexta-type) allatostatin prepro-hormone from Drosophila melanogaster. Biochem. Biophys. Res. Commun. 2001;282(1):124–30. doi: 10.1006/bbrc.2001.4565. [DOI] [PubMed] [Google Scholar]
- 41.Kramer SJ, Toschi A, Miller CA, Kataoka H, Quistad GB, Li JP, Carney RL, Schooley DA. Identification of an allatostatin from the tobacco hornworm Manduca sexta. Proc. Natl. Acad. Sci. U.S.A. 1991;88(21):9458–62. doi: 10.1073/pnas.88.21.9458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Veenstra JA. Isolation and structure of corazonin, a cardioactive peptide from the American cockroach. FEBS Lett. 1989;250(2):231–4. doi: 10.1016/0014-5793(89)80727-6. [DOI] [PubMed] [Google Scholar]
- 43.Nassel DR. Neuropeptides in the nervous system of Drosophila and other insects: multiple roles as neuromodulators and neurohormones. Prog. Neurobiol. 2002;68(1):1–84. doi: 10.1016/s0301-0082(02)00057-6. [DOI] [PubMed] [Google Scholar]
- 44.McCormick J, Nichols R. Spatial and temporal expression identify dromyosuppressin as a brain-gut peptide in Drosophila melanogaster. J. Comp. Neurol. 1993;338(2):278–88. doi: 10.1002/cne.903380210. [DOI] [PubMed] [Google Scholar]
- 45.Merte J, Nichols R. Drosophila melanogaster FMRFamide-containing peptides: redundant or diverse functions. Peptides. 2002;23(1):209–20. doi: 10.1016/s0196-9781(01)00598-8. [DOI] [PubMed] [Google Scholar]
- 46.Kaminski S, Orlowski E, Berry K, Nichols R. The effects of three Drosophila melanogaster myotropins on the frequency of foregut contractions differ. J. Neurogenet. 2002;16(2):125–34. doi: 10.1080/01677060213156. [DOI] [PubMed] [Google Scholar]
- 47.Duttlinger A, Berry K, Nichols R. The different effects of three Drosophila melanogaster dFMRFamide-containing peptides on crop contractions suggest these structurally related peptides do not play redundant functions in gut. Peptides. 2002;23(11):1953–7. doi: 10.1016/s0196-9781(02)00179-1. [DOI] [PubMed] [Google Scholar]
- 48.Nichols R. Isolation and structural characterization of Drosophila TDVDHVFLRFamide and FMRFamide-containing neural peptides. J. Mol. Neurosci. 1992;3(4):213–8. doi: 10.1007/BF03380141. [DOI] [PubMed] [Google Scholar]
- 49.Dockray GJ. Cholecystokinins in rat cerebral cortex: identification, purification and characterization by immunochemical methods. Brain Res. 1980;188(1):155–65. doi: 10.1016/0006-8993(80)90564-8. [DOI] [PubMed] [Google Scholar]
- 50.McDonald TJ, Jornvall H, Nilsson G, Vagne M, Ghatei M, Bloom SR, Mutt V. Characterization of a gastrin releasing peptide from porcine non-antral gastric tissue. Biochem. Biophys. Res. Commun. 1979;90(1):227–33. doi: 10.1016/0006-291x(79)91614-0. [DOI] [PubMed] [Google Scholar]
- 51.Nichols R, Schneuwly SA, Dixon JE. Identification and characterization of a Drosophila homologue to the vertebrate neuropeptide cholecystokinin. J. Biol. Chem. 1988;263(25):12167–70. [PubMed] [Google Scholar]
- 52.Nachman RJ, Holman GM, Cook BJ. Active fragments and analogs of the insect neuropeptide leucopyrokinin: structure-function studies. Biochem. Biophys. Res. Commun. 1986;137(3):936–42. doi: 10.1016/0006-291x(86)90315-3. [DOI] [PubMed] [Google Scholar]
- 53.Hewes RS, Snowdeal EC, III, Saitoe M, Taghert PH. Functional redundancy of FMRFamide-related peptides at the Drosophila larval neuromuscular junction. J. Neurosci. 1998;18(18):7138–51. doi: 10.1523/JNEUROSCI.18-18-07138.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Benveniste RJ, Taghert PH. Cell type-specific regulatory sequences control expression of the Drosophila FMRF-NH2 neuropeptide gene. J. Neurobiol. 1999;38(4):507–20. doi: 10.1002/(sici)1097-4695(199903)38:4<507::aid-neu7>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 55.Nambu JR, Murphy-Erdosh C, Andrews PC, Feistner GJ, Scheller RH. Isolation and characterization of a Drosophila neuropeptide gene. Neuron. 1988;1(1):55–61. doi: 10.1016/0896-6273(88)90209-7. [DOI] [PubMed] [Google Scholar]
- 56.Schneider LE, Taghert PH. Isolation and characterization of a Drosophila gene that encodes multiple neuropeptides related to Phe-Met-Arg-Phe-NH2 (FMRFamide) Proc. Natl. Acad. Sci. U.S.A. 1988;85(6):1993–7. doi: 10.1073/pnas.85.6.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Landon Cleavage at aspartyl-prolyl bonds. Methods Enzymol. 1977;47:145–9. doi: 10.1016/0076-6879(77)47017-4. [DOI] [PubMed] [Google Scholar]
- 58.Roos C. GenBank. 1999. Drosophila melanogaster hug gene for hugin, exons 1 to 3. [Google Scholar]
- 59.Taghert PH, Veenstra JA. Drosophila neuropeptide signaling. Adv. Genet. 2003;49:1–65. doi: 10.1016/s0065-2660(03)01001-0. [DOI] [PubMed] [Google Scholar]
- 60.Schoofs L, Holman GM, Nachman R, Proost P, Van Damme J, De Loof A. Isolation, identification and synthesis of locustapyrokinin II from Locusta migratoria, another member of the FXPRL-amide peptide family. Comp. Biochem. Physiol., Part C. 1993;106(1):103–9. doi: 10.1016/0742-8413(93)90260-r. [DOI] [PubMed] [Google Scholar]
- 61.Meng X, Wahlstrom G, Immonen T, Kolmer M, Tirronen M, Predel R, Kalkkinen N, Heino TI, Sariola H, Roos C. The Drosophila hugin gene codes for myostimulatory and ecdysis-modifying neuropeptides. Mech. Dev. 2002;117(12):5–13. doi: 10.1016/s0925-4773(02)00175-2. [DOI] [PubMed] [Google Scholar]
- 62.Skaer NJ, Nassel DR, Maddrell SH, Tublitz NJ. Neurochemical fine tuning of a peripheral tissue: peptidergic and aminergic regulation of fluid secretion by Malpighian tubules in the tobacco hawkmoth M. sexta. J. Exp. Biol. 2002;205(Pt 13):1869–80. doi: 10.1242/jeb.205.13.1869. [DOI] [PubMed] [Google Scholar]
- 63.Terhzaz S, O’Connell FC, Pollock VP, Kean L, Davies SA, Veenstra JA, Dow JA. Isolation and characterization of a leucokinin-like peptide of Drosophila melanogaster. J. Exp. Biol. 1999;202(Pt 24):3667–76. doi: 10.1242/jeb.202.24.3667. [DOI] [PubMed] [Google Scholar]
- 64.Wen T, Parrish CA, Xu D, Wu Q, Shen P. Drosophila neuropeptide F and its receptor, NPFR1, define a signaling pathway that acutel modulates alcohol sensitivity. Proc. Natl. Acad. Sci. U.S.A. 2005;102(6):2141–6. doi: 10.1073/pnas.0406814102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dierick HA, Greenspan RJ. Serotonin and neuropeptide F have opposite modulatory effects on fly aggression. Nat. Genet. 2007;39(5):678–82. doi: 10.1038/ng2029. [DOI] [PubMed] [Google Scholar]
- 66.Vanden Broeck J. Neuropeptides and their precursors in the fruitfly, Drosophila melanogaster. Peptides. 2001;22(2):241–54. doi: 10.1016/s0196-9781(00)00376-4. [DOI] [PubMed] [Google Scholar]
- 67.Lee G, Bahn JH, Park JH. Sex- and clock-controlled expression of the neuropeptide F gene in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 2006;103(33):12580–5. doi: 10.1073/pnas.0601171103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rao KR, Riehm JP. Pigment-dispersing hormones. Ann. N.Y. Acad. Sci. 1993;680:78–88. doi: 10.1111/j.1749-6632.1993.tb19676.x. [DOI] [PubMed] [Google Scholar]
- 69.Ewer J, Frisch B, Hamblen-Coyle MJ, Rosbash M, Hall JC. Expression of the period clock gene within different cell types in the brain of Drosophila adults and mosaic Analysis of these cells’ influence on circadian behavioral rhythms. J. Neurosci. 1992;12(9):3321–49. doi: 10.1523/JNEUROSCI.12-09-03321.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lin Y, Stormo GD, Taghert PH. The neuropeptide pigment-dispersing factor coordinates pacemaker interactions in the Drosophila circadian system. J. Neurosci. 2004;24(36):7951–7. doi: 10.1523/JNEUROSCI.2370-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Park JH, Hall JC. Isolation and chronobiological Analysis of a neuropeptide pigment-dispersing factor gene in Drosophila melanogaster. J. Biol. Rhythms. 1998;13(3):219–28. doi: 10.1177/074873098129000066. [DOI] [PubMed] [Google Scholar]
- 72.Renn SC, Park JH, Rosbash M, Hall JC, Taghert PH. A pdf neuropeptide gene mutation and ablation of PDF neurons each cause severe abnormalities of behavioral circadian rhythms in Drosophila. Cell. 1999;99(7):791–802. doi: 10.1016/s0092-8674(00)81676-1. [DOI] [PubMed] [Google Scholar]
- 73.Schoofs L, Veelaert D, Vanden Broeck J, De Loof A. Peptides in the locusts, Locusta migratoria and Schistocerca gregaria. Peptides. 1997;18(1):145–56. doi: 10.1016/s0196-9781(96)00236-7. [DOI] [PubMed] [Google Scholar]
- 74.Loi PK, Tublitz NJ. Sequence and expression of the CAPA/ CAP2b gene in the tobacco hawkmoth, Manduca sexta. J. Exp. Biol. 2004;207(Pt 21):3681–91. doi: 10.1242/jeb.01186. [DOI] [PubMed] [Google Scholar]
- 75.Broeck JV. Insect G protein-coupled receptors and signal transduction. Arch. Insect Biochem. Physiol. 2001;48(1):1–12. doi: 10.1002/arch.1054. [DOI] [PubMed] [Google Scholar]
- 76.Terhzaz S. Drosophila melanogaster neuropeptide IFamide preproprotein mRNA, complete cds. GenBank. 2001 [Google Scholar]
- 77.Janssen I, Schoofs L, Spittaels K, Neven H, Vanden Broeck J, Devreese B, Van Beeumen J, Shabanowitz J, Hunt DF, De Loof A. Isolation of NEB-LFamide, a novel myotropic neuropeptide from the grey fleshfly. Mol. Cell. Endocrinol. 1996;117(2):157–65. doi: 10.1016/0303-7207(95)03746-2. [DOI] [PubMed] [Google Scholar]
- 78.Terhzaz S, Rosay P, Goodwin SF, Veenstra JA. The neuropeptide SIFamide modulates sexual behavior in Drosophila. Biochem. Biophys. Res. Commun. 2007;352(2):305–10. doi: 10.1016/j.bbrc.2006.11.030. [DOI] [PubMed] [Google Scholar]
- 79.Siviter RJ, Coast GM, Winther AM, Nachman RJ, Taylor CA, Shirras AD, Coates D, Isaac RE, Nassel DR. Expression and functional characterization of a Drosophila neuropeptide precursor with homology to mammalian preprotachykinin A. J. Biol. Chem. 2000;275(30):23273–80. doi: 10.1074/jbc.M002875200. [DOI] [PubMed] [Google Scholar]
- 80.Uttenweiler-Joseph S, Moniatte M, Lagueux M, Van Dorsselaer A, Hoffmann JA, Bulet P. Differential display of peptides induced during the immune response of Drosophila: a matrix-assisted laser desorption ionization time-of-flight mass spectrometry study. Proc. Natl. Acad. Sci. U.S.A. 1998;95(19):11342–7. doi: 10.1073/pnas.95.19.11342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22(3):451–61. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- 82.Wang X, Bo J, Bridges T, Dugan KD, Pan TC, Chodosh LA, Montell DJ. Analysis of cell migration using whole-genome expression profiling of migratory cells in the Drosophila ovary. Dev. Cell. 2006;10(4):483–95. doi: 10.1016/j.devcel.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 83.Zitnan D, Sehnal F, Bryant PJ. Neurons producing specific neuropeptides in the central nervous system of normal and pupariation-delayed Drosophila. Dev. Biol. 1993;156(1):117–35. doi: 10.1006/dbio.1993.1063. [DOI] [PubMed] [Google Scholar]
- 84.Wegener C, Herbert Z, Eckert M, Predel R. The periviscerokinin (PVK) peptide family in insects: evidence for the inclusion of CAP(2b) as a PVK family member. Peptides. 2002;23(4):605–11. doi: 10.1016/s0196-9781(01)00665-9. [DOI] [PubMed] [Google Scholar]
- 85.Siegmund T, Korge G. Innervation of the ring gland of Drosophila melanogaster. J. Comp. Neurol. 2001;431(4):481–91. doi: 10.1002/1096-9861(20010319)431:4<481::aid-cne1084>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 86.Nichols R, Lim IA. Spatial and temporal immunocytochemical analysis of drosulfakinin (Dsk) gene products in the Drosophila melanogaster central nervous system. Cell Tissue Res. 1996;283(1):107–16. doi: 10.1007/s004410050518. [DOI] [PubMed] [Google Scholar]
- 87.Schneider LE, Roberts MS, Taghert PH. Cell type-specific transcriptional regulation of the Drosophila FMRFamide neuropeptide gene. Neuron. 1993;10(2):279–91. doi: 10.1016/0896-6273(93)90318-l. [DOI] [PubMed] [Google Scholar]
- 88.Schneider LE, Sun ET, Garland DJ, Taghert PH. An immunocytochemical study of the FMRFamide neuropeptide gene products in Drosophila. J. Comp. Neurol. 1993;337(3):446–60. doi: 10.1002/cne.903370308. [DOI] [PubMed] [Google Scholar]
- 89.Helfrich-Forster C. The period clock gene is expressed in central nervous system neurons which also produce a neuropeptide that reveals the projections of circadian pacemaker cells within the brain of Drosophila melanogaster. Proc. Natl. Acad. Sci. U.S.A. 1995;92(2):612–6. doi: 10.1073/pnas.92.2.612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Choi MY, Rafaeli A, Jurenka RA. Pyrokinin/PBAN-like peptides in the central nervous system of Drosophila melanogaster. Cell Tissue Res. 2001;306(3):459–65. doi: 10.1007/s00441-001-0467-x. [DOI] [PubMed] [Google Scholar]
- 91.Melcher C, Pankratz MJ. Candidate gustatory interneurons modulating feeding behavior in the Drosophila brain. PLoS Biol. 2005;3(9):e305. doi: 10.1371/journal.pbio.0030305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Nassel DR, Shiga S, Wikstrand EM, Rao KR. Pigment-dispersing hormone-immunoreactive neurons and their relation to serotonergic neurons in the blowfly and cockroach visual system. Cell Tissue Res. 1991;266(3):511–23. doi: 10.1007/BF00318593. [DOI] [PubMed] [Google Scholar]
- 93.Nichols R. FMRFamide-related peptides and serotonin regulate Drosophila melanogaster heart rate: mechanisms and structure requirements. Peptides. 2006;27(5):1130–7. doi: 10.1016/j.peptides.2005.07.032. [DOI] [PubMed] [Google Scholar]
- 94.Rosoff ML, Burglin TR, Li C. Alternatively spliced transcripts of the flp-1 gene encode distinct FMRFamide-like peptides in Caenorhabditis elegans. J. Neurosci. 1992;12(6):2356–61. doi: 10.1523/JNEUROSCI.12-06-02356.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Amara SG, Jonas V, Rosenfeld MG, Ong ES, Evans RM. Alternative RNA processing in calcitonin gene expression generates mRNAs encoding different polypeptide products. Nature. 1982;298(5871):240–4. doi: 10.1038/298240a0. [DOI] [PubMed] [Google Scholar]
- 96.Park D, Veenstra JA, Park JH, Taghert PH. Mapping peptidergic cells in Drosophila: where DIMM fits in. PLoS One. 2008;3(3):e1896. doi: 10.1371/journal.pone.0001896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Emeson RB, Morabito MV. Food fight: the NPY-serotonin link between aggression and feeding behavior. Sci STKE. 2005;2005(277):pe12. doi: 10.1126/stke.2772005pe12. [DOI] [PubMed] [Google Scholar]
- 98.Hokfelt T, Lundberg JM, Schultzberg M, Johansson O, Skirboll L, Anggard A, Fredholm B, Hamberger B, Pernow B, Rehfeld J, Goldstein M. Cellular localization of peptides in neural structures. Proc. R. Soc. London, Ser. B. 1980;210(1178):63–77. doi: 10.1098/rspb.1980.0119. [DOI] [PubMed] [Google Scholar]
- 99.Rotzinger S, Bush DE, Vaccarino FJ. Cholecystokinin modulation of mesolimbic dopamine function: regulation of motivated behaviour. Pharmacol. Toxicol. 2002;91(6):404–13. doi: 10.1034/j.1600-0773.2002.910620.x. [DOI] [PubMed] [Google Scholar]
- 100.Vaccarino FJ, Rankin J. Nucleus accumbens cholecystokinin (CCK) can either attenuate or potentiate amphetamine-induced locomotor activity: evidence for rostral-caudal differences in accumbens CCK function. Behav. Neurosci. 1989;103(4):831–6. [PubMed] [Google Scholar]
- 101.Cole SH, Carney GE, McClung CA, Willard SS, Taylor BJ, Hirsh J. Two functional but noncomplementing Drosophila tyrosine decarboxylase genes: distinct roles for neural tyramine and octopamine in female fertility. J. Biol. Chem. 2005;280(15):14948–55. doi: 10.1074/jbc.M414197200. [DOI] [PubMed] [Google Scholar]
- 102.Certel SJ, Savella MG, Schlegel DC, Kravitz EA. Modulation of Drosophila male behavioral choice. Proc. Natl. Acad. Sci. U.S.A. 2007;104(11):4706–11. doi: 10.1073/pnas.0700328104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hoyer SC, Eckart A, Herrel A, Zars T, Fischer SA, Hardie SL, Heisenberg M. Octopamine in male aggression of Drosophila. Curr. Biol. 2008;18(3):159–67. doi: 10.1016/j.cub.2007.12.052. [DOI] [PubMed] [Google Scholar]
- 104.Scholz H. Influence of the biogenic amine tyramine on ethanol-induced behaviors in Drosophila. J. Neurobiol. 2005;63(3):199–214. doi: 10.1002/neu.20127. [DOI] [PubMed] [Google Scholar]
- 105.Biemann K. Sequencing of peptides by tandem mass spectrometry and high-energy collision-induced dissociation. Methods Enzymol. 1990;193:455–79. doi: 10.1016/0076-6879(90)93433-l. [DOI] [PubMed] [Google Scholar]
- 106.Huesmann GR, Cheung CC, Loi PK, Lee TD, Swiderek KM, Tublitz NJ. Amino acid sequence of CAP2b, an insect cardioacceleratory peptide from the tobacco hawkmoth Manduca sexta. FEBS Lett. 1995;371(3):311–4. doi: 10.1016/0014-5793(95)00929-4. [DOI] [PubMed] [Google Scholar]
- 107.Davies SA, Stewart EJ, Huesmann GR, Skaer NJ, Maddrell SH, Tublitz NJ, Dow JA. Neuropeptide stimulation of the nitric oxide signaling pathway in Drosophila melanogaster Malpighian tubules. Am. J. Physiol. 1997;273(2 Pt 2):R823–7. doi: 10.1152/ajpregu.1997.273.2.R823. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.