SUMMARY
The prion protein (PrP) is essential for the pathogenesis of prion disease. PrP has been detected in the cytosol of neurons and transgenic mice expressing PrP in the cytosol (cyPrP) under a pan-neuronal promoter developed rapid cerebellar granule neuron degeneration. Yet, it remains unclear whether cyPrP is capable to cause toxicity in other neuronal populations. Here, we report that transgenic mice expressing cyPrP in the forebrain neurons developed behavioral abnormalities including clasping and hyperactivity. These mice had a reduced thickness in cortex and developed astrogliosis in hippocampal and cortical regions. Moreover, cyPrP in these mice was recognized by the A11 anti-oligomer antibody and was associated with the hydrophobic lipid core of membranes, indicating that cyPrP oligomer caused membrane perturbation contributes to cyPrP neurotoxicity. Together, our results clearly revealed that cyPrP is able to cause toxicity in different neuronal populations, supporting a role of cyPrP in PrP-mediated neurodegenerative disorders.
Keywords: Prion, PrP, forebrain neurotoxicity, oligomer, membrane perturbation
INTRODUCTION
Prion diseases are a group of neurodegenerative disorders characterized by the conversion of normal prion protein (PrPC) to a pathogenic conformation (PrPSc) [1–4]. Previous studies established that the expression of PrPC is indispensable for prion disease [5–7]. Moreover, these studies also indicate that an alteration in PrPC metabolism contributes to neurotoxicity.
Besides its normal cell surface localization, PrP has been detected in the cytosol of neurons in wild-type animals [8]. The cytosolic appearance of PrP has been attributed to retro-translocation of misfolded PrP from the endoplasmic reticulum (ER) [9–11], impaired import into ER due to the inefficiency of PrP signal sequences [12], or preemptive quality control in the ER [13, 14]. We previously reported that transgenic mice expressing PrP in the cytosol (cyPrP) under its own pan-neuronal promoter developed rapid cerebellar granule neuron degeneration [15]. The lack of detectable toxicity in other brain regions led to the question of whether cyPrP toxicity could affect other neurons. Moreover, the controversy over cyPrP neurotoxicity is further deepened by conflicting results from cell culture studies, from toxic [12, 15–17], to not toxic [18, 19], or even protecting certain types of cell against Bax induced apoptosis [20, 21]. In prion disease, the neurotoxicity involves different neuronal populations [22–25]. Should cyPrP play any role in the pathogenesis of prion disease, it must be able to cause toxicity in other neuronal populations in an in vivo setting.
In this study, we generated transgenic mice expressing cyPrP under the control of a tetracycline responsive promoter, in which the transgene expression requires the binding of tetracycline-transactivator (tTA) to the promoter in the absence of tetracycline (known as the “Tet-OFF system”). We crossed inducible cyPrP transgenic mice with mice expressing tTA under the control of calcium-calmodulin-dependent kinase II (CamKII) promoter [26], creating double transgenic mice expressing cyPrP in the forebrain. This strategy avoided the rapid cyPrP-evoked cerebellar degeneration, allowing us to determine whether cyPrP is toxic to other neurons after an extended period of time. Here, we report that cyPrP indeed causes toxicity in forebrain neurons. In addition, cyPrP is recognized by the A11 anti-oligomer antibody and binds to the hydrophobic lipid core of membranes, which supports the hypothesis that oligomeric cyPrP perturbs lipid membranes and causes neurotoxicity.
MATERIAL AND METHODS
Generation of transgenic mice
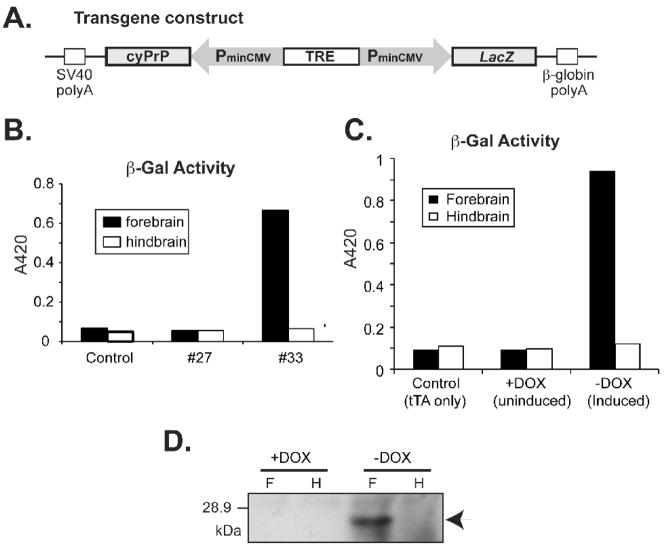
Purified DNA fragment containing cyPrP coding sequences under the regulation of a tetracycline-responsive bi-directional promoter (designated as: pBI-cyPrP-Gal, Fig. 1A) was microinjected into fertilized FVB/N mouse oocytes. Pups were screened for founders by PCR of tail DNA with two pairs of primers. The β-Gal Primers amplify a sequence within the β-galactosidase gene. The PrP primers amplify a sequence between cyPrP and the minimal CMV promoter (PminCMV, Fig. 1A).
Fig. 1. Characterization of tetracycline-inducible transgenic mice expressing cyPrP in the forebrain.
(A) The pBI-cyPrP-Gal construct. (B) Double transgenic mice (tTA+/−; pBI-cyPrP-Gal+/−) from two transgenic lines (#27 and #33) and a control mouse (tTA+/−; pBI-cyPrP-Gal−/−) were switched to dox-free food at 6 weeks of age and sacrificed when 8 weeks old. The expression of the β-Gal gene was monitored by measuring β-Gal activity in forebrain and hindbrain lysates as indicated. (C) Beta-Gal activity was measured in forebrain and hindbrain lysates of double and single transgenic littermates fed with or without dox as indicated. (D) The expression of cyPrP was measured by immunoblot analysis with 3F4 antibody. F, forebrain; H, hindbrain.
The β-Gal primers are: BI 1806, 5′ GACGCCATCCACGCTGTTTTGACCTCC3′; BI 2722A, 5′ CACGCAACTCGCCGCACATCTGAACTT3′.
The PrP primers are: BI459, 5′AACCTCCCACACCTCCCCCTGAACCTG3′ BI944A, 5′GGCAACGACTGGGAGGACCGCTACTAC3′
Determination of transgene dosage with radioactive PCR
PCR reaction was carried out in 10 μl volume with ~ 40ng tail DNA, PrP primers, and mouse cyclophilin primers (as internal control) in 20mM Tris pH 8.4, 50mM KCl, 2% Formamide, 0.2mM dNTP, 1μM each of the primers, 0.5 units of Taq polymerase, and 1μCi α-P32-dCTP. After 20 cycles, half of the sample was loaded on a 4% polyacrylamide denaturing gel and subjected to electrophoresis. Results were quantified by a Storm PhosphorImager System (Molecular Dynamics).
Behavior analysis
Fourteen double transgenic mice (tTA+/−; pBI-cyPrP-Gal+/−) and 11 single transgenic control mice (tTA−/−; pBI-cyPrP-Gal+/−) from two transgenic lines, which went through the same transgene induction regime of switching food containing doxycycline (dox-food) (Bio-Serv) to regular mouse food at 6 weeks of age, were kept at the mouse behavior testing facility at Ohio State University for more than a month before conducting the test. Two double transgenic mice died during the course of testing and all available mice were subjected to full panel of tests as previously described [27].
The open field test was conducted in a test chamber that was enclosed in a sound and light attenuating cabinet and consisted of a 60cm3 clear Plexiglas arena lined with corncob bedding. The arena was surrounded by a series of infrared lights for tracking mouse movement in three dimensions. Between each test, the chamber was rinsed thoroughly with a 70% ethanol solution and the bedding was changed. Each test session was 30 min in duration and the results were generated by the PAS software package (San Diego Instruments, San Diego, CA, USA). The total locomotor activity (number of beam breaks), percentage of activity in the periphery versus the center of the arena, and the total number of rears served as the dependent measures.
For the tail pinch test, the mouse was pinched at approx 2 cm from the base of tail and scored as: 0, no reaction; 1, look back, pull forward or light squeal; 2, twitch (muscle tension); 3, aggressive or vocal response. For the toe pinch test, the mouse was pinched at the middle hind left toe and scored as: 0, no reaction; 1, slight withdrawal; 2, moderate withdrawal (not brisk); 3, brisk, rapid withdrawal; 4, very rapid, repeated withdrawal.
For tail suspension test, the mouse was suspended ~30 cm above the surface of a table for 15 seconds with each 2-second clasping counted as 1 point. Eleven double transgenic mice (tTA+/−; pBI-cyPrP-Gal+/−) and 11 single transgenic control mice (tTA−/−; pBI-cyPrP-Gal+/−) that went through the same induction regime of switching dox-food to regular mouse food at the age of 6 weeks were subjected to the test.
Membrane association, solubility and immunoblot analysis
The cyPrP membrane association analysis was performed as previously described [28]. Briefly, post-nuclear supernatants containing 100 μg of protein were separated on a discontinuous iodixanol gradient [28, 29] and 200 μl fractions were collected. For membrane extractions, 300 μg post-nuclear supernatant was centrifuged at 346,000g for 30 minutes at 4°C. The pellet was re-suspended in a solution containing 0.5M NaHCO3, pH 11, or 1M KCl plus 10mM NaOH and subjected to the iodixanol gradient separation. The solubility analysis was performed as previously described [28] except that the centrifugation of induced homozygous transgenic mice was carried out at 100,000g for 1 hour at 4°C. Subcellular fractionation of neuronal membranes was performed as previously described [28, 30]. The presence of cyPrP and synaptophysin were determined by immunoblot analysis with the 3F4 anti-PrP antibody (Covance) and an anti-synaptophysin antibody (Sigma). Quantification was performed on a Storm PhosphorImager System (Molecular Dynamics).
Histological analysis
Hematoxylin & Eosin staining (H&E) was performed with Harris Hematoxylin (Sigma) and Eosin Y (Fisher). Immunohistochemistry was performed as previously described (27). CyPrP was detected with a mouse 3F4 antibody (Signet) at 1:1000. Rabbit anti GFAP antibody (DAKO) was used at 1:4000. Antigen Amplify system (prohisto lab) was used as manufacturer suggested to increase sensitivity of detection. Chicken anti-neurofilament heavy chain (NF-H) antibody (EnCor Biotech) was used at 1:1000, and donkey anti chicken secondary antibody (Jackson ImmunoResearch Laboratories) was used at 1:500. ABC and DAB kits were purchased from the Vector Lab.
For immunofluorescence staining, cyPrP was detected with 3F4 antibody (4mg/ml, Covance) at 1:1500. Rabbit anti-calnexin antibody (Stressgen) was used at 1:1000. Alexa Fluor 488 goat anti-mouse IgG (1:500) and Alexa Fluor 594 goat anti-rabbit IgG (1:500) (Invitrogen) were used at as secondary antibodies. Antigen Amplify system (prohisto lab) was used to increase sensitivity of detection. Incubations with primary and secondary antibodies were carried out at 4°C for 48 and 24 hours, respectively. After washing twice with PBS, cells were incubated with 100 ng/ml 4,6-diamidino-2-phenylindole (DAPI) (Sigma) in PBS for 10 min and washed three times with PBS. Coverslips were mounted on glass slides, and staining was visualized with an Olympus IX81 inverted microscope.
Beta-galactosidase (β-Gal) staining and activity
For staining, brain hemispheres were rinsed with ice-cold PBS pH 7.8, fixed at 4°C for 1 hour in 2% paraformaldehyde, 0.2% glutaldehyde, 0.02% NP-40, and 0.01% deoxycholate. After three PBS rinses, brain hemispheres were stained at 37°C for 4 hours with X-Gal solution (5mM KFeCN, 2mM MgCl2, 0.02% NP-40, 0.01% Deoxycholate, 1mg/ml X-GAL), and post-fixed with 4% paraformaldehyde for 2 hours.
For activity, 100μg of brain lysates were incubated at 37°C for 20 to 180 minutes with 1ml LacZ solution (60mM Na2HPO4, 40mM NaH2PO4 pH 6.95, 10mMKCl, 1mM MgSO4; and freshly added 50mM β-mercaptoethanol) and 200μl substrate solution (60mM Na2HPO4, 40mM NaH2PO4 pH 6.95; and freshly added 2mg/ml o-Nitrophenyl β-D-Glactopyranoside). The reaction was stopped by adding 0.5 ml of 1M Na2CO3 and OD was measured at 420 nm.
Detection of cyPrP oligomers
Mouse forebrains were homogenized in Mg2+ and Ca2+ free PBS (1:10 (w/v)). To prepare lysates for immunoprecipitation, stock solutions were added to 100μl homogenate to reach final concentrations of 10mM Phosphate buffer, pH7.4, 150mM NaCl, 1% NP-40, 1% Deoxycholate, 0.1% SDS, and 1X complete protease inhibitors (Roche) in a final volume of 150μl. The lysates were kept on ice for 10 minutes, followed by 10 minute sonication in a cup-hold sonicator and pre-cleared by rotating at room temperature for 40 minutes in the presence of 20 μl of Protein G magnetic beads (Dynabeads, Invitrogen). The pre-cleared lysates were rotated at 4°C overnight in the presence of 7 μg of the A11 anti-oligomer antibody (BioSource, Invitrogen), followed by incubation with 40 μl of Protein G magnetic beads at room temperature for 40 minutes. The beads were then washed and eluted according to manufacturer suggested protocol. Samples were subjected to SDS-PAGE and immunoblot analysis.
RESULTS
Characterization of tetracycline-responsive transgenic mice expressing cyPrP
We generated transgenic mice expressing murine cyPrP under the regulation of a tetracycline-responsive bi-directional promoter. This promoter, as shown in figure 1A, contains a tetracycline-responsive element (TRE) flanked by two identical minimal CMV promoters (PminCMV) driving cyPrP and β-Gal expression respectively. The transgene expression is regulated by the tTA transcription factor of the “Tet-off system”. The widely used 3F4 epitope, an epitope derived from hamster PrP, was engineered in murine cyPrP, allowing us to readily distinguish cyPrP from endogenous murine PrPC.
Six transgenic founders were identified and all of them were crossed with FVB/N mice expressing tTA under the control of the CamKII promoter [26]. The pregnant mice were supplied with dox-food to suppress transgene expression. Once double transgenic offspring (tTA+/−; pBI-cyPrP-Gal+/−) were identified, dox-food was removed at 6 weeks of age and the expression of cyPrP and β-gal was monitored after 2 weeks of induction. Using a β-Gal activity assay, we identified two lines of mice (line 5 and 33) expressing transgene at different levels. Figure 1B shows that β-gal activity was detected in the forebrain of a double transgenic mouse from line 33, but not in a double transgenic mouse from line 27 that does not express the β-Gal reporter gene. Similarly, no β-gal activity was detected in a control single transgenic mouse containing only the tTA transgene. The whole brain β-gal staining revealed that the β-gal expression was largely confined to the forebrain (Supplemental Fig. S1), consistent with CamKII promoter-driven tTA expression pattern [26]. Transgene expression was regulated by dox and no β-gal activity or cyPrP expression was detected in double transgenic mice fed with dox-food (Fig. 1C and D). When dox was removed, cyPrP and β-gal were expressed and the expression was almost exclusively in the forebrain (Fig. 1C and D). The equal loading of the gel (Fig. 1D) was verified by total protein stain of the blot (data not shown).
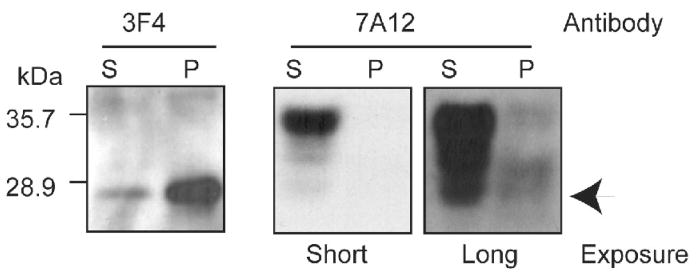
To determine the aggregation status of cyPrP in these mice, we separated forebrain detergent lysates prepared from induced double transgenic mice into supernatant and pellet fractions. The majority of cyPrP was detected in the pellet fraction (Fig. 2, left panel). The same blot was re-probed with 7A12 antibody that recognizes both endogenous PrP and cyPrP transgene [31]. Clearly, most of the heterogeneously glycosylated endogenous PrP was soluble and appeared in the supernatant (Fig. 2, right panels). Notably, the aggregated cyPrP could not be reliably detected with 7A12 antibody, suggesting a low cyPrP level compared to endogenous PrP. In these induced mice that are heterozygous for the cyPrP transgene, cyPrP was sensitive to proteinase K (PK) digestion (data not shown).
Fig. 2. Characterization of CyPrP in the inducible transgenic mice.
Forebrain detergent lysate from an induced heterozygous transgenic mouse were separated into supernatant (S) and pellet (P) fractions, and the presence of cyPrP was determined by immunoblot analysis with the 3F4 anti-PrP antibody. The same blot was re-probed with 7A12 antibody to determine total PrP. Arrow points at the expected position of cyPrP.
Mice expressing cyPrP in the forebrain developed behavioral abnormalities
When transgene expression was suppressed by feeding mice with dox-food, no difference was noticed between double transgenic mice (tTA+/−; pBI-cyPrP-Gal+/−) and control mice (pBI-cyPrP-Gal only, tTA only or wild-type mice). When cyPrP expression was induced by switching from dox-food to regular diet, behavioral phenotypes emerged. Inducing cyPrP expression for 2 weeks led to clasping, a common neurodegenerative phenotype, in both transgenic line 5 and 33 (Fig. 3A).
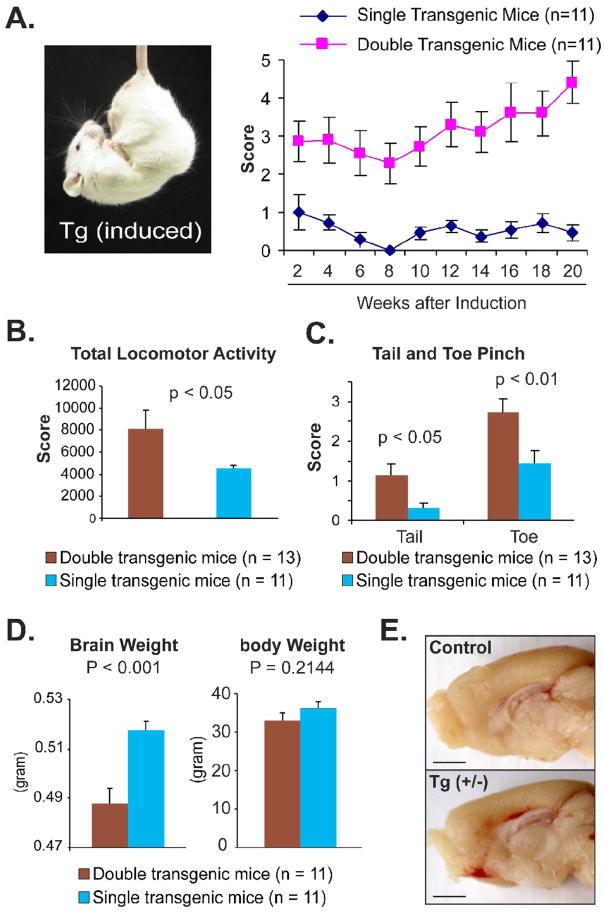
Fig. 3. Forebrain toxicity in induced transgenic mice expressing cyPrP.
Pregnant mice and pups were supplied with Dox food until 6 weeks of age and then switched to normal food to induce transgene expression in double transgenic mice (tTA+/−; pBI-cyPrP-Gal +/−). Among 11 double transgenic mice, 8 mice were from line 33 and 5 mice were from line 5. Single transgenic littermates (tTA−/−; pBI-cyPrP-Gal+/−) that went through the same induction regime were used as controls. (A) Picture and score of tail suspension test. Error bars represent standard error and the differences are statistically significant, p<0.05 at 2 weeks and p<0.01 at remaining time points. (B) Total locomotor activity as indicated by the number of beam breaks. (C) Scores of tail and toe pinch. (D) The brain and body weight of induced heterozygous double transgenic mice and their single transgenic littermates. (E) A representative pair of double transgenic mouse (Tg(+/−)) and single transgenic littermate (control) showing the difference in the cortex thickness. Bars represent 1 mm.
To determine other behavioral abnormalities, double transgenic mice (both line 5 and 33) and their single transgenic control littermates were induced and subjected to a battery of behavior tests. The open field test revealed that induced double transgenic mice had a significantly increased locomotor activity compared to control mice (Fig. 3B), which was consistent with our daily observation that many induced transgenic mice developed stereotypical circling. The significance of the open field test results might be compromised somewhat by a small number of induced double transgenic mice that had obviously reduced locomotor activity and remained inactive. The observation of hyper-and hypoactive mice in both transgenic line 5 and 33 ruled out the possibility that this phenotype results from the random transgene integration. Nonetheless, the induced transgenic mice as a whole group had a statistically significant increase in locomotor activity.
The hyperactivity of these mice interfered with the interpretation of other behavior tests. For example, induced double transgenic mice had reduced numbers of rears and showed a preference for the light chamber during the light-dark preference test (data not shown). Although the differences were statistically significant, the fact that several induced transgenic mice were continuously circling during the testing period, rather than exploring the arena, precludes a conclusive interpretation of the results.
In contrast, a significant difference in their response to tail and toe pinch was observed between control and cyPrP expressing mice, with induced double transgenic mice reacting more violently (Fig. 3C). This phenotype may have contributed to the higher tendency for double transgenic mice, both males and females, to initiate fight with their littermates, which was observed during our daily monitoring.
When these mice were sacrificed, we compared the brain weight of double transgenic mice to that of single transgenic mice (tTA−/−; pBI-cyPrP-Gal+/−), which had gone through the same induction regime. The brains of cyPrP expressing double transgenic mice were significantly lighter compared to control mice, while there is no significant difference in body weight between these two groups (Fig. 3D). When these brains were dissected sagittally, a noticeably thinner cortex was observed in induced double transgenic mice (Fig. 3E).
Neurodegeneration in induced cyPrP homozygous mice
In order to determine whether an increased cyPrP dosage leads to a more severe toxicity, we generated homozygous double transgenic mice from transgenic line 33. Heterozygous double transgenic mice (tTA+/−; pBI-cyPrP-Gal+/−) were mated with single transgenic mice (tTA−/−; pBI-cyPrP-Gal+/−), and the pregnant females were separated and remained on regular diet. Thus, the transgene expression was never suppressed in these pups. We compared the phenotype among littermates that were without cyPrP transgene (wild-type), with tTA and a single copy of cyPrP transgene (heterozygous, tTA+/−; pBI-cyPrP-Gal+/−), or with two copies of cyPrP transgene (homozygous, tTA+/−; pBI-cyPrP-Gal+/+).
Although both heterozygous and homozygous mice developed stereotypical circling and clasping phenotype, the homozygous transgenic mice were much smaller compared to wild-type or heterozygous transgenic littermates (Fig. 4A), which was noticeable from 14 days after birth. No obvious difference in food intake was observed, and the body size difference was not obvious among heterozygous mice (tTA+/−; pBI-cyPrP-Gal+/−), wild-type mice, and single transgenic mice containing only tTA transgene (tTA+/−; pBI-cyPrP-Gal−/−) or one or two copies of the pBI-cyPrP-Gal transgene (tTA−/−; pBI-cyPrP-Gal+/− or tTA−/−; pBI-cyPrP-Gal+/+), suggesting that the body size difference is due to the toxic effect of higher amount of cyPrP. In accordance with this notion, the tapering of cortex appeared to be more severe in homozygous double transgenic mice (Fig. 4B).
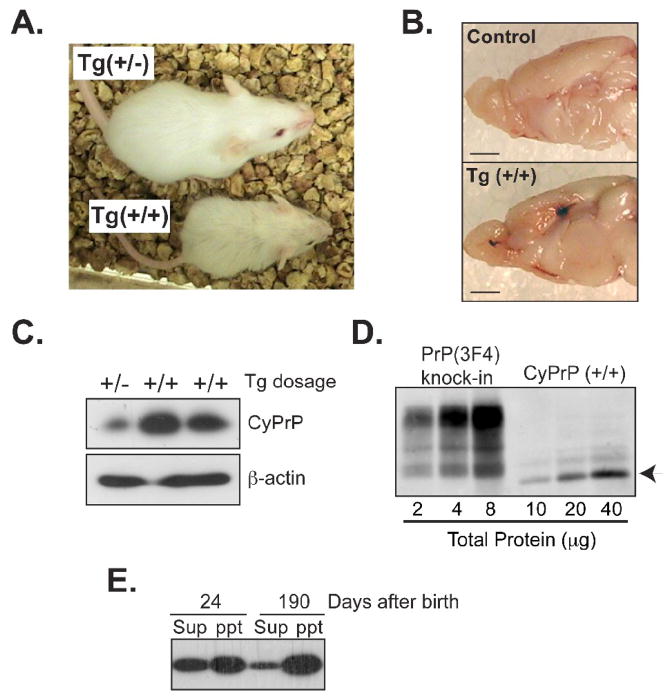
Fig. 4. More severe toxicity associated with homozygous transgenic mice (tTA +/−, pBI-cyPrP-Gal+/+).
(A) Homozygous (+/+) and heterozygous (+/−) transgenic littermates at 4 weeks of age. The transgene expression was never suppressed in these mice and they were always kept in the same cage since birth. (B) Brain images of an induced homozygous transgenic mouse (Tg (+/+)) and its single transgenic littermate (control) sacrificed at 6 months of age. Bars represent 1 mm. (C) CyPrP in the forebrain of induced hetero- (+/−) or homozygous (+/+) mice were detected by immunoblot analysis with the 3F4 antibody. Equal loading was verified by re-probing the same blot with an antibody against β-actin. (D) Serial dilutions of forebrain lysates of an induced homozygous transgenic mouse and a knock-in mouse in which endogenous PrP was replaced by mouse PrP with the 3F4 epitope. The total protein concentration was indicated and PrP was detected by immunoblot analysis with the 3F4 antibody. Arrow points at cyPrP. (E) Forebrain lysates of homozygous transgenic mice were separated into supernatant (sup) and pellet (ppt) fractions by a 1-hour 100,000g centrifugation. The presence of cyPrP was determined by immunoblot analysis with the 3F4 antibody.
The cyPrP level in homozygous mice was about 3 fold higher compared to heterozygous littermates (Fig. 4C and Supplemental Fig. S2), which is higher than what would be predicted based on gene dosage, indicating that, once above certain levels, more cyPrP escapes degradation. However, the amount of cyPrP in homozygous mice was still a small fraction of endogenous PrPC. Using serial dilutions of forebrain homogenates, we compared the cyPrP level in homozygous mice with that of knock-in mice expressing wild-type PrP with the 3F4 epitope (Jackson, W. and Lindquist, S. unpublished data) (Fig. 4D). Phosphorimager quantification revealed that the cyPrP level was less than 10% of the endogenous PrP.
Surprisingly, even though the majority of cyPrP was aggregated in aged homozygous mice (Fig. 4E and Supplemental Fig. S3), a significant portion of cyPrP was soluble in 24 days old homozygous transgenic mice (Fig. 4E and Supplemental Fig. S4), suggesting a higher aggregation tendency of cyPrP in aged animals. PK digestion analysis revealed a slight increase in cyPrP PK-resistance in both young and old homozygous mice. A 15 kDa cyPrP fragment was detected after a 1 hour 2μg/ml PK digestion at 37°C, whereas endogenous PrP remained PK sensitive (Supplemental Fig. S5). Although the PK-resistance is much lower compared to pathogenic PrPSc form, the increase of PK-resistance in a specific cyPrP fragment may indicate the formation of ordered aggregates.
Collectively, these results suggest that a small amount of cyPrP is sufficient to cause neurotoxicity and the toxicity is dependent on the level of cyPrP.
Neurotoxicity in cortex and hippocampus
Histology analysis revealed that there is no overt brain structure abnormality in hetero- or homozygous mice in which transgene expression was never suppressed (data not shown), indicating that the levels of cyPrP expression in these mice did not affect brain development. CyPrP and various neuronal markers were stained to determine the relationship between cyPrP expression and neurotoxicity. Similar to reported expression pattern of CamKII promoter driven tTA [32], the cyPrP was detected in hippocampus and cortex by the 3F4 antibody (compare Fig. 5A and B). Using immunofluorescence staining, we detected cyPrP in the processes and cell body of hippocampal neurons (compare Fig. 5C and 5D), while no nuclear accumulation or formation of perinuclar aggresome was detected (Fig. 5D). Moreover, we did not observe any extracellular cyPrP accumulation in either young or aged mice, indicating that cyPrP neurotoxicity is cell autonomous.
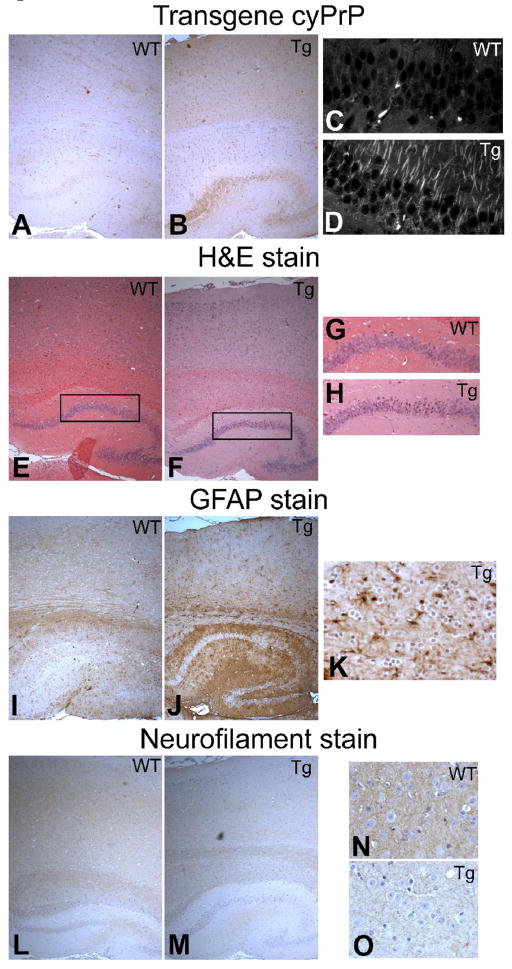
Fig. 5. Neurotoxicity in the cortical and hippocampal regions.
(A and B) CyPrP in wild-type and transgenic mouse brains were stained with 3F4 anti-PrP antibody (brown). (C and D) Immunofluorescence staining of cyPrP in hippocampal CA1 region of wild-type and transgenic mice as indicated. (E and F)H&E staining of the cortical and hippocampal regions of wild-type and transgenic mice brains. (G and H) High magnification images of figure D and E as indicated by the inserts. (I and J) Cortical and hippocampal regions of wild-type and transgenic brains stained with an anti-GFAP antibody (brown). (K) A high magnification image of transgenic mouse cortical region stained with an anti-GFAP antibody (brown). (L and M) Brains of wild-type and transgenic mice were stained with an anti-neurofilament heavy chain antibody (brown). (N and O) High magnification images of wild-type and transgenic mouse cortical regions stained with an anti-neurofilament heavy chain antibody (brown). In all images, nuclei were counter stained with hematoxylin (blue). Images were taken using an Olympus IX81 inverted microscope and the settings were exactly the same for images in the same group. WT, wild-type mouse; Tg, cyPrP-expressing double transgenic mouse.
More condensed nuclei were observed in cortical and hippocampal neurons of cyPrP expressing transgenic mice (compare Fig. 5E and F, Fig 5G and H), and Glial fibrillary acidic protein (GFAP) staining revealed astrogliosis in both regions (compare Fig. 5I and 5J, Fig. 5K is a high magnification of transgenic mouse cortical region). In addition, a reduced neurofilament staining was observed in the cortical and hippocampal regions of cyPrP expressing transgenic mice (compare Fig. 5L and M), which was more evident under a high magnification (compare Fig. 5N and O). Together, these results revealed that the neurotoxicity is in the cortical and hippocampal regions, supporting the relationship between cyPrP expression and neurotoxicity.
A portion of cyPrP is associated with the hydrophobic lipid core of membranes
To determine whether cyPrP in these mice are associated with lipid membranes, we performed iodixanol gradient analysis, in which the forebrain homogenates were loaded at the bottom of the gradient and membrane-bound PrP would migrate to the top. Our result indicated that a portion of cyPrP in the double transgenic mice was membrane-bound, migrating to top fractions similar to GPI-anchored endogenous PrP (Fig. 6A), while the cytosolically localized GAPDH (glyceraldehydes 3-phosphate dehydrogenase) remained at the bottom.
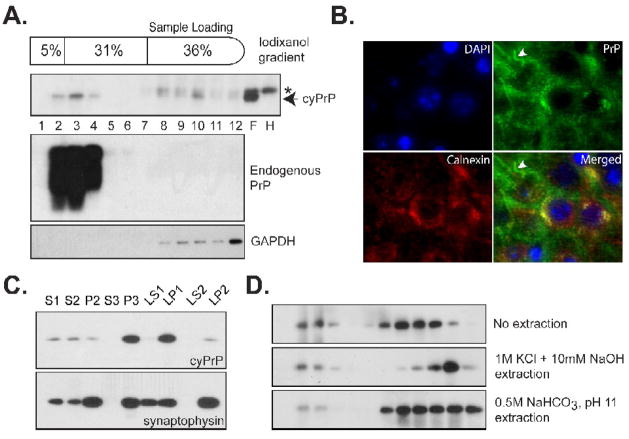
Fig. 6. The interaction between cyPrP and lipid membranes.
(A) Post-nuclear supernatant prepared from the forebrain of an induced heterozygous transgenic mouse (pBI-cyPrP-Gal+/−; tTA+/−) was separated by the iodixanol gradient. The presence of cyPrP in each fraction was detected by the 3F4 antibody. The same blot was re-probed by POM1 anti-PrP antibody to identify membrane associated endogenous PrP, and an anti-GAPDH antibody to identify cytosolic GAPDH protein. Number represents fractions from top to the bottom. F, forebrain; H, hindbrain; Arrow indicates the position of cyPrP; Asterisk indicates a non-specific reactive band that was present in both forebrain and hindbrain lysates. (B) CA1 region of a transgenic mouse brain was stained with antibodies against cyPrP (green) and ER membrane protein calnexin (red). Nuclei were stained with DAPI (blue). Yellow color in the merged image indicates the co-localization between calnexin and cyPrP in the cell body. White arrowhead points to cyPrP in the neuronal process that is not co-localized with calnexin. (C) The presence of cyPrP in different neuronal subcellular membranes. CyPrP and synaptophysin were detected by immunoblot analysis, and the total protein of each fraction is 10μg. (D) Post-nuclear supernatant prepared from an induced homozygous transgenic mouse were subjected to no extraction, extraction with 1M KCl plus 10mM NaOH or with 0.5M NaHCO3, pH 11, and then separated by the iodixanol gradient. CyPrP was detected by immunoblot analysis with 3F4 antibody.
The cyPrP’s membrane association was supported by co-staining cyPrP with calnexin, an ER membrane protein. We found that a significant portion of cyPrP in the cell body was co-localized with calnexin (Fig. 6B), while no co-localization was detected for cyPrP in neuronal processes (Fig. 6B, indicated by a white arrowhead). The possible cyPrP’s association with plasma membrane was not clearly detected using this method, which may be attributed to reasons that only a portion of cyPrP is membrane associated (Fig. 6A and D) and/or that plasma membrane associated proteins are difficult to stain in paraffin-embedded sections. Nevertheless, the co-localization of cyPrP with calnexin is in accordance with result of gradient analysis, that is, a portion of cyPrP is associated with membranes.
We also analyzed which subcellular membrane cyPrP is associated with in these mice by separating neuronal membranes according to a standard protocol [28, 30] (Fig. 6C). S1 and P2 represent post-nuclear supernatant and crude membranes respectively, P3 is enriched with light membranes and microsomes, LP1 represents synaptosomal membranes, and LP2 represents a synaptic vesicle-enriched fraction [30, 33, 34]. Notably, cyPrP was enriched in microsomal (P3) and synaptosomal (LP1) membrane fractions (Fig. 6C, top panel). In contrast to neuronal membrane protein synaptophysin, very little cyPrP was detected in the synaptic vesicle enriched LP2 fraction (Fig. 6C). These results suggest that cyPrP in these mice are associated with plasma and ER membranes.
To determine whether cyPrP in these mice was associated with the hydrophobic lipid core of membranes, we extracted membranes with a buffer containing 1 M KCl plus 10 mM NaOH. As shown in Figure 6D, a portion of cyPrP remained membrane-associated after the extraction (Fig. 6D, middle panel). A more stringent condition, extracting membranes with 0.5M NaHCO3 at pH 11, was also performed and, still, some cyPrP remained membrane-bound (Fig. 6D, bottom panel). High salt and alkaline extraction condition will remove all the peripheral membrane associated proteins from lipid bilayer. Moreover, we previously showed that aggregated cyPrP in the forebrain of straight transgenic mice remained at the bottom fractions after extraction [28], which suggests that the migration of cyPrP to top of the gradient is not due to cyPrP aggregation caused density change. Thus, these results indicate that cyPrP is associated with the hydrophobic lipid core of membranes in the inducible transgenic mice, which is in agreement with our hypothesis that cyPrP caused membrane perturbation leads to neurotoxicity [28].
CyPrP is recognized by the A11 anti-oligomer antibody
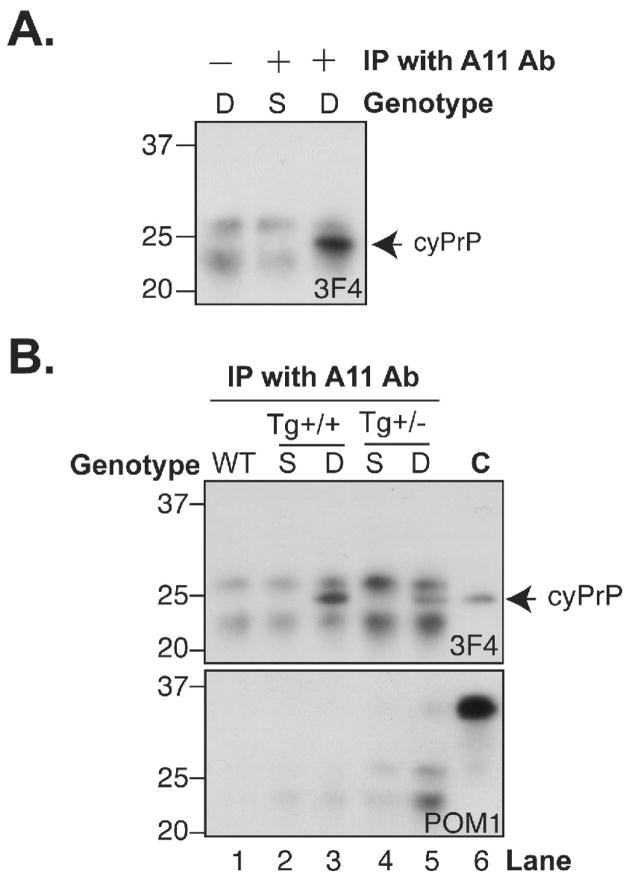
Recent studies have suggested that a common ordered oligomeric species adopted by numerous amyloidogenic proteins often happens on the amyloid fiber formation pathway [35, 36]. Instead of mature amyloid fibers, the soluble oligomeric species permeabilizes lipid bilayer and causes cytotoxicity [37, 38]. To determine whether cyPrP forms ordered oligomer, we used protein G-coated magnetic beads together with an anti-oligomer antibody (A11 antibody), which specifically recognizes oligomer formed by various amyloidogenic proteins including PrP [35], to pull up cyPrP oligomers present in the forebrain lysates. Notably, cyPrP was pulled up by the A11 antibody and detected by immunoblot analysis with the 3F4 anti-PrP antibody (Fig. 7A, lane 3). When the same experiment was performed without adding the A11 antibody, cyPrP was not detected (Fig. 7A, lane 1), ruling out the possibility of non-specific binding. Neither was another control in which a single transgenic littermate containing only pBI-cyPrP-Gal transgene was used (Fig. 7A, lane 2), supporting that the band detected by the 3F4 antibody was indeed cyPrP.
Fig. 7. CyPrP forms ordered amyloid oligomer structure.
(A) Forebrain lysates prepared from induced double transgenic mice (D; pBI-cyPrP-Gal+/+; tTA+/−) or single transgenic mice (S; pBI-cyPrP-Gal+/+; tTA−/−) were subjected to immunoprecipitation with or without the anti-oligomer A11 antibody as indicated, and the presence of cyPrP was detected by immunoblot analysis with the 3F4 antibody (3F4). (B) Same as those in A. WT, a wild-type mouse; Tg(+/+) represents induced mice that were homozygous for pBI-cyPrP-Gal transgene; Tg(+/−) represents induced mice that were heterozygous for pBI-cyPrP-Gal transgene; C represents a control sample in which forebrain lysate of an induced homozygous transgenic mouse was directly subject to immunoblot analysis. As indicated, the presence of cyPrP was detected by immunoblot analysis with 3F4 antibody, and the same blot was re-probed using POM1 antibody to detect endogenous PrP.
To compare the amount of cyPrP oligomer in the induced homozygous and heterozygous transgenic mice, the same experiment was repeated using forebrain lysates prepared from induced homozygous or heterozygous transgenic mice (Fig. 7B, top panel). Our result showed that the amount of cyPrP oligomer was higher in the homozygous mice (Fig. 7B, top panel, compare lane 3 and 5). Since the age of mice used in this analysis was comparable, 15 and 16 months respectively, this difference is most likely not due to the age of animals. Instead, the formation of amyloid oligomer appears to be dependant on the cyPrP levels in the brain, which also correlates with the more severe toxicity seen in homozygous mice.
The SDS-PAGE migration pattern of cyPrP pulled up by the A11 antibody (Fig. 7B, top panel, lane 3 and 5) was exactly the same as cyPrP in the forebrain lysates detected directly by immunoblot analysis (Fig. 7B, top panel, lane 6). In addition, when the same blot was re-probed by the POM1 anti-PrP antibody that recognizes all PrPs [39], almost no endogenous PrP was detected in the pull-up samples (Fig. 7B, bottom panel). Since the input of the pull-up experiment was 50 times greater than that used in the control (Fig. 7B, bottom panel, lane 6), the lack of endogenous PrP detection in the pull-up samples supported the specificity of this analysis. Together, these results indicate that, in vivo, cyPrP forms ordered amyloid oligomer and the amount of cyPrP oligomer appeared to be associated with the severity of neurotoxicity.
DISCUSSION
Our results of inducible transgenic mice provided several novel findings of cyPrP neurotoxicity. First, we have clearly showed that cyPrP evoked toxicity is not restricted to cerebellar granule neurons. In the inducible mice, it causes toxicity in different forebrain neuronal populations. Second, a portion of cyPrP can be recognized by the A11 anti-oligomer antibody, indicating that it forms ordered amyloid oligomer structure. Third, our results indicate that cyPrP is not invariably aggregated in vivo. When the amount of cyPrP is above certain threshold, such as in homozygous transgenic mice, a large portion of it remains soluble in young animals (Fig. 4E and S4). When cyPrP expression level is below the threshold, such as in heterozygous transgenic mice, the majority of it forms aggregate regardless of mouse age (Fig. 2 and S3).
Neurodegeneration in prion disease occurs in different brain regions, and thereby our finding that cyPrP causes toxicity in different neuronal populations is critical for it to be considered as a legitimate candidate for the neurotoxic species in prion disease. Since the CamKII promoter is neuron specific [26, 40] and the fact that we did not observed extracellular accumulation of cyPrP, the phenotypes observed here likely result from cell autonomous cyPrP toxicity. The phenotype in the inducible cyPrP-expressing mice appeared to be milder compared to what was observed in the cerebellum [15], which could be attributed to reasons such as a higher resistance of forebrain neurons to cyPrP toxicity, different mouse background, or the mosaic expression pattern of the CamKII promoter [26, 40]. Nonetheless, our results clearly revealed that cyPrP is capable to cause forebrain neurotoxicity, which supports the hypothesis that it may contribute to neurotoxicity in prion disease.
The neurotoxic mechanism of prion disease remains unclear. Studies of several apoptotic cell death pathways including Bax, Bcl2 and caspase-12 concluded that these pathways do not contribute significantly to the pathogenic process [41–43], revealing the complexity of neurotoxic mechanism in prion disease. In cyPrP expressing transgenic mice, neither spongiosis nor PrPSc was detected. This phenomenon might be due to the low cyPrP expression levels in the inducible (this study) and the straight transgenic mice [15, 28], which is presumably due to the high neurotoxicity of cyPrP. On the other hand, it is also possible that the lack of spongiosis or PrPSc is a characteristic of cyPrP neurotoxicity. Even in this case, cyPrP caused neurotoxicity may still play a role in the pathogenesis of prion disease. It is well known that different forms of prion disease differ in clinical manifestation and pathology [2, 22–24], and it is possible that different pathogenic mechanisms may account for different forms or different stages of prion diseases. CyPrP induced toxicity, alone or as one of the toxic components, could be involved in various forms of prion disease, but its involvement in inherited prion disease seems more appealing. Several disease-associated PrP mutants have been shown to increase the retro-translocation of PrP to the cytosol [9, 11, 44, 45]. In addition, the duration of the inherited prion disease is generally long [2, 22–24], classic PrPSc was not detected in some human cases [24, 25, 46] or in animal models [47–49], spongiosis is not evident in some cases [22, 24], and brain atrophy has been reported [46].
One of the interesting findings in this study is the recognition of cyPrP by the A11 antibody, indicating that cyPrP may form ordered oligomeric structures in vivo. Soluble oligomers have been implicated as the major neurotoxic species in a variety of neurodegenerative diseases [35]. The formation of soluble cyPrP oligomer may explain the peculiar relationship between cyPrP aggregation and neurotoxicity observed in this study. The homozygous mice differ from their heterozygous littermates in that the cyPrP level was higher and a large portion of cyPrP remained soluble in 24-day old mice, suggesting that, in vivo, a higher cyPrP level does not simply lead to a high degree of aggregation. The increased cyPrP solubility could be due to that the intra-neuronal environment in 24-day old mice is unfavorable for cyPrP to form large aggregates. Alternatively, the cyPrP aggregation may require its interaction with other intracellular factors that might be limited in neurons of mice at this age. More importantly, the increased solubility of cyPrP in 24-day old homozygous mice appears to correlate with the progression of neurotoxic phenotype. The most striking difference between homozygous and heterozygous mice is the smaller body size in homozygous mice, which progresses most rapidly from 14 days to 4 weeks after birth. After that, the body size of homozygous mice remains smaller compared to their heterozygous littermates, but it does not appear to progress any further. Further study is required to determine whether this phenotype is related to higher levels of soluble cyPrP oligomers.
Our results are also in accordance with that a low level of cyPrP is sufficient to cause neurotoxicity and that the cyPrP neurotoxicity correlates with its interaction with the hydrophobic lipid core of membranes [15, 28]. The neurotoxicity associated with such a small amount of cyPrP is likely due to its interference with certain vital physiological processes. Because neurons generally contain long processes and membrane integrity is critical for their physiological functions, membrane perturbation would be a plausible explanation for the impairment of their functionality and viability. The observations that cyPrP forms ordered oligomeric species that are known to disrupt lipid bilayer [37, 38], and that the cyPrP neurotoxicity correlates with its interaction with the hydrophobic lipid core of membranes are in line with this hypothesis. The interaction between cyPrP and the hydrophobic lipid core of membranes does not necessarily mean that cyPrP acquires a transmembrane topology. More likely, it becomes an integral monotopic protein like cyclooygenase, which only penetrate one leaflet of the lipid bilayer. Oligomerization of cyPrP in this topological form will likely result in a disruption of lipid bilayer homeostasis. The enrichment of cyPrP in microsomal and synaptosomal fractions is consistent with our previous finding that cyPrP in cultured cells are associated with ER and plasma membranes [28]. Because of the high calcium concentrations in ER and extracellular space, permeability alteration in ER and plasma membranes could potentially lead to a disruption of cytosolic calcium homeostasis, which may ultimately lead to neurotoxicity.
It is conceivable that cyPrP, which is in a completely foreign cytoplasmic environment, may cause toxicity via different pathways. A recent study showed that an oligomeric PrP species, recognized by the anti-amyloid oligomer antibody and present in partially purified PrPSc, reduced proteasome activity by inhibiting the 20S proteasome catalytic core [50]. We analyzed proteasome activity (including caspase-like, chymotrypsin-like, and trypsin-like protease activities) in the inducible transgenic mice at various ages and did not find any significant difference between cyPrP-expressing and control mice (data not shown). However, this negative result does not rule out the possibility that cyPrP oligomer may compromise proteasome activity since total forebrain lysates were used in our assay. In the inducible mice, the expression of cyPrP is determined by the CamKII promoter, which only expresses in a subset of forebrain neurons [26, 51]. Moreover, total forebrain lysates contain not only neurons but also a large number of other types of cells that may interfere with proteasome activity analysis. In fact, the peculiar increase of cyPrP levels during aging (Supplemental Fig. S2) may actually indicate a decrease in proteasome activity. More detailed studies, such as crossing our mice with ubiquitin-GFP reporter mice [50], are required to determine whether the cyPrP oligomer compromises the ubiquitin-proteasome degradation system.
Collectively, our results reveal that a low level of cyPrP is sufficient to cause toxicity in different neuronal populations. Since cyPrP was detected in cells expressing endogenous wild-type PrP [52] and in neurons of wild-type mice [8], the cyPrP neurotoxicity may play a role in various PrP-mediated neurodegenerative disorders.
Supplementary Material
Acknowledgments
We thank Drs. Walker Jackson and Susan Lindquist for providing a 3F4 tagged PrP knock-in mouse brain and Dr. Man-Sun Sy for providing the 7A12 antibody. This study was supported by grants from the Ellison Medical Foundation (JM), a Pfizer/AFAR Innovations in Aging Research Grant (JM), National Institutes of Health (NIH) Grants R01NS060729 (JM), MH 57535 and P30 NS045758 (RJN) and National Science Foundation Grant IOS 04-16897 (RJN).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Prusiner SB. Prions. Proc Natl Acad Sci U S A. 1998;95:13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aguzzi A, Sigurdson C, Heikenwaelder M. Molecular mechanisms of prion pathogenesis. Annu Rev Pathol. 2008;3:11–40. doi: 10.1146/annurev.pathmechdis.3.121806.154326. [DOI] [PubMed] [Google Scholar]
- 3.Caughey B, Baron GS. Prions and their partners in crime. Nature. 2006;443:803–810. doi: 10.1038/nature05294. [DOI] [PubMed] [Google Scholar]
- 4.Collinge J, Clarke AR. A general model of prion strains and their pathogenicity. Science. 2007;318:930–936. doi: 10.1126/science.1138718. [DOI] [PubMed] [Google Scholar]
- 5.Bueler H, Aguzzi A, Sailer A, Greiner RA, Autenried P, Aguet M, Weissmann C. Mice devoid of PrP are resistant to scrapie. Cell. 1993;73:1339–1347. doi: 10.1016/0092-8674(93)90360-3. [DOI] [PubMed] [Google Scholar]
- 6.Brandner S, Isenmann S, Raeber A, Fischer M, Sailer A, Kobayashi Y, Marino S, Weissmann C, Aguzzi A. Normal host prion protein necessary for scrapie-induced neurotoxicity. Nature. 1996;379:339–343. doi: 10.1038/379339a0. [DOI] [PubMed] [Google Scholar]
- 7.Mallucci G, Dickinson A, Linehan J, Klohn PC, Brandner S, Collinge J. Depleting neuronal PrP in prion infection prevents disease and reverses spongiosis. Science. 2003;302:871–874. doi: 10.1126/science.1090187. [DOI] [PubMed] [Google Scholar]
- 8.Mironov A, Jr, Latawiec D, Wille H, Bouzamondo-Bernstein E, Legname G, Williamson RA, Burton D, DeArmond SJ, Prusiner SB, Peters PJ. Cytosolic prion protein in neurons. J Neurosci. 2003;23:7183–7193. doi: 10.1523/JNEUROSCI.23-18-07183.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jin T, Gu Y, Zanusso G, Sy M, Kumar A, Cohen M, Gambetti P, Singh N. The chaperone protein BiP binds to a mutant prion protein and mediates its degradation by the proteasome. J Biol Chem. 2000;275:38699–38704. doi: 10.1074/jbc.M005543200. [DOI] [PubMed] [Google Scholar]
- 10.Yedidia Y, Horonchik L, Tzaban S, Yanai A, Taraboulos A. Proteasomes and ubiquitin are involved in the turnover of the wild-type prion protein. Embo J. 2001;20:5383–5391. doi: 10.1093/emboj/20.19.5383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma J, Lindquist S. Wild-type PrP and a mutant associated with prion disease are subject to retrograde transport and proteasome degradation. Proc Natl Acad Sci U S A. 2001;98:14955–14960. doi: 10.1073/pnas.011578098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rane NS, Yonkovich JL, Hegde RS. Protection from cytosolic prion protein toxicity by modulation of protein translocation. Embo J. 2004;23:4550–4559. doi: 10.1038/sj.emboj.7600462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kang SW, Rane NS, Kim SJ, Garrison JL, Taunton J, Hegde RS. Substrate-specific translocational attenuation during ER stress defines a pre-emptive quality control pathway. Cell. 2006;127:999–1013. doi: 10.1016/j.cell.2006.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rane NS, Kang SW, Chakrabarti O, Feigenbaum L, Hegde RS. Reduced translocation of nascent prion protein during ER stress contributes to neurodegeneration. Dev Cell. 2008;15:359–370. doi: 10.1016/j.devcel.2008.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ma J, Wollmann R, Lindquist S. Neurotoxicity and neurodegeneration when PrP accumulates in the cytosol. Science. 2002;298:1781–1785. doi: 10.1126/science.1073725. [DOI] [PubMed] [Google Scholar]
- 16.Rambold AS, Miesbauer M, Rapaport D, Bartke T, Baier M, Winklhofer KF, Tatzelt J. Association of Bcl-2 with misfolded prion protein is linked to the toxic potential of cytosolic PrP. Mol Biol Cell. 2006;17:3356–3368. doi: 10.1091/mbc.E06-01-0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grenier C, Bissonnette C, Volkov L, Roucou X. Molecular morphology and toxicity of cytoplasmic prion protein aggregates in neuronal and non-neuronal cells. J Neurochem. 2006;97:1456–1466. doi: 10.1111/j.1471-4159.2006.03837.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fioriti L, Dossena S, Stewart LR, Stewart RS, Harris DA, Forloni G, Chiesa R. Cytosolic prion protein (PrP) is not toxic in N2a cells and primary neurons expressing pathogenic PrP mutations. J Biol Chem. 2005;280:11320–11328. doi: 10.1074/jbc.M412441200. [DOI] [PubMed] [Google Scholar]
- 19.Crozet C, Vezilier J, Delfieu V, Nishimura T, Onodera T, Casanova D, Lehmann S, Beranger F. The truncated 23–230 form of the prion protein localizes to the nuclei of inducible cell lines independently of its nuclear localization signals and is not cytotoxic. Mol Cell Neurosci. 2006;32:315–323. doi: 10.1016/j.mcn.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 20.Roucou X, Guo Q, Zhang Y, Goodyer CG, LeBlanc AC. Cytosolic Prion Protein Is Not Toxic and Protects against Bax-mediated Cell Death in Human Primary Neurons. J Biol Chem. 2003;278:40877–40881. doi: 10.1074/jbc.M306177200. [DOI] [PubMed] [Google Scholar]
- 21.Jodoin J, Laroche-Pierre S, Goodyer CG, LeBlanc AC. Defective retrotranslocation causes loss of anti-Bax function in human familial prion protein mutants. J Neurosci. 2007;27:5081–5091. doi: 10.1523/JNEUROSCI.0957-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Montagna P, Gambetti P, Cortelli P, Lugaresi E. Familial and sporadic fatal insomnia. Lancet Neurol. 2003;2:167–176. doi: 10.1016/s1474-4422(03)00323-5. [DOI] [PubMed] [Google Scholar]
- 23.Gambetti P, Kong Q, Zou W, Parchi P, Chen SG. Sporadic and familial CJD: classification and characterisation. Br Med Bull. 2003;66:213–239. doi: 10.1093/bmb/66.1.213. [DOI] [PubMed] [Google Scholar]
- 24.Collins S, McLean CA, Masters CL. Gerstmann-Straussler-Scheinker syndrome, fatal familial insomnia, and kuru: a review of these less common human transmissible spongiform encephalopathies. J Clin Neurosci. 2001;8:387–397. doi: 10.1054/jocn.2001.0919. [DOI] [PubMed] [Google Scholar]
- 25.Kovacs GG, Trabattoni G, Hainfellner JA, Ironside JW, Knight RS, Budka H. Mutations of the prion protein gene phenotypic spectrum. J Neurol. 2002;249:1567–1582. doi: 10.1007/s00415-002-0896-9. [DOI] [PubMed] [Google Scholar]
- 26.Mansuy IM, Winder DG, Moallem TM, Osman M, Mayford M, Hawkins RD, Kandel ER. Inducible and reversible gene expression with the rtTA system for the study of memory. Neuron. 1998;21:257–265. doi: 10.1016/s0896-6273(00)80533-4. [DOI] [PubMed] [Google Scholar]
- 27.Young KA, Berry ML, Mahaffey CL, Saionz JR, Hawes NL, Chang B, Zheng QY, Smith RS, Bronson RT, Nelson RJ, Simpson EM. Fierce: a new mouse deletion of Nr2e1; violent behaviour and ocular abnormalities are background-dependent. Behav Brain Res. 2002;132:145–158. doi: 10.1016/s0166-4328(01)00413-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang X, Wang F, Arterburn L, Wollmann R, Ma J. The interaction between cytoplasmic prion protein and the hydrophobic lipid core of membrane correlates with neurotoxicity. J Biol Chem. 2006;281:13559–13565. doi: 10.1074/jbc.M512306200. [DOI] [PubMed] [Google Scholar]
- 29.Wang F, Yang F, Hu Y, Wang X, Jin C, Ma J. Lipid interaction converts prion protein to a PrPSc-like proteinase K-resistant conformation under physiological conditions. Biochemistry. 2007;46:7045–7053. doi: 10.1021/bi700299h. [DOI] [PubMed] [Google Scholar]
- 30.Nishiyama A, Endo T, Takeda S, Imamura M. Identification and characterization of epsilon-sarcoglycans in the central nervous system. Brain Res Mol Brain Res. 2004;125:1–12. doi: 10.1016/j.molbrainres.2004.01.012. [DOI] [PubMed] [Google Scholar]
- 31.Yin S, Pham N, Yu S, Li C, Wong P, Chang B, Kang SC, Biasini E, Tien P, Harris DA, Sy MS. Human prion proteins with pathogenic mutations share common conformational changes resulting in enhanced binding to glycosaminoglycans. Proc Natl Acad Sci U S A. 2007;104:7546–7551. doi: 10.1073/pnas.0610827104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Santacruz K, Lewis J, Spires T, Paulson J, Kotilinek L, Ingelsson M, Guimaraes A, DeTure M, Ramsden M, McGowan E, Forster C, Yue M, Orne J, Janus C, Mariash A, Kuskowski M, Hyman B, Hutton M, Ashe KH. Tau suppression in a neurodegenerative mouse model improves memory function. Science. 2005;309:476–481. doi: 10.1126/science.1113694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dunah AW, Standaert DG. Dopamine D1 receptor-dependent trafficking of striatal NMDA glutamate receptors to the postsynaptic membrane. J Neurosci. 2001;21:5546–5558. doi: 10.1523/JNEUROSCI.21-15-05546.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Onofri F, Giovedi S, Vaccaro P, Czernik AJ, Valtorta F, De Camilli P, Greengard P, Benfenati F. Synapsin I interacts with c-Src and stimulates its tyrosine kinase activity. Proc Natl Acad Sci U S A. 1997;94:12168–12173. doi: 10.1073/pnas.94.22.12168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kayed R, Head E, Thompson JL, McIntire TM, Milton SC, Cotman CW, Glabe CG. Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis. Science. 2003;300:486–489. doi: 10.1126/science.1079469. [DOI] [PubMed] [Google Scholar]
- 36.Shorter J, Lindquist S. Hsp104 catalyzes formation and elimination of self-replicating Sup35 prion conformers. Science. 2004;304:1793–1797. doi: 10.1126/science.1098007. [DOI] [PubMed] [Google Scholar]
- 37.Kayed R, Sokolov Y, Edmonds B, McIntire TM, Milton SC, Hall JE, Glabe CG. Permeabilization of lipid bilayers is a common conformation-dependent activity of soluble amyloid oligomers in protein misfolding diseases. J Biol Chem. 2004;279:46363–46366. doi: 10.1074/jbc.C400260200. [DOI] [PubMed] [Google Scholar]
- 38.Demuro A, Mina E, Kayed R, Milton SC, Parker I, Glabe CG. Calcium dysregulation and membrane disruption as a ubiquitous neurotoxic mechanism of soluble amyloid oligomers. J Biol Chem. 2005;280:17294–17300. doi: 10.1074/jbc.M500997200. [DOI] [PubMed] [Google Scholar]
- 39.Polymenidou M, Stoeck K, Glatzel M, Vey M, Bellon A, Aguzzi A. Coexistence of multiple PrPSc types in individuals with Creutzfeldt-Jakob disease. Lancet Neurol. 2005;4:805–814. doi: 10.1016/S1474-4422(05)70225-8. [DOI] [PubMed] [Google Scholar]
- 40.Chandrasekaran K, Hazelton JL, Wang Y, Fiskum G, Kristian T. Neuron-specific conditional expression of a mitochondrially targeted fluorescent protein in mice. J Neurosci. 2006;26:13123–13127. doi: 10.1523/JNEUROSCI.4191-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Steele AD, Hetz CH, Yi CH, Jackson WS, Borkowski AW, Yuan J, Wollmann RH, Lindquist S. Prion Pathogenesis is Independent of Caspase-12. Prion. 2007;1:1–5. doi: 10.4161/pri.1.4.5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Steele AD, King OD, Jackson WS, Hetz CA, Borkowski AW, Thielen P, Wollmann R, Lindquist S. Diminishing apoptosis by deletion of Bax or overexpression of Bcl-2 does not protect against infectious prion toxicity in vivo. J Neurosci. 2007;27:13022–13027. doi: 10.1523/JNEUROSCI.3290-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Coulpier M, Messiaen S, Hamel R, Fernandez de Marco M, Lilin T, Eloit M. Bax deletion does not protect neurons from BSE-induced death. Neurobiol Dis. 2006;23:603–611. doi: 10.1016/j.nbd.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 44.Mishra RS, Bose S, Gu Y, Li R, Singh N. Aggresome formation by mutant prion proteins: the unfolding role of proteasomes in familial prion disorders. J Alzheimers Dis. 2003;5:15–23. doi: 10.3233/jad-2003-5103. [DOI] [PubMed] [Google Scholar]
- 45.Zanusso G, Petersen RB, Jin T, Jing Y, Kanoush R, Ferrari S, Gambetti P, Singh N. Proteasomal degradation and N-terminal protease resistance of the codon 145 mutant prion protein. J Biol Chem. 1999;274:23396–23404. doi: 10.1074/jbc.274.33.23396. [DOI] [PubMed] [Google Scholar]
- 46.Yamada M, Itoh Y, Inaba A, Wada Y, Takashima M, Satoh S, Kamata T, Okeda R, Kayano T, Suematsu N, Kitamoto T, Otomo E, Matsushita M, Mizusawa H. An inherited prion disease with a PrP P105L mutation: clinicopathologic and PrP heterogeneity. Neurology. 1999;53:181–188. doi: 10.1212/wnl.53.1.181. [DOI] [PubMed] [Google Scholar]
- 47.Hsiao KK, Scott M, Foster D, Groth DF, DeArmond SJ, Prusiner SB. Spontaneous neurodegeneration in transgenic mice with mutant prion protein [see comments] Science. 1990;250:1587–1590. doi: 10.1126/science.1980379. [DOI] [PubMed] [Google Scholar]
- 48.Chiesa R, Piccardo P, Ghetti B, Harris DA. Neurological illness in transgenic mice expressing a prion protein with an insertional mutation. Neuron. 1998;21:1339–1351. doi: 10.1016/s0896-6273(00)80653-4. [DOI] [PubMed] [Google Scholar]
- 49.Nazor KE, Kuhn F, Seward T, Green M, Zwald D, Purro M, Schmid J, Biffiger K, Power AM, Oesch B, Raeber AJ, Telling GC. Immunodetection of disease-associated mutant PrP, which accelerates disease in GSS transgenic mice. Embo J. 2005;24:2472–2480. doi: 10.1038/sj.emboj.7600717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kristiansen M, Deriziotis P, Dimcheff DE, Jackson GS, Ovaa H, Naumann H, Clarke AR, van Leeuwen FW, Menendez-Benito V, Dantuma NP, Portis JL, Collinge J, Tabrizi SJ. Disease-associated prion protein oligomers inhibit the 26S proteasome. Mol Cell. 2007;26:175–188. doi: 10.1016/j.molcel.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 51.Jerecic J, Single F, Kruth U, Krestel H, Kolhekar R, Storck T, Kask K, Higuchi M, Sprengel R, Seeburg PH. Studies on conditional gene expression in the brain. Ann N Y Acad Sci. 1999;868:27–37. doi: 10.1111/j.1749-6632.1999.tb11271.x. [DOI] [PubMed] [Google Scholar]
- 52.Wang X, Wang F, Sy MS, Ma J. Calpain and other cytosolic proteases can contribute to the degradation of retro-translocated prion protein in the cytosol. J Biol Chem. 2005;280:317–325. doi: 10.1074/jbc.M410649200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.