Abstract
The ability to derive neural progenitors, differentiated neurons and glial cells from human embryonic stem cells (hESCs) with high efficiency holds promise for a number of clinical applications. However, investigating the temporal events is crucial for defining the underlying mechanisms that drive this process of differentiation along different lineages. We carried out quantitative proteomic profiling using a multiplexed approach capable of analyzing eight different samples simultaneously to monitor the temporal dynamics of protein abundance as human embryonic stem cells differentiate into motor neurons or astrocytes. Using this approach, a catalog of ~1,200 proteins along with their relative quantitative expression patterns was generated. The differential expression of the large majority of these proteins has not previously been reported or studied in the context of neural differentiation. As expected, two of the widely used markers of pluripotency - alkaline phosphatase (ALPL) and LIN28 - were found to be downregulated during differentiation while S-100 and tenascin C were upregulated in astrocytes. Neurofilament 3 protein, doublecortin and CAM kinase-like 1 and nestin proteins were upregulated during motor neuron differentiation. We identified a number of proteins whose expression was largely confined to specific cell types - embryonic stem cells, embryoid bodies and differentiating motor neurons. For example, glycogen phosphorylase (PYGL) and fatty acid binding protein 5 (FABP5) were enriched in ESCs while beta spectrin (SPTBN5) was highly expressed in embryoid bodies. Karyopherin, heat shock 27 kDa protein 1 and cellular retinoic acid binding protein 2 (CRABP2) were upregulated in differentiating motor neurons but were downregulated in mature motor neurons. We validated some of the novel markers of the differentiation process using immunoblotting and immunocytochemical labeling. To our knowledge, this is the first large scale temporal proteomic profiling of human stem cell differentiation into neural cell types highlighting proteins with limited or undefined roles in neural fate.
Keywords: proteomic profiling, embryonic stem cells, motor neurons, astrocytes, iTRAQ
Introduction
Neural differentiation of embryonic stem cells (ESCs) has become a prolific area of stem cell research. This can be attributed to molecular embryologic studies of the developing nervous system which have resulted in major discoveries defining the mechanisms underlying developmental processes and because ESCs readily differentiate into neural lineages in vitro 1, 2. Coupled with the therapeutic potential of these cells, the ability to efficiently produce pure populations of various neural subtypes is a promising goal, especially for therapeutic purposes. However, the use of hESCs for generating cell types appropriate for cell-based interventions is dependent on progress in defining the mechanisms controlling their fate into different subtypes including motor neurons and astrocytes as well as developing distinct biomarkers for their isolation3, 4. Indeed, several methods to achieve neural differentiation have been reported whereby mitotically-active, nestin-positive progenitor cells are obtained and can be induced to differentiate into neurons, astrocytes, and immature oligodendrocytes in a manner similar to normal fetal development5. However, the characterization of these populations during differentiation is limited to only a few biomarkers and the mechanisms that regulate their differentiation are largely unknown.
While there have been a number of studies comparing the transcriptome of human ESC-derived neuronal 5-8 and glial cells 9, only a limited number of studies have examined their individual proteomes 10-13 Changes at the transcript level do not necessarily correlate with protein abundance.14,15 Proteomics has emerged as a powerful direct method of simultaneous identification and quantitation of proteins. Quantitative proteomics can provide temporal information through the use of multiplexing by incorporating stable isotopes into proteins and/or peptides either in vivo or in vitro. An in vivo method, stable isotope labeling with amino acids in cell culture (SILAC), can be used for simultaneous analysis of up to 5 states. A popular in vitro labeling method is the use of iTRAQ reagents for labeling peptides. Recently, a 4-plex iTRAQ labeling strategy was used to study stem cell differentiation that demonstrated the use of multiple reaction monitoring to validate the quantitation data13. This study showed the utility of protein specific peptide quantitation because relative quantitation of highly homologous proteins can be misrepresented in conventional LC-MS/MS experiments.
More recently, an 8-plex iTRAQ reagent has become available for multiplexed analysis16, 17. This newer generation of reagents is similar to 4-plex reagents and is compatible with platforms used for 4-plex iTRAQ based analysis. We chose this system for a temporal analysis of neural differentiation because it could be used to compare several time points in a single experiment. By studying the progression of neural development from human ESCs, our study aims at providing information regarding the expression of key proteins, which may control the fate of ESCs into motor neurons and astrocytes or may serve to `mark' certain states during the differentiation process. This preliminary study reports the expression profile of ~1,200 proteins during motor neuron and astrocyte differentiation, comparable to the number previously reported in undifferentiated human ESCs.10 Subsequently, a subset of proteomics data was validated using western blot and immunocytochemical analysis.
Materials and Methods
Cell Culture
Human embryonic stem cells (hESC) (H1; WA01) were purchased from Wicell (Wisconsin). Undifferentiated H1 cells were cultured on irradiated mouse embryonic fibroblasts (MEFs) in DMEM/F12 supplemented with 20% KnockOut serum (Invitrogen), 3.5μl BME, 2 mM glutamax, 2 mM nonessential amino acid and 4 ng/mL basic fibroblast growth factor (FGF-2) for 5 days. H1 cells were detached using 0.05% trypsin/EDTA for 5 minutes at 37° C then neutralized using trypsin neutralizing solution (Lonza). H1 cells were routinely subcultured every 5 to 7 days and karyotyped prior to placing them in a differentiation scheme.
Human ESC Differentiation
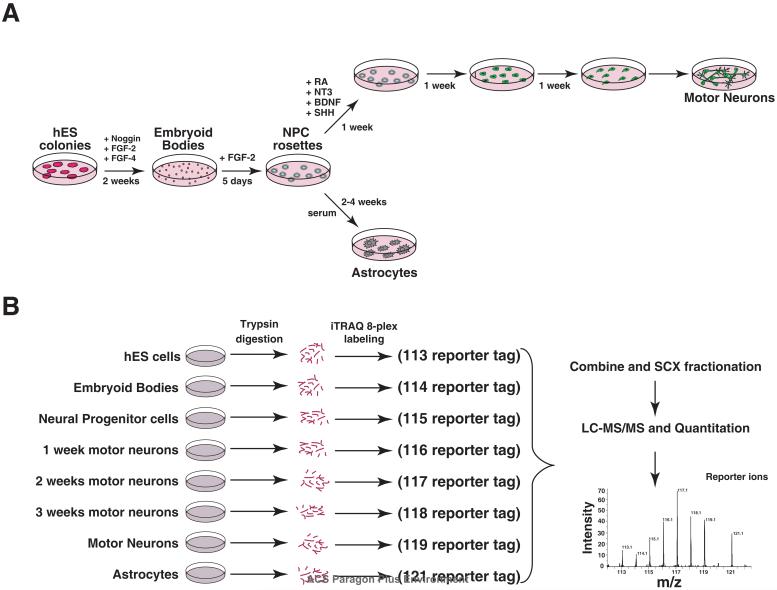
The protocol for deriving cells used in this differentiation study is illustrated in Figure 1. Briefly, hESCs were first trypsinized and grown for 5 days on matrigel with feeder-conditioned media. Once colonies formed, 1 mg/ml collagenase in Dulbecco's PBS (DPBS) was used to produce aggregates (or embryoid bodies) which were then cultured in suspension with N2B27 medium 18 supplemented with 200 ng/ml noggin, 20 ng/ml FGF-2, and 20 ng/ml FGF-4 (R&D Systems). This media was changed every 2 days. After 10-14 days the embryoid bodies (EBs) were plated on matrigel-coated plates and grown in N2B27 supplemented with 20 ng/ml FGF-2. After 5 days, neural rosettes appeared consisting of neural progenitor cells. Motor neuron and astrocyte differentiation was then induced from the neural progenitors (NPs). For motor neuron differentiation, NPs were plated in N2B27 media supplemented with 1 μM RA (Sigma), 10 ng/ml NT3 (R&D Systems), 10 ng/ml BDNF (R&D Systems) and 200 ng/ml sonic hedgehog (R&D Systems). Astrocyte differentiation was induced from NP cultures using N2B27 media supplemented with 10% fetal calf serum. Differentiation at each stage was confirmed with known markers of neural induction using standard immunocytochemical methods. Markers included those expressed by neural progenitors including nestin, islet-1, LIM1, FOXA2 and PAX6; those specific to neurons such as tubulin beta 3 (TUJ1) and motor neuron and pancreas homeobox 1 (Hb9) as well as markers for astrocytes including glial fibrillary acidic protein (GFAP) and S100 calcium binding protein (S-100). Conversely, loss of pluripotency was monitored by the loss of expression of OCT4 and alkaline phosphatase expression.
Figure 1. Experimental strategy for quantitative proteomics analysis.
A. Outline of the protocol for differentiating hES cells along neural lineages. The cells were grown in N2B27 media on tissue culture plastic coated with matrigel. B. A schematic of the quantitative proteomic analysis by labeling with 8-plex iTRAQ reagents followed by strong cation exchange (SCX) based fractionation and liquid chromatography tandem mass spectrometry (LC-MS/MS)
Immunocytochemistry
Antibodies and the concentrations used are summarized in Supplementary Table 1. At all stages except EBs, immunocytochemistry was employed while EBs were frozen in O.C.T. freezing compound (TissueTek), cut into 5μm sections and placed on slides (ProbeOn Plus, Fisher Scientific). In all cases, cells were fixed in 4% paraformaldehyde for 10 min and antibodies diluted in 15% goat serum in DPBS and incubated with the sample for 1 hr at room temperature. Antibodies were then detected by using fluorescently-labeled secondary antibodies (1:200 dilution; Molecular Probes) in 15% goat serum in DPBS for another hour at room temperature. Nuclei were visualized using DAPI (Sigma). Negative controls were performed with secondary antibodies only and with the appropriate sera or ascites fluid.
Fluorescent images were captured using a Nikon Eclipse E800 microscope (Nikon, Inc., Melville, NY). Alexa Fluor 594 fluorescence was detected using a G2ERHOD 541-551 nm excitation filter, a 575 nm dichroic mirror and a barrier filter with a band width of 590. Alexa Fluor 488 was detected using a FITC excitation filter, a 505 nm dichroic mirror and a barrier filter with a band width of 515-555 nm. DAPI was detected using a standard DAPI/Hoechst filter set, UV 2E/C 340380 nm excitation filter, 400 nm dichroic mirror, and a barrier filter with a band width of 435-485 nm. Barrier filters were manufactured by Chroma, Inc. (Burlington, VT). Images were captured with a Photometrics 20 MHz cooled interlined CCD camera and imported into Metamorph software, v.6.2 (Universal Imaging Corp).
Western Blotting
Samples containing 1 × 106 cells were prepared using concentrated Laemmli buffer and heated for 5 min at 100°C. Equal amounts of protein lysates (20μg) were loaded onto reducing 4-12% Bis-Tris Midigels (NUPAGE Novex) for size fractionation and transferred onto PVDF membrane for western blot analysis. Visualization was performed by chemiluminescence detection using the ECL Plus detection system per manufacturer's instruction (Amersham Biosciences).
8-plex iTRAQ Labeling
Whole cell lysates were prepared from hESCs at 8 different time points during neural differentiation. These time points included (1) undifferentiated hESCs, (2) embryoid bodies grown in suspension after 15 days, (4) Cells grown from EBS on matrigel after 5 days forming neural rosettes, a hallmark of neural progenitor cells (NPs), (5-7) NPs after one, two or three weeks of growth in culture conditions favoring the induction of motor neurons (MN) or (8) astrocytes.
Cells were rinsed in ice cold PBS for 3 times, lysed in 0.5% sodium dodecyl sulfate and sonicated for 3min at 30 rpm. The amount of samples was normalized based on protein concentration (Lowry's method). Peptides from each sample were then differentially-labeled using iTRAQ 8-plex reagent (Applied Biosystems, Cat. No. 4390812) according to manufacturer's instructions. Briefly, 60 μg of each sample from ESC, EB, NP, week 1 MN, week 2 MN, week 3 MN and astrocytes were treated with 2 μl of reducing agent (tris(2-carboxyethyl) phosphine (TCEP)) at 60°C for 1 hr followed by addition of 1 μl of cysteine blocking reagent, methyl methanethiosulfonate (MMTS) and kept for 10 minutes at room temperature. The pH of the samples was maintained between 7.5-8.0 by adding 20 μl of 0.5 M triethylammonium bicarbonate. Protein samples were digested using sequencing grade trypsin (Promega) (1:15) for 12 hr at 37°C. Peptides from each sample were then placed in a final volume of 30 μl and labeled with one of the 8 iTRAQ reagents in 60 μl of isopropanol at room temperature. After 2 hours, iTRAQ labeling reactions were terminated by adding 100 μl water to each sample and the corresponding samples were combined and organic solvent was evaporated using a Speedvac. The pH was adjusted to 3.0 using 100 mM phosphoric acid and then diluted to 1 ml in strong cation exchange (SCX) solvent A (10 mM potassium phosphate buffer, pH 2.85, in 25% acetonitrile). Combined mixtures of iTRAQ labeled tryptic digests were fractionated using strong cation exchange chromatography on a PolySULFOETHYL A column (PolyLC, Columbia, MD) (300Å, 5μm, 100 × 2.1 mm) using an Agilent 1100 HPLC system containing a binary pump, UV detector and a fraction collector. Fractionation of peptides (0.2 ml fraction) was carried out by a linear gradient between solvent A and solvent B (solvent A, 350 mM KCl, pH 2.85). The fractions were completely dried and reconstituted in 40μl of 0.2% formic acid and stored at -80°C until LC-MS/MS analysis.
Liquid chromatography and tandem mass spectrometry (LC-MS/MS)
Tandem mass spectrometry analysis of iTRAQ labeled peptides was carried out on a quadrupole time-of-flight mass spectrometer (QSTAR/pulsar, Applied Biosystems). Peptide fractions from SCX chromatography were further separated by reversed-phase liquid chromatography (RP-LC) (Agilent 1100 system) interfaced with a mass spectrometer. RP-LC system consisted of a desalting column (75μm × 3 cm, C18 material 5-10μm, 120Å) and an analytical column (75μm × 10 cm, C18 material 5μm, 120Å) with a nanoflow solvent delivery. Electrospray source was fitted with an emitter tip 8μm (New Objective, Woburn, MA) and maintained at 900 V ion spray voltage. Peptide samples (40μl) were loaded onto a trap column in 0.1% formic acid, 5% acetonitrile for 15 min and LC-MS/MS data was acquired by online analysis of peptides eluted in an acetonitrile in 0.1% formic acid (5-40%) gradient for 30 min with a flow rate of 300 nl/min. Using Analyst v 1.1 (Applied Biosystems), MS/MS data were acquired by targeting three most abundant ions in the scan range of m/z 400 to 1200 Da and those ions selected were excluded from MS/MS for 45s. Unlike the non-labeled peptides, twenty percent higher collision energy was applied during MS/MS scans of iTRAQ labeled peptides. In addition, we generated a technical replicate by labeling the samples again with iTRAQ and analyzed them as described above except that “IDA Extension II,” a script in Analysis software which defines the peptides to be selected for fragmentation based on a user selected ion count threshold, was enabled for selection of low abundant peptides (nearest to threshold ion count >70 with the count every 2 cycle) for fragmentation. This option was used to increase the coverage of the proteome as well as to obtain replicate measurements.
Data analysis and relative quantitation of proteins
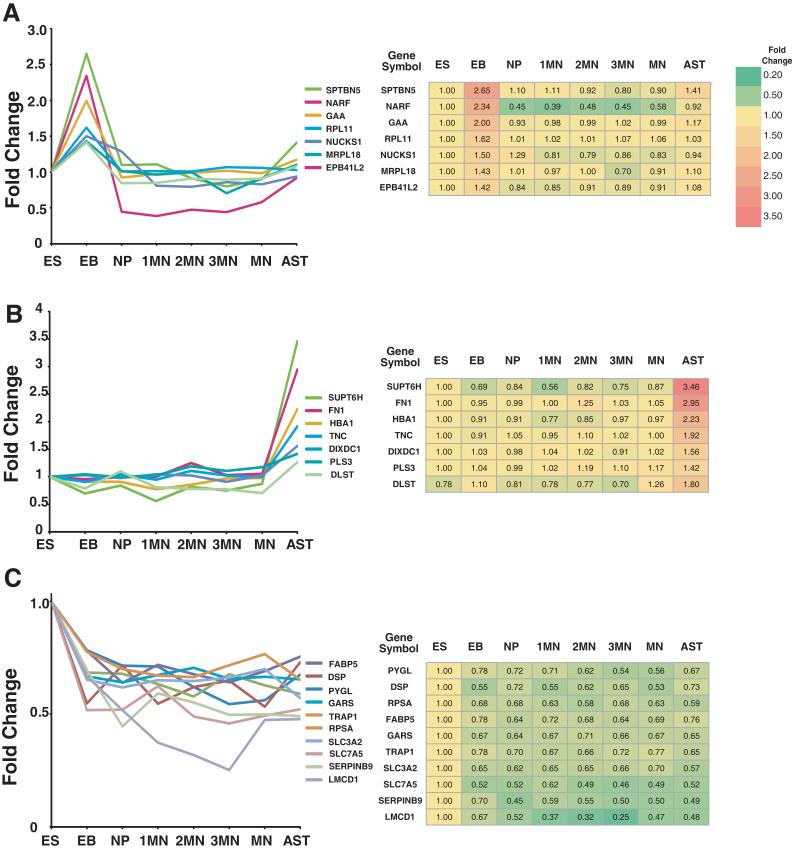
The analysis and documentation of peptide and protein identification was carried out in accordance with Molecular and Cellular Proteomics guidelines. Analyst raw data files were uploaded to ProteinPilot software v2.0.1 (Applied Biosystems) and searched against human RefSeq database version 26. A reverse database was also generated and searched to determine the false discovery rate (FDR) in this analysis. ProteinPilot uses Paragon algorithm for peptide identification and ProGroup algorithm subsequently processes the searched results. The LC-MS/MS analyses were combined and relative abundance of proteins calculated based on individual peptide ratios. Search parameters included iTRAQ labeling at N-terminus and lysine residues, cysteine modification by methyl methanethiosulfonate (MMTS), methionine oxidation and digestion by trypsin. Isoform specific identification of proteins was carried out by selecting peptides distinct to each form, which excludes all shared peptides from quantitation. Proteins identified with >95% confidence or Protscore >1.3 were used for further analysis. Protein identification was based on ProtScore unused score criteria (Pro Group Algorithm, ProteinPilot software). The “Unused” ProtScore is a measurement of all the peptide evidence for a protein that is not better explained by a higher ranking protein. It is the true indicator of protein confidence. Identification and quantitation of a protein is reported for unique peptides with “unused” confidence threshold (ProtScore) >1.3%. For illustrating proteins showing temporal patterns of regulation shown in figures 4 and 5, proteins that exhibited trends of progressive upregulation, progressive downregulation, upregulation during differentiation but downregulation in mature motor neurons and up regulation in hESCs, embryoid bodies and astrocytes were selected manually. The extent of fold-change was not considered while selecting proteins for these expression patterns. In cases where quantitative data was available from both replicates, the ratios were taken from the experiment with a higher score for protein identification.
Figure 4. Temporal patterns of differentially expressed proteins.
Proteins were selected based on the temporal patterns of expression during differentiation. A. Proteins that show a high expression in early stages of motor neuron differentiation, B. Proteins showing progressive increase during motor neuron differentiation, C. Proteins showing progressive decrease during motor neuron differentiation. The fold-changes from the iTRAQ experiments are indicated in the corresponding tables.
Figure 5. Proteins showing high expression in specific cell types.
Sets of proteins showing high expression levels in embryoid bodies (EBs) (A), astrocytes (B) or embryonic stem cells (ES) (C) are shown A. The fold-changes from the iTRAQ experiments are indicated in the corresponding tables.
Results
Embryonic stem cell differentiation into motor neuron and astrocytes
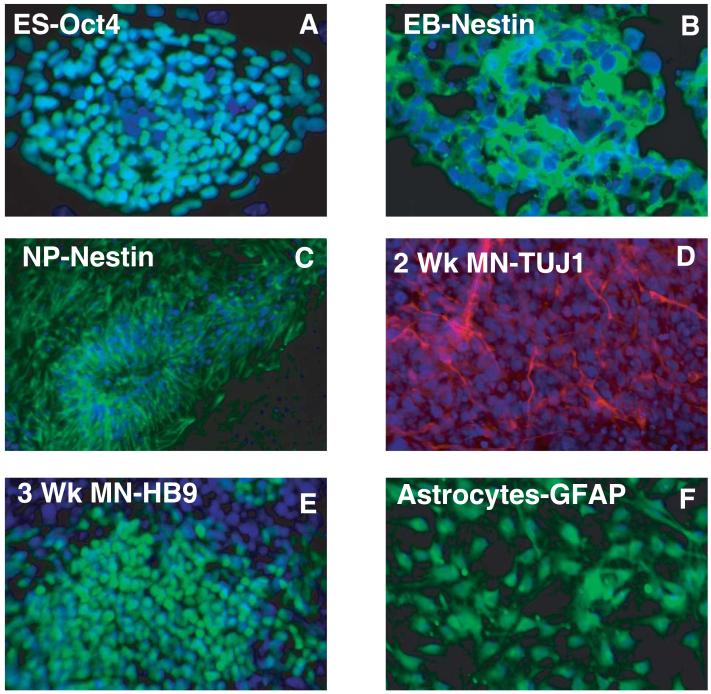
Human stem cells were subjected to well established motor neuron and astrocytes differentiation protocols as illustrated in Figure 1 19. In this study, changes in the proteome were measured at sequential steps in the derivation of hESCs into postmitotic motor neurons and astrocytes. These steps represent distinct stages of differentiation and include early neural progenitors (NPs) produced during embryoid body formation, neural-rosette forming NPs produced in monolayer culture followed by subsequent populations induced to either progress into pre- and postmitotic motor neurons or into mature astrocytes. Figure 2 shows the immunocytochemical staining of different cell types using classic markers. Undifferentiated hESCs were monitored for morphological integrity and expression of OCT4 (POU5F1: POU domain class 5, transcription factor 1) (Figure 2A) and alkaline phosphatase, two well established markers of pluripotency using immunocytochemistry.4, 20 These two markers were used to monitor the loss of pluripotency throughout the differentiation procedure. No expression of either marker was detected by day 10 of embryoid body formation (data not shown). The purity of each population was measured by immocytochemical staining using cell stage specific markers nestin (NPs), glial fibrillary acidic protein (GFAP; astrocytes), tubulin beta 3 (TUJ1; NPs and premitotic motor neurons) and motor neuron and pancreas homeobox 1 (HB9; postmitotic motor neurons) in addition to cell specific morphological changes. At each stage cells were collected for immunocytochemical and proteomic analysis. By day 10, over 95% of the cells form embryoid bodies expressing nestin (Figure 2B) with a concomitant lack of expression of OCT4 and alkaline phosphatase. Neural progenitors in monolayers were almost all nestin positive and over 70% of the cells TUJ1 positive (Figure 2C). As expected TUJ1 was not detected in EBs (data not shown) demonstrating further differentiation along neural lineage at this stage. After 1 week of motor neural induction, >95% of the cells were TUJ1 positive (Figure 2D) and after the third week >65% of the cells expressed the homeobox protein, HB9, the marker specific for mature postmitotic motor neurons (Figure 2E). In contrast over 90% of the cells expressed GFAP, a marker of astrocytes three weeks after induction with fetal calf serum (Figure 2F).
Figure 2. Immunocytochemical characterization of the differentiation process.
Indirect immunofluorescence labeling of different cell types was carried out using Alexa Fluor 594 or Alexa Fluor 488 conjugated secondary antibodies. A. Oct4 expression in undifferentiated hESC colony. B. Nestin positive staining of a cryosection through 10-day differentiating embryoid body (EB) cultured under neural inducing culture conditions. C. Neural rosette formation of nestin-positive cells cultured in monolayer from EBs shown in B. D. Neuronal marker staining of tubulin beta 3 (TUJ1) in 2-wk derived motor neuron cultures and E. HB9 expression in postmitotic motor neurons derived from 3-wk culture conditions favoring neuronal derivation. F. GFAP staining of astrocytes.
iTRAQ labeling and tandem mass spectrometry
We have carried out a replicate analysis using 8-plex iTRAQ. The protein lysates from each population were used for iTRAQ labeling followed by SCX chromatography (Figure 1B). A total of 32 SCX fractions were analyzed by reversed phase LC-MS/MS as explained in the methods section. In all, 52,306 MS/MS spectra were searched and quantitated. The first 8-plex labeling experiment yielded 620 protein identifications (ProtScore >1.3 implies 95% confidence). We then repeated the iTRAQ labeling and analyzed using alternating low abundant and high abundant ions dependent scan method in tandem mass spectrometry (see methods). Using this second approach, we identified 1,079 proteins (ProtScore >1.3 implies 95% confidence) with an overlap of 448 proteins from the first experiment. A total of 1,251 proteins were considered and proteins (n=66) without quantitation ratios were excluded from further analysis. 1,185 non-redundant proteins were considered confidently identified with quantitation ratios. The calculated false discovery rate based on search of a reverse database was <1.3%. The complete list of all protein identifications from the two experiments along with the confidence scores is provided in Supplementary table 2. The list of identified peptides and the corresponding quantitation data is provided as Supplementary table 3.
Quantitative proteomics reveals temporal regulation of proteins during neural differentiation
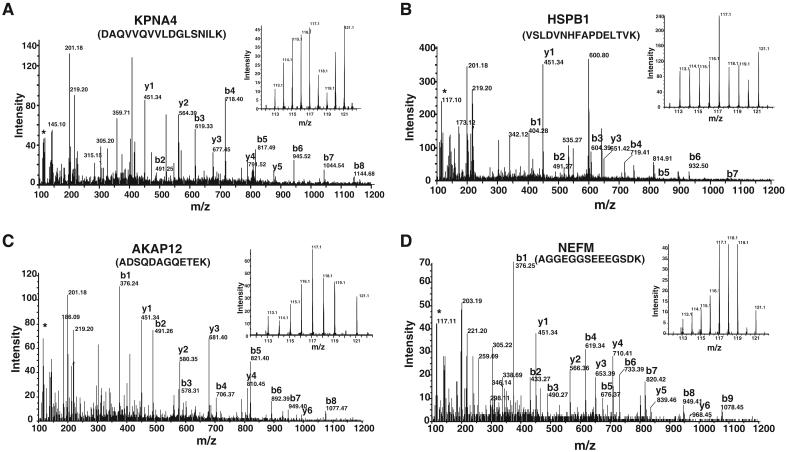
This multiplexed quantitative proteomic approach allowed us to precisely obtain a profile of expression patterns of proteins in a detailed time course analysis of neural differentiation. A number of patterns of expression were observed and representative MS/MS spectra of peptides from 4 proteins - KPNA4, HSPB1, AKAP12 and NEFM - are shown in Figure 3 with each showing a characteristic profile. The inset in each panel shows the relative abundance of each peptide during the 8 time points during differentiation.
Figure 3. MS/MS spectra of iTRAQ labeled peptides.
MS/MS spectra of peptide from representative differentially expressed proteins identified in this study. A. Karyopherin alpha 4 (KPNA4) B. Heat shock protein binding 1 (HSPB1) C. A kinase (PRKA) anchor protein (gravin) 12 (AKAP12); and, D. Neurofilament-3. The inset shows the reporter ions obtained during MS/MS that were used for quantitation.
To illustrate, we manually grouped the proteins into classes that exhibited different trends of expression at various stages of differentiation. These patterns include increase in expression levels during motor neuron induction (1 to 3 week) but decreasing in mature motor neurons (Fig. 4A), proteins showing a progressive increase during the induction of motor neurons (Fig. 4B), progressive decrease in their expression during induction of motor neurons (Fig. 4C). The category shown in Figure 4A includes proteins such as cytokeratin 8 (KRT8), keratin 18 (KRT18), heat shock 27 kDa protein 1 (HSPB1) and Karyopherin alpha 4 (KPNA4). The levels of cytokeratin 8 and cytokeratin 18 (KRT8 and KRT18) were found to increase after formation of NPs with highest levels at 2 week time point for motor neuron differentiation.13, 21 HSPB1 showed high levels during motor neuron differentiation but was also upregulated in astrocytes, which is consistent with an earlier report demonstrating its expression in astrocytes in a murine model of amyotrophic lateral sclerosis (ALS).22 Karyopherin alpha 4, a nuclear transporter, was also found to be upregulated during MN differentiation. This protein has been shown to regulate neural cell fate in differentiating mouse ESCs.23 The dynamics of expression for HSPB1 and KPNA4 protein were also verified by Western blot analysis (Figure 6) and immunocytochemistry (Figure 7). Other proteins in this category include cellular retinoic acid binding 2 (CRABP2), which is consistent with its selective role in retinoic acid induction of MN differentiation.24 The category shown in figure 4B includes proteins such as doublecortin and CAM kinase-like 1 (DCLK1) and neurofilament 3 (NEFM). The iTRAQ analysis showed neurofilament 3 to be upregulated at 2 and 3 weeks after motor neuron induction as well as in mature motor neurons, which was confirmed by Western blot analysis (Figure 6) and immunocytochemical analysis (Figure 7). The role of NEFM and its expression has been extensively studied in neuronal differentiation and growth and its functional importance in motor neurons. The proteins showing the pattern illustrated in Figure 4C include alkaline phosphatase (ALPL) and DNA (cytosine-5-)-methyltransferase 3 beta (DNMT3B). DNMT3B has been studied in mouse ESCs with a potential role in OCT4 and NANOG regulation of pluripotency and development.25, 26-28
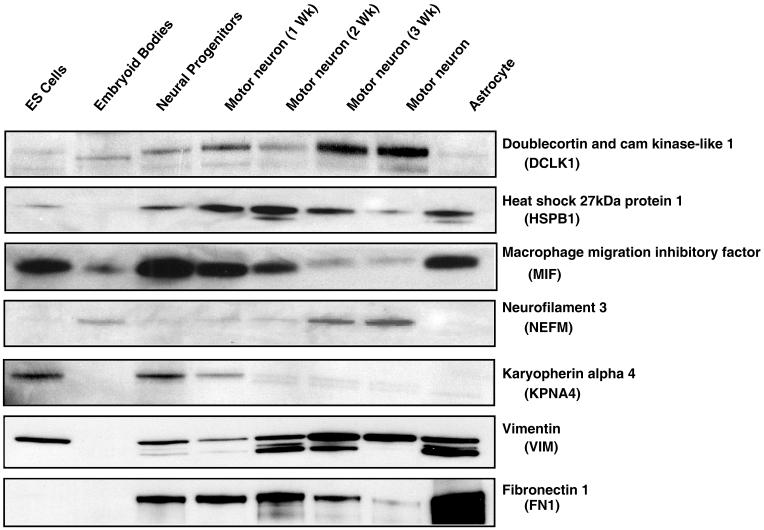
Figure 6. Western blot validation.
Cell lysates from different stages of differentiation as indicated were tested for expression of a select set of molecules against which antibodies were commercially available.
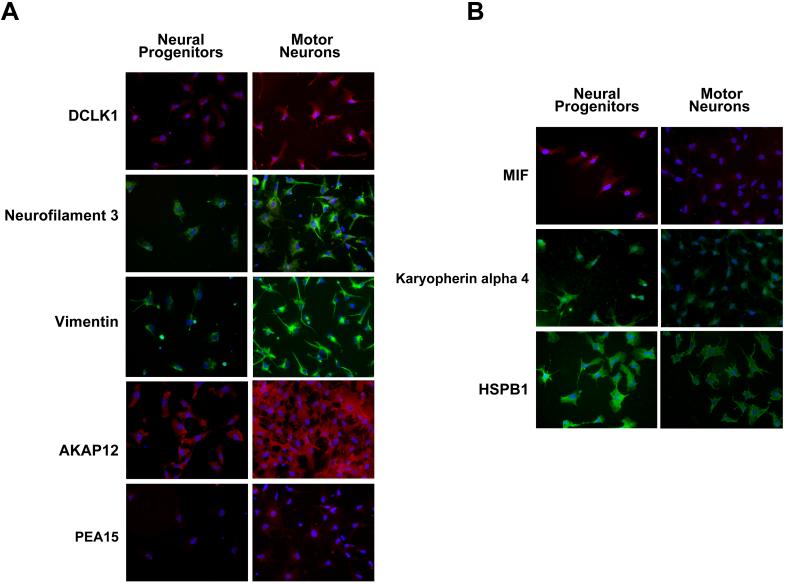
Figure 7. Immunocytochemical analysis of differentially expressed proteins.
Panel A shows an increase in expression of five proteins in hESC-derived motor neurons as compared to neural progenitors. Panel B shows a decrease in expression of three proteins in hESC-derived motor neurons compared to neural progenitors. The nuclei in each case are visualized using DAPI (blue).
We also grouped proteins, which show high expression in specific cell types such as EBs (Fig. 5A), astrocytes (Fig. 5B) and undifferentiated hESCs (Fig. 5C). Proteins expressed during astrocytic differentiation were compared between NPs and GFAP-positive astrocytes. Figure 5B comprises the group of proteins overexpressed in astrocytes including tenascin C (TNC) and fibronectin-1 (FN1). Tenacin C is expressed prominently in cerebral cortex of the developing CNS 29 and is known to play an important role in cortical function 30 and is required for astrocytes proliferation.31 Fibronectin, although known as a mesenchymal stem cell marker,32 is also highly expressed in astrocytes.33 High expression of VIM observed in this study is keeping with earlier reports describing it as a marker of reactive astrocytes34,35 (Fig. 4B, Fig 6 and Fig 7). Additionally, we observed a progressive upregulation of VIM during motor neuron formation, which has not been previously described (Fig 4B). Figure 5A shows proteins upregulated only in embryoid bodies, which include spectrin beta (SPTBN5). Figure 5C shows proteins showing high expression in ESCs with respect to all other neural differentiation stages. Examples of proteins in this category are solute carrier family proteins SLC3A2, SLC7A5 as well as glycogen phosphorylase (PYGL). SLC3A2, a membrane protein interacts with integrins to support RHOA-driven contractility, can control the capacity of cells to assemble a fibronectin matrix.36 SLC7A5 belong to a group of tyrosine transporters with multiples transmembrane domains. It has been shown to be overexpressed in obese mice compared to wild type.37 In this study, we found many novel proteins with respect to their association with neuronal differentiation and further studies are necessary to understand their exact roles in this process.
Among other proteins that showed significant patterns, several histone proteins (HIST1H1C, HIST2H2AB, and HIST1H1E) showed overexpression in neural progenitor cells and early motor neurons. This is consistent with earlier reports showing upregulation of histone proteins in differentiating neural precursor cells. 38, 39 It must be pointed out that many of these transient dynamic changes observed in this study would be missed with a two state analysis such as differential gel electrophoresis or double isotope labeling methods. This is clear from Figure 4A which shows proteins that are upregulated at specific times during commitment of neural progenitors into the neuronal lineage.
Western blot validation of proteomic analysis
Western blots were performed for 7 differentially regulated proteins that are either known to be expressed at different time points or in a specific cell type or they were based on novel proteins selected from the proteomics data set (Figure 6). The known markers of differentiation included neurofilament 3, and vimentin, which showed similar results in both western blot and immunocytochemical analyses. Among the novel proteins validated, a positive correlation was observed with the proteomic analysis for DCLK1, HSPB1, MIF and KPNA4. Fibronectin 1 was detected in the proteomic analyses, which is known to be expressed by pluripotent stem cells and astrocytes. These known controls demonstrated a decrease in expression as illustrated by the decrease in fibronectin 1 levels during motor neuron developmental lineage along with a concomitant increase in astrocytes. Interestingly, while a decrease in expression was noted in MIF during human neural differentiation, the opposite effect has been reported in retinoic acid-treated cells on day 14 during mouse ESC neural induction 40 suggesting that there might be species-specific differences in this process.
Immunocytochemical validation of proteins detected by iTRAQ analyses
Proteins were selected from the iTRAQ analyses based on their known or potential role in neurogenesis for further validation. Figure 7A demonstrates an increase in the expression of DCLK1, neurofilament 3, vimentin, AKAP12 and PEA15 in motor neurons when compared to neural progenitor cells. Three of these (DCLK1, neurofilament 3 and vimentin) were validated by Western blotting as shown in Figure 6. Figure 7B shows immunocytochemical staining of HSPB1, KPNA4 and MIF, all of which show results similar to what was observed by western blotting in Figure 6. Overall, our validation by immunocytochemical analysis shows good concordance with results obtained from western blotting and the iTRAQ data.
Discussion
Mass spectrometry based quantitative proteomics using isotope labeling is a powerful approach for global screening of differentially expressed proteins. An accompanying manuscript (Molina et. al. JPR This issue) describes the development and application of a 5-plex in vivo labeling method using SILAC for studying the differentiation of preadipocytes to adipocytes.41 In the present study, we chose an in vitro labeling approach to simultaneously analyze 8 different stages during the differentiation of human ES cells along neural lineages. In addition to the standard data dependent acquisition, we took advantage of the ability of the QSTAR mass spectrometer to carry out a directed MS/MS analysis of peptide ions with a lower ion count selected by the user. This allowed us to increase the coverage of peptides and proteins especially for those of lower abundance such as transcription factors and kinases. The magnitude of change in the expression level of the large majority of proteins that do show temporal differences in their expression levels is not high (<3 fold). This is important because such small differences are difficult to be reliably analyzed by semi-quantitative proteomic approaches such as label-free peptide counting that have been used to study the proteome of ES cells previously.10
Many proteins found to be regulated in ES cell differentiation may have potential role in regulation of neurogenesis and pluripotency. We identified two known pluripotent markers of undifferentiated ESCs - LIN 28 and alkaline phosphatase. Although the three well-established regulators of pluripotency, OCT4, NANOG, and SOX2, were not identified in these analyses, this is to be expected, as many transcription factors are difficult to identify from whole cell lysates owing to their low abundance. However, several potential regulators of pluripotency were found. For instance, two transcription factors, HMGA1 and SIN3A were downregulated during differentiation. HMGA1 is a protooncogene involved in cell proliferation, differentiation, and development with demonstrated effects in mouse ESC lympho-hematopoietic differentiation 42 while the transcriptional co-repressor SIN3 homolog A (SIN3A) is overexpressed in ESCs and ECCs.43 Another protein, HSPB1 (also known as heat shock protein 27) has been shown to be expressed in undifferentiated mouse ESCs and during very early stages of differentiation. 21 HSPB1 was found to upregulated in 1 to 3 weeks of motor neuron differentiation in both iTRAQ and western blot analysis. Conversely, developmental pluripotent protein-4 (DPPA4) was found to be downregulated in neural progenitor cells, motor neurons at different stages and astrocytes as expected from developmental models. 44
Yocum et al. carried out a quantitative proteomic analysis in which they compared early neuroectodermal induction by noggin to mesodermal induction by BMP4, an antagonist of noggin.13 In that study, BG01 hESCs were grown on 0.1% gelatin coated dishes for 2 days and then induced with 1000 ng/ml noggin for a week. In the present study, H1 hESCs were grown on matrigel for 5 days and collagenase treated to obtain embryoid bodies before inducing with 200 ng/ml noggin in presence of 20 ng/ml FGF-2 and 20 ng/ml FGF-4 for 2 weeks. Although there are differences in the cells and growth conditions used in these studies, we compared the quantitative data from these studies to examine the overlap. Out of 55 differentially expressed proteins upon noggin treatment from the Yocum et al study, 39 were detected in our study (Supplementary Table 4). Almost half of these 39 proteins exhibited changes in the same direction in both studies. However, none of the 34 proteins (Supplementary table 2) that were found to be differentially expressed in neural progenitors (showing 1.5 fold change in either direction) in the current study were reported as differentially expressed by Yocum et al. This lack of congruence in these two datasets could be because the time points at which cells were harvested for proteomic analysis in the two studies were different although the cells were at a somewhat similar stage of differentiation. Another factor is that our approach allowed identification of lower abundant proteins resulting in identification of 1,251 proteins as against 603 proteins from Yocum et al. study.
In quantitative proteomics use of isobaric tags is becoming popular as it allows simultaneous quantitation of protein levels and data is considered reliable even at subtle changes in relative abundance. However, one of the limitations of large-scale quantitative proteomic analysis is that it is difficult to capture all the specific proteins of our interest among large number of proteins identified. Particularly, in iTRAQ labeling methods, after combining the labeled samples, peptides derived from many different states can dominate over those that are specifically found in one or two states thereby limiting their identification in regular data dependent acquisition of highly abundant ions. For example, many transcription-related proteins found in this study (SUPT16H, BTF3, POU6F2) were expressed at the same levels in eight different stages. In contrast, OCT4,NANOG, and SOX2 transcription factors were among those that were not identified in this study. Since these proteins are expected to be high only in undifferentiated cells, peptides from these proteins become diluted upon mixing the labeled peptides from several stages of differentiation. Although our low abundant ion scan method employed in this study enabled us to find a greater number of differentially expressed proteins, it is by chance that these proteins were not detected among thousands of low abundant proteins. Further, repeated mass spectrometry analysis and/or subcellular fractionation prior to mass spectrometry could be used to carry out a more comprehensive analysis. We would like to point out that we have not validated a large number of proteins that could turn out to be exciting and valuable markers of the differentiation process. For instance, solute carrier protein 3 member 2 (SLC3A2) is a cell surface molecule that is somewhat specific to hESCs, and can potentially be used for isolation purposes. Finally, many of the proteins identified in this study have not been previously described in the context of neural stem cell differentiation and to facilitate dissemination of this dataset we have made it available through “Human Proteinpedia” where the peptide and protein identifications can be visualized in the context of individual proteins 45
Supplementary Material
Acknowledgements
This work was supported by a grant from the Maryland Stem Cell Research Fund, State of Maryland (2007-MSCRFE-0137-01) to C.L.K., J.D.G. and A.P. and an NIH Roadmap grant “Technology Center for Networks and Pathways” (U54 RR 020839) to A.P.
Footnotes
Supporting Information Supporting Information Available: This material is available free at http://pubs.acs.org.
References
- 1.Smukler SR, Runciman SB, Xu S, van der Kooy D. Embryonic stem cells assume a primitive neural stem cell fate in the absence of extrinsic influences. J Cell Biol. 2006;172(1):79–90. doi: 10.1083/jcb.200508085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vallier L, Rugg-Gunn PJ, Bouhon IA, Andersson FK, Sadler AJ, Pedersen RA. Enhancing and diminishing gene function in human embryonic stem cells. Stem Cells. 2004;22(1):2–11. doi: 10.1634/stemcells.22-1-2. [DOI] [PubMed] [Google Scholar]
- 3.Thomson JA. Embryonic stem cell lines derived from human blastocysts. 1998. [comment][erratum appears in Science 1998 Dec 4; 282(5395): 1827] [DOI] [PubMed] [Google Scholar]
- 4.Reubinoff BE, Pera MF, Fong CY, Trounson A, Bongso A. Embryonic stem cell lines from human blastocysts: somatic differentiation in vitro. Nat Biotechnol. 2000;18(4):399–404. doi: 10.1038/74447. [DOI] [PubMed] [Google Scholar]
- 5.Shin S, Sun Y, Liu Y, Khaner H, Svant S, Cai J, Xu QX, Davidson BP, Stice SL, Smith AK, Goldman SA, Reubinoff BE, Zhan M, Rao MS, Chesnut JD. Whole genome analysis of human neural stem cells derived from embryonic stem cells and stem and progenitor cells isolated from fetal tissue. Stem Cells. 2007;25(5):1298–306. doi: 10.1634/stemcells.2006-0660. [DOI] [PubMed] [Google Scholar]
- 6.Cheng L, Sung MT, Cossu-Rocca P, Jones TD, MacLennan GT, De Jong J, Lopez-Beltran A, Montironi R, Looijenga LH. OCT4: biological functions and clinical applications as a marker of germ cell neoplasia. J Pathol. 2007;211(1):1–9. doi: 10.1002/path.2105. [DOI] [PubMed] [Google Scholar]
- 7.Lee H, Shamy GA, Elkabetz Y, Schofield CM, Harrsion NL, Panagiotakos G, Socci ND, Tabar V, Studer L. Directed differentiation and transplantation of human embryonic stem cell-derived motoneurons. Stem Cells. 2007;25(8):1931–9. doi: 10.1634/stemcells.2007-0097. [DOI] [PubMed] [Google Scholar]
- 8.Wu H, Xu J, Pang ZP, Ge W, Kim KJ, Blanchi B, Chen C, Sudhof TC, Sun YE. Integrative genomic and functional analyses reveal neuronal subtype differentiation bias in human embryonic stem cell lines. Proc Natl Acad Sci U S A. 2007;104(34):13821–6. doi: 10.1073/pnas.0706199104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhong JF, Song Y, Du J, Gamache C, Burke KA, Lund BT, Weiner LP. Gene regulation networks related to neural differentiation of hESC. Gene Expr. 2007;14(1):23–34. doi: 10.3727/000000007783991781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Van Hoof D, Passier R, Ward-Van Oostwaard D, Pinkse MW, Heck AJ, Mummery CL, Krijgsveld J. A quest for human and mouse embryonic stem cell-specific proteins. Mol Cell Proteomics. 2006;5(7):1261–73. doi: 10.1074/mcp.M500405-MCP200. [DOI] [PubMed] [Google Scholar]
- 11.Van Hoof D, Mummery CL, Heck AJ, Krijgsveld J. Embryonic stem cell proteomics. Expert Rev Proteomics. 2006;3(4):427–37. doi: 10.1586/14789450.3.4.427. [DOI] [PubMed] [Google Scholar]
- 12.Baharvand H, Hajheidari M, Ashtiani SK, Salekdeh GH. Proteomic signature of human embryonic stem cells. Proteomics. 2006;6(12):3544–9. doi: 10.1002/pmic.200500844. [DOI] [PubMed] [Google Scholar]
- 13.Yocum AK, Gratsch TE, Leff N, Strahler JR, Hunter CL, Walker AK, Michailidis G, Omenn GS, O'Shea KS, Andrews PC. Coupled global and targeted proteomics of human embryonic stem cells during induced differentiation. Mol Cell Proteomics. 2008;7(4):750–67. doi: 10.1074/mcp.M700399-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ideker T, Thorsson V, Ranish JA, Christmas R, Buhler J, Eng JK, Bumgarner R, Goodlett DR, Aebersold R, Hood L. Integrated genomic and proteomic analyses of a systematically perturbed metabolic network. Science. 2001;292(5518):929–34. doi: 10.1126/science.292.5518.929. [DOI] [PubMed] [Google Scholar]
- 15.Tian GW, Mohanty A, Chary SN, Li S, Paap B, Drakakaki G, Kopec CD, Li J, Ehrhardt D, Jackson D, Rhee SY, Raikhel NV, Citovsky V. High-throughput fluorescent tagging of full-length Arabidopsis gene products in planta. Plant Physiol. 2004;135(1):25–38. doi: 10.1104/pp.104.040139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Choe L, D'Ascenzo M, Relkin NR, Pappin D, Ross P, Williamson B, Guertin S, Pribil P, Lee KH. 8-plex quantitation of changes in cerebrospinal fluid protein expression in subjects undergoing intravenous immunoglobulin treatment for Alzheimer's disease. Proteomics. 2007;7(20):3651–60. doi: 10.1002/pmic.200700316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ow SY, Cardona T, Taton A, Magnuson A, Lindblad P, Stensjo K, Wright PC. Quantitative shotgun proteomics of enriched heterocysts from Nostoc sp. PCC 7120 using 8-plex isobaric peptide tags. J Proteome Res. 2008;7(4):1615–28. doi: 10.1021/pr700604v. [DOI] [PubMed] [Google Scholar]
- 18.Bottenstein JE, Sato GH. Growth of a rat neuroblastoma cell line in serum-free supplemented medium. Proc Natl Acad Sci U S A. 1979;76(1):514–7. doi: 10.1073/pnas.76.1.514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sterneckert JL, Hill CM, Palmer R, Gearhart JD. Bone morphogenetic proteins produced by cells within embryoid bodies inhibit ventral directed differentiation by Sonic Hedgehog. Cloning Stem Cells. 2005;7(1):27–34. doi: 10.1089/clo.2005.7.27. [DOI] [PubMed] [Google Scholar]
- 20.Draper JS, Pigott C, Thomson JA, Andrews PW. Surface antigens of human embryonic stem cells: changes upon differentiation in culture. J Anat. 2002;200(Pt 3):249–58. doi: 10.1046/j.1469-7580.2002.00030.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Winger QA, Guttormsen J, Gavin H, Bhushan F. Heat shock protein 1 and the mitogen-activated protein kinase 14 pathway are important for mouse trophoblast stem cell differentiation. Biol Reprod. 2007;76(5):884–91. doi: 10.1095/biolreprod.106.056820. [DOI] [PubMed] [Google Scholar]
- 22.Fukada Y, Yasui K, Kitayama M, Doi K, Nakano T, Watanabe Y, Nakashima K. Gene expression analysis of the murine model of amyotrophic lateral sclerosis: studies of the Leu126delTT mutation in SOD1. Brain Res. 2007;1160:1–10. doi: 10.1016/j.brainres.2007.05.044. [DOI] [PubMed] [Google Scholar]
- 23.Yasuhara N, Shibazaki N, Tanaka S, Nagai M, Kamikawa Y, Oe S, Asally M, Kamachi Y, Kondoh H, Yoneda Y. Triggering neural differentiation of ES cells by subtype switching of importin-alpha. Nat Cell Biol. 2007;9(1):72–9. doi: 10.1038/ncb1521. [DOI] [PubMed] [Google Scholar]
- 24.Wrage PC, Tran T, To K, Keefer EW, Ruhn KA, Hong J, Hattangadi S, Trevino I, Tansey MG. The neuro-glial properties of adipose-derived adult stromal (ADAS) cells are not regulated by Notch 1 and are not derived from neural crest lineage. PLoS ONE. 2008;3(1):e1453. doi: 10.1371/journal.pone.0001453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li JY, Pu MT, Hirasawa R, Li BZ, Huang YN, Zeng R, Jing NH, Chen T, Li E, Sasaki H, Xu GL. Synergistic function of DNA methyltransferases Dnmt3a and Dnmt3b in the methylation of Oct4 and Nanog. Mol Cell Biol. 2007;27(24):8748–59. doi: 10.1128/MCB.01380-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu YL, Guo YS, Xu L, Wu SY, Wu DX, Yang C, Zhang Y, Li CY. Alternation of neurofilaments in immune-mediated injury of spinal cord motor neurons. Spinal Cord. 2008 doi: 10.1038/sc.2008.90. [DOI] [PubMed] [Google Scholar]
- 27.Perrot R, Berges R, Bocquet A, Eyer J. Review of the Multiple Aspects of Neurofilament Functions, and their Possible Contribution to Neurodegeneration. Mol Neurobiol. 2008 doi: 10.1007/s12035-008-8033-0. [DOI] [PubMed] [Google Scholar]
- 28.Brady ST. Motor neurons and neurofilaments in sickness and in health. Cell. 1993;73(1):1–3. doi: 10.1016/0092-8674(93)90151-f. [DOI] [PubMed] [Google Scholar]
- 29.Crossin KL, Hoffman S, Tan SS, Edelman GM. Cytotactin and its proteoglycan ligand mark structural and functional boundaries in somatosensory cortex of the early postnatal mouse. Dev Biol. 1989;136(2):381–92. doi: 10.1016/0012-1606(89)90264-9. [DOI] [PubMed] [Google Scholar]
- 30.Irintchev A, Rollenhagen A, Troncoso E, Kiss JZ, Schachner M. Structural and functional aberrations in the cerebral cortex of tenascin-C deficient mice. Cereb Cortex. 2005;15(7):950–62. doi: 10.1093/cercor/bhh195. [DOI] [PubMed] [Google Scholar]
- 31.Ikeshima-Kataoka H, Saito S, Yuasa S. Tenascin-C is required for proliferation of astrocytes in primary culture. In Vivo. 2007;21(4):629–33. [PubMed] [Google Scholar]
- 32.Witusik M, Piaskowski S, Hulas-Bigoszewska K, Zakrzewska M, Gresner SM, Azizi SA, Krynska B, Liberski PP, Rieske P. Successful elimination of non-neural cells and unachievable elimination of glial cells by means of commonly used cell culture manipulations during differentiation of GFAP and SOX2 positive neural progenitors (NHA) to neuronal cells. BMC Biotechnol. 2008;8:56. doi: 10.1186/1472-6750-8-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kose N, Asashima T, Muta M, Iizasa H, Sai Y, Terasaki T, Nakashima E. Altered expression of basement membrane-related molecules in rat brain pericyte, endothelial, and astrocyte cell lines after transforming growth factor-beta1 treatment. Drug Metab Pharmacokinet. 2007;22(4):255–66. doi: 10.2133/dmpk.22.255. [DOI] [PubMed] [Google Scholar]
- 34.Chan-Ling T, Chu Y, Baxter L, Weible Ii M, Hughes S. In vivo characterization of astrocyte precursor cells (APCs) and astrocytes in developing rat retinae: Differentiation, proliferation, and apoptosis. Glia. 2008 doi: 10.1002/glia.20733. [DOI] [PubMed] [Google Scholar]
- 35.Pekny M, Wilhelmsson U, Bogestal YR, Pekna M. The role of astrocytes and complement system in neural plasticity. Int Rev Neurobiol. 2007;82:95–111. doi: 10.1016/S0074-7742(07)82005-8. [DOI] [PubMed] [Google Scholar]
- 36.Feral CC, Zijlstra A, Tkachenko E, Prager G, Gardel ML, Slepak M, Ginsberg MH. CD98hc (SLC3A2) participates in fibronectin matrix assembly by mediating integrin signaling. J Cell Biol. 2007;178(4):701–11. doi: 10.1083/jcb.200705090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim JY, Wu Y, Smas CM. Characterization of ScAP-23, a new cell line from murine subcutaneous adipose tissue, identifies genes for the molecular definition of preadipocytes. Physiol Genomics. 2007;31(2):328–42. doi: 10.1152/physiolgenomics.00206.2006. [DOI] [PubMed] [Google Scholar]
- 38.Salim K, Kehoe L, Minkoff MS, Bilsland JG, Munoz-Sanjuan I, Guest PC. Identification of differentiating neural progenitor cell markers using shotgun isobaric tagging mass spectrometry. Stem Cells Dev. 2006;15(3):461–70. doi: 10.1089/scd.2006.15.461. [DOI] [PubMed] [Google Scholar]
- 39.Scaturro M, Cestelli A, Castiglia D, Nastasi T, Di Liegro I. Posttranscriptional regulation of H1 zero and H3.3B histone genes in differentiating rat cortical neurons. Neurochem Res. 1995;20(8):969–76. doi: 10.1007/BF00970744. [DOI] [PubMed] [Google Scholar]
- 40.Sarkar SA, Sharma RP. Expression of selected apoptosis related genes, MIF, IGIF and TNF alpha, during retinoic acid-induced neural differentiation in murine embryonic stem cells. Cell Struct Funct. 2002;27(2):99–107. doi: 10.1247/csf.27.99. [DOI] [PubMed] [Google Scholar]
- 41.Molina H, Yang Y, Ruch T, Kim JW, Mortensen P, Otto T, Nalli A, Tang QQ, Lane MD, Chaerkady R, Pandey A. Temporal Profiling of the Adipocyte Proteome during Differentiation Using a Five-Plex SILAC Based Strategy. J Proteome Res. 2008 doi: 10.1021/pr800650r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Battista S, Pentimalli F, Baldassarre G, Fedele M, Fidanza V, Croce CM, Fusco A. Loss of Hmga1 gene function affects embryonic stem cell lympho-hematopoietic differentiation. Faseb J. 2003;17(11):1496–8. doi: 10.1096/fj.02-0977fje. [DOI] [PubMed] [Google Scholar]
- 43.Liang J, Wan M, Zhang Y, Gu P, Xin H, Jung SY, Qin J, Wong J, Cooney AJ, Liu D, Songyang Z. Nanog and Oct4 associate with unique transcriptional repression complexes in embryonic stem cells. Nat Cell Biol. 2008;10(6):731–9. doi: 10.1038/ncb1736. [DOI] [PubMed] [Google Scholar]
- 44.Maldonado-Saldivia J, van den Bergen J, Krouskos M, Gilchrist M, Lee C, Li R, Sinclair AH, Surani MA, Western PS. Dppa2 and Dppa4 are closely linked SAP motif genes restricted to pluripotent cells and the germ line. Stem Cells. 2007;25(1):19–28. doi: 10.1634/stemcells.2006-0269. [DOI] [PubMed] [Google Scholar]
- 45.Mathivanan S, Ahmed M, Ahn NG, Alexandre H, Amanchy R, Andrews PC, Bader JS, Balgley BM, Bantscheff M, Bennett KL, Bjorling E, Blagoev B, Bose R, Brahmachari SK, Burlingame AS, Bustelo XR, Cagney G, Cantin GT, Cardasis HL, Celis JE, Chaerkady R, Chu F, Cole PA, Costello CE, Cotter RJ, Crockett D, DeLany JP, De Marzo AM, DeSouza LV, Deutsch EW, Dransfield E, Drewes G, Droit A, Dunn MJ, Elenitoba-Johnson K, Ewing RM, Van Eyk J, Faca V, Falkner J, Fang X, Fenselau C, Figeys D, Gagne P, Gelfi C, Gevaert K, Gimble JM, Gnad F, Goel R, Gromov P, Hanash SM, Hancock WS, Harsha HC, Hart G, Hays F, He F, Hebbar P, Helsens K, Hermeking H, Hide W, Hjerno K, Hochstrasser DF, Hofmann O, Horn DM, Hruban RH, Ibarrola N, James P, Jensen ON, Jensen PH, Jung P, Kandasamy K, Kheterpal I, Kikuno RF, Korf U, Korner R, Kuster B, Kwon MS, Lee HJ, Lee YJ, Lefevre M, Lehvaslaiho M, Lescuyer P, Levander F, Lim MS, Lobke C, Loo JA, Mann M, Martens L, Martinez-Heredia J, McComb M, McRedmond J, Mehrle A, Menon R, Miller CA, Mischak H, Mohan SS, Mohmood R, Molina H, Moran MF, Morgan JD, Moritz R, Morzel M, Muddiman DC, Nalli A, Navarro JD, Neubert TA, Ohara O, Oliva R, Omenn GS, Oyama M, Paik YK, Pennington K, Pepperkok R, Periaswamy B, Petricoin EF, Poirier GG, Prasad TS, Purvine SO, Rahiman BA, Ramachandran P, Ramachandra YL, Rice RH, Rick J, Ronnholm RH, Salonen J, Sanchez JC, Sayd T, Seshi B, Shankari K, Sheng SJ, Shetty V, Shivakumar K, Simpson RJ, Sirdeshmukh R, Siu KW, Smith JC, Smith RD, States DJ, Sugano S, Sullivan M, Superti-Furga G, Takatalo M, Thongboonkerd V, Trinidad JC, Uhlen M, Vandekerckhove J, Vasilescu J, Veenstra TD, Vidal-Taboada JM, Vihinen M, Wait R, Wang X, Wiemann S, Wu B, Xu T, Yates JR, Zhong J, Zhou M, Zhu Y, Zurbig P, Pandey A. Human Proteinpedia enables sharing of human protein data. Nat Biotechnol. 2008;26(2):164–7. doi: 10.1038/nbt0208-164. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.