Abstract
Krüppel-like factor 4 (KLF4) is an epithelially enriched, zinc finger-containing transcription factor, the expression of which is associated with growth arrest. Constitutive expression of KLF4 inhibits G1/S transition of the cell cycle but the manner by which it accomplishes this effect is unclear. To better understand the biochemical function of KLF4, we identified its target genes using cDNA microarray analysis in an established human cell line containing inducible KLF4. RNA extracted from induced and control cells were hybridized differentially to microarray chips containing 9600 human cDNAs. In all, 84 genes with significantly increased expression and 107 genes with significantly reduced expression due to KLF4 induction were identified. The affected genes are sorted to several clusters on the basis of functional relatedness. A major cluster belongs to genes involved in cell-cycle control. Within this cluster, many up-regulated genes are inhibitors of the cell cycle and down-regulated genes are promoters of the cell cycle. Another up-regulated gene cluster includes nine keratin genes, of which seven are located in a specific region on chromosome 12. The results indicate that KLF4 is involved in the control of cell proliferation and does so by eliciting changes in expression of numerous cell-cycle regulatory genes in a concerted manner. Furthermore, KLF4 regulates expression of a group of epithelial-specific keratin genes in a manner consistent with a potential locus control region function.
Keywords: KLF4, cDNA microarray, transcriptional profiling, cell cycle, epithelial differentiation
Introduction
The mammalian gut epithelium is a dynamic system in which cell proliferation, occurring primarily in the crypt, is carefully balanced with differentiation and death of epithelial cells located outside the crypt.1– 4 Other epithelial tissues such as the skin exhibit this characteristic.5–7 The homeostasis can occasionally become perturbed, leading to disease states such as cancer.8,9 Previous studies have identified a number of key factors in regulating proliferation and differentiation of the gut epithelial cells. Examples include members of the Wnt pathway of regulators such as β-catenin10,11 and TCF-4,12,13 and transcription factors such as CDX214–16 and PDX.17 However, the exact mechanism by which activities of these regulatory proteins are orchestrated to control intestinal epithelial proliferation and differentiation is not clearly defined.
Krüppel is a zinc finger-containing transcription factor in Drosophila melanogaster that has a crucial function in controlling embryogenesis.18 A large number of mammalian genes exhibit sequence homology to the DNA-binding domain of Krüppel. Among these, a group, called the Krüppel-like factors (KLFs), constitutes a particularly close family.19 – 23 The prototype gene in this group is erythroid Krüppel-like factor (EKLF or KLF1), which has an essential function in regulating erythropoiesis.24 – 26 Two additional KLFs, lung Krüppel-like factor (LKLF or KLF227,28) and gut-enriched Krüppel-like factor (GKLF or KLF429,30), are related to KLF1 more closely than any other members of the KLF family. In addition to sequence homology, structural-functional studies cluster these three genes into a subfamily of KLFs.31
KLF4 was identified originally on the basis of sequence similarity to another zinc finger-containing transcription factor, zif 268 or EGR1.29,32 Subsequent studies indicate that its expression is enriched in epithelial tissues such as the gut and skin.29,30,33 Moreover, the KLF4 mRNA is found primarily in the post-mitotic, terminally differentiated epithelial cells in these organs.29,30 In vitro, expression of KLF4 is associated with the growth-arrested process.29 Conditions that increase its expression include serum deprivation, contact inhibition, and DNA damage.29,34 Conversely, expression of KLF4 is decreased in conditions that involve increased proliferation such as neoplasm of the intestinal tract35,36 and in cancer cell lines.37 Moreover, forced expression of KLF4 in cells by either transient or stable transfection inhibits DNA synthesis.29,38 These studies suggest that KLF4 may have an important function in regulating cell proliferation.
We showed recently that induction in expression of KLF4 in response to DNA damage is dependent largely on the tumor suppressor p53.34 Following DNA damage, KLF4 binds to a specific region in the promoter of the gene encoding a key cell-cycle inhibitor, the cyclin-dependent kinase inhibitor p21WAF1/Cip1,39,40 resulting in the activation of this promoter. The ability of p53 to activate the p21WAF1/Cip1 promoter is dependent on KLF4 and the two proteins synergize to regulate p21WAF1/Cip1 expression.34 Using a stably transfected cell line in which an inducible promoter controls expression of KLF4, we showed that induction of KLF4 results in an arrest in the transition from the G1 to the S phase of the cell cycle.41 More recently, using a combination of genetic and biochemical approaches, we showed that KLF4 is essential in mediating the G1/S cell-cycle effect of p53 as a consequence of DNA damage.42 These studies demonstrate an essential role for KLF4 in cell-cycle control.
In addition to its documented effect on cell proliferation, KLF4 has been shown to be important in regulating tissue differentiation, as indicated by gene knockout studies. In one example, KLF4 is shown to be required for establishing the barrier function of the skin, as measured by penetration of external dyes and rapid loss of body fluids in newborn mice null for KLF4.43 In another, KLF4 is shown to be essential for the terminal differentiation of goblet cells in the colon.44 These studies demonstrate an important aspect of the role of KLF4 in controlling in vivo differentiation of specific epithelial functions.
In order to further understand the biochemical mechanism by which KLF4 regulates cell proliferation and differentiation, we initiated an effort to identify its target genes using a genomic approach. In the current study, we used the previously established inducible cell system for KLF4 and performed cDNA microarray analysis to establish the transcriptional program regulated by KLF4. This effort led to the identification of two groups of genes, one activated and the other inhibited, by KLF4. Furthermore, these genes can be sorted to a limited number of clusters on the basis of functional similarity, one of which is a group of cell-cycle regulators. Another interesting cluster in the genes up-regulated by KLF4 contains a large number of keratin genes, which are epithelial-specific. Moreover, at least seven of the identified keratin genes are located in the same region of the same chromosome. These findings suggest that KLF4 may have a dual function. One is to regulate the cell cycle by coordinating the expression of numerous cell-cycle regulators. The other is the coordinated expression of a certain family of differentiation and epithelial-specific genes in a manner that is suggestive of a locus control region (LCR) function.
Results
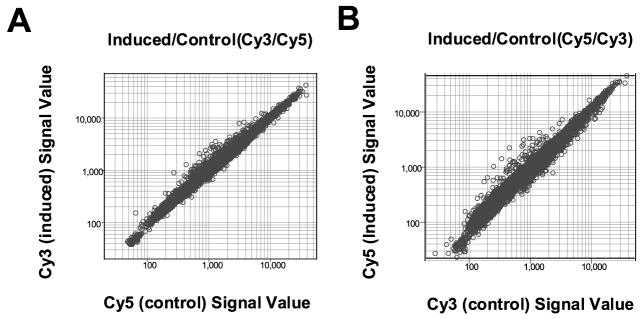
To identify potential target genes of KLF4, we treated EcR-RKO/KLF4 cells with 5 μM ponasterone A or vehicle alone for 24 hours and compared the expression profiles between the two treatment conditions using cDNA microarrays. Previous studies indicate that treatment of EcR-RKO/KLF4 cells with ponasterone A results in a significant induction in KLF4 expression, which is accompanied by G1/S cell-cycle arrest.41 In one experiment (experiment A), mRNAs from induced cells were reverse-transcribed and labeled with Cy3-fluorescence dye, while those from control cells were labeled with Cy5 dye. In a second experiment (experiment B), the dyes were reversed. Following hybridization to the Unigene cDNA microarray that contained 9600 human genes, the signal intensity for each of the labeled cDNAs from induced cells was measured and plotted against that from control cells for each experiment (Figure 1). In experiment A (Figure 1(A)), genes whose expression was induced by treatment with ponasterone A are found to the left of diagonal axis. Similarly, genes whose expression was decreased by treatment with ponasterone A are found to the right of the diagonal axis. Results of experiment B, in which the Cy3 and Cy5 dyes were reversed, are shown in Figure 1(B).
Figure 1.
Results of cDNA micro-array hybridization. EcR-RKO/KLF4 cells were treated with 5 μM ponasterone A (PA) or vehicle alone for 24 hours before subject to mRNA isolation. In experiment A, mRNAs from induced and control cells were reverse-transcribed and labeled with Cy3 and Cy5 fluorescence dye, respectively. The dyes were reversed in experiment B. Labeled cDNAs from induced and control cells were then combined and hybridized to the human cDNA microarray chip as described in Experimental Procedures. The signal value for each cDNA in the treated and control cells was measured and plotted in the Figure for both experiments A and B.
Figure 2 is a scatter plot analysis of balanced differential expression values for each of the genes on the microarray between experiments A and B. A change in expression of >1.7-fold (either increased or decreased) is considered significant, based on the recommendation of the manufacturer (>99.5% tolerance intervals for >99% of the elements on the microarray).45 As expected, the majority of genes is clustered in the center of Figure 2 and represents those whose expression were unchanged after treatment with ponasterone A in the two experiments. A total of 84 genes had a differential expression value of >1.7-fold between induced and control cells in both experiments and are considered up-regulated by KLF4. Among these, 20 genes had values greater than twofold, with intestinal alkaline phosphatase (ALPI) having the largest change at 4.9-fold. A total of 107 genes had a differential expression value of >1.7-fold between control and induced cells in both experiments and are considered down-regulated. In all, 24 genes had a value of at least twofold when control cells were compared to induced cells, with KIA (proliferation related Ki-67 antigen) being the highest with a 2.99-fold change.
Figure 2.
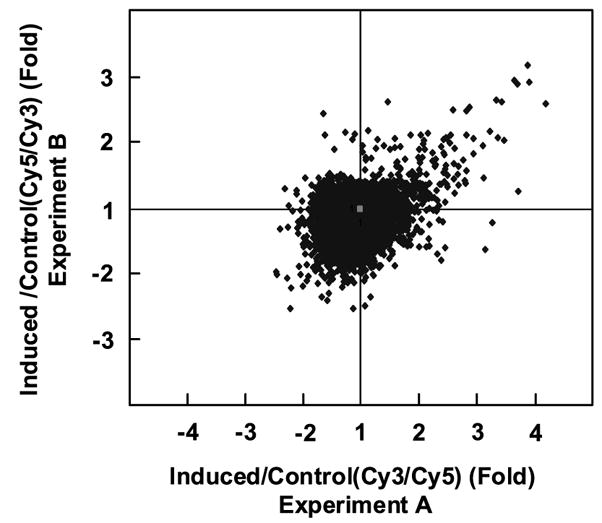
Scatter plot analysis of microarray hybridization results from experiments A and B. The signal value for each cDNA from induced cells was compared to that from control cells and expressed as a balanced differential expression value. Shown is a scatter plot of balanced differential expression values for all the genes in experiments A and B. Genes in the center of the plot have a value of 1 and are unchanged in their expression by treatment with ponasterone A. Genes in the right upper quadrants are up-regulated and those in the lower left quadrants are down-regulated in both experiments.
The group of genes up-regulated by KLF4 was further analyzed on the basis of functional similarity using the DRAGON database†. As shown in Table 1, these genes can be clustered into several groups, as genes involved in metabolism, cell-cycle control, immune response, structural integrity, signal transduction, translation control, and others. Notable examples include intestinal alkaline phosphatase (ALPI), p57Kip2, p21WAF1/Cip1, and numerous keratin (KRT) genes. Table 2 shows the identities of the group of genes down-regulated by KLF4. As in the case of up-regulated genes, the down-regulated genes can be clustered into several functional groups, including cell-cycle control, signal transduction, malignancy, transport, DNA replication and relaxation, mitochondrial processes, and others. Among the down-regulated genes, the group of cell cycle-related genes particularly stands out, as they represent primarily genes that are important in progression of the cell cycle with examples such as KIA, cyclin D1, CDC2/CDK1 and RAD21.
Table 1.
Gene clusters up-regulated by KLF4
| Symbol | Gene name | Symbol | Gene name |
|---|---|---|---|
| Metabolism | Structural | ||
| ALPI | Alkaline phosphatase, intestinal | KRT18 | Keratin 18 |
| ALPP | Alkaline phosphatase, placental | KRT13 | Keratin 13 |
| CTBS | Di-N-acetyl-chitobiase | VIL2 | Villin 2 (ezrin) |
| RDP | Dipeptidase 1, renal | MCM | Methylmalonyl coenzyme A mutase |
| PRODH | Proline dehydrogenase 1 | KRTHB1 | Keratin, hair, basic 1 |
| CAMK1 | Calmodulin-dep. protein kinase 1 | RTN2C | Reticulon 2C |
| AGPAT2 | 1-Acylglycerol-3-phosphate O-acyltransferase 2 | SNL | Singed-like (fascin homolog, sea urchin) |
| PON | Paraoxinase 1 | KRTHB3 | Keratin, hair, basic 3 |
| AAT | Antitrypsin serine (or cysteine) proteinase inhibitor, clade A (α-1 antiproteinase, antitrypsin), member 1 | KRT8 | Keratin 8 |
| ASS | Argininosuccinate synthetase | KRT2A | Keratin 2A |
| DHCR7 | 7-Dehydrocholesterol reductase | KRT6A | Keratin 6A |
| B4GALT1 | UDP-Gal: β GlcNAc β 1,4- galactosyltransferase 1 | KRT7 | Keratin 7 |
| GCNT3 | Glucosaminyl (N-acetyl) transferase 3, mucin-type | KRT15 | Keratin 15 |
| KLK1 | Kallikrein 1 | DSG2 | Desmoglein 2 |
| CSTB | Cystatin B (stefin B) | VIM | Vimentin |
| Cell cycle control | Signal transduction | ||
| p57/Kip2 | Cyclin-dependent kinase inhibitor 1C | MT1L | Metallothionein 1L |
| IGFBP6 | Insulin-like growth factor binding protein 6 | ADSF | Adipose specific 2 |
| p21/Cip1 | Cyclin-dependent kinase inhibitor 1A | GNAI2B | Guanine nucleotide binding protein, α inhibiting activity 2 |
| SFN | Stratifin (14-3-3 sigma) | GPRK5 | G protein-coupled receptor kinase 5 |
| PGGT1B | Protein geranyl-geranyltransferase type I β | CRIP2 | Cysteine-rich protein 2 |
| PPFIA3 | Protein tyrosine phosphatase, receptor type, f polypeptide, interacting protein, α 3 | MT1E | Metallothionein 1E |
| UFD2 | Ubiquitination factor E4B | EST | Highly similar to metallothionein-IB |
| Immune response | Translation | ||
| HBZ | Hemoglobin, zeta | TCEA2 | Transcription elongation factor A (SII), 2 |
| ENTPD2 | Ectonucleoside triphosphate diphosphohydrolase 2 | RNA helicase | RNA helicase-related protein |
| HCG II | H. sapiens HCG II mRNA | EST | Moderately similar to integrase |
| HBG1 | Hemoglobin, γA | ||
| ARHH | Ras homolog gene family, member H | Other | |
| FUT1 | Fucosyltransferase 1 | CDNA | cDNA FLJ13155, human |
| NK4 | Natural killer cell transcript 4 | CDNA | cDNA FLJ11658, human |
| PVRL1 | Poliovirus receptor-related 1 | CYP4F1-like | Rat CYP4F1-like |
| FTL | Ferritin, light polypeptide |
Table 2.
Gene clusters down-regulated by KLF4
| Symbol | Gene name | Symbol | Gene name |
|---|---|---|---|
| Cell cycle | Malignancy | ||
| KIA | Proliferation related Ki-67 antigen | CBFB | Core-binding factor, β subunit |
| FOXM1 | Forkhead box M1 | PURB | Purine-rich element binding protein B |
| CENPE | Centromere protein E | TXNIP | Thioredoxin interacting protein |
| THBS1 | Thrombospondin 1 | STK15 | Serine/threonine kinase 15 |
| JAG1 | Jagged 1 | TTK | TTK protein kinase |
| MAD2L1 | Mitotic arrest deficient-like 1 | ZWINTAS | ZW10 interactor anti-sense |
| CDC2/CDK1 | Cell division cycle 2/cyclin-dependent kinase 1 | BMI-1 | B lymphoma Mo-MLV insertion region |
| BUB1 | Budding uninhibited by benzimidazoles 1 homolog B | ALDOA | Aldolase A, fructose-bisphosphate |
| RAD21 | RAD21 homolog (S. pombe) | PPP2CA | Protein phosphatase 2A, catalytic, alpha |
| Wee1 | WEE1 + homolog (S. pombe) | PTEN | Phosphatase and tensin homolog |
| RRM1 | Ribonucleotide reductase M1 polypeptide | DEK | DEK oncogene (DNA binding) |
| TMPO | Thymopoietin | BAGE | B melanoma antigen |
| KNSL6 | Kinesin-like 2 | NUP98 | Nucleoporin 98kD |
| MCM2 | Minichromosome maintenance deficient 2 | ZNF220 | Zinc finger protein 220 |
| CCND1 | Cyclin D1 | ||
| HAT1 | Histone acetyltransferase 1 | ||
| XPO1 | Exportin 1 (CRM1 homolog, yeast) | ||
| NEDD5 | Neural precursor cell expressed, developmentally down-regulated 5 | ||
| KAP1 | KRAB-associated protein 1 | ||
| Signaling | Transport | ||
| NPR3 | Atrionatriuretic peptide receptor, type C | MAN1B1 | Mannosidase, α, class 1B, member 1 |
| KIAA0008 | KIAA0008 gene product | SLC12A2 | Solute carrier family 12, member 2 |
| VRK1 | VRK1 | SLC4A3 | Solute carrier family 4, member 3 |
| F2 | Coagulation factor II (thrombin) receptor | SEC24C | SEC24 related gene family, member C |
| CALCRL | Calcitonin, receptor-like | COPE | Coatomer protein complex, subunit epsilon |
| IDE | Insulin-degrading enzyme | SORT1 | Sortilin 1 |
| ARL6 | ADP-ribosylation factor-like 6 interacting protein | KPNA2 | Karyopherin alpha 2 |
| NESP55 | Neuroendocrine secretory protein 55 | SLC15A1 | Solute carrier family 15, member 1 |
| FLJ10955 | Hypothetical protein FLJ10955 | ||
| GUCYB3 | Guanylate cyclase 1, soluble, beta 3 | ||
| FZD1 | Frizzled (Drosophila) homolog 1 | ||
| Mitochondria | DNA-repair/relaxation | ||
| HMT1L1 | hnRNP methyltransferase-like 1 | TOP2 | Topoisomerase (DNA) II, alpha |
| CDNA FLJ13016 | Moderately similar to Rattus norvegicus | FEN1 | Flap structure-specific endonuclease 1 |
| fis | mRNA for SECIS binding protein 2 | ||
| ART4 | ADP-ribosyltransferase 4 | H2AX | H2A histone family, member X |
| Protein phospha tase 1D | Protein phosphatase 1D magnesium-dependent, δ isoform | HEX1 | Exonuclease 1 |
| CDNA FLJ10293 | Clone NT2RM1000280, highly similar to vacuolar ATP synthase subunit D | H2AFZ | H2A histone family, member Z |
| MpV17 | Transgene, murine homolog, glomerulosclerosis | ||
| MTMR7 | Myotubularin-related protein 7 | ||
| CLPP | Caseinolytic protease, ATP-dependent, proteolytic subunit, E. coli homolog | ||
| MTND2 | NADH-ubiquinone oxidoreductase, subunit ND2 | ||
| IDO | Indoleamine-pyrrole 2,3 dioxygenase | ||
| EIF3S5 | Eukaryotic translation initiation factor 3, subunit 5 | ||
| ERCC6 | Excision repair cross-complementing rodent repair deficiency, group 6 | ||
| Metabolism | DNA-binding/transcription | ||
| OCLN | Occludin | TERF1 | Telomeric repeat binding factor (NIMA-interacting) 1 |
| PAPSS2 | 3′-Phosphoadenosine 5′-phosphosulfate synthase 2 | DBP | D site of albumin promoter (albumin D-box) binding protein |
| CTH, gammalyase | Cystathionine γ-lyase | ATF2 | Activating transcription factor 2 |
| RANBP2L1 | RAN binding protein 2-like 1 | NR1D2 | Nuclear receptor subfamily 1, group D, member 2 |
| PCTK2 | PCTAIRE protein kinase 2 | SF3B3 | Splicing factor 3B, subunit 3 |
| PRKAA2 | Protein kinase, AMP-activated, α 2 catalytic | ||
| Adhesion | Other/EST | ||
| MMP10 | Matrix metalloproteinase 10 (stromelysin 2) | ESTs | ESTs |
| CEACAM6 | Carcinoembryonic antigen-related cell adhesion molecule 6 | Incyte EST | Incyte EST |
| LAMC2 | Laminin, γ 2 | ALDH7A1 | Aldehyde dehydrogenase 7 family, member A1 |
| LUM | Lumican | P311 protein | P311 protein |
| MMP13 | Matrix metalloproteinase 13 (collagenase 3) | RNA binding motif protein | RNA binding motif protein, X chromosome |
| MMP10 | Matrix metalloproteinase 10 (stromelysin 2) | Incyte EST | Incyte EST |
| PRKAR2A | PRKAR2A | ||
| KIAA0074 protein | KIAA0074 protein |
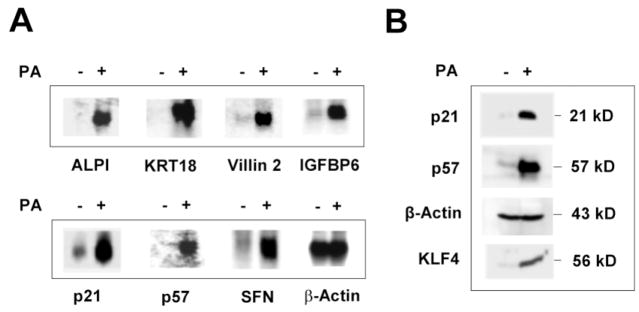
To verify the results of the microarray experiments, we performed Northern blot analysis of RNA isolated from EcR-RKO/KLF4 cells treated with ponasterone A or vehicle alone for 24 hours using several cDNA probes encoding genes that were shown to be up-regulated by KLF4 in Table 1. As seen in Figure 3(A), each of the seven genes tested showed significant induction in the mRNA levels in treated cells as compared to control. These genes represent three clusters of up-regulated genes involved in metabolism (ALPI), structural integrity (KRT18 and villin 2), and cell-cycle control (p21WAF1/Cip1, p57Kip2, IGFBP6, and SFN). It should be noted that each of the cell-cycle control genes tested here has been shown to be an inhibitor of the cell cycle.40,46– 49 To further verify the authenticity of the microarray experiments, we performed Western blot analysis of proteins isolated from treated and control cells. As seen in Figure 3(B), the levels of both p21WAF1/Cip1 and p57Kip2 proteins were elevated significantly, as was that of KLF4. In contrast, the level of the loading control, β-actin, remained the same.
Figure 3.
Verification of expression levels of up-regulated genes identified by microarrays. (A) Total RNA was isolated from EcR-RKO/KLF4 cells treated for 24 hours with ponasterone A (PA) (+) or vehicle alone (−) treated and analyzed by Northern blot analysis using cDNA probes encoding some of the up-regulated genes identified by microarray analysis. ALPI, intestinal alkaline phosphatase; KRT18, keratin 18; IGFBP6, insulin-like growth factor binding protein 6; SFN, stratifin or 14-3-3σ. β-Actin was used as a loading control. (B) Proteins were extracted from treated or control cells and analyzed by Western blot analysis using antibodies directed against p21WAF1/Cip1, p57Kip2, β-actin, and KLF4.
We verified the expression levels of some of the down-regulated genes obtained from the micro-array experiments by Northern blot analysis (Figure 4). As shown, all seven selected genes showed decreased levels of expression when cells were treated with ponasterone A. All seven belong to the cell-cycle group, and have been shown to control the progression of the cell cycle at various checkpoints.50 – 59
Figure 4.
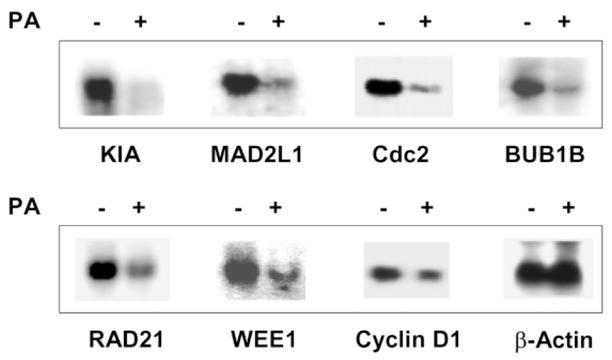
Verification of expression levels of down-regulated genes identified by microarrays. Total RNA was isolated from EcR-RKO/KLF4 cells treated for 24 hours with ponasterone A (PA) (+) or vehicle alone (−) treated and analyzed by Northern blot analysis using cDNA probes encoding some of the down-regulated genes identified by microarray analysis. See Table 2 for abbreviations. β-Actin was used as a loading control.
To further examine the kinetics of induction of several of the up-regulated genes as a result of the inducible expression of KLF4, we performed Northern blot analysis using RNA isolated from EcR-RKO/KLF4 cells that had been treated with ponasterone A for various periods of time (Figure 5). As shown, of the five genes tested, the induction of p21WAF1/Cip1 and IGFBP6 was relatively early, beginning around four hours following treatment. In contrast, the levels of transcript for ALPI rose relatively late. These results suggest that the time-courses of induction of the up-regulated genes by KLF4 fall into different groups.
Figure 5.
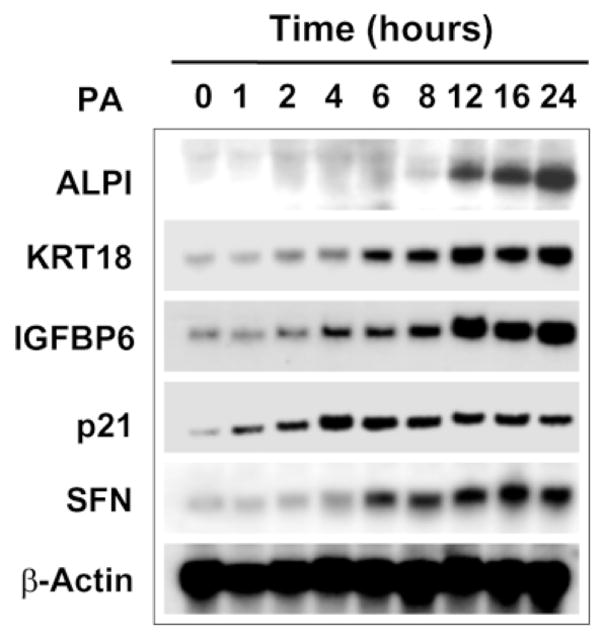
Time + course of expression of up-regulated gene during induction of KLF4RNA was isolated from EcR-RKO/KLF4 cells treated with ponasterone A for the time indicated and probed by Northern blot analysis using cDNA encoding the genes in the Figure. β-Actin was used as a loading control.
Among the genes up-regulated by KLF4 (Table 1), we noticed that there are a large number of genes encoding a structural protein, keratin. As keratin is a major constituent protein of the epithelial tissues,60 – 62 we analyzed the group of keratin genes that are up-regulated by KLF4 on the basis of sequence homology. Figure 6(A) shows that all nine of the keratin genes up-regulated by KLF4 are closely related, in addition to several other structural proteins such as vimentin (VIM), villin 2 or ezrin (VIL2/p81), desmoglein 2 (DSG2), and singed-like (SNL/p55). It is of note that many of the up-regulated keratin genes are located on the same region of chromosome 12. In human, there are a total of 20 genes encoding keratin, which are distributed on chromosomes 12 and 17.63 – 65 Of the 12 keratin genes that are on the chip and located on chromosome 12, seven are up-regulated by KLF4 (Figure 6(B)). In contrast, of the seven keratin genes on the chip and located on chromosome 17, only two are up-regulated by KLF4. These results suggest that KLF4 is involved in the coordinated induction of a family of keratin genes located on a similar region of chromosome 12.
Figure 6.
A cluster of keratin and keratin-related genes is up-regulated by KLF4. (A) The sequence-relatedness of the group of keratin and keratin-related genes that are up-regulated by KLF4. Shown are also the chromosomal locations for each of the genes. KRT, keratin; VIM, vimentin; VIL2/p81, villin 2 or eczrin; DSG2, desmoglein 2; SNL/p55, singed-like. (B) The up-regulated keratin genes are sorted on the basis of their chromosomal localization.
Discussion
In the present study, we report the expression profiling of the transcriptional program controlled by KLF4 using cDNA microarrays. Our study identified 84 and 107 genes whose expression are significantly up- and down-regulated, respectively, upon the inducible expression of KLF4 in a stably transfected cell line. These results are derived after duplication of the experiments by dye reversal and extensive data filtering. Further, expression of many of the target genes was verified by Northern blot analysis and, on occasion, Western blot analysis. Examples are provided in Figure 3 for the up-regulated genes and in Figure 4 for the down-regulated genes. In these two groups of genes, there is a general agreement in fold-changes of expression between the microarray experiments and densitometric tracings of mRNA levels (Table 3). Combining the 14 genes, the Pearson correlation coefficient r between Northern and microarray data is 0.61. Of these, KRT18, VIL2 and CDC2 have the most discrepant correlation between microarray and Northern data. If these three genes are excluded, the Pearson correlation coefficient r is raised to 0.94, indicating an excellent correlation in the remaining 11 genes. We verified the protein levels of two genes, p21WAF1/Cip1 and p57Kip2, and they correlate well with the microarray and the Northern data (Table 3).
Table 3.
Comparison of expression ratios between microarray experiments and Northern blot analysis
| Up-regulated genes | Down-regulated genes | ||||||
|---|---|---|---|---|---|---|---|
| Gene | Microarraysa | Northern blotsb | Western blotsb | Gene | Microarraysc | Northern blotsd | |
| ALPI | 4.89 | 3.2 | KIA | 2.99 | 2.6 | ||
| P57 | 4.08 | 3.0 | 4 | MAD2L1 | 2.12 | 1.9 | |
| KRT18 | 3.33 | 5 | CDC2 | 2.09 | 2.8 | ||
| IGFBP6 | 3.23 | 2.4 | BUB1B | 2.06 | 1.8 | ||
| Villin2 | 2.94 | 3.9 | RAD21 | 2.06 | 1.9 | ||
| P21 | 2.22 | 2.3 | 3 | WEE1 | 2.01 | 1.9 | |
| SFN | 2.13 | 2 | Cyclin D1 | 1.70 | 1.6 | ||
Numbers indicate means of fold-induction of two experiments.
Fold-induced by densitometric tracing.
Means of fold-decreased of two experiments.
Fold-decreased by densitometric tracing.
The present study was designed to identify target genes of KLF4 after 24 hours of its inducible expression. We therefore identified genes whose expression are either maximally or near-maximally activated or suppressed by KLF4 at this particular time. The time-course Northern experiments for some of the up-regulated genes in Figure 5 support this claim. However, there are notable differences in the kinetics of activation for the group of genes tested. For example, ALPI is activated relatively late in the process, whereas p21WAF1/Cip1 is activated relatively early. We presume that our study failed to identify some of the target genes that a re activated early following induction of KLF4 but the expression levels subsequently fall back to baseline at 24 hours. These potential “immediate-early” genes can be identified when RNA isolated from earlier time-points are included in the microarray experiments in a manner similar to a previous study that examined the transcriptional program in the response of human fibroblasts to serum.66 We presume that such studies may identify additional KLF4 target genes that are regulatory in nature and that they may mediate the effect of KLF4.
KLF4 was identified initially as a gene whose expression accompanies growth arrest.29 Conditions such as contact inhibition and serum deprivation stimulated KLF4 expression.29 Further, its in vivo pattern of expression mirrors that seen in vitro. Thus, KLF4 mRNA is found primarily in terminally differentiated and mitotically inert epithelial cells of the intestine29 and epidermis.30,43 Constitutive expression of KLF4 results in the inhibition of DNA synthesis29 and colony formation.38 A recent study using the inducible RKO cell line described here demonstrated that the inducible expression of KLF4 results in an arrest in the cell cycle at the transition phase between G1 and S, and that this arrest is accompanied by the activation of expression of p21WAF1/Cip1,41 a potent suppressor of cell proliferation.39 Moreover, KLF4 is induced as a consequence of DNA damage in a p53-dependent manner, and this induction results in the transcriptional activation of p21WAF1/Cip1 due to a direct effect of KLF4 on a specific cis-element in the p21WAF1/Cip1 promoter.34 These studies indicate that p21WAF1/Cip1 is an important downstream mediator of KLF4. Our present study confirmed these observations and provided independent evidence that p21WAF1/Cip1 is a target gene of KLF4. Moreover, the group of target genes up-regulated by KLF4 includes additional genes that have been proven to encode inhibitors of cell proliferation, including p57Kip2,46,47 IGFBP6,48 and SFN (stratifin).49 These findings illustrate a mechanism by which KLF4 suppresses proliferation by activating a group of cell-cycle inhibitors.
Studies have indicated that KLF4 is an inhibitor of DNA synthesis,29 and that this effect is likely mediated at the G1/S transition of the cell cycle with a resultant arrest at this checkpoint following KLF4 induction.41 The results of our current study show that both p21WAF1/Cip1 and p57Kip2 are prominent target genes of KLF4 and are likely the explanation for its cell-cycle effect. Both p21WAF1/Cip140 and p57Kip247 are potent inhibitors of cyclin-dependent kinases (Cdks) in the G1 phase of the cell cycle. It is of interest to note that a similar G1 Cdk inhibitor, p27Kip1, is regulated primarily at a post-translational level,67,68 which may be the reason that its mRNA level was unchanged after KLF4 induction in the current study (data not shown).
In addition to up-regulating several inhibitors of the cell cycle, KLF4 suppresses several genes that are positive regulators of the cell cycle. Examples include cyclin D1, CDC2/CDK1 (cell division cycle/cyclin-dependent kinase 1), KIA (proliferation related Ki-67 antigen), and MCM2 (minichromosome maintenance deficient 2). Many of these proteins are known to promote the cell cycle, some at the G1/S phase. Cyclin D1 is one of several D-type cyclins that couples extracellular signals to the biochemical machinery that governs progression through the G1 phase of the mammalian cell division cycle.69 Cyclin D1 assembles with cyclin-dependent kinases CDK4 and CDK6 to form holoenzymes that facilitate exit from G1 by phosphorylating key substrates, including the retinoblastoma protein. Previous studies showed that the inhibitory effect of KLF4 on cyclin D1 expression is exerted at a transcriptional level.70 Similarly, CDC2/CDK1 is responsible for controlling the transition from the G1 to the S phase, and from the G2 to the M phase of the cell cycle.71 In addition, both KIA50 and MCM272 are shown to be associated with cell proliferation. MCM2, in particular, is absolutely necessary for the initiation of DNA replication.72 Thus, the concomitant increase of inhibitors and decrease of promoters of G1 to S phase progression upon induction of KLF4 expression is likely to be the reason that KLF4 causes the previously observed G1/S block.41
Another group of cell cycle-related genes that are suppressed by KLF4 are components of the kinetochore. This group includes CENPE (centromere protein E), MAD2L1 (mitotic arrest deficient 2-like 1), BUB1B (budding uninhibited by benzimidazoles 1 homolog B), and RAD21. These proteins are required for the execution of the mitotic checkpoint in the cell cycle.51,53,73,74 Although the significance of the down-regulation of these kinetochore-associated genes by KLF4 is not clear, it is possible that a consequence is mitotic arrest, which is the case when CENPE expression becomes suppressed.73 Two additional lines of evidence link KLF4 to the G2/M phase of the cell cycle. One is its ability to down-regulate STK15 (serine/threonine kinase 5), a serine/threonine kinase mainly functioning in the G2/M phase75 and whose amplification was shown to cause centrosome amplification, aneuploidy and transformation.76 The other is the finding that KLF4 activates expression of SFN (stratifin or 14-3-3 σ), an inhibitor of G2/M progression of the cell cycle following DNA damage.49,77 It is of interest to note that DNA damage-induced activation of SFN expression is dependent on p53,49 as is KLF4.34 The lone exception to the notion that KLF4 inhibits the cell cycle by activating or suppressing cell-cycle genes with opposite effects is its ability to down-regulated WEE1, which, like SFN, is an inhibitor of G2/M progression in response to DNA damage.78 Whether WEE1 may be activated by KLF4 at a time other than the 24 hours tested in this study is unclear.
The in vivo expression of KLF4 is found primarily in epithelial tissues, including the intestine29,30 and epidermis.30,43 Moreover, the KLF4 mRNA is found mostly in the post-mitotic, terminally differentiated epithelial cells.29,30,43 These findings suggest that KLF4 may play a role in the expression of differentiation-dependent genes. Indeed, KLF4 has been shown to trans-activate the promoters of several epithelial enriched genes such as CYP1A1,79 laminin α 3A,80 keratin 19,81 and keratin 4.82 Our current study identified additional genes that are targets of KLF4 and which are enriched in epithelial cells. They include ALPI, VIL2 or ezrin, DSG2 (desmoglein 2), and numerous genes encoding keratins (Table 1). These findings suggest that KLF4, in addition to function as a cell cycle regulator, is involved in regulating expression of differentiation-specific genes.
Evidence supporting the role of KLF4 in regulating differentiation of epithelial tissues is demonstrated by gene targeting experiments of Klf4 that confirmed the effect of Klf4 on terminal differentiation of two epithelial cell types, epidermal keratinocytes43 and colonic goblet cells.44 Klf4-null (−/−) mice died shortly after birth due to a loss of barrier function of the skin.43 Here, the late-stage differentiation structures of the skin, including the cornified envelope, are selectively perturbed. Accompanying Klf4 deletion is an altered expression of Sprr2a, which encodes a cornified envelope protein that functions as a marker of keratinocyte differentiation.83 In our experiments, expression of SPRR2A was unchanged upon KLF4 induction (data not shown). This may be due to the fact that the RKO cell line used in the present study is intestinal, rather than epidermal in origin.
A more recent study examined the effect of Klf4 knockout on the intestine.44 It was shown that Klf4-null mice have a 90% reduction in the number of goblet cells in the colon with an altered expression of the gene encoding a goblet cell-specific marker, MUC2.44 It was concluded that Klf4 plays a crucial role in the in vivo differentiation of colonic goblet cells. The cDNA encoding MUC2 was not included in the chip and whether its expression is controlled by KLF4 therefore could not be determined. However, cDNA encoding GCNT3 (Glucosaminyl (N-acetyl) transferase 3, mucin-type), a colonic epithelial enriched mucin,84 was included in the chip and its level of expression was increased by 1.9-fold upon KLF4 induction (Table 1). These findings suggest a potential mechanism by which KLF4 may regulate goblet cell differentiation by activating mucin gene expression.
A particularly interesting group among the genes up-regulated by KLF4 is one that encodes structural proteins (Table 1). Most of these structural proteins are involved in the formation of intermediate filaments including keratins, villin 2 and vimentin. Of particular interest is the activation of nine keratin genes by KLF4 (Figure 6). Keratins constitute intermediate filaments and are markers of epithelial differentiation.61 There are 20 keratin genes in the human genome and they are divided into two classes: type I and type II. Type I keratins are relatively low in molecular mass and acidic, whereas type II keratins are larger and more basic. Filament formation usually requires heterodimerization of keratins in pairs consisting of one type I and one type II polypeptide. The two types of keratin genes are located in clusters on two separate chromosomes; type I on chromosome 17 and type II on chromosome 12.63 It is of interest to note that the majority (seven out of nine) of keratin genes activated by KLF4 are located on chromosome 12 (Figure 6). These findings raise the possibility of the presence of a locus control region (LCR)85 in this keratin gene cluster that is regulated by KLF4 in a manner reminiscent of the regulation of the globin gene LCR by KLF1.86
In summary, using expression profiling, we identified groups of genes that are up-regulated or down-regulated upon the inducible expression of KLF4 in a colonic epithelial cell line, RKO. Functional and sequence clusterings of these genes reveal that they may mediate some, if not most, of the previously reported biochemical functions of KLF4, including inhibition of cell proliferation and promotion of terminal differentiation. The identification of the type II keratin gene cluster as a site of regulation by KLF4 raises the possibility that KLF4 may have an LCR function. Further exploration of the mechanism by which KLF4 regulates its target genes, including the keratin gene cluster, may reveal additional information on how KLF4 regulates cellular proliferation and differentiation.
Experimental Procedures
Cell culture
The inducible cell system for KLF4 has been described.41 Briefly, a colon cancer cell line, RKO, was stably transfected with a plasmid encoding the insect hormone receptor, ecdysone receptor (EcR) and the retinoid X receptor (RXR), and a second plasmid containing KLF4 and green fluorescence protein (GFP) under the control of an ecdysone receptor response element.41 This cell line, abbreviated EcR-RKO/KLF4, was maintained in DMEM (GIBCO, Gaithersburg, MD) supplemented with heat-inactivated 10% (v/v) fetal calf serum (Hyclone), 2 mM L-glutamine, 10 mM Hepes (pH 7.2), 100 units/ml of penicillin, 100 μg/ml of streptomycin (GIBCO), and 150 μg/ml of Zeocin for selection (Invitrogen, CA) in a 37 °C environment with 5% (v/v) CO2-in-air. Upon reaching 80% confluence, cells were treated with 5 μM ponasterone A (PA), an ecdysone analogue, for various periods of time. To control for the experiment, the vehicle, ethanol, was added for the same periods of time.
RNA preparation and cDNA microarray analysis
Total RNA was isolated from EcR-RKO/KLF4 cells treated with ponasterone A or vehicle alone for 24 hours using Trizol™ (Invitrogen). Poly(A)-containing RNA was then purified using the Oligotex mRNA midi-kit (Qiagen, Valencia, CA) and quantified using the Ribo-Green™ RNA quantification kit (Molecular Probes, Eugene, OR). Complementary DNAs were then generated from mRNAs obtained from treated and control cells and labeled with Cy3 and Cy5 fluorescent dyes, respectively, in one experiment and with the dyes in reverse in a second experiment. Fluorescent-labeled cDNAs were hybridized to the Human Unigene™ version 1.33 microarrays (IncyteGenomics, Palo Alto, CA), which contained cDNAs from 9600 unique human genes. Data were analyzed using the GemTools™ 2.5.0 software and expressed as a balanced differential†. Genes that were up-regulated and down-regulated (defined as >1.7-fold induced or suppressed) were filtered using Partek Pro (Partek, Inc., St. Charles, MO) and S-Plus™ (Insightful Corp., Seattle, WA). In addition, the genes were grouped by their function into gene clusters using the DRAGON database for human genes (http://pevsnerlab.kennedykrieger.org/dragon.htm), the Pfam database‡, and the Swiss-Prot database§. The structural group of genes that were up-regulated was clustered using the MegAlign program within the LaserGene™ software package by DNASTAR (Madison, WI). The phylogenetic tree of the up-regulated structural group/intermediate filament group was done using ClustalW multiple alignment87 and the Blosum Series protein weight matrix.88
Northern blot analysis
Complementary cDNA clones encoding the various genes analyzed by Northern blots were purchased from Research Genetics (Huntsville, AL). cDNA clones for p21WAF1/Cip1 and cyclin D1 were gifts from Dr B. Vogelstein (Johns Hopkins Oncology Center, Baltimore, MD) and for intestinal alkaline phosphatase (ALPI) from Dr R. Hodin (Harvard Medical School, Boston, MA). β-Actin cDNA probe was purchased from GIBCO. All cDNA clones were verified by sequencing at the Emory DNA Core Facility. The probes were radioactively labeled using the Random Primer Labeling kit (Roche). Total RNA (20 μg) prepared from ponasterone A-treated or control EcR-RKO/KLF4 cells at various time-points up to 24 hours was resolved by electrophoresis in 1.2% (w/v) agarose gels containing 2.4 M formaldehyde and transblotted onto nylon membranes (Hybond-N; Amersham). Hybridization and washing were performed under high-stringency conditions using radioactively labeled cDNA probes.
Western blot analysis
Western blot analyses were performed using standard procedures. In brief, the concentrations of proteins from treated or untreated cells were measured with a spectrophotometer (Beckman) using BioRad Protein Assay with bovine serum albumin (BSA) as a standard. Total proteins (40 μg) were dissolved in loading buffer (60 mM Tris–HCl (pH 6.8), 2% (w/v) SDS, 100 mM dithiothreitol, 0.01% (w/v) bromphenol blue), heated at 100 °C for three minutes, and loaded onto an SDS/polyacrylamide gel in running buffer containing 25 mM Tris–HCl (pH 8.3), 250 mM glycine, 0.1% SDS. At the completion of electrophoresis, proteins were transferred electrophoretically to a nitrocellulose membrane, which was then immunoblotted sequentially with primary antibodies followed by a secondary antibody (horseradish peroxidase-conjugated goat anti-rabbit IgG; Santa Cruz). The rabbit antiserum against KLF4 was as described.29 Anti-serum directed against p21WAF1/Cip1, p57Kip2 and β-actin was purchased from Santa Cruz. All blots were visualized with enhanced chemiluminescence from Amersham.
Acknowledgments
We thank Drs B. Vogelstein and R. Hodin for providing reagents. This work was supported, in part, by grants from the National Institutes of Health (DK52230 and CA84197). V.W.Y. is a Georgia Cancer Coalition Distinguished Cancer Scholar. E.M.W. is supported, in part, by the Postdoctoral Research and Education Program (PREP) (GM00680).
Abbreviations
- ALPI
intestinal alkaline phosphatase
- BUB1B
budding uninhibited by benzimidazoles 1 homolog B
- CENPE
centromere protein E
- CDC2/CDK1
cell division cycle 2/cyclin-dependent kinase 1
- Cdk
cyclin-dependent kinase
- EcR
ecdysone receptor
- IGFBP6
insulin-like growth factor binding protein 6
- KIA
proliferation-related Ki-67 antigen
- KRT
keratin
- KLF
Krüppel-like factor
- MAD2L1
mitotic arrest deficient 2-like 1
- MCM2
minichromosome maintenance deficient 2
- PA
ponasterone A
- SFN
stratifin
- STK15
serine/threonine kinase 5
Footnotes
References
- 1.Marshman E, Booth C, Potten CS. The intestinal epithelial stem cell. BioEssays. 2002;24:91–98. doi: 10.1002/bies.10028. [DOI] [PubMed] [Google Scholar]
- 2.Booth C, Potten CS. Gut instincts: thoughts on intestinal epithelial stem cells. J Clin Invest. 2000;105:1493–1499. doi: 10.1172/JCI10229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Simon TC, Gordon JI. Intestinal epithelial cell differentiation: new insights from mice, flies and nematodes. Curr Opin Genet Dev. 1995;5:577–586. doi: 10.1016/0959-437x(95)80026-3. [DOI] [PubMed] [Google Scholar]
- 4.Gordon JI, Hermiston ML. Differentiation and self-renewal in the mouse gastrointestinal epithelium. Curr Opin Cell Biol. 1994;6:795–803. doi: 10.1016/0955-0674(94)90047-7. [DOI] [PubMed] [Google Scholar]
- 5.Fuchs E, Raghavan S. Getting under the skin of epidermal morphogenesis. Nature Rev Genet. 2002;3:199–209. doi: 10.1038/nrg758. [DOI] [PubMed] [Google Scholar]
- 6.Watt FM. Stem cell fate and patterning in mammalian epidermis. Curr Opin Genet Dev. 2001;11:410–417. doi: 10.1016/s0959-437x(00)00211-2. [DOI] [PubMed] [Google Scholar]
- 7.Eckert RL, Crish JF, Robinson NA. The epidermal keratinocyte as a model for the study of gene regulation and cell differentiation. Physiol Rev. 1997;77:397–424. doi: 10.1152/physrev.1997.77.2.397. [DOI] [PubMed] [Google Scholar]
- 8.Kinzler KW, Vogelstein B. Lessons from hereditary colorectal cancer. Cell. 1996;87:159–170. doi: 10.1016/s0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- 9.Vogelstein B, Kinzler KW. The multistep nature of cancer. Trends Genet. 1993;9:138–141. doi: 10.1016/0168-9525(93)90209-z. [DOI] [PubMed] [Google Scholar]
- 10.Wong MH, Huelsken J, Birchmeier W, Gordon JI. Selection of multipotent stem cells during morphogenesis of small intestinal crypts of Lieberkuhn is perturbed by stimulation of Lef-1/beta-catenin signaling. J Biol Chem. 2002;277:15843–15850. doi: 10.1074/jbc.M200184200. [DOI] [PubMed] [Google Scholar]
- 11.Mariadason JM, Bordonaro M, Aslam F, Shi L, Kuraguchi M, Velcich A, Augenlicht LH. Down-regulation of beta-catenin TCF signaling is linked to colonic epithelial cell differentiation. Cancer Res. 2001;61:3465–3471. [PubMed] [Google Scholar]
- 12.Korinek V, Barker N, Moerer P, van Donselaar E, Huls G, Peters PJ, Clevers H. Depletion of epithelial stem-cell compartments in the small intestine of mice lacking Tcf-4. Nature Genet. 1998;19:379–383. doi: 10.1038/1270. [DOI] [PubMed] [Google Scholar]
- 13.Barker N, Huls G, Korinek V, Clevers H. Restricted high level expression of Tcf-4 protein in intestinal and mammary gland epithelium. Am J Pathol. 1999;154:29–35. doi: 10.1016/S0002-9440(10)65247-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Silberg DG, Swain GP, Suh ER, Traber PG. Cdx1 and cdx2 expression during intestinal development. Gastroenterology. 2000;119:961–971. doi: 10.1053/gast.2000.18142. [DOI] [PubMed] [Google Scholar]
- 15.Lorentz O, Duluc I, Arcangelis AD, Simon-Assmann P, Kedinger M, Freund JN. Key role of the Cdx2 homeobox gene in extracellular matrix-mediated intestinal cell differentiation. J Cell Biol. 1997;139:1553–1565. doi: 10.1083/jcb.139.6.1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suh E, Traber PG. An intestine-specific homeobox gene regulates proliferation and differentiation. Mol Cell Biol. 1996;16:619–625. doi: 10.1128/mcb.16.2.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Larsson LI, Madsen OD, Serup P, Jonsson J, Edlund H. Pancreatic-duodenal homeobox 1—role in gastric endocrine patterning. Mech Dev. 1996;60:175–184. doi: 10.1016/s0925-4773(96)00609-0. [DOI] [PubMed] [Google Scholar]
- 18.Schuh R, Aicher W, Gaul U, Cote S, Preiss A, Maier D, et al. A conserved family of nuclear proteins containing structural elements of the finger protein encoded by Krüppel, a Drosophila segmentation gene. Cell. 1986;47:1025–1032. doi: 10.1016/0092-8674(86)90817-2. [DOI] [PubMed] [Google Scholar]
- 19.Bieker JJ. Krüppel-like factors: three fingers in many pies. J Biol Chem. 2001;276:34355–34358. doi: 10.1074/jbc.R100043200. [DOI] [PubMed] [Google Scholar]
- 20.Black AR, Black JD, Azizkhan-Clifford J. Sp1 and Krüppel-like factor family of transcription factors in cell growth regulation and cancer. J Cell Physiol. 2001;188:143–160. doi: 10.1002/jcp.1111. [DOI] [PubMed] [Google Scholar]
- 21.Dang DT, Pevsner J, Yang VW. The biology of the mammalian Krüppel-like family of transcription factors. Int J Biochem Cell Biol. 2000;32:1103–1121. doi: 10.1016/s1357-2725(00)00059-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Turner J, Crossley M. Mammalian Krüppel-like transcription factors: more than just a pretty finger. Trends Biochem Sci. 1999;24:236–240. doi: 10.1016/s0968-0004(99)01406-1. [DOI] [PubMed] [Google Scholar]
- 23.Philipsen S, Suske G. A tale of three fingers: the family of mammalian Sp/XKLF transcription factors. Nucl Acids Res. 1999;27:2991–3000. doi: 10.1093/nar/27.15.2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller IJ, Bieker JJ. A novel, erythroid cell-specific murine transcription factor that binds to the CACCC element and is related to the Krüppel family of nuclear proteins. Mol Cell Biol. 1993;13:2776–2786. doi: 10.1128/mcb.13.5.2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perkins AC, Sharpe AH, Orkin SH. Lethal beta-thalassaemia in mice lacking the erythroid CACCC-transcription factor EKLF. Nature. 1995;375:318–322. doi: 10.1038/375318a0. [DOI] [PubMed] [Google Scholar]
- 26.Nuez B, Michalovich D, Bygrave A, Ploemacher R, Grosveld F. Defective haematopoiesis in fetal liver resulting from inactivation of the EKLF gene. Nature. 1995;375:316–318. doi: 10.1038/375316a0. [DOI] [PubMed] [Google Scholar]
- 27.Anderson KP, Kern CB, Crable SC, Lingrel JB. Isolation of a gene encoding a functional zinc finger protein homologous to erythroid Krüppel-like factor: identification of a new multigene family. Mol Cell Biol. 1995;15:5957–5965. doi: 10.1128/mcb.15.11.5957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wani MA, Conkright MD, Jeffries S, Hughes MJ, Lingrel JB. cDNA isolation, genomic structure, regulation, and chromosomal localization of human lung Krüppel-like factor. Genomics. 1999;60:78–86. doi: 10.1006/geno.1999.5888. [DOI] [PubMed] [Google Scholar]
- 29.Shields JM, Christy RJ, Yang VW. Identification and characterization of a gene encoding a gut-enriched Krüppel-like factor expressed during growth arrest. J Biol Chem. 1996;271:20009–20017. doi: 10.1074/jbc.271.33.20009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garrett-Sinha LA, Eberspaecher H, Seldin MF, de Crombrugghe B. A gene for a novel zinc-finger protein expressed in differentiated epithelial cells and transiently in certain mesenchymal cells. J Biol Chem. 1996;271:31384–31390. doi: 10.1074/jbc.271.49.31384. [DOI] [PubMed] [Google Scholar]
- 31.Shields JM, Yang VW. Two potent nuclear localization signals in the gut-enriched Krüppel-like factor define a subfamily of closely related Krüppel proteins. J Biol Chem. 1997;272:18504–18507. doi: 10.1074/jbc.272.29.18504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Christy BA, Lau LF, Nathans D. A gene activated in mouse 3T3 cells by serum growth factors encodes a protein with “zinc finger” sequences. Proc Natl Acad Sci USA. 1988;85:7857–7861. doi: 10.1073/pnas.85.21.7857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jenkins TD, Opitz OG, Okano J, Rustgi AK. Transactivation of the human keratin 4 and Epstein–Barr virus ED-L2 promoters by gut-enriched Krüppel-like factor. J Biol Chem. 1998;273:10747–10754. doi: 10.1074/jbc.273.17.10747. [DOI] [PubMed] [Google Scholar]
- 34.Zhang W, Geiman DE, Shields JM, Dang DT, Mahatan CS, Kaestner KH, et al. The gut-enriched Krüppel-like factor (Krüppel-like factor 4). mediates the transactivating effect of p53 on the p21WAF1/Cip1 promoter. J Biol Chem. 2000;275:18391–18398. doi: 10.1074/jbc.C000062200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dang DT, Bachman KE, Mahatan CS, Dang LH, Giardiello FM, Yang VW. Decreased expression of the gut-enriched Krüppel-like factor gene in intestinal adenomas of multiple intestinal neoplasia mice and in colonic adenomas of familial adenomatous polyposis patients. FEBS Letters. 2000;476:203–207. doi: 10.1016/s0014-5793(00)01727-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ton-That H, Kaestner KH, Shields JM, Mahatanankoon CS, Yang VW. Expression of the gut-enriched Krüppel-like factor gene during development and intestinal tumorigenesis. FEBS Letters. 1997;419:239–243. doi: 10.1016/s0014-5793(97)01465-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dang DT, Mahatan CS, Dang LH, Agboola IA, Yang VW. Expression of the gut-enriched Krüppel-like factor (Krüppel-like factor 4) gene in the human colon cancer cell line RKO is dependent on CDX2. Oncogene. 2001;20:4884–4890. doi: 10.1038/sj.onc.1204645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Geiman DE, Ton-That H, Johnson JM, Yang VW. Transactivation and growth suppression by the gut-enriched Krüppel-like factor (Krüppel-like factor 4) are dependent on acidic amino acid residues and protein–protein interaction. Nucl Acids Res. 2000;28:1106–1113. doi: 10.1093/nar/28.5.1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.El-Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM, et al. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 40.Harper JW, Adami GR, Wei N, Keyomarsi K, Elledge SJ. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- 41.Chen X, Johns DC, Geiman DE, Marban E, Dang DT, Hamlin G, Sun R, Yang VW. Krüppel-like factor 4 (gut-enriched Krüppel-like factor). inhibits cell proliferation by blocking G1/S progression of the cell cycle. J Biol Chem. 2001;276:30423–30428. doi: 10.1074/jbc.M101194200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yoon HS, Chen X, Yang VW. Krüppel-like factor 4 mediates p53-dependent G1/S cell cycle arrest in response to DNA damage. J Biol Chem. 2003 doi: 10.1074/jbc.M211027200. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Segre JA, Bauer C, Fuchs E. Klf4 is a transcription factor required for establishing the barrier function of the skin. Nature Genet. 1999;22:356–360. doi: 10.1038/11926. [DOI] [PubMed] [Google Scholar]
- 44.Katz JP, Perreault N, Goldstein BG, Lee CS, Labosky PA, Yang VW, Kaestner KH. The zinc-finger transcription factor Klf4 is required for terminal differentiation of goblet cells in the colon. Development. 2002;129:2619–2628. doi: 10.1242/dev.129.11.2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yue H, Eastman PS, Wang BB, Minor J, Doctolero MH, Nuttall RL, et al. An evaluation of the performance of cDNA microarrays for detecting changes in global mRNA expression. Nucl Acids Res. 2001;29:e41. doi: 10.1093/nar/29.8.e41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Matsuoka S, Edwards MC, Bai C, Parker S, Zhang P, Baldini A, et al. p57KIP2, a structurally distinct member of the p21CIP1 Cdk inhibitor family, is a candidate tumor suppressor gene. Genes Dev. 1995;9:650–662. doi: 10.1101/gad.9.6.650. [DOI] [PubMed] [Google Scholar]
- 47.Lee MH, Reynisdottir I, Massague J. Cloning of p57KIP2, a cyclin-dependent kinase inhibitor with unique domain structure and tissue distribution. Genes Dev. 1995;9:639–649. doi: 10.1101/gad.9.6.639. [DOI] [PubMed] [Google Scholar]
- 48.Kato M, Ishizaki A, Hellman U, Wernstedt C, Kyogoku M, Miyazono K, et al. A human keratinocyte cell line produces two autocrine growth inhibitors, transforming growth factor-beta and insulin-like growth factor binding protein-6, in a calcium- and cell density-dependent manner. J Biol Chem. 1995;270:12373–12379. doi: 10.1074/jbc.270.21.12373. [DOI] [PubMed] [Google Scholar]
- 49.Hermeking H, Lengauer C, Polyak K, He TC, Zhang L, Thiagalingam S, et al. 14-3-3 sigma is a p53-regulated inhibitor of G2/M progression. Mol Cell. 1997;1:3–11. doi: 10.1016/s1097-2765(00)80002-7. [DOI] [PubMed] [Google Scholar]
- 50.Schluter C, Duchrow M, Wohlenberg C, Becker MHG, Key G, Flad HD, Gerdes J. The cell proliferation-associated antigen of antibody Ki-67: a very large, ubiquitous nuclear protein with numerous repeated elements, representing a new kind of cell cycle-maintaining proteins. J Cell Biol. 1993;123:513–522. doi: 10.1083/jcb.123.3.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li Y, Benezra R. Identification of a human mitotic checkpoint gene: hsMAD2. Science. 1996;274:246–248. doi: 10.1126/science.274.5285.246. [DOI] [PubMed] [Google Scholar]
- 52.Lee MG, Nurse P. Complementation used to clone a human homologue of the fission yeast cell cycle control gene cdc2. Nature. 1987;327:31–35. doi: 10.1038/327031a0. [DOI] [PubMed] [Google Scholar]
- 53.Cahill DP, Lengauer C, Yu J, Riggins GJ, Willson JKV, Markowitz SD, et al. Mutations of mitotic checkpoint genes in human cancers. Nature. 1998;392:300–303. doi: 10.1038/32688. [DOI] [PubMed] [Google Scholar]
- 54.Davenport JW, Fernandes ER, Harris LD, Neale GAM, Goorha R. The mouse mitotic checkpoint gene bub1b, a novel bub1 family member, is expressed in a cell cycle-dependent manner. Genomics. 1999;55:113–117. doi: 10.1006/geno.1998.5629. [DOI] [PubMed] [Google Scholar]
- 55.Hoque MT, Ishikawa F. Human chromatid cohesin component hRad21 is phosphorylated in M phase and associated with metaphase centromeres. J Biol Chem. 2001;276:5059–5067. doi: 10.1074/jbc.M007809200. [DOI] [PubMed] [Google Scholar]
- 56.McKay MJ, Troelstra C, van der Spek P, Kanaar R, Smit B, Hagemeijer A, et al. Sequence conservation of the rad21 Schizosaccharomyces pombe DNA double-strand break repair gene in human and mouse. Genomics. 1996;36:305–315. doi: 10.1006/geno.1996.0466. [DOI] [PubMed] [Google Scholar]
- 57.Heald R, McLoughlin M, McKeon F. Human wee1 maintains mitotic timing by protecting the nucleus from cytoplasmically activated Cdc2 kinase. Cell. 1993;74:463–474. doi: 10.1016/0092-8674(93)80048-j. [DOI] [PubMed] [Google Scholar]
- 58.Hinds PW, Dowdy SF, Eaton EN, Arnold A, Weinberg RA. Function of a human cyclin gene as an oncogene. Proc Natl Acad Sci. 1994;91:709–713. doi: 10.1073/pnas.91.2.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xiong Y, Connelly T, Futcher B, Beach D. Human D-type cyclin. Cell. 1991;65:691–699. doi: 10.1016/0092-8674(91)90100-d. [DOI] [PubMed] [Google Scholar]
- 60.Steinert PM, Roop DR. Molecular and cellular biology of intermediate filaments. Ann Rev Biochem. 1988;57:593–625. doi: 10.1146/annurev.bi.57.070188.003113. [DOI] [PubMed] [Google Scholar]
- 61.Fuchs E. Keratins as biochemical markers of epithelial differentiation. Trends Genet. 1988;4:277–281. doi: 10.1016/0168-9525(88)90169-2. [DOI] [PubMed] [Google Scholar]
- 62.Sun TT, Eichner R, Nelson WG, Tseng SC, Weiss RA, Jarvinen M, Woodcock-Mitchell J. Keratin classes: molecular markers for different types of epithelial differentiation. J Invest Dermatol. 1983;81:109s–115s. doi: 10.1111/1523-1747.ep12540831. [DOI] [PubMed] [Google Scholar]
- 63.Yoon SJ, LeBlanc-Straceski J, Ward D, Krauter K, Kucherlapati R. Organization of the human keratin type II gene cluster at 12q13. Genomics. 1994;24:502–508. doi: 10.1006/geno.1994.1659. [DOI] [PubMed] [Google Scholar]
- 64.Ceratto N, Dobkin C, Carter M, Jenkins E, Yao XL, Cassiman JJ, et al. Human type I cyto-keratin genes are a compact cluster. Cytogenet Cell Genet. 1997;77:169–174. doi: 10.1159/000134566. [DOI] [PubMed] [Google Scholar]
- 65.Milisavljevic V, Freedberg IM, Blumenberg M. Close linkage of the two keratin gene clusters in the human genome. Genomics. 1996;34:134–138. doi: 10.1006/geno.1996.0252. [DOI] [PubMed] [Google Scholar]
- 66.Iyer VR, Eisen MB, Ross DT, Schuler G, Moore T, Lee JC, et al. The transcriptional program in the response of human fibroblasts to serum. Science. 1999;283:83–87. doi: 10.1126/science.283.5398.83. [DOI] [PubMed] [Google Scholar]
- 67.Toyoshima H, Hunter T. p27, a novel inhibitor of G1 cyclin-Cdk protein kinase activity, is related to p21. Cell. 1994;78:67–74. doi: 10.1016/0092-8674(94)90573-8. [DOI] [PubMed] [Google Scholar]
- 68.Hengst L, Reed SI. Translational control of p27Kip1 accumulation during the cell cycle. Science. 1996;271:1861–1864. doi: 10.1126/science.271.5257.1861. [DOI] [PubMed] [Google Scholar]
- 69.Sherr CJ. D-type cyclins. Trends Biochem Sci. 1995;20:187–190. doi: 10.1016/s0968-0004(00)89005-2. [DOI] [PubMed] [Google Scholar]
- 70.Shie JL, Chen ZY, Fu M, Pestell RG, Tseng CC. Gut-enriched Krüppel-like factor represses cyclin D1 promoter activity through Sp1 motif. Nucl Acids Res. 2000;28:2969–2976. doi: 10.1093/nar/28.15.2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Doree M, Galas S. The cyclin-dependent protein kinases and the control of cell division. FASEB J. 1994;8:1114–1121. doi: 10.1096/fasebj.8.14.7958616. [DOI] [PubMed] [Google Scholar]
- 72.Tye BK. MCM proteins in DNA replication. Annu Rev Biochem. 1999;68:649–686. doi: 10.1146/annurev.biochem.68.1.649. [DOI] [PubMed] [Google Scholar]
- 73.Yao X, Abrieu A, Zheng Y, Sullivan KF, Cleveland DW. CENP-E forms a link between attachment of spindle microtubules to kinetochores and the mitotic checkpoint. Nature Cell Biol. 2000;2:484–491. doi: 10.1038/35019518. [DOI] [PubMed] [Google Scholar]
- 74.Hoque MT, Ishikawa F. Human chromatid cohesin component hRad21 is phosphorylated in M phase and associated with metaphase centromeres. J Biol Chem. 2001;276:5059–5067. doi: 10.1074/jbc.M007809200. [DOI] [PubMed] [Google Scholar]
- 75.Shindo M, Nakano H, Kuroyanagi H, Shirasawa T, Mihara M, Gilbert DJ, et al. cDNA cloning, expression, subcellular localization, and chromosomal assignment of mammalian aurora homologues, aurora-related kinase (ARK) 1 and 2. Biochem Biophys Res Commun. 1998;244:285–292. doi: 10.1006/bbrc.1998.8250. [DOI] [PubMed] [Google Scholar]
- 76.Zhou H, Kuang J, Zhong L, Kuo W, Gray JW, Sahin A, et al. Tumour amplified kinase STK15/BTAK induces centrosome amplification, aneuploidy and transformation. Nature Genet. 1998;20:189–193. doi: 10.1038/2496. [DOI] [PubMed] [Google Scholar]
- 77.Chan TA, Hermeking H, Langauer C, Kinzler KW, Vogelstein B. 14-3-3 sigma is required to prevent mitotic catastrophe after DNA damage. Nature. 1999;401:616–620. doi: 10.1038/44188. [DOI] [PubMed] [Google Scholar]
- 78.Raleigh JM, O’Connell MJ. The G(2) DNA damage checkpoint targets both Wee1 and Cdc25. J Cell Sci. 2000;113:1727–1736. doi: 10.1242/jcs.113.10.1727. [DOI] [PubMed] [Google Scholar]
- 79.Zhang W, Shields JM, Sogawa K, Fujii-Kuriyama Y, Yang VW. The gut-enriched Krüppel-like factor suppresses the activity of the CYP1A1 promoter in an Sp1-dependent fashion. J Biol Chem. 1998;273:17917–17925. doi: 10.1074/jbc.273.28.17917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Miller KA, Eklund EA, Peddinghaus ML, Cao Z, Fernandes N, Turk PW, et al. Krüppel-like factor 4 regulates laminin alpha 3A expression in mammary epithelial cells. J Biol Chem. 2001;276:42863–42868. doi: 10.1074/jbc.M108130200. [DOI] [PubMed] [Google Scholar]
- 81.Brembeck FH, Rustgi AK. The tissue-dependent keratin 19 gene transcription is regulated by GKLF/KLF4 and Sp1. J Biol Chem. 2000;275:28230–28239. doi: 10.1074/jbc.M004013200. [DOI] [PubMed] [Google Scholar]
- 82.Okano J, Opitz OG, Nakagawa H, Jenkins TD, Friedman SL, Rustgi AK. The Krüppel-like transcriptional factors Zf9 and GKLF coactivate the human keratin 4 promoter and physically interact. FEBS Letters. 2000;473:95–100. doi: 10.1016/s0014-5793(00)01468-x. [DOI] [PubMed] [Google Scholar]
- 83.Gibbs S, Fijneman R, Wiegant J, van Kessel AG, van De Putte P, Backendorf C. Molecular characterization and evolution of the SPRR family of keratinocyte differentiation markers encoding small proline-rich proteins. Genomics. 1993;16:630–637. doi: 10.1006/geno.1993.1240. [DOI] [PubMed] [Google Scholar]
- 84.Yeh JC, Ong E, Fukuda M. Molecular cloning and expression of a novel beta-1,6-N-acetyl-glucosaminyltransferase that forms core 2, core 4, and I branches. J Biol Chem. 1999;274:3215–3221. doi: 10.1074/jbc.274.5.3215. [DOI] [PubMed] [Google Scholar]
- 85.Fraser P, Grosveld F. Locus control regions, chromatin activation and transcription. Curr Opin Cell Biol. 1998;10:361–365. doi: 10.1016/s0955-0674(98)80012-4. [DOI] [PubMed] [Google Scholar]
- 86.Tewari R, Gillemans N, Wijgerde M, Nuez B, von Lindern M, Grosveld F, Philipsen S. Erythroid Krüppel-like factor (EKLF) is active in primitive and definitive erythroid cells and is required for the function of 5′HS3 of the beta-globin locus control region. EMBO J. 1998;17:2334–2341. doi: 10.1093/emboj/17.8.2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucl Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Henikoff S, Henikoff JG. Performance evaluation of amino acid substitution matrices. Proteins: Struct Funct Genet. 1993;17:49–61. doi: 10.1002/prot.340170108. [DOI] [PubMed] [Google Scholar]