SUMMARY
The DevR transcriptional switch that defines the response of Mycobacterium tuberculosis to the lack of oxygen is now well established and likely helps the bacteria shift to a state of persistence. The M. tuberculosis two component signal transduction system (TCS), DevR-DevS, implicated in this transition to latency, is differentially expressed in H37Ra and H37Rv strains. Despite originating from the H37 ancestral strain, H37Ra and H37Rv have significant differences in their growth, physiology, and virulence. To further dissect the role of DevR in growth adaptive processes of M. tuberculosis, we investigated the hypoxic response of the avirulent H37Ra strain. Our results show that the DevR-DevS TCS in H37Ra is responsive to hypoxia and capable of target gene regulation, indicating similar DevR-DevS signaling pathways in H37Ra and H37Rv. A key finding of this study was the constitutive expression of the Rv3134c-devR-devS operon and a subset of sentinel DevR regulated genes in aerobic cultures of H37Ra but not H37Rv grown in Dubos Tween Albumin medium. Asparagine and/or catabolites of asparagine metabolism were implicated in aerobic induction of the DevR-DevS TCS in H37Ra. This is the first report of medium-specific constitutive expression of the DevR regulon in an avirulent strain and suggests a potential role for metabolite(s) in activation of the DevR-DevS TCS.
Keywords: M. tuberculosis H37Ra, DevR, constitutive aerobic expression, hypoxia, asparagine metabolism
INTRODUCTION
Tuberculosis continues its global rampage with the number of deaths exceeding 2 million people per year, making it the leading cause of mortality from a single bacterial pathogen. In recent years, cumulative data from transcriptomic, proteomic, and metabolomic profiling of Mycobacterium tuberculosis have enabled us to begin to understand the mechanisms involved in mycobacterial adaptation and survival in host cells. In parallel, comparative genomics between virulent and avirulent strains allow insights into mechanisms crucial for latency and persistence of M. tuberculosis. The recently annotated genome of H37Ra (GenBank accession no. CP000611), an avirulent counterpart of M. tuberculosis H37Rv 1 promises to provide new perspectives into the virulence regulation of tubercle bacilli.
Studies from various laboratories have revealed differences between H37Rv and H37Ra in cell membrane proteins and lipid metabolism,2 cell wall methyl branched lipids,3 culture filtrate proteins,4 a transcription regulator,5 PPE proteins,6 and the devR-devS TCS.7 Of particular interest, the devR-devS TCS was found to be differentially expressed in H37Rv versus H37Ra.8
The DevR response regulator (also called DosR) is a key factor in the metabolic shift-down during bacterial adaptation to oxygen limitation, low concentrations of nitric oxide (NO) or reactive nitrogen intermediates (RNI) and carbon monoxide (CO).9–16 A DevR regulon of ~ 50 genes is implicated in this transcriptional re-programming of M. tuberculosis. While there are considerable data on gene expression under hypoxic stress in H37Rv, we know little about the response of the H37Ra strain. In agreement with our previous observation,17 comparison of the annotated H37Ra genome sequence with that of H37Rv (GenBank accession no. AL123456) did not reveal any differences in devR-devS loci and since these genes were expressed at lower levels in H37Ra, we sought to determine whether differential DevR expression compromised the transcriptional response of H37Ra to hypoxia.
The Wayne model of dormancy18, 19 has contributed significantly to our understanding of mycobacterial gene expression under hypoxia.9,11, 20 Although the model uses Dubos broth base supplemented with Albumin-Dextrose-Saline (Dubos-ADS), commonly referred to as Dubos Tween Albumin medium (DTA) for culturing bacteria under hypoxia, several laboratories have used Middlebrook 7H9 base supplemented with 7H9-ADS and 0.05% Tween 80 (7H9T) or 7H9T plus 0.2% glycerol (7H9TG). Media constituents can play a critical role in defining the bacterial transcriptome and physiology. Pertinent to this is the idea that transition of M. tuberculosis into a state of non-replicative persistence could also be triggered by nutrient starvation.21 Therefore, in view of (1) the disparity in use of different kinds of media in gene expression studies and (2) consideration that the devR-devS TCS was initially identified and characterized from bacteria cultured in Kirchener’s and Middlebrook 7H9T media,7, 8 we investigated the effects of various media (7H9T, 7H9TG and DTA) on expression of the devR-devS genes, in virulent and avirulent strains of M. tuberculosis.
MATERIALS AND METHODS
A detailed description of all methods and protocols is available as Supplemental Data.
Bacterial strains and culture conditions
M. tuberculosis strains H37Rv (American Type Culture Collection, ATCC No. 25618) and H37Ra (ATCC No. 25177, obtained from Dr. Richard F. Silver, Case Western Reserve University, OH) were the test strains used in this study. Middlebrook 7H9 and Dubos broth base were made according to the manufacturer’s instructions (Difco). The detailed compositions of various media and supplements used in this study can be found in Supplementary Tables S1 and S2. The different carbon and nitrogen sources present in the two core media used in this study, 7H9T and DTA are listed in Table 1. For aerobic growth, mycobacterial cultures were harvested at logarithmic phase for RNA and protein extraction. For hypoxic growth, late logarithmic phase cultures of mycobacteria were shifted to hypoxia as previously described 22 followed by RNA isolation at 24 hr and 48 hr post hypoxia shift.
TABLE 1.
Carbon and Nitrogen sources in 7H9T and DTA media
| Broth base | Supplements | Acronym | Nitrogen source | Carbon source |
|---|---|---|---|---|
| Middlebrook 7H9 | 7H9-ADSa, Tween 80 | 7H9T | ammonium sulfate, L-glutamate and ferric ammonium citrate | glucose |
| Dubos | Dubos-ADSa | DTA | pancreatic digest of casein, L- asparagine and ferric ammonium citrate | glucose |
Compositions of ADS supplements for 7H9 and Dubos bases can be found in Supplementary Table S2.
RNA isolation and quantitative Reverse Transcriptase-PCR (qRT-PCR)
RNA was isolated from M. tuberculosis cultures using TRI reagent (Ambion Inc., TX) according to the manufacturer’s protocol. Briefly, cells were lysed using a bead beater in a 1:1 mixture of 0.1 mm zirconium-silica beads and TRI reagent, followed by chloroform extraction. RNA was reverse transcribed to cDNA that served as template for amplification by gene-specific primers (Supplementary Table S3). The 16S rRNA gene was used for normalizing expression before calculating the fold change between test and control samples. Statistical significance between means from three independent experiments was determined with the one-way variance test ANOVA using GraphPad Prism 5.0 (GraphPad software, CA).
Protein isolation and Western analysis
Total protein from logarithmic phase cultures of M. tuberculosis was isolated by lysing cell pellets in a bead beater. Immunoblotting was done using rabbit anti-HspX and anti-DevR antibodies as previously described.22 Semi-quantitative measurement of DevR expression was done by densitometric analysis of the scanned blot using Alpha Ease FC software (Alpha Innotech Corp, CA).
RESULTS
Culture media modulate aerobic expression of the Rv3134c-devR-devS operon and DevR regulon
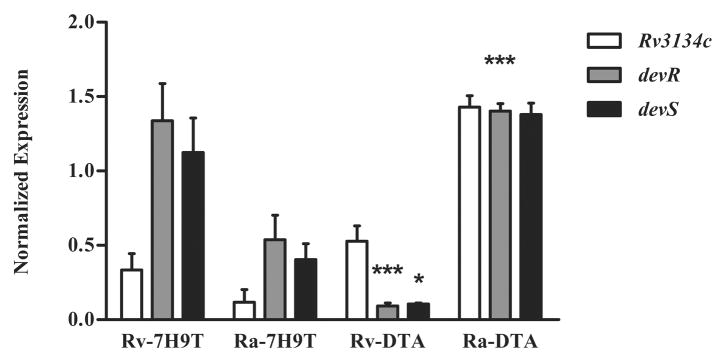
The devR and devS genes are co-transcribed as an operon with the upstream gene, Rv3134c.8 The expression of the Rv3134c-devR-devS operon in exponential cultures of H37Ra and H37Rv grown in various media was measured using qRT-PCR. No difference was observed in the pattern of aerobic expression of devR between H37Ra and H37Rv strains in 7H9T and 7H9TG medium (data not shown). Thus, all further comparisons of gene expression were performed between bacteria cultured in 7H9T and DTA media. qRT-PCR analyses of this operon revealed interesting differences between H37Ra and H37Rv strains (Figure 1). First, in 7H9T medium, devR operon expression was lower in H37Ra compared to H37Rv, as observed previously.8 Conversely, in DTA medium, the operon was significantly upregulated in H37Ra compared to H37Rv (P value < 0.001, Figure 1). Second, in H37Rv, switching culture medium from 7H9T to DTA was accompanied by a significant decrease in devR and devS gene expression (P value < 0.001 and 0.05, respectively) but not that of Rv3134c (Figure 1). On the basis of these observations, we hypothesized that media constituents can modulate aerobic expression of this operon in H37Ra and H37Rv. In subsequent experiments, we focused on characterizing the constitutive expression of devR-devS TCS in H37Ra cultivated in DTA medium.
Figure 1. Differential aerobic expression of Rv3134c-devR-devS operon in M. tuberculosis cultivated in 7H9T and DTA media.
qRT-PCR analyses of Rv3134c-devR-devS operon during exponential growth of H37Rv (Rv) and H37Ra (Ra) in 7H9T and DTA media. The normalized expression of these genes with respect to expression of 16S rRNA is presented as mean ± standard deviations (SD) of three independent experiments. *** represents P < 0.001 for the differences in expression of the operon between H37Ra and H37Rv in DTA medium. The decrease in the expression of devR (***, P < 0.001) and devS (*, P < 0.05) genes in H37Rv grown in DTA versus 7H9T medium is significant.
To analyze the effect of devR induction on target gene regulation during aerobic growth of H37Ra in DTA medium, we measured transcript levels for six genes in the DevR regulon namely, hspX, fdxA, narK2, Rv3130c, Rv1738 and Rv2626c. All of these are known to be strongly induced under hypoxia and exposure to NO.11, 14 Significant upregulation was observed for all 6 target genes in H37Ra but not in H37Rv grown in DTA medium (Figure 2A; black bars). In contrast, none of these genes was induced in H37Ra grown in 7H9T (Figure 2A; white bars). Overall, induction of the DevR regulon in aerobic cultures of H37Ra in DTA medium suggests that H37Ra but not H37Rv is responsive to a medium-specific signal.
Figure 2. Aerobic expression of DevR and a subset of sentinel DevR-regulated genes in M. tuberculosis grown in 7H9T and DTA media.
(A) Fold induction of devR, hspX, narK2, fdxA, Rv1738, Rv2626c, and Rv3130c genes in H37Ra grown aerobically in 7H9T (white bars) and DTA (black bars) media with respect to H37Rv (baseline expression set to 1.0). *** represents P < 0.001, for the differences in expression between H37Ra and H37Rv strains in DTA medium. (B) Western blot analyses of M. tuberculosis H37Ra and H37Rv grown in 7H9T (Lanes 1 and 2) and DTA (Lanes 3 and 4) media using anti-DevR and anti-HspX antibodies. Lane 5 is purified HspX protein. The levels of DevR in H37Ra cultivated in 7H9T (RaT) and DTA (RaDTA) media are reported as percentage expression with respect to H37Rv (RvT or RvDTA, fixed at 100%) based on densitometric measurement of band intensities (below Figure 2B).
Aerobic expression of DevR and α-crystallin-like protein HspX in M. tuberculosis
Western blotting analysis was performed to determine whether the elevated levels of devR and hspX transcripts in DTA-grown H37Ra were accompanied by an increase in protein levels. Based on densitometric measurement of the band intensities, DevR in H37Ra was ~ 13% lower in 7H9T (Figure 2B; Lane 1) while in DTA medium it was ~ 43% higher (Figure 2B; Lane 3) compared to H37Rv (100%) cultured in the respective media. Likewise, HspX was significantly induced in H37Ra grown in DTA medium (Figure 2B; Lane 3). Note that though DevR expression was observed in aerobic cultures of H37Ra and H37Rv grown in 7H9T and DTA media, HspX induction was evident only in H37Ra grown in DTA medium.
L-Asparagine and/or asparagine catabolism: a metabolic signal for DevR induction
To understand the media-specific effect on aerobic expression of the DevR regulon, media compositions were examined. The different carbon and nitrogen sources in 7H9T and DTA media are listed in Table 1. Assimilation of L-asparagine, the major nitrogen source in DTA medium, is significantly different in H37Ra and H37Rv.23, 24 We, therefore sought to determine if L-asparagine in DTA medium was responsible for the increased aerobic expression of the DevR regulon in H37Ra. Involvement of L-asparagine in the DevR regulon induction was established by three experiments.
First, M. tuberculosis L-asparaginase (AnsA) is stereospecific for its substrate L-asparagine although it can bind to D-asparagine.23 If L-asparagine utilization through AnsA plays a key role in induction of the DevR regulon, then competitive inhibition of AnsA by D-asparagine should abrogate the response. Accordingly, H37Ra was cultured in DTA medium supplemented with a 20-fold excess of D-asparagine (DTA+Dasn). qRT-PCR (Figure 3A) and western blot analyses (Figure 3B; Lanes 3 and 4) revealed that devR was not induced in DTA+Dasn medium. Likewise, HspX expression was significantly reduced (Figure 3B; Lanes 3 and 4). This implicated L-asparagine or asparagine metabolism in DevR regulon induction in DTA-grown H37Ra. Second, glycerol is reported to inhibit AnsA activity in mycobacteria25, 26, hence the effect of glycerol supplementation on aerobic expression of devR was assessed. In the presence of glycerol, devR induction was not observed in H37Ra or H37Rv (DTA+G, Figure 3A). Immunoblotting indicated minimal expression of DevR and HspX in H37Ra grown in DTA+G medium (Figure 3B; Lane 5). Third, in 7H9T medium supplemented with L-asparagine (7H9T+Lasn), the DevR regulon was significantly upregulated in aerobic cultures of H37Ra (Figure 3C). No upregulation was observed in H37Rv (data not shown). Taken together, these data suggest a role for L-asparagine in aerobic induction of the DevR regulon in H37Ra.
Figure 3. Asparagine: A metabolic signal for aerobic induction of the DevR regulon in M. tuberculosis H37Ra.
(A) devR expression in H37Rv (Rv, black bars), H37Ra (Ra, gray bars) grown in DTA medium with (hatched lines, DTA+G) or without glycerol (solid, DTA) and in H37Ra grown in DTA supplemented with 300 mM D-asparagine (vertical lines, DTA+Dasn). *** represents P < 0.001. (B) Western blot analyses of DevR and HspX expression in 30 μg (Lanes 1 and 3) and 45 μg (Lanes 2 and 4) of H37Ra cell lysates prepared from DTA and DTA+Dasn media respectively. Lane 5 represents cell lysate of H37Ra grown in DTA+G medium (C) Fold induction of devR, devS, Rv3134c, hspX, narK2 and Rv3130c genes in H37Ra grown in 7H9T medium with L-asparagine (7H9T+Lasn) with respect to 7H9T control (set at 1.0).
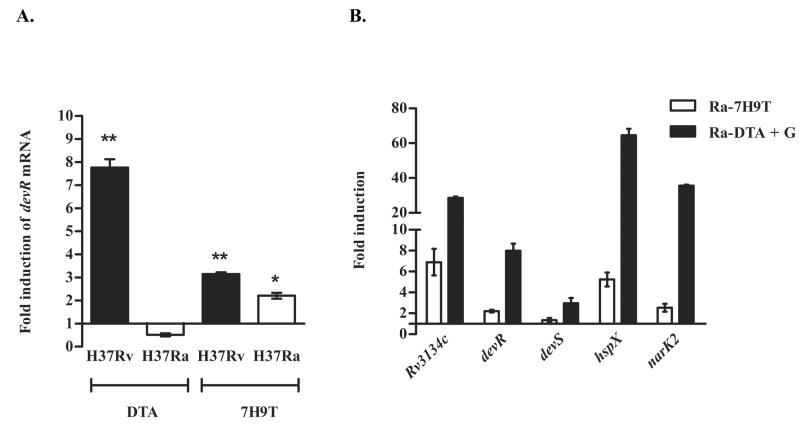
Hypoxic response of M. tuberculosis H37Ra
The hypoxic response of M. tuberculosis was assessed in cultures subjected to 24 hr hypoxic stress as described in Methods. In DTA medium, devR transcripts were induced in H37Rv, however no induction was observed in H37Ra (Figure 4A). Interestingly, in 7H9T medium, hypoxia-induced transcription of devR was similar in both H37Ra and H37Rv (Figure 4A). Similar results were obtained at 48 hr time point (data not shown). Furthermore, when H37Ra grown in DTA+G medium was subjected to hypoxia, a robust induction of DevR regulon genes was observed (~ 27-, 8- and 3-fold induction of Rv3134c, devR and devS genes, respectively; P < 0.001, Figure 4B). Our results indicate that glycerol is inhibitory to devR induction in DTA-grown H37Ra under aerobic conditions (Figure 3A) but not during hypoxia. The apparent lack of devR induction in hypoxia-adapted H37Ra cultured in DTA medium is ascribed to constitutively high expression of devR transcripts under aerobic conditions (Figure 1) and not to a dysfunction of the TCS.
Figure 4. Hypoxic response of M. tuberculosis H37Rv and H37Ra.
Fold induction at 24 hr hypoxia of (A) devR expression in H37Rv (black bars) and H37Ra (white bars) cultured in DTA and 7H9T media and of (B) Rv3134c, devR, devS, hspX and narK2 genes in H37Ra grown in 7H9T (white bars) and DTA+G (black bars) media with respect to their aerobic controls (set at 1.0). **, * represents P < 0.01 and 0.05 respectively for devR induction in H37Rv and H37Ra in the respective media compared to their aerobic controls. The hypoxic induction of the DevR regulon in DTA+G media is significant (P < 0.001).
DISCUSSION
The DevR regulon of M. tuberculosis is activated by hypoxia, NO and CO.9–16 These signals are likely to be relevant for dormancy adaptation in vivo. Hence, it is of significant interest to understand the intricacies of the DevR-DevS regulatory cascade. The DevR-mediated hypoxia response has not been characterized in H37Ra. Since H37Ra is closely related to H37Rv, any aberration of the hypoxia response in H37Ra could contribute to its attenuation. Our findings here establish that DevR in H37Ra is a hypoxia-responsive regulator capable of activating downstream target genes in a medium-specific manner (Figure 4). The constitutive aerobic expression of devR in DTA (Figure 1) masked the hypoxic induction that was noted in DTA+G medium (Figure 4). While this manuscript was being written, Lee et al., reported upregulation of devR (dosR) transcripts under dormancy-like conditions in H37Ra 5, though media effects were not investigated.
A remarkable observation of the present study was the differential modulation of aerobic expression of Rv3134c-devR-devS operon in H37Ra and H37Rv in response to culture media (Figure 1). While switching from 7H9T to DTA medium, Rv3134c-devR-devS genes showed significantly high aerobic expression in H37Ra whereas a decrease in the level of devR-devS, but not Rv3134c transcripts, was noted in H37Rv (Figure 1). Transcriptional regulation of Rv3134c-devR-devS genes is characterized by the presence of multiple aerobic transcription start sites and hypoxia-inducible promoters 27, 28 that could potentially affect the expression of individual genes of this operon. Variations in transcriptional activity of this operon were recently observed in M. bovis BCG cultivated under aerobic, hypoxic and nutrient stress conditions.28
The observations made in the present study do not correlate with the findings of Gao et al.2 in which no differences were detected in the expression of Rv3134c-devR-devS genes in H37Ra cultured in various media. One possible explanation is their use of DTA medium containing glycerol (DTA+G), which was shown in the present study to abrogate devR gene induction. The devR-devS genes were originally identified on the basis of differential expression between H37Rv and H37Ra strains cultured in Kirchener’s and 7H9T media.7, 8 Those observations can now be understood based on the present findings: the low level of devR expression in H37Ra cultured in Kirchener’s medium can be attributed to the effect of glycerol on asparagine metabolism. Repression of the key enzyme, AnsA by glycerol has been suggested to occur in mycobacteria 25, 26 and shown in Pseudomonas aeroginosa.29 Although the underlying mechanism of glycerol-mediated inhibition of devR-devS aerobic expression is not understood, our observations raise questions about the global changes in gene expression by glycerol, a carbon source widely used in growth of mycobacteria.
Aerobic expression of a TCS normally responsive to hypoxia and NO, both absent during aerobic growth, suggested three possibilities: (1) that the cells were experiencing an overall artificial anaerobic environment in spite of surplus oxygen, (2) that conditions mimicking hypoxia were created by poor uptake of oxygen and/or (3) that generation of an endogenous signal distinct from oxygen was responsible for the induction of the TCS. The first two possibilities were ruled out in view of a robust growth of H37Ra in DTA medium and no up-regulation of cydA, a marker for anaerobic respiration30 (data not shown). High level of HspX in H37Ra cultivated in DTA medium in spite of basal level expression of DevR in cell lysates of H37Rv and H37Ra in 7H9T and DTA medium favored the third possibility (Figure 2B). As phosphorylation of DevR is required for hspX upregulation in M. tuberculosis,31 it is tempting to speculate that an inducing signal was generated during cultivation of H37Ra in DTA medium resulting in hspX induction. Further analysis of medium-specific induction implicated asparagine in DTA medium as a key component required for the aerobic activation of DevR-DevS TCS in H37Ra (Figure 3).
Much of the data regarding asparagine metabolism in mycobacteria comes from biochemical studies. The first step in utilization of asparagine by bacterial cells is its deamidation into aspartic acid and ammonia by L-asparaginase (AnsA). M. tuberculosis H37Ra and H37Rv possess AnsA activity having optimum pH of 9.0. A second AnsA activity with an optimum pH of 9.6 was reported exclusively in H37Ra.23 However, comparative analyses of the H37Ra and H37Rv genomes revealed the presence of one identical gene (ansA/MRA_1550/Rv1538c) encoding AnsA. Instances of a single gene giving rise to multiple isoenzymes exist in mycobacteria: for example, M. smegmatis ask gene encoding aspartokinase gives rise to three different isoenzymes.32 Whether AnsA in M. tuberculosis is subjected to differential regulation is yet to be explored. Additionally, in H37Ra but not H37Rv, an aspartotransferase enzyme catalyzing the conversion of asparagine to aspartohydroxamic acid 24 has been described. However, the genetic basis of such an activity is unknown.
While it is possible that the aforementioned differences in asparagine metabolism are responsible for the aerobic induction of the DevR regulon in H37Ra cultured in DTA medium, however a dysregulation of the Rv3134c-devR-devS operon in H37Ra cannot be excluded. Comparative genome analyses of H37Ra and H37Rv have revealed several mutations within coding and putative promoter regions in H37Ra.33 Of particular interest is a transversion (A-T) mutation in the putative promoter region of H37Ra sigC sigma factor (MRA_2083) that resulted in higher level sigC expression in cultures of H37Ra versus H37Rv.33 In H37Rv, putative promoter −10 and −35 elements having matches for SigC sigma factor were identified upstream of the transcriptional start sites of Rv3134c and hspX. 27, 31 Furthermore, hspX expression, along with three other members of the DevR regulon, namely, pfkB, Rv2028c and Rv2004c were repressed in an M. tuberculosis sigC strain 34 suggesting a possible role for SigC in DevR regulon expression. However, in the context of asparagine-mediated effects, further investigations are imperative to decipher the underlying link between DevR, asparagine metabolism and sigma factor C in H37Ra.
The constitutive expression of the devR-devS TCS in H37Ra grown in DTA medium underscores the importance of media constituents in defining the transcriptional signatures of the bacterium. These effects are particularly relevant in the present realm of microarray analyses, wherein use of different kinds of media by investigators complicates inter-laboratory comparisons of data. The differences observed in our studies when glycerol was used as an additional carbon source in DTA medium highlight this point. In view of our results several questions arise. First, what is the underlying basis of asparagine-mediated aerobic induction of a dormancy regulator? Second, how does this induction affect growth of H37Ra and third, why is this phenomenon not observed with H37Rv grown under similar conditions? We are currently addressing these questions and anticipate that these insights will strengthen our understanding of the complex mechanisms of signaling and virulence in mycobacteria.
Supplementary Material
Acknowledgments
This work was supported by US Public Health Service grant AI046428 from the National Institutes of Health awarded to J.E.C.-C. We are grateful to Richard F. Silver for the generous gift of M. tuberculosis H37Ra strain. Purified 16 kD HspX protein was obtained from Colorado State University as part of NIH, NIAID Contract No. HHSN266200400091C, entitled “Tuberculosis Vaccine Testing and Research Materials”. We are also thankful to Illah Joshi and Arpita Bose for providing the DevR and HspX antibodies respectively. We gratefully acknowledge Shelley Haydel for critical review of the manuscript and fellow colleagues for helpful suggestions and discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Steenken W, Jr, Oatway W, Jr, Petroff SA. Biological Studies of the Tubercle Bacillus: III. Dissociation and pathogenicity of the R and S variants of the human tubercle bacillus (H37) J Exp Med. 1934;60:515–40. doi: 10.1084/jem.60.4.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gao Q, Kripke K, Arinc Z, Voskuil MI, Small P. Comparative expression studies of a complex phenotype: cord formation in Mycobacterium tuberculosis. Tuberculosis. 2004;84:188–96. doi: 10.1016/j.tube.2003.12.013. [DOI] [PubMed] [Google Scholar]
- 3.Cardona PJ, Soto CY, Martin C, Giquel B, Agusti G, Andreu N, Guirado E, Sirakova T, Kolattukudy P, Julian E, Luquin M. Neutral-red reaction is related to virulence and cell wall methyl-branched lipids in Mycobacterium tuberculosis. Microbes Infect. 2006;8:183–90. doi: 10.1016/j.micinf.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 4.He XY, Zhuang YH, Zhang XG, Li GL. Comparative proteome analysis of culture supernatant proteins of Mycobacterium tuberculosis H37Rv and H37Ra. Microbes Infect. 2003;5:851–56. doi: 10.1016/s1286-4579(03)00179-5. [DOI] [PubMed] [Google Scholar]
- 5.Lee JS, Krause R, Schreiber J, Mollenkopf HJ, Kowall J, Stein R, Jeon BY, Kwak JY, Song MK, Patron JP, Jorg S, Roh K, Cho SN, Kaufmann SH. Mutation in the transcriptional regulator PhoP contributes to avirulence of Mycobacterium tuberculosis H37Ra strain. Cell Host Microbe. 2008;3:97–103. doi: 10.1016/j.chom.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 6.Rindi L, Lari N, Garzelli C. Search for genes potentially involved in Mycobacterium tuberculosis virulence by mRNA differential display. Biochem Biophys Res Commun. 1999;258:94–101. doi: 10.1006/bbrc.1999.0591. [DOI] [PubMed] [Google Scholar]
- 7.Kinger AK, Tyagi JS. Identification and cloning of genes differentially expressed in the virulent strain of Mycobacterium tuberculosis. Gene. 1993;131:113–17. doi: 10.1016/0378-1119(93)90678-v. [DOI] [PubMed] [Google Scholar]
- 8.Dasgupta N, Kapur V, Singh KK, Das TK, Sachdeva S, Jyothisri K, Tyagi JS. Characterization of a two-component system, devR-devS, of Mycobacterium tuberculosis. Tuber Lung Dis. 2000;80:141–59. doi: 10.1054/tuld.2000.0240. [DOI] [PubMed] [Google Scholar]
- 9.Muttucumaru DG, Roberts G, Hinds J, Stabler RA, Parish T. Gene expression profile of Mycobacterium tuberculosis in a non-replicating state. Tuberculosis. 2004;84:239–46. doi: 10.1016/j.tube.2003.12.006. [DOI] [PubMed] [Google Scholar]
- 10.Ohno H, Zhu G, Mohan VP, Chu D, Kohno S, Jacobs WR, Jr, Chan J. The effects of reactive nitrogen intermediates on gene expression in Mycobacterium tuberculosis. Cell Microbiol. 2003;5:637–48. doi: 10.1046/j.1462-5822.2003.00307.x. [DOI] [PubMed] [Google Scholar]
- 11.Park HD, Guinn KM, Harrell MI, Liao R, Voskuil MI, Tompa M, Schoolnik GK, Sherman DR. Rv3133c/dosR is a transcription factor that mediates the hypoxic response of Mycobacterium tuberculosis. Mol Microbiol. 2003;48:833–43. doi: 10.1046/j.1365-2958.2003.03474.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sherman DR, Voskuil MI, Schnappinger D, Liao R, Harrell MI, Schoolnik GK. Regulation of the Mycobacterium tuberculosis hypoxic response gene encoding alpha -crystallin. Proc Natl Acad Sci USA. 2001;98:7534–39. doi: 10.1073/pnas.121172498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Timm J, Post FA, Bekker LG, Walther GB, Wainwright HC, Manganelli R, Chan WT, Tsenova L, Gold B, Smith I, Kaplan G, McKinney JD. Differential expression of iron-, carbon-, and oxygen-responsive mycobacterial genes in the lungs of chronically infected mice and tuberculosis patients. Proc Natl Acad Sci USA. 2003;100:14321–26. doi: 10.1073/pnas.2436197100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Voskuil MI, Schnappinger D, Visconti KC, Harrell MI, Dolganov GM, Sherman DR, Schoolnik GK. Inhibition of respiration by nitric oxide induces a Mycobacterium tuberculosis dormancy program. J Exp Med. 2003;198:705–13. doi: 10.1084/jem.20030205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumar A, Deshane JS, Crossman DK, Bolisetty S, Yan BS, Kramnik I, Agarwal A, Steyn AJ. Heme oxygenase-1-derived carbon monoxide induces the Mycobacterium tuberculosis dormancy regulon. J Biol Chem. 2008;283:18032–9. doi: 10.1074/jbc.M802274200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shiloh MU, Manzanillo P, Cox JS. Mycobacterium tuberculosis senses host-derived carbon monoxide during macrophage infection. Cell Host Microbe. 2008;3:323–30. doi: 10.1016/j.chom.2008.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sharma D, Tyagi JS. The value of comparative genomics in understanding mycobacterial virulence: Mycobacterium tuberculosis H37Ra genome sequencing -a worthwhile endeavour. J Biosci. 2007;32:185–89. doi: 10.1007/s12038-007-0018-z. [DOI] [PubMed] [Google Scholar]
- 18.Wayne LG. Synchronized replication of Mycobacterium tuberculosis. Infect Immun. 1977;17:528–30. doi: 10.1128/iai.17.3.528-530.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wayne LG, Hayes LG. An in vitro model for sequential study of shiftdown of Mycobacterium tuberculosis through two stages of nonreplicating persistence. Infect Immun. 1996;64:2062–69. doi: 10.1128/iai.64.6.2062-2069.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Voskuil MI, Visconti KC, Schoolnik GK. Mycobacterium tuberculosis gene expression during adaptation to stationary phase and low-oxygen dormancy. Tuberculosis. 2004;84:218–27. doi: 10.1016/j.tube.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 21.Betts JC, Lukey PT, Robb LC, McAdam RA, Duncan K. Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol Microbiol. 2002;43:717–31. doi: 10.1046/j.1365-2958.2002.02779.x. [DOI] [PubMed] [Google Scholar]
- 22.Saini DK, Malhotra V, Dey D, Pant N, Das TK, Tyagi JS. DevR-DevS is a bonafide two-component system of Mycobacterium tuberculosis that is hypoxia-responsive in the absence of the DNA-binding domain of DevR. Microbiology. 2004;150:865–75. doi: 10.1099/mic.0.26218-0. [DOI] [PubMed] [Google Scholar]
- 23.Jayaram HN, Ramakrishnan R, Vaidyanathan CS. L-asparaginases from Mycobacterium tuberculosis strains H37Rv and H37Ra. Arch Biochem Biophys. 1968;126:165–74. doi: 10.1016/0003-9861(68)90570-5. [DOI] [PubMed] [Google Scholar]
- 24.Jayaram HN, Ramakrishnan T, Vaidyanathan CS. Aspartotransferase from Mycobacterium tuberculosis H37Ra. Ind J Biosci. 1969;6:106–10. [Google Scholar]
- 25.Segal W, Edwards BS. Carbohydrate and amino acid metabolism and control. In: Kubica GP, Wayne LG, editors. The Mycobacteria: A Source Book. New York and Basel: Microbiology series; 1984. pp. 575–94. [Google Scholar]
- 26.Masood R, Sharma YK, Venkitasubramanian TA. Metabolism of Mycobacteria. J Biosci. 1985;7:421–31. [Google Scholar]
- 27.Bagchi G, Chauhan S, Sharma D, Tyagi JS. Transcription and autoregulation of the Rv3134c-devR-devS operon of Mycobacterium tuberculosis. Microbiology. 2005;151:4045–53. doi: 10.1099/mic.0.28333-0. [DOI] [PubMed] [Google Scholar]
- 28.Rodriguez JG, Burbano CS, Nunez C, Gonzalez CE, Zambrano MM, Garcia MJ, Del Portillo P. Rv3134c/devR/devS operon of Mycobacterium bovis BCG is differentially transcribed under “in vitro” stress conditions. Tuberculosis. 2008;88:273–82. doi: 10.1016/j.tube.2007.11.011. [DOI] [PubMed] [Google Scholar]
- 29.Geckil H, Gencer S, Ates B, Ozer U, Uckun M, Yilmaz I. Effect of Vitreoscilla hemoglobin on production of a chemotherapeutic enzyme, L-asparaginase, by Pseudomonas aeruginosa. Biotechnol J. 2006;1:203–08. doi: 10.1002/biot.200500048. [DOI] [PubMed] [Google Scholar]
- 30.Shi L, Sohaskey CD, Kana BD, Dawes S, North RJ, Mizrahi V, Gennaro ML. Changes in energy metabolism of Mycobacterium tuberculosis in mouse lung and under in vitro conditions affecting aerobic respiration. Proc Natl Acad Sci USA. 2005;102:15629–34. doi: 10.1073/pnas.0507850102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chauhan S, Tyagi JS. Cooperative binding of phosphorylated DevR to upstream sites is necessary and sufficient for activation of the Rv3134c-devRS operon in Mycobacterium tuberculosis: Implication in the induction of DevR target genes. J Bacteriol. 2008;190:4301–12. doi: 10.1128/JB.01308-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sritharan V, Wheeler PR, Ratledge C. Metabolism of aspartate in Mycobacterium smegmatis. Eur J Biochem. 1989;180:587–93. doi: 10.1111/j.1432-1033.1989.tb14685.x. [DOI] [PubMed] [Google Scholar]
- 33.Zheng H, Lu L, Wang B, Pu S, Zhang X, Zhu G, Shi W, Zhang L, Wang H, Wang S, Zhao G, Zhang Y. Genetic basis of virulence attenuation revealed by comparative genomic analysis of Mycobacterium tuberculosis strain H37Ra versus H37Rv. PLoS ONE. 2008;3(6):e2375. doi: 10.1371/journal.pone.0002375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun R, Converse PJ, Ko C, Tyagi S, Morrison NE, Bishai WR. Mycobacterium tuberculosis ECF sigma factor sigC is required for lethality in mice and for the conditional expression of a defined gene set. Mol Microbiol. 2004;52:25–38. doi: 10.1111/j.1365-2958.2003.03958.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.