Abstract
A family of RNA m5C methyl transferases (MTases) containing over 55 members in eight subfamilies has been identified recently by an iterative search of the genomic sequence databases by using the known 16S rRNA m5C 967 MTase, Fmu, as an initial probe. The RNA m5C MTase family contained sequence motifs that were highly homologous to motifs in the DNA m5C MTases, including the ProCys sequence that contains the essential Cys catalyst of the functionally similar DNA-modifying enzymes; it was reasonable to assign the Cys nucleophile to be that in the conserved ProCys. The family also contained an additional conserved Cys residue that aligns with the nucleophilic catalyst in m5U54 tRNA MTase. Surprisingly, the mutant of the putative Cys catalyst in the ProCys sequence was active and formed a covalent complex with 5-fluorocytosine-containing RNA, whereas the mutant at the other conserved Cys was inactive and unable to form the complex. Thus, notwithstanding the highly homologous sequences and similar functions, the RNA m5C MTase uses a different Cys as a catalytic nucleophile than the DNA m5C MTases. The catalytic Cys seems to be determined, not by the target base that is modified, but by whether the substrate is DNA or RNA. The function of the conserved ProCys sequence in the RNA m5C MTases remains unknown.
Keywords: fluorocytosine, S-adenosyl-l-methionine, Fmu, ProCys
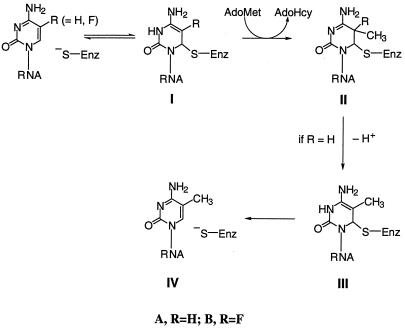
Enzymes that catalyze 5-methylation (or hydroxymethylation) of pyrimidines use the thiol of a Cys residue to attack the 6 position of the heterocycle to activate the 5 position toward the one-carbon transfer (1, 2). Scheme S1 depicts the mechanism for methylation of cytosine nucleotides. Subsequent to one-carbon transfer, the 5-proton of the 5,6-dihydropyrimidine intermediate IIA is removed, and β-elimination provides the product and free enzyme. The covalent 5,6-dihydropyrimidine intermediate IIA has been identified by biochemical studies as well as by structures of stable covalent complexes formed between enzymes and 5-fluoropyrimidine substrate analogs that act as mechanism-based inhibitors (e.g., Scheme S1, IIB). In the latter, the stable carbon–fluorine bond in IIB prevents β-elimination of the enzyme and hence provides stability to the complexes.
Scheme 1.
Proposed mechanism for methylation of cytosine nucleotides.
The Escherichia coli fmu gene product Fmu was shown recently to be the 16S rRNA m5C967 methyl transferase (MTase; ref. 3). Shortly thereafter, use of the Fmu sequence as a probe to search available genomic databases revealed a family of more than 55 putative RNA m5C MTases (4).
Members of the RNA m5C MTase family contain six signature motifs that are highly homologous to motifs in the DNA m5C MTases in which functions have been assigned from structural studies (5, 6). One of the RNA m5C MTase sequence motifs, designated motif IV, has the completely conserved ProCys (residues 324–325 in Fmu) that contains the Cys catalyst in most enzymes that transfer one-carbon units to the 5 position of pyrimidines; these include DNA m5C MTase, dCMP hydroxymethylase, thymidylate synthase, and dUMP hydroxymethylase. For such enzymes, the only reported case of the catalytic Cys not being contained within a conserved ProCys dipeptide is with m5U54 tRNA MTase (7). This enzyme does not contain the ProCys sequence and uses an unrelated Cys residue in a SerCys sequence of motif VI as the catalytic nucleophile (8). In the DNA m5C MTase family, motif VI is associated with binding to the substrate Cyt via a conserved Glu that is also found in the RNA m5C MTases. Interestingly, the RNA m5C MTase family, but not the DNA m5C MTase family, also contained the second conserved Cys residue in motif VI (375Cys in Fmu). Because all of the RNA m5C MTases contained the ProCys dipeptide in motif IV and were highly homologous to the well characterized, functionally similar DNA m5C MTases, it was reasonable to assign the Cys nucleophile to be that in the conserved ProCys of motif IV (4).
In the present work, we show that, contrary to our expectations, the Cys of the ProCys dipeptide in Fmu is not the catalytic nucleophile in RNA m5C methylation; rather, the conserved Cys in motif VI serves this function.
Materials and Methods
Materials.
Plasmids pWK1 and pWK1.3 used for preparation of E. coli 16S rRNA and the 56-mer corresponding to nucleotides 927–982 of 16S rRNA, respectively, have been reported (3). T7 RNA polymerase was prepared and purified as described (9). [3H-Me]S-Adenosyl-l-methionine (AdoMet) (79 Ci/mmol; 1 Ci = 37 GBq) was from Amersham Pharmacia, and 5-fluorocytidine (FCyd) was from ICN. All other materials were the highest purity available from commercial sources.
5-Fluorocytidine-5′-Triphosphate (FCTP).
FCyd (400 mg, 1.5 mmol) was converted to FCTP as described for other nucleosides (10). The reaction mixture was quenched with 40 ml of 0.2 M NH4HCO3 and adjusted to pH 7.0 with 0.5 M NaOH. The solution was extracted with 5 × 50 ml of CH2Cl2, and FCTP was purified by DE52 (Whatman) column chromatography (2.5 cm × 20 cm) by using a 1.0-liter linear gradient of 50–200 mM NH4HCO3; the eluant was monitored at 260 nm and collected in 8-ml fractions. Fractions containing FCTP eluting at ≈200 mM NH4HCO3 were evaporated in vacuo, and the residue was dissolved in 2 ml of 100 mM Tris⋅HCl (pH 8.0); concentration measurements assumed the same extinction coefficient as that for FCyd (ref. 11; ɛ = 8,060 at 281 nm; pH 7.0) and indicated a yield of 0.13 mmol (8.5% from FCyd). Electrospray MS gave an m/z ratio of 500 (M-H) as predicted. 1H NMR (400 MHz, 2H2O) gave peaks at δ 8.08 (d, 1 H, J = 6.8 Hz, 6-H), 5.92 (m, 1 H, 1′-H), and 4.20–4.40 (m, 5 H, ribose). The UV spectrum (in 10 mM KPi, pH 7.0) gave λmax values of 244 and 284 nm.
RNA Preparation.
Unmodified 16S rRNA, the 56-mer corresponding to nucleotides 927–982 of 16S rRNA, and 5-fluorocytosine (FC) RNA 56-mer were synthesized by runoff transcription of Bsu36I-linearized pWK1 or linearized pWK1.3 as described (3). For FC RNAs, FCTP replaced CTP. Purification and quantitation of RNA concentration were performed as described (12).
Mutagenesis.
pET-FmuC325A and pET-FmuC375A were prepared by mutagenesis of pET-Fmu with the QuickChange site-directed mutagenesis kit (Stratagene) by using two complementary primers: 5′-GATGCGCCTGCTAGCGCAACCGGTGTGATTCG for pET-FmuC325A and 5′-GGTCTATGCCACCGCTAGCGTGTTACCGG for pET-FmuC375A. In each, an Ala codon GCT (bold face) replaced the wild-type TGT, and a silent mutation was introduced at the next codon to create a diagnostic NheI site (underlined). Clones carrying mutant plasmids were identified by restriction enzyme digestion, and DNA sequences were confirmed.
Purification of Fmu Mutants.
Protein expression and purification of Fmu C325A and C375A were as described for Fmu (3), with addition of hydroxyapatite chromatography. Cells from 1-liter cultures were processed through DEAE-Sepharose chromatography (3), and fractions containing Fmu were loaded on a hydroxyapatite column (2 × 5 cm) equilibrated with buffer B [10 mM KPi, pH 6.8/0.5 mM EDTA/5 mM DTT/10% (vol/vol) glycerol]. The column was washed with 250 ml of buffer B and eluted with a 400-ml linear gradient of 0–0.5 M KPi in buffer B. Fractions containing Fmu mutants were pooled, concentrated, and desalted with a Centriprep concentrator (Amicon, Beverly, MA).
Enzyme Assays.
The methylation assay was performed as described (3). For 16S rRNA, 2 μM RNA, 20 μM [3H-Me]AdoMet (7.9 Ci/mmol), and 0.25 μM Fmu were used. For the 56-mer corresponding to nucleotides 927–982 of 16S rRNA, 20 μM RNA, 50 μM [3H-Me]AdoMet (4.0 Ci/mmol), and 0.5 μM were used.
Binding Assays.
The 56-mer corresponding to nucleotides 927–982 of 16S rRNA was labeled at the 3′ end with (5′-32P)pCp by using T4 RNA ligase (13) and purified by 7 M urea/10% PAGE. Mixtures (20 μl) containing 5 nM 56-mer (8 × 103 cpm) and varying concentrations (5–500 nM) of Fmu, Fmu-C325A, or Fmu-C375A in methylation buffer were incubated at 15°C for 60 min and assayed for protein–RNA complexes by nitrocellulose filtration with filtration efficiency of 50% (14). The apparent dissociation constants (Kd) for the 56-mer obtained by a nonlinear least-squares fit of the data to nonlinear least-squares fit of the data to RNAbound/RNAtotal = 1/(1 + Kapp/Etotal) (14) were 0.22 μM for Fmu, 0.20 μM for Fmu C325A, and 0.27 μM for Fmu-375A.
SDS/PAGE of Fmu RNA CH3 Complexes.
A solution (10 μl) containing 20 μM 56-mer FC RNA (nucleotides 927–982 of 16S rRNA), 50 μM AdoMet (or [3H-Me]AdoMet at 4.0 Ci/mmol), 1 unit/ml RNase inhibitor, and 2 μM enzyme in methylation buffer was incubated at room temperature for 30 min, denatured in boiling water with 2× loading buffer for 10 min, and then electrophoresed on SDS/12% PAGE with a 4% stacking gel. Gels were stained for protein with Coomassie blue. For radioactivity detection, gels were soaked in Amplify (Amersham Pharmacia) for 15 min, dried under vacuum, and analyzed by exposure to Biomax films (Kodak).
Results and Discussion
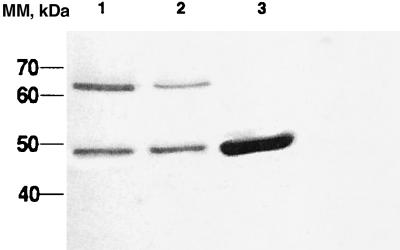
We have previously shown that Fmu catalyzes the AdoMet-dependent methylation at C967 of in vitro synthesized 16S rRNA and, albeit at reduced rates, smaller analogs of 16S rRNA such as the 56-mer corresponding to nucleotides 927–982 of 16S rRNA containing the target Cyd residue (3). Herein, we show that, in the presence of AdoMet, Fmu forms a covalent adduct with the 56-mer 16S rRNA substrate analog that contains FC in place of C residues. Evidence for the covalent protein–RNA complex is as follows. The complex formed is stable on heating in SDS and on SDS/PAGE (Fig. 1). On SDS/PAGE, the complex migrates more slowly (63 kDa) than the free enzyme (47 kDa) and can be visualized after staining protein with Coomassie blue or RNA with ethidium bromide. On treatment with nuclease P1, the mobility of the complex increases to 55 kDa, consistent with partial digestion of the RNA in the complex. Further, the formation of the complex requires AdoMet and is labeled with tritium when [3H-Me]AdoMet is used as methyl donor (Fig. 1). Together with what is known of the reaction of 5-fluorinated substrates of analogous enzymes such as m5C DNA MTases and thymidylate synthase, these data indicate that the structure of the Fmu FC RNA covalent complex covalent is as shown in IIB, Scheme S1. Here, there is covalent adduct formation between a thiol of the enzyme and carbon-6 of the target FC residue as well as methylation of carbon 5 of the pyrimidine.
Figure 1.
SDS/PAGE of Fmu and its complexes with FC RNA. (A) Coomassie-blue-stained gels of Fmu and FC RNA (lane 1); Fmu, FC RNA, and [3H-Me]AdoMet (lane 2); and nuclease P1 digest of covalent complex from lane 2 (lane 3). (B) Autoradiogram of the same gel. MM, molecular mass.
Based on analogy with the well studied DNA m5C MTases, 325Cys of Fmu was the most likely candidate to serve as the catalytic nucleophile; however, this expectation was unproven, and there is a second conserved Cys at residue 375 that is completely conserved in RNA m5C MTases. To identify the catalytic Cys of Fmu and assign function or lack thereof of the second conserved Cys, we prepared the Cys-to-Ala mutations at each of the two conserved Cys residues—the Cys residue of the 324ProCys dipeptide in motif IV and that in the 374ThrCys dipeptide of motif VI. The two mutants, as well as Fmu, were shown to bind to the 56-mer 16S rRNA analog with Kd values of 0.20–0.27 μM by using a direct binding assay (Table 1), indicating that no gross alteration of structure resulted from the mutations. Unexpectedly, we found that the Cys325Ala mutant was catalytically active, showing one-third to one-fourth the activity of the wild-type enzyme (Table 1). The Cys325Ala mutant also formed a stable AdoMet-dependent covalent complex with the 56-mer RNA containing FC in place of C (Fig. 2). In contrast, the Cys375Ala mutant was catalytically inactive on both full-length and 56-mer substrates (Table 1) and did not form a covalent complex with the 56-mer RNA substrate analog containing FC in place of C (Fig. 2). To ensure that no sample mix-up was made, each of the mutants was prepared in isolation of the other; for each, the DNA sequence was again verified, and the protein was isolated and reanalyzed; the results were as described above.
Table 1.
Activity and binding of Fmu and mutants
| Fmu | Activity, nmol⋅min−1⋅mg−1
|
Kd, μM 56-mer | |
|---|---|---|---|
| 16S RNA | 56-mer | ||
| Wild type | 37.2 | 75.1 | 0.22 |
| C325A | 12.8 | 18.9 | 0.20 |
| C375A | <0.1 | <0.1 | 0.27 |
Substrates and ligands were in vitro synthesized 16S rRNA and the 56-mer corresponding to nucleotides 927–982 of 16S rRNA.
Figure 2.
SDS/PAGE of FC RNA complexed to wild-type Fmu (lane 1), Fmu C325A (lane 2), and Fmu C375A (lane 3). Proteins were visualized by staining with Coomassie blue. MM, molecular mass.
Thus, unlike the m5C DNA MTases that use the ProCys thiol in motif IV as a catalyst, the m5C RNA MTase Fmu uses the thiol of the conserved 375Cys in motif VI as the catalyst. Interestingly, although 375Cys has no counterpart in the m5C DNA MTases, it is completely conserved in the RNA m5U MTases (8) and corresponds to the nucleophilic catalyst in m5U54 tRNA MTase (7). In effect, the m5C RNA MTases represent hybrids of the m5C DNA MTases and the m5U RNA MTases in that they contain the conserved Cys of motif IV found in the former and the conserved Cys of motif VI found in the latter (Fig. 3). However, the amino acid sequences in motifs VI of the RNA and DNA m5C MTases cannot be aligned adequately to map the catalytic 375Cys of Fmu to a known crystal structure. For example, when motif VI of Fmu is aligned to that of mHhaI without insertions or deletions, the 375Cys incorrectly aligns to 113Pro of HhaI, which is some 24 Å away from its active site Cys. Assuming that other RNA m5C MTases are as Fmu, we conclude that the Cys nucleophile used for m5C methylation of nucleic acids is not universally contained within the ProCys sequence of motif IV but rather is determined by whether the substrate is DNA or RNA. That is, for DNA substrates, the catalyst is the Cys residue of the conserved ProCys dipeptide in motif IV, whereas, for RNA substrates, it is the conserved Cys in motif VI.
Figure 3.
Sequence motifs of representative DNA and RNA MTases; bold face residues are conserved in subfamilies. TrmA is m5U54 tRNA MTase of E. coli. Fmu-ecoli, Fmu-bacsu, and Fmu-haein are Fmu proteins from E. coli, Bacillus subtilis, and Haemophilus influenzae, respectively. Met2-ecoli and mtbf-bacsu are two DNA m5C MTases from E. coli and B. subtilis.
It seems unlikely that the ProCys dipeptide of the m5C RNA MTases would be completely conserved if it did not have some important role. Interestingly, one of the putative m5C RNA MTases, Nop2p, is also an essential nucleolar protein of Saccharomyces cerevisiae involved in large ribosomal subunit assembly (15). Mutation of the motif IV ProCys of Nop2p to ProAla results in loss of cell viability, but mutation of the Cys in motif VI does not (16). In context of what has been described herein, this result suggests that the putative RNA methylation function of Nop2p is not essential, whereas that of the ProCys sequence is. Further, ProCys motifs are essential in some nucleotide-modifying enzymes that do not catalyze one-carbon transfers to pyrimidines. These include double-stranded RNA adenosine deaminases (17), the RNA Cyt deaminase responsible for apolipoprotein B RNA editing activity (18), and the Ada suicide DNA-repair protein from E. coli and related proteins (19). It seems reasonable to conclude that the ProCys dipeptide is sufficient but not necessary for methylation of pyrimidines in nucleic acids and that cysteines within conserved ProCys sequences may be involved in other important as-yet unidentified functions.
Acknowledgments
We thank Dr. X. R. Gu for performing the binding assay. This work was supported by U.S. Public Health Service Grant GM51232 (to D.V.S.) from the National Institutes of Health.
Abbreviations
- FC
5-fluorocytosine
- FCyd
5-fluorocytidine
- FCTP
5-fluorocytidine-5′-triphosphate
- MTase
methyl transferase
- AdoMet
S-adenosyl-l-methionine
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Ivanetich K M, Santi D V. Prog Nucleic Acid Res Mol Biol. 1992;42:127–156. doi: 10.1016/s0079-6603(08)60575-9. [DOI] [PubMed] [Google Scholar]
- 2.Carreras C W, Santi D V. Annu Rev Biochem. 1995;64:721–762. doi: 10.1146/annurev.bi.64.070195.003445. [DOI] [PubMed] [Google Scholar]
- 3.Gu X R, Gustafsson C, Ku J, Yu M, Santi D V. Biochemistry. 1999;38:4053–4057. doi: 10.1021/bi982364y. [DOI] [PubMed] [Google Scholar]
- 4.Reid R, Greene P J, Santi D V. Nucleic Acids Res. 1999;27:3138–3145. doi: 10.1093/nar/27.15.3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheng X. Annu Rev Biophys Biomol Struct. 1995;24:293–318. doi: 10.1146/annurev.bb.24.060195.001453. [DOI] [PubMed] [Google Scholar]
- 6.Cheng X D. Curr Opin Struct Biol. 1995;5:4–10. doi: 10.1016/0959-440x(95)80003-j. [DOI] [PubMed] [Google Scholar]
- 7.Kealey J T, Santi D V. Biochemistry. 1991;30:9724–9728. doi: 10.1021/bi00104a022. [DOI] [PubMed] [Google Scholar]
- 8.Gustafsson C, Reid R, Greene J G, Santi D V. Nucleic Acids Res. 1996;24:3756–3762. doi: 10.1093/nar/24.19.3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ellinger T, Ehricht R. BioTechniques. 1998;24:718–720. doi: 10.2144/98245bm03. [DOI] [PubMed] [Google Scholar]
- 10.Sakthivel K, Barbas C F., III Angew Chem Int Ed Engl. 1998;37:2872–2875. doi: 10.1002/(SICI)1521-3773(19981102)37:20<2872::AID-ANIE2872>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 11.Wempen I, Duschinsky R, Kaplan L, Fox J J. J Am Chem Soc. 1961;83:4755–4766. [Google Scholar]
- 12.Gu X R, Ofengand J, Santi D V. Biochemistry. 1994;33:2255–2261. doi: 10.1021/bi00174a036. [DOI] [PubMed] [Google Scholar]
- 13.England T E, Bruce A G, Uhlenbeck O C. Methods Enzymol. 1980;65:65–74. doi: 10.1016/s0076-6879(80)65011-3. [DOI] [PubMed] [Google Scholar]
- 14.Gu X R, Santi D V. Biochemistry. 1992;31:10295–10302. doi: 10.1021/bi00157a017. [DOI] [PubMed] [Google Scholar]
- 15.Hong B, Brockenbrough J S, Wu P, Aris J P. Mol Cell Biol. 1997;17:378–388. doi: 10.1128/mcb.17.1.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.King M, Ton D, Redman K L. Biochem J. 1999;337:29–35. [PMC free article] [PubMed] [Google Scholar]
- 17.Lai F, Drakas R, Nishikura K. J Biol Chem. 1995;270:17098–17105. doi: 10.1074/jbc.270.29.17098. [DOI] [PubMed] [Google Scholar]
- 18.Navaratnam N, Bhattacharya S, Fujino T, Patel D, Jarmuz A L, Scott J. Cell. 1995;81:187–195. doi: 10.1016/0092-8674(95)90328-3. [DOI] [PubMed] [Google Scholar]
- 19.Friedberg E C, Walker G C, Siede W. DNA Repair and Mutagenesis. Washington DC: Am. Soc. Microbiol.; 1995. [Google Scholar]