Abstract
Molecular controls over the development of the exceptional neuronal subtype diversity of the cerebral cortex are now beginning to be identified. The initial subtype fate decision early in the life of a neuron, and the malleability of this fate when the balance of key postmitotic signals is modified, reveals not only that a neuron is deterministically set on a general developmental path at its birth, but also that this program must be precisely executed during postmitotic differentiation. Here, we show that callosal projection neurons (CPN) and subcerebral projection neurons (subcerebral PN) in layer V of the neocortex share aspects of molecular identity after their birth that are progressively resolved during differentiation. The LIM-homeodomain–related genes Lmo4 and Clim1 are initially expressed by both CPN and subcerebral PN in layer V, and only during mid to late differentiation does expression of Lmo4 and Clim1 become largely segregated into distinct neuronal subtypes. This progressive postmitotic resolution of molecular identity reveals similarities and possibly shared evolutionary origin between layer V CPN and subcerebral PN, and provides insight into how and when these neuronal subtypes achieve their distinct identities during cortical development.
Keywords: callosal, corticospinal, cortical development, Lmo4, Clim1, Ldb2, subcerebral, subtype identity
Introduction
A striking feature of the extraordinary neuronal diversity of the 6-layered architecture of the mammalian neocortex is the precision and reproducibility with which this heterogeneity is organized and assembled. Birth-dating experiments revealed the inside-out pattern of corticogenesis, as early born neurons occupy deep layers and later-born neurons occupy superficial layers (Angevine and Sidman 1961; Rakic 1974). Laminar fate, however, does not dictate neuronal identity, as each layer contains many distinct neuronal subtypes, each with their own distinct molecular “biography” (Shaywitz and Melton 2005) of cell-intrinsic and cell-extrinsic controls. Though major progress has been made in identifying molecular signals that control broad aspects of neuronal development in the forebrain (Guillemot et al. 2006), only recently have specific critical controls over excitatory cortical neuron subtype specification and differentiation begun to be characterized (Supplementary Fig. 1) (Arlotta et al. 2005; Molyneaux et al. 2005, 2007; Chen, Rasin, et al. 2005; Chen, Schaevitz, et al. 2005; Alcamo et al. 2008; Britanova et al. 2008; Chen et al. 2008; Fishell & Hanashima 2008; Lai et al. 2008; Joshi et al. 2008; Kwan et al. 2008; Leone et al. 2008). The elucidation of such molecular programs responsible for the generation of distinct subtypes of neocortical neurons and their integration into long-distance and local circuitry provides substantial insight into cortical organization, function, and evolution, and promises to shed light onto how neuronal diversity arises throughout the central nervous system.
Layer V of the murine neocortex provides special insight into neuronal identity bifurcation decisions in that it contains 2 broad classes of projection neurons (PN), born at the same time from the same proliferative ventricular zone (VZ): 1) subcerebral PN, whose axons exit the cortex and descend through the internal capsule toward targets in the brainstem and spinal cord (this broad neuronal class includes multiple subtypes, such as corticospinal motor neurons (CSMN) located rostrally, and corticotectal projection neurons (CTPN) located caudally); and 2) callosal projection neurons (CPN), whose axons extend to the contralateral hemisphere (this broad neuronal class includes the subset of intratelencephalic corticostriatal projection neurons (CStrPNi), most of which project to both the contralateral cortex and the contralateral striatum). The developmental biology of all of these diverse neuronal subtypes is highly clinically relevant (Pasinelli and Brown 2006; Sohur et al. 2006; Minshew and Williams 2007; Molyneaux et al. 2007).
Previous work in our lab identified developmentally regulated genes that constitute a program of combinatorial molecular genetic controls over the specification and differentiation of subtypes of cortical PN. Retrogradely labeled CSMN, CTPN, and CPN were purified via fluorescence activated cell sorting, and their gene expression was compared by microarray at key stages of differentiation (embryonic day [E] 18, postnatal day [P] 3, P6, and P14) (Arlotta et al. 2005). From this work, a number of critical subcerebral PN developmental controls were characterized, including Ctip2 (Arlotta et al. 2005; Arlotta et al. 2008), Fezf2 (Chen, Rasin, et al. 2005; Chen, Schaevitz, et al. 2005; Molyneaux et al. 2005; Chen et al. 2008), Sox5 (Kwan et al. 2008; Lai et al. 2008), and Bhlhb5 (Joshi et al. 2008). This work also revealed a set of LIM-homeodomain (LIM-HD)–related genes that are expressed preferentially in either subcerebral PN or CPN. These genes are of significant interest, as LIM-HD transcription factors and their regulators, LIM only (LMO) proteins and cofactor-of-LIM proteins (CLIM, also known as Nuclear LIM interactor, NLI, or LIM domain binding, LDB, proteins) are centrally involved in multiple aspects of neuronal specification, differentiation, and axon pathfinding (Lundgren et al. 1995; Hobert and Westphal 2000; Kania et al. 2000; Segawa et al. 2001; Kashani et al. 2006; Lee et al. 2008). Clim1 (also known as Ldb2), exhibits predominantly subcerebral PN expression that decreases postnatally, whereas both Lmo4, whose gene product interacts with CLIM1 and CLIM2 (Kashani et al. 2006), and Lhx2, exhibit CPN-specific expression postnatally (Arlotta et al. 2005) (Supplementary Fig. 2A–C). In addition, Lmo4 is expressed in retrogradely labeled CPN and excluded from labeled subcerebral PN at late stages of neuronal differentiation (Arlotta et al. 2005) (Fig. 3D,E).
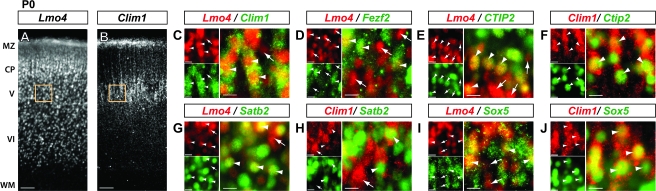
Figure 2.
By intermediate stages of layer V neuronal differentiation, Lmo4 and Clim1 expression becomes partially restricted to distinct cortical PN subtypes. Coronal mouse brain sections showing protein localization by immunocytochemistry. (C-J) Boxed regions in A and B are representative locations of high magnification images. (A) At P0, Lmo4 is expressed in subsets of neurons throughout all neocortical layers, in contrast to (B) Clim1 expression, which is restricted to cortical layer V. (C) Lmo4 and Clim1 are still extensively coexpressed at P0 (arrowheads), though neurons that only express Lmo4 are apparent (arrow). (D) Lmo4 continues to be coexpressed with Fezf2 at P0 (arrowheads), though many Lmo4-expressing neurons lack Fezf2 expression (arrows). (E and F) Lmo4 is extensively coexpressed with Ctip2 at P0 (E; arrowheads), with some exceptions (E; arrows), while Clim1 is extensively coexpressed with Ctip2 (F; arrowheads). (G and H) Lmo4 is extensively coexpressed with Satb2 at P0 (G; arrowheads), with rare exceptions (G; arrow), while Clim1 is expressed by many neurons, both coexpressed with (H; arrowheads) and expressed independently from Satb2 (H; arrow). (I and J) Lmo4 and Clim1 are both extensively coexpressed with Sox5 (arrowheads), though some Lmo4-expressing neurons do not express Sox5 (I; arrows). MZ: marginal zone; CP: cortical plate; V-VI: neocortical layers V-VI; WM: white matter. Scale bars: (A and B) 50mm, (C-J) 10mm.
Here, we find that cortical PN of layer V progressively adopt this distinct and complementary Lmo4 and Clim1 expression. During early differentiation, Lmo4 and Clim1 are expressed in both presumptive subcerebral PN and CPN in layer V, colocalizing with a number of key subtype identity-controlling transcription factors. During mid to late differentiation, this overlapping expression gradually diminishes, and Lmo4 and Clim1 adopt their largely cell type–specific expression pattern in CPN and subcerebral PN, respectively. Only small subsets of neurons maintain overlap. The progressive sharpening of molecular expression boundaries highlights important postmitotic events that occur during the sequential acquisition of distinct cortical PN subtype identities, and suggests important transcriptional regulatory roles of these LIM-HD–related genes during neuronal differentiation.
Materials and Methods
Mice
Clim1+/lacZ mice were obtained from MMRRC (University of California at Davis, CA; 011733-UCD) (GeneID: 16826). Fezf2+/lacZ mice were generated by Hirata et al. (2004) (Fezf2 GeneID 54713). Clim1+/lacZ mice were backcrossed into a pure C57BL/6 background. Fezf2+/lacZ mice were bred on a pure C57BL/6 background. The day of vaginal plug detection was designated as E0.5. The day of birth was designated as P0. All mouse studies were approved by the Massachusetts General Hospital IACUC, and were performed in accordance with institutional and federal guidelines.
Immunocytochemistry
Brains were fixed and stained using standard methods (Fricker-Gates et al. 2002). Primary antibodies and dilutions were used as follows: goat anti-LMO4, 1:100 (Santa Cruz Biotech, Santa Cruz, CA); rabbit anti-βgal, 1:3000 (MP Biomedicals, Solon, OH); goat anti-βgal, 1:2500 (Biogenesis, Brentwood, NH); rat anti-CTIP2, 1:1000 (Abcam, Cambridge, MA); rabbit anti-SOX5, 1:500 (Santa Cruz Biotech); goat anti-SOX5, 1:500 (Santa Cruz Biotech); rabbit anti-SATB2, 1:1000, gift of V. Tarabykin; mouse anti-SATB2, 1:100 (Abcam). Appropriate secondary antibodies were from the Molecular Probes Alexa series (Invitrogen, Carlsbad, CA).
Retrograde PN Labeling
CPN were labeled via injection of FluoroGold (FG) at P3 into the contralateral cortex, as previously described (Fricker-Gates et al. 2002; Arlotta et al. 2005). All injections were performed using a Vevo 770 ultrasound backscatter microscopy system (VisualSonics, Toronto, Canada) to visualize the injection site. Mice were perfused for analysis at P6.
Microscopy and Image Analysis
Tissue sections were viewed on a Nikon E1000 microscope equipped with an X-Cite 120 illuminator (EXFO, Ontario, Canada) and cooled CCD camera, and images were collected and analyzed with Volocity image analysis software (Version 4.0.1; Improvision Inc, Waltham, MA). Images were optimized for size, color, and contrast using Photoshop 7.0 (Adobe, San Jose, CA).
Results
During Early Differentiation, Lmo4 and Clim1 are Expressed in both Subcerebral PN and CPN in Layer V
By E15.5, VZ progenitors have already given rise to layer V PN, which have migrated radially into the cortical plate and begun early differentiation, whereas neurons that will occupy more superficial layers are concurrently being born. At this early stage of development, immunocytochemical analysis reveals that Lmo4 and Clim1 are both expressed postmitotically in the cortical plate (Fig. 1A,B) in an overlapping subset of neurons (Fig. 1C; arrowheads), whereas neither is expressed in the VZ. Heterozygote Clim1+/lacZ transgenic mice, in which Clim1-expressing neurons can be identified by βgal expression, seen as βgal-filled cytoplasm surrounding the nucleus, were used to characterize Clim1 expression.
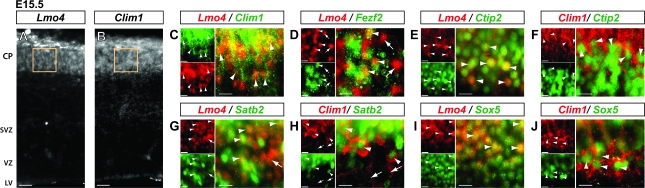
Figure 1.
At early stages of neocortical neuronal differentiation, Lmo4 and Clim1 are coexpressed in the CP. Coronal mouse brain sections showing protein localization by immunocytochemistry. Heterozygote Clim1+/lacZ transgenic mice, which express βgal in all Clim1-expressing neurons, were used to characterize Clim1 expression, which appears as βgal-filled cytoplasm surrounding the nucleus. (A and B) Lmo4 and Clim1 are broadly expressed in the CP at E15.5. (C–J) Boxed regions in A and B are representative locations of layer V high magnification images. (C) At E15.5, Lmo4 and Clim1 are expressed in overlapping neuronal populations in the CP (arrowheads). (D–F) Lmo4 and Clim1 are expressed at E15.5 in many neurons that express recently identified, central molecular controls over subcerebral PN development: Fezf2 (D; arrowheads), with some exceptions (D; arrows); and Ctip2 (E and F; arrowheads). (G and H) Lmo4 and Clim1 are also coexpressed at E15.5 with Satb2, a recently identified molecular control over CPN development (arrowheads), though some other neurons expressing Lmo4 or Clim1 do not express Satb2 (arrows). (I and J) Lmo4 and Clim1 are extensively coexpressed at E15.5 with Sox5, a recently identified, central molecular control over corticofugal PN development (arrowheads). CP: cortical plate; SVZ: subventricular zone; VZ: ventricular zone; LV: lateral ventricle. Scale bars: (A and B) 25 μm, (C–J) 10 μm.
Fezf2, a transcription factor that is required for the specification of subcerebral PN identity, is normally expressed by a subset of VZ progenitors and by differentiating subcerebral PN from the time of their birth (Chen, Rasin, et al. 2005; Chen, Schaevitz, et al. 2005; Molyneaux et al. 2005; Chen et al. 2008). Using heterozygote Fezf2+/lacZ transgenic mice, in which all neurons that express Fezf2 simultaneously express cytoplasmic βgal, we find that a subset of cortical plate neurons that express Fezf2 also express Lmo4 (Fig. 1D; arrowheads), indicating that at least a subpopulation of neurons destined to project subcerebrally coexpress Lmo4 early in their development. Previous work demonstrated that loss of Fezf2 expression leads to a total loss of strong Clim1 expression in layer V, due to the absence of subcerebral PN (Molyneaux et al. 2005) (Supplementary Fig. 2D), strongly suggesting that E15.5 Clim1-expressing neurons in the cortical plate coexpress Fezf2, and that Fezf2 function is required for subcerebral PN Clim1 expression.
Ctip2 is a transcription factor expressed postmitotically in subcerebral PN from the earliest stages of their development; it acts downstream of Fezf2 (Molyneaux et al. 2005; Chen et al. 2008), and is required for proper subcerebral PN differentiation, axonal fasciculation, and pathfinding (Arlotta et al. 2005; Lai et al. 2008). Although at E13.5 Ctip2 is briefly coexpressed with Satb2 in presumptive CPN in the CP, by E15.5 Ctip2 expression in these neurons is largely eliminated due to Satb2 repression (Alcamo et al. 2008; Britanova et al. 2008). We find that a large number of Ctip2-expressing neurons in layer V coexpress Lmo4 at E15.5, further indicating that many presumptive subcerebral PN express Lmo4 during early differentiation (Fig. 1E; arrowheads). In addition, we find that, as expected, Ctip2-expressing neurons coexpress Clim1, visible as CTIP2+ nuclei surrounded by βgal+ cytoplasm when the plane of imaging cuts through the nucleus (Fig. 1F; arrowheads).
Satb2 is a transcription factor expressed postmitotically in CPN from the earliest stages of their development, and is required for proper CPN specification and differentiation (Alcamo et al. 2008; Britanova et al. 2008). We find that, at E15.5, at least a subpopulation of Satb2-expressing presumptive CPN coexpress both Lmo4 (Fig. 1G; arrowheads) and Clim1 (Fig. 1H; arrowheads), indicating that, at this early stage, Clim1 is expressed in both presumptive CPN and subcerebral PN populations.
Sox5 is expressed postmitotically in corticofugal PN, including subcerebral PN in layer V, from their earliest stages of development, and is necessary for the appropriate sequential generation of the deep layer corticofugal PN subtypes (Lai et al. 2008). We find extensive coexpression of Sox5 and Lmo4 in the cortical plate at E15.5 (Fig. 1I; arrowheads), further indicating early expression of Lmo4 in subcerebral PN. Additionally, as expected, many Sox5-expressing presumptive subcerebral PN coexpress Clim1 (Fig. 1J; arrowheads).
At Intermediate Stages of Differentiation, Lmo4 and Clim1 Expression Partially Segregates into Distinct Cortical PN Subtypes
By P0, essentially all cortical PN have been born, the layers have more fully segregated, and PN subpopulations in layer V have entered intermediate stages of their differentiation, including axon elongation to their appropriate callosal or subcerebral targets. At this stage of development, Lmo4 is expressed in subsets of neurons broadly throughout the cortical layers (Fig. 2A), whereas Clim1 is restricted to neurons in layer V (Fig. 2B). Lmo4 and Clim1 continue to be extensively coexpressed in layer V (Fig. 2C; arrowheads), though some neurons express Lmo4 and not Clim1 (Fig. 2C; arrow), and vice versa, strongly suggesting that the expression of these genes has begun to be segregated into distinct neuronal subpopulations.
At P0, many neurons that express Fezf2 continue to coexpress Lmo4 (Fig. 2D; arrowheads), yet a large population expresses Lmo4 and not Fezf2 (Fig. 2D; arrows; Supplementary Fig. 3A), strongly suggesting that, by P0, Lmo4 expression has become partially excluded from the subcerebral PN population. As mentioned earlier, the dramatic reduction of Clim1 expression in layer V of Fezf2−/− mice (Supplementary Fig. 2D) indicates that the Clim1-expressing populations at P0 also express Fezf2.
At P0, we find that Ctip2 is expressed in many of the Lmo4-expressing neurons (Fig. 2E; arrowheads), and absent from many others (Fig. 2E; arrows; Supplementary Fig. 3B). As expected, at this stage, essentially all Ctip2-expressing neurons coexpress Clim1 (Fig. 2F; arrowheads; Supplementary Fig. 3C). Lmo4 and Satb2 are extensively coexpressed in CPN of layer V (Fig. 2G; arrowheads; Supplementary Fig. 3D), whereas Clim1 continues to be expressed in a subpopulation of Satb2-expressing neurons (Fig. 2H; arrowheads), though many Clim1-expressing neurons do not express Satb2 (Fig. 2H; arrow; Supplementary Fig. 3E). At P0, Lmo4 is still expressed in a large number of Sox5-expressing neurons (Fig. 2I; arrowheads), suggesting that many corticofugal PNs still express Lmo4, though many Lmo4-expressing neurons lack Sox5 expression (Fig. 2I; arrows). As expected, we find extensive coexpression of Clim1 and Sox5 in layer V subcerebral PN (Fig. 2J; arrowheads).
At Later Stages of Differentiation, Expression of Lmo4 and Clim1 Delineate Distinct Layer V PN Subtypes
By P6, layer V PN have more fully differentiated, and their axons have reached their respective targets. At this age, Lmo4 continues to be expressed in subsets of neurons broadly across cortical layers (Fig. 3A), whereas Clim1 expression remains confined to a subset of neurons in layer V (Supplementary Fig. 2D). We find that Lmo4 and Clim1 are expressed in largely distinct layer V neuronal populations (Fig. 3C; arrows), though some colocalization remains (Fig. 3C; arrowheads), indicating that, while largely segregated, a subpopulation of neurons maintains coexpression into later differentiation, possibly reflecting ongoing neuronal maturation at P6, or alternatively, highlighting a distinct subpopulation of layer V PN.
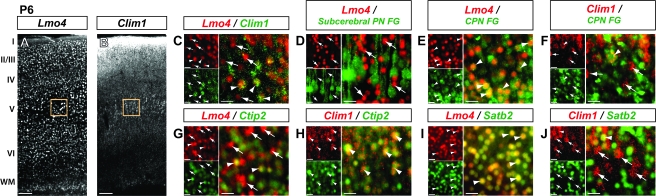
Figure 3.
By late stages of layer V neuronal differentiation, Lmo4 and Clim1 expression is largely segregated into CPN and subcerebral PN subtypes, respectively. Coronal mouse brain sections showing protein localization by immunocytochemistry. (A) At P6, Lmo4 is widely expressed in all cortical layers, whereas (B) Clim1 is exclusively expressed in cortical layer V (immunocytochemical background fluorescence is visible in upper layers). (C–J) Boxed regions in A and B are representative locations of high magnification images. (C) At P6, a large proportion of Lmo4-expressing neurons do not express Clim1 (arrows), although a subpopulation of Lmo4 and Clim1 coexpressing neurons exist (arrowheads). (D–F) Lmo4 expression is excluded from FG-labeled subcerebral PN (D; arrows), but it is extensively expressed in CPN (E; arrowheads), and Clim1 expression is absent from CPN (F; arrows), with rare but distinct exceptions (F; arrowhead). (G and H) Lmo4 is largely excluded from Ctip2-expressing neurons (G; arrows), with some exceptions (G; arrowheads), whereas Clim1 is extensively coexpressed with Ctip2 (H; arrowheads). (I and J) Lmo4-expressing neurons coexpress Satb2 (I; arrowheads), whereas Satb2 expression is largely excluded from Clim1-expressing neurons (J; arrows), with occasional exceptions (J; arrowhead). (D) Adapted from (Arlotta et al. 2005). I–VI: neocortical layers I–VI; WM: white matter. Scale bars: (A and B) 100 μm, (C–J) 20 μm.
Because axonal projections have reached their destinations by P6, analysis of connectivity can be performed via retrograde labeling from axonal targets. Previous FG retrograde labeling experiments from the spinal cord demonstrated that Lmo4 expression is excluded from subcerebral PN (Arlotta et al. 2005) (Fig. 3D; arrows). By retrogradely labeling from the contralateral cortex at P3 and examining the cortex at P6, we find that nearly all Lmo4-expressing neurons are CPN (Fig. 3E; arrowheads), whereas nearly all CPN do not express Clim1, or express it at very low levels, with rare but distinct exceptions (Fig. 3F; arrowhead).
At this age, Lmo4 expression is excluded from the majority of Ctip2-expressing neurons (Fig. 3G; arrows; Supplementary Fig. 3F), whereas Clim1 is expressed in nearly all Ctip2-expressing neurons (Fig. 3H; arrowheads; Supplementary Fig. 3G). Essentially all Satb2-expressing neurons of layer V express Lmo4 (Fig. 3I; arrowheads; Supplementary Fig. 3H), whereas Satb2 and Clim1 expression is mostly mutually exclusive (Fig. 3J; arrows; Supplementary Fig. 3I), with rare exceptions (Fig. 3J; arrowhead). By P6, Sox5 no longer exhibits specific corticofugal PN expression (Lai et al. 2008) and was, therefore, not examined.
Taken together, our data verify our previous microarray analysis identifying Lmo4 and Clim1 expression late in differentiation as essentially confined to CPN and subcerebral PN, respectively. However, we find this not to be the case earlier in differentiation, when presumptive CPN and subcerebral PN express both Lmo4 and Clim1, demonstrating that cortical PN molecular identity is progressively refined throughout embryonic and postnatal neuronal differentiation.
Discussion
The postmitotic acquisition and execution of neuronal identity is an emerging theme in cortical development (Alcamo et al. 2008; Britanova et al. 2008; Fishell and Hanashima 2008; Joshi et al. 2008; Lai et al. 2008). Although cortical neuron subtypes acquire much of their prospective identity at the time of their birth (McConnell and Kaznowski 1991; Molyneaux et al. 2007), key postmitotic molecular controls execute the exquisitely precise, temporally regulated refinement of identity during the sequential generation of distinct cortical PN subtypes. The recent characterization of the postmitotic functions of critical transcriptional regulators, including Sox5 in controlling the sequential generation of deep layer corticofugal PN subtypes (Kwan et al. 2008; Lai et al. 2008); Satb2 in controlling CPN identity (Alcamo et al. 2008; Britanova et al. 2008); and Bhlhb5 in controlling the acquisition of appropriate sensori-motor areal identity (Joshi et al. 2008), highlights the delicate balance of multiple key postmitotic cell-intrinsic signals during the execution and acquisition of neuronal identity, as well as the plasticity of this identity when the expression of these factors is altered. This postmitotic malleability strongly suggests that, for at least a phase of early postmitotic differentiation, maturing cortical PN subtypes share many fate executing molecular controls, able to implement multiple developmental paths. Here, we identify that expression of the LIM domain only protein LMO4 and the LIM-HD cofactor CLIM1 overlaps in presumptive layer V subcerebral PN and CPN during early development. This early coexpression exemplifies this early period of shared molecular identity, before progressively resolving into distinct subtypes during mid to late differentiation (Fig. 4K).
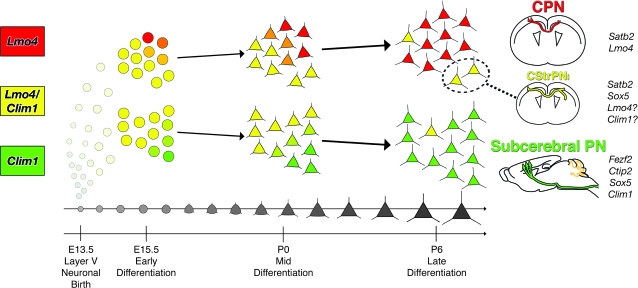
Figure 4.
Model illustrating the initial coexpression and progressive segregation by late differentiation of Lmo4 and Clim1 expression into layer V CPN and subcerebral PN, respectively. The small subpopulation that continues to coexpress Lmo4, Clim1, and other molecular controls over both CPN and subcerebral PN development might represent the CStrPNi subpopulation of CPN. This model is consistent with either a scenario where fate is specified premitotically and executed and refined postmitotically, or one where fate is specified predominantly postmitotically (see Discussion).
Previous work from our lab identified LMO4 and CLIM1 as 2 LIM domain-related proteins that, at late stages of PN differentiation, are nearly exclusively expressed by CPN or by subcerebral PN, respectively (Arlotta et al. 2005) (Supplementary Fig. 2A,B). However, the earlier progressive refinement of this expression into cortical neuron subtypes has not previously been described.
At E15.5, during early layer V cortical PN differentiation, Lmo4 is initially coexpressed with the critical postmitotic molecular determinant of CPN identity, Satb2, in a subpopulation of neurons. However, at this stage, Lmo4 is also broadly coexpressed with known molecular controls over subcerebral PN development, Fezf2, Ctip2, and Sox5. Reciprocally, many Clim1-expressing neurons coexpress these molecular controls over subcerebral PN development, but also coexpress Satb2 and Lmo4. As differentiation progresses, the overlapping expression of Lmo4 and Clim1 is progressively segregated into layer V CPN and subcerebral PN populations, respectively. By P6, at late stages of differentiation when distinct neuronal subtype axonal circuitries are established, the expression of Lmo4 and Clim1 becomes largely mutually exclusive; at this stage, Lmo4 expression is largely segregated to Satb2-expressing CPN, whereas Clim1 expression is largely confined to Ctip2-expressing subcerebral PN.
Discussion of the postmitotic acquisition of identity raises 2 important hypotheses regarding the distinction between the specification of fate, and the subsequent execution of these fate programs. At least 2 major models can describe the progressive refinement of molecular identity that we observe: 1) CPN and subcerebral PN are specified before they are born, and although their molecular identities (as defined by the small group of genes that have been elucidated in layer V subtype development; e.g., Fezf2, Ctip2, Sox5, Satb2) might overlap very early on in their development, each postmitotic neuron is already specified and needs only the appropriate combination of downstream signals to execute this identity, leading to acquisition of precise phenotype. In this case, 2 distinct pools of postmitotic neurons give rise to layer V CPN and subcerebral PN, though their molecular identities are only progressively refined. 2) The identity of CPN and subcerebral PN is not entirely specified at the time of birth, and specific combinations of postmitotic signals are needed to fully specify the fate of these neurons. In this case, CPN and subcerebral PN arise from one common pool of postmitotic neurons whose overlapping molecular profiles are indicative of their not yet fully specified identity. We support the first of these hypotheses, given work that strongly suggests that a neuron's identity is specified by the time it is born (McConnell and Kaznowski 1991; Molyneaux et al. 2005, 2007), whereas many critical postmitotic signals, including Sox5 (Kwan et al. 2008; Lai et al. 2008), Satb2 (Alcamo et al. 2008; Britanova et al. 2008), and Bhlhb5 (Joshi et al. 2008) are needed at appropriate times and with appropriate levels of expression to properly execute the acquisition of precise subtype identity.
During this postmitotic execution of fate, several plausible models could specifically explain the progressive refinement of Lmo4 and Clim1 expression in layer V. One model is that only one presumptive subpopulation, CPN or subcerebral PN, coexpresses Lmo4 and Clim1 during early differentiation, and these coexpressing neurons eventually downregulate the expression of one gene, invariably acquiring a single subtype fate. Though this model is theoretically possible, we propose an alternative model in which many presumptive CPN and subcerebral PN express both Lmo4 and Clim1 before eventually segregating into either one developmental path or the other (Fig. 4K). In support of this model, we find that, at E15.5, during early differentiation, Lmo4 is coexpressed with Fezf2, known to be specifically expressed in presumptive subcerebral PN, and Clim1 is coexpressed with Satb2, a critical transcription factor in CPN development. Taken together, these data indicate that many newly born CPN and subcerebral PN share expression of Lmo4 and Clim1, and, therefore, the expression of either of these genes is not restricted by presumptive fate boundaries during early cortical development. Only at later postmitotic stages of differentiation does expression of Lmo4 and Clim1 distinguish between cortical PN subtypes.
Interestingly, we find that this segregation of expression during postnatal development is not absolute. At P6, a small subset of neurons continue to coexpress Lmo4 and Clim1 (Fig. 3C; arrowheads); Lmo4 and Ctip2 (Fig. 3G; arrowheads); Clim1 and Satb2 (Fig. 3J; arrowhead); and a small subpopulation of retrogradely labeled CPN express Clim1 (Fig. 3F; arrowhead). Supporting these findings, previous work demonstrated that Clim1 is expressed at low levels in at least a subpopulation of postnatal CPN, as indicated by microarray analysis (Supplementary Fig. 2A) (note that CPN expression, although much lower than subcerebral PN expression, still has a substantial normalized intensity). Further, analysis of the cortex of Fezf2−/− mice at P6, in the absence of subcerebral PN, demonstrates that Clim1 is still expressed by some neurons in layer V at relatively low levels (Supplementary Fig. 2D; arrow).
One possible explanation for the persistent overlapping expression of these CPN and subcerebral PN-associated genes in a small number of neurons late in differentiation is that these dual CLIM+/LMO4+ neurons comprise a unique subpopulation that share characteristics of both PN subtypes. CStrPNi are a subset of CPN, many located in layer V, which project both to the contralateral hemisphere and to the contralateral striatum. Our data suggest that the small subset of neurons in which Clim1 and Lmo4 are coexpressed with CPN and subcerebral PN-specific transcription factors may be this CStrPNi subpopulation, whose axonal identity shares characteristics of both CPN, which cross the midline as they project toward cortical targets, and subcerebral PN, which project away from the cortex. CStrPNi also share molecular characteristics of both CPN and subcerebral PN, including expression of both Sox5 and Satb2 (U.S.S., unpublished data). This combination of anatomical and molecular characteristics of both CPN and subcerebral PN in a single neuronal population may reflect the likely closer evolutionary relationship of layer V subcerebral PN to CPN in layer V than to the more superficially located layer II/III CPN, based on their shared time of birth and on the more ancient origin of deep layer relative to superficial layer PNs (Molnar et al. 2006; Lai et al. 2008). Additional analysis of the molecular controls over the specification and differentiation of this unique cortical PN subpopulation will elucidate aspects of their molecular identity that are unique, and those that are shared with their neighboring PN subtypes.
Given the refinement of Lmo4 and Clim1 expression patterns that temporally coincide with neuronal subtype differentiation, and their known developmental functions in other systems, these are promising genes to play a functionally significant role in progressively segregating and executing the molecular identity of CPN and subcerebral PN. The critical developmental biological roles of many LIM-HD proteins suggest the likelihood that Clim and Lmo genes, by modulating LIM-HD protein transcriptional activity, might function directly in the differentiation of their respective cortical PN subtypes. A number of LIM-HD transcription factors function centrally during telencephalic development, including Lhx2 during both the patterning of the dorsal telencephalon and cortical hem (Bulchand et al. 2001; Monuki et al. 2001) and later in cortical development and connectivity (Padmanabhan et al., unpublished data), and Lhx6 during the migration and specification of interneuron subtypes (Alifragis et al. 2004; Liodis et al. 2007). CLIM cofactors associate with LIM-HD transcription factors to potentiate or repress their transcriptional activity (Jurata and Gill 1997; Thaler et al. 2002; Bhati et al. 2008), whereas LMO proteins, which contain a LIM domain but not a DNA-binding domain, compete with CLIM proteins for binding to LIM-HD transcription factors, thereby interfering with LIM-HD:CLIM complex formation and function (Milan et al. 1998; Milan and Cohen 1999; van Meyel et al. 1999).
LMO4 functions during spinal cord development to suppress V2 interneuron gene expression in spinal motor neurons by disrupting assembly of the LHX3:CLIM transcriptional complex (Lee et al. 2008). Similarly, by directly interacting with CLIM1 (Kashani et al. 2006), LMO4 might selectively inhibit CLIM1:LIM-HD complex formation in developing CPN or subcerebral PN. These LIM-HD interacting partners might be subtype specific, as is the case with Lhx2, which is specifically expressed in CPN at early and intermediate stages of cortical development (Arlotta et al. 2005) (Supplementary Fig. 2C). Alternatively, LMO4 and CLIM1 might interact with a LIM-HD transcription factor expressed by both neuronal populations, and subtype-specific differentiation could be conferred by unique interaction partners within each subtype. Additionally, LMO4 and CLIM1 might regulate transcription of subtype-specific genes independently of interaction with LIM-HD transcription factors, as has been previously described during Drosophila development (Torigoi et al. 2000). Loss- and gain-of-function characterization of Lmo4 and Clim1 at distinct stages of corticogenesis might further elucidate important functions and interaction partners during PN subtype development.
Only recently have molecular controls over the specification and differentiation of distinct cortical neuron subtypes begun to be identified. It is becoming apparent that a delicate and precise balance of postmitotic signals is critical for executing fate specification programs set in place at the birth of individual neurons. The progressive refinement and distinction of Lmo4 and Clim1 expression in cortical PN subtypes of layer V highlights the dynamic and likely functional nature of this postmitotic molecular sharpening of boundaries, providing insight into how neuronal subtype identity and neuronal diversity emerges.
Supplementary Material
Supplementary material can be found at: http://www.cercor.oxfordjournals.org/
Funding
National Institutes of Health (NS49553 and NS45523; additional infrastructure supported by NS41590); Harvard Stem Cell Institute; Spastic Paraplegia Foundation; Travis Roy Foundation; Massachusetts Spinal Cord Injury Research Fund; Jane and Lee Seidman Fund for CNS Research; Emily and Robert Pearlstein Fund for Nervous System Repair to J.D.M.; National Institutes of Health predoctoral NRSA (F31 NS060421) partially supported E.A.; National Institutes of Health predoctoral NRSA (F31 NS063516) partially supported S.J.S.; Harvard Stem Cell Institute Internship Program partially supported G.C.; and Cerebral Palsy Research Fund and fellowship from the Lefler Center for the Study of Neurodegenerative Disorders partially supported U.S.S.
Supplementary Material
Acknowledgments
We thank L. Pasquina, D. Schuback, E. Sievert, A. Wheeler, and T. Yamamoto for superb technical assistance; E. Sievert for valuable artistic assistance on Fig. 4; Drs P. Arlotta and B. Molyneaux for helpful discussions and input; Drs M. Hibi and V. Tarabykin for generous sharing of mice and antibodies; and current and past members of our laboratory for helpful suggestions. Conflict of Interest: None declared.
References
- Alcamo EA, Chirivella L, Dautzenberg M, Dobreva G, Farinas I, Grosschedl R, McConnell SK. Satb2 regulates callosal projection neuron identity in the developing cerebral cortex. Neuron. 2008;57:364–377. doi: 10.1016/j.neuron.2007.12.012. [DOI] [PubMed] [Google Scholar]
- Alifragis P, Liapi A, Parnavelas JG. Lhx6 regulates the migration of cortical interneurons from the ventral telencephalon but does not specify their GABA phenotype. J Neurosci. 2004;24:5643–5648. doi: 10.1523/JNEUROSCI.1245-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angevine JB, Jr, Sidman RL. Autoradiographic study of cell migration during histogenesis of cerebral cortex in the mouse. Nature. 1961;192:766–768. doi: 10.1038/192766b0. [DOI] [PubMed] [Google Scholar]
- Arlotta P, Molyneaux BJ, Chen J, Inoue J, Kominami R, Macklis JD. Neuronal subtype-specific genes that control corticospinal motor neuron development in vivo. Neuron. 2005;45:207–221. doi: 10.1016/j.neuron.2004.12.036. [DOI] [PubMed] [Google Scholar]
- Arlotta P, Molyneaux BJ, Jabaudon D, Yoshida Y, Macklis JD. Ctip2 controls the differentiation of medium spiny neurons and the establishment of the cellular architecture of the striatum. J Neurosci. 2008;28:622–632. doi: 10.1523/JNEUROSCI.2986-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhati M, Lee C, Nancarrow AL, Lee M, Craig VJ, Bach I, Guss JM, Mackay JP, Matthews JM. Implementing the LIM code: the structural basis for cell type-specific assembly of LIM-homeodomain complexes. EMBO J. 2008;27:2018–2029. doi: 10.1038/emboj.2008.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britanova O, de Juan Romero C, Cheung A, Kwan KY, Schwark M, Gyorgy A, Vogel T, Akopov S, Mitkovski M, Agoston D, et al. Satb2 is a postmitotic determinant for upper-layer neuron specification in the neocortex. Neuron. 2008;57:378–392. doi: 10.1016/j.neuron.2007.12.028. [DOI] [PubMed] [Google Scholar]
- Bulchand S, Grove EA, Porter FD, Tole S. LIM-homeodomain gene Lhx2 regulates the formation of the cortical hem. Mech Dev. 2001;100:165–175. doi: 10.1016/s0925-4773(00)00515-3. [DOI] [PubMed] [Google Scholar]
- Chen B, Schaevitz LR, McConnell SK. Fezl regulates the differentiation and axon targeting of layer 5 subcortical projection neurons in cerebral cortex. Proc Natl Acad Sci USA. 2005;102:17184–17189. doi: 10.1073/pnas.0508732102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Wang SS, Hattox AM, Rayburn H, Nelson SB, McConnell SK. The Fezf2-Ctip2 genetic pathway regulates the fate choice of subcortical projection neurons in the developing cerebral cortex. Proc Natl Acad Sci USA. 2008;105:11382–11387. doi: 10.1073/pnas.0804918105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JG, Rasin MR, Kwan KY, Sestan N. Zfp312 is required for subcortical axonal projections and dendritic morphology of deep-layer pyramidal neurons of the cerebral cortex. Proc Natl Acad Sci USA. 2005;102:17792–17797. doi: 10.1073/pnas.0509032102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishell G, Hanashima C. Pyramidal neurons grow up and change their mind. Neuron. 2008;57:333–338. doi: 10.1016/j.neuron.2008.01.018. [DOI] [PubMed] [Google Scholar]
- Fricker-Gates RA, Shin JJ, Tai CC, Catapano LA, Macklis JD. Late-stage immature neocortical neurons reconstruct interhemispheric connections and form synaptic contacts with increased efficiency in adult mouse cortex undergoing targeted neurodegeneration. J Neurosci. 2002;22:4045–4056. doi: 10.1523/JNEUROSCI.22-10-04045.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillemot F, Molnar Z, Tarabykin V, Stoykova A. Molecular mechanisms of cortical differentiation. Eur J Neurosci. 2006;23:857–868. doi: 10.1111/j.1460-9568.2006.04626.x. [DOI] [PubMed] [Google Scholar]
- Hirata T, Suda Y, Nakao K, Narimatsu M, Hirano T, Hibi M. Zinc finger gene fez-like functions in the formation of subplate neurons and thalamocortical axons. Dev Dyn. 2004;230:546–556. doi: 10.1002/dvdy.20068. [DOI] [PubMed] [Google Scholar]
- Hobert O, Westphal H. Functions of LIM-homeobox genes. Trends Genet. 2000;16:75–83. doi: 10.1016/s0168-9525(99)01883-1. [DOI] [PubMed] [Google Scholar]
- Joshi PS, Molyneaux BJ, Feng L, Xie X, Macklis JD, Gan L. Bhlhb5 regulates the postmitotic acquisition of area identities in layers II-V of the developing neocortex. Neuron. 2008;60:258–272. doi: 10.1016/j.neuron.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurata LW, Gill GN. Functional analysis of the nuclear LIM domain interactor NLI. Mol Cell Biol. 1997;17:5688–5698. doi: 10.1128/mcb.17.10.5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kania A, Johnson RL, Jessell TM. Coordinate roles for LIM homeobox genes in directing the dorsoventral trajectory of motor axons in the vertebrate limb. Cell. 2000;102:161–173. doi: 10.1016/s0092-8674(00)00022-2. [DOI] [PubMed] [Google Scholar]
- Kashani AH, Qiu Z, Jurata L, Lee SK, Pfaff S, Goebbels S, Nave KA, Ghosh A. Calcium activation of the LMO4 transcription complex and its role in the patterning of thalamocortical connections. J Neurosci. 2006;26:8398–8408. doi: 10.1523/JNEUROSCI.0618-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan KY, Lam MM, Krsnik Z, Kawasawa YI, Lefebvre V, Sestan N. SOX5 postmitotically regulates migration, postmigratory differentiation, and projections of subplate and deep-layer neocortical neurons. Proc Natl Acad Sci USA. 2008;105:16021–16026. doi: 10.1073/pnas.0806791105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai T, Jabaudon D, Molyneaux BJ, Azim E, Arlotta P, Menezes JR, Macklis JD. SOX5 controls the sequential generation of distinct corticofugal neuron subtypes. Neuron. 2008;57:232–247. doi: 10.1016/j.neuron.2007.12.023. [DOI] [PubMed] [Google Scholar]
- Lee S, Lee B, Joshi K, Pfaff SL, Lee JW, Lee SK. A regulatory network to segregate the identity of neuronal subtypes. Dev Cell. 2008;14:877–889. doi: 10.1016/j.devcel.2008.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leone DP, Srinivasan K, Chen B, Alcamo E, McConnell SK. The determination of projection neuron identity in the developing cerebral cortex. Curr Opin Neurobiol. 2008;18:28–35. doi: 10.1016/j.conb.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liodis P, Denaxa M, Grigoriou M, Akufo-Addo C, Yanagawa Y, Pachnis V. Lhx6 activity is required for the normal migration and specification of cortical interneuron subtypes. J Neurosci. 2007;27:3078–3089. doi: 10.1523/JNEUROSCI.3055-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundgren SE, Callahan CA, Thor S, Thomas JB. Control of neuronal pathway selection by the Drosophila LIM homeodomain gene apterous. Development. 1995;121:1769–1773. doi: 10.1242/dev.121.6.1769. [DOI] [PubMed] [Google Scholar]
- McConnell SK, Kaznowski CE. Cell cycle dependence of laminar determination in developing neocortex. Science. 1991;254:282–285. doi: 10.1126/science.254.5029.282. [DOI] [PubMed] [Google Scholar]
- Milan M, Cohen SM. Regulation of LIM homeodomain activity in vivo: a tetramer of dLDB and apterous confers activity and capacity for regulation by dLMO. Mol Cell. 1999;4:267–273. doi: 10.1016/s1097-2765(00)80374-3. [DOI] [PubMed] [Google Scholar]
- Milan M, Diaz-Benjumea FJ, Cohen SM. Beadex encodes an LMO protein that regulates Apterous LIM-homeodomain activity in Drosophila wing development: a model for LMO oncogene function. Genes Dev. 1998;12:2912–2920. doi: 10.1101/gad.12.18.2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minshew NJ, Williams DL. The new neurobiology of autism: cortex, connectivity, and neuronal organization. Arch Neurol. 2007;64:945–950. doi: 10.1001/archneur.64.7.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnar Z, Metin C, Stoykova A, Tarabykin V, Price DJ, Francis F, Meyer G, Dehay C, Kennedy H. Comparative aspects of cerebral cortical development. Eur J Neurosci. 2006;23:921–934. doi: 10.1111/j.1460-9568.2006.04611.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molyneaux BJ, Arlotta P, Hirata T, Hibi M, Macklis JD. Fezl is required for the birth and specification of corticospinal motor neurons. Neuron. 2005;47:817–831. doi: 10.1016/j.neuron.2005.08.030. [DOI] [PubMed] [Google Scholar]
- Molyneaux BJ, Arlotta P, Menezes JR, Macklis JD. Neuronal subtype specification in the cerebral cortex. Nat Rev Neurosci. 2007;8:427–437. doi: 10.1038/nrn2151. [DOI] [PubMed] [Google Scholar]
- Monuki ES, Porter FD, Walsh CA. Patterning of the dorsal telencephalon and cerebral cortex by a roof plate-Lhx2 pathway. Neuron. 2001;32:591–604. doi: 10.1016/s0896-6273(01)00504-9. [DOI] [PubMed] [Google Scholar]
- Pasinelli P, Brown RH. Molecular biology of amyotrophic lateral sclerosis: insights from genetics. Nat Rev Neurosci. 2006;7:710–723. doi: 10.1038/nrn1971. [DOI] [PubMed] [Google Scholar]
- Rakic P. Neurons in rhesus monkey visual cortex: systematic relation between time of origin and eventual disposition. Science. 1974;183:425–427. doi: 10.1126/science.183.4123.425. [DOI] [PubMed] [Google Scholar]
- Segawa H, Miyashita T, Hirate Y, Higashijima S, Chino N, Uyemura K, Kikuchi Y, Okamoto H. Functional repression of Islet-2 by disruption of complex with Ldb impairs peripheral axonal outgrowth in embryonic zebrafish. Neuron. 2001;30:423–436. doi: 10.1016/s0896-6273(01)00283-5. [DOI] [PubMed] [Google Scholar]
- Shaywitz DA, Melton DA. The molecular biography of the cell. Cell. 2005;120:729–731. doi: 10.1016/j.cell.2005.03.002. [DOI] [PubMed] [Google Scholar]
- Sohur US, Emsley JG, Mitchell BD, Macklis JD. Adult neurogenesis and cellular brain repair with neural progenitors, precursors and stem cells. Philos Trans R Soc Lond B Biol Sci. 2006;361:1477–1497. doi: 10.1098/rstb.2006.1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaler JP, Lee SK, Jurata LW, Gill GN, Pfaff SL. LIM factor Lhx3 contributes to the specification of motor neuron and interneuron identity through cell-type-specific protein-protein interactions. Cell. 2002;110:237–249. doi: 10.1016/s0092-8674(02)00823-1. [DOI] [PubMed] [Google Scholar]
- Torigoi E, Bennani-Baiti IM, Rosen C, Gonzalez K, Morcillo P, Ptashne M, Dorsett D. Chip interacts with diverse homeodomain proteins and potentiates bicoid activity in vivo. Proc Natl Acad Sci USA. 2000;97:2686–2691. doi: 10.1073/pnas.050586397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Meyel DJ, O'Keefe DD, Jurata LW, Thor S, Gill GN, Thomas JB. Chip and apterous physically interact to form a functional complex during Drosophila development. Mol Cell. 1999;4:259–265. doi: 10.1016/s1097-2765(00)80373-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.