Abstract
During mammalian corticogenesis a series of transient cell layers establish laminar architectonics. The preplate, which forms from the earliest-generated neurons, separates into the marginal zone and subplate layer. To provide a systematic screen for genes involved in subplate development and function, we screened lines of transgenic mice, generated using bacterial artificial chromosome methodology (GENSAT Project), to identify transgenic lines of mice that express the enhanced green fluorescent protein (EGFP) reporter in preplate neurons destined for the subplate. Gene expression profiling of RNA purified from EGFP-positive neurons identified over 200 genes with enriched expression in future subplate neurons. Major classes of subplate-enriched genes included genes involved in transcriptional processes, cortical development, cell and axon motility, protein trafficking and steroid hormone signaling. Additionally, we identified 10 genes related to degenerative diseases of the cerebral and cerebellar cortex. Cre recombinase–based fate mapping of cells expressing Phosphodiesterase 1c (Pde1c) revealed beta-galactosidase positive cells in the ventricular zone, as well as the subplate, suggesting that subplate neurons and cortical projection neurons may be derived from common progenitors. These experiments therefore reveal genetic markers, which identify subplate neurons from the earliest stages of their development, and genes with enriched expression in subplate neurons during early stages of corticogenesis.
Keywords: cortical development, subplate neurons, fate mapping, gene expression
Introduction
The development of the mammalian neocortex involves the formation of a series of transient layers, resulting from temporal waves of radial and tangential migrations of neuronal precursors from their germinal zones. The preplate is formed by the earliest-generated neurons, which are born in the dorsal forebrain between embryonic day 10.5 (E10.5) and E12.5 in the mouse (Caviness 1982; Wood et al. 1992; Del Rio et al. 2000; Hevner et al. 2003). Radially migrating, later-born neurons from the ventricular zone “split” the preplate, segregating the 2 major classes of preplate neurons to positions above and below the cortical plate. Cajal–Retzius neurons remain in the most superficial cortical layer, the marginal zone, whereas subplate neurons descend into a deeper layer below the cortical plate neurons. The prevailing hypothesis is that subplate neurons pattern the connectivity in cortical circuits during early periods of cortical development (Ghosh et al. 1990; Allendoerfer and Shatz 1994; Marin-Padilla 1998). Beginning in the preplate stage, neurons destined for the subplate extend axons that pioneer both corticothalamic projections (McConnell et al. 1989; De Carlos and O'Leary 1992) and projections to the contralateral cortex via the corpus callosum and anterior commissure (Jacobs et al. 2007). Later in development, subplate neurons are crucial for guiding thalamocortical axons to their layer IV targets (Allendoerfer and Shatz 1994).
Relatively few analyses of the regulatory pathways involved in the early differentiation and specification of subplate neurons have been reported (Zhou et al. 1999; Hevner et al. 2001; Shinozaki et al. 2002; Lai et al. 2008). Although antigen markers are currently used to identify subplate neurons (e.g., Map2, CSPG, Tbr1), the expression of these markers is not restricted to subplate neurons. Golli promoter–driven transgenes, which specifically identify subplate neurons (Jacobs et al. 2007; Landry et al. 1998) are transiently expressed in Cajal–Retzius neurons at the preplate stage, precluding analysis of gene expression at early embryonic time points (Xie et al. 2002). The goal of this study was to provide a systematic screen to identify genes and pathways that are enriched in subplate neurons during the period of early cortical development when the subplate is thought to establish a template for the cortical circuitry. To identify genes expressed in mouse preplate and subplate neurons, we screened lines of transgenic mice created by the GENSAT Project (Gong et al. 2003, 2007). The GENSAT Project uses bacterial artificial chromosome (BAC) transgenic methodology to achieve stable expression of the enhanced green fluorescent protein (EGFP) reporter under the control of the native locus. In tissue sections of the developing neocortex of transgenic mice, it is possible to track the movements of individual EGFP-positive subplate neurons (or their precursor preplate neurons) and monitor patterns of axon outgrowth and connectivity. In the present study, we analyzed 2 lines in detail: Tg(Pde1C-EGFP) mouse embryos, in which the EGFP transgene labeled all preplate neurons, and Tg(Girk4-EGFP) mouse embryos, where EGFP expression was restricted to Cajal–Retzius neurons. Because Cajal–Retzius neurons segregate into the marginal zone during cortical plate formation, gene expression profiles of these 2 transgenic lines provided an experimental approach to identify genes expressed in preplate neurons destined for the subplate. Analysis of gene expression profiles of FAC-sorted EGFP-positive cells from these 2 lines identified 229 genes expressed in future subplate neurons. This set of genes included genes involved cortical development, cell and axon motility, signaling pathways (receptors, G-proteins, second messenger systems), protein trafficking, steroid hormone signaling, and central nervous system (CNS) degenerative diseases of the cerebral and cerebellar cortex. We also identified 2 novel markers that specifically identify the future subplate subpopulation at the preplate stage. This study is a critical first step in identifying the regulatory pathways and interactions involved in the role of the subplate in the earliest stages of corticogenesis.
Materials and Methods
Animals
BAC-EGFP and BAC-Cre recombinase transgenic lines were maintained on the Swiss–Webster background in the Rockefeller University animal facility. Experiments were carried out in accordance with the Principles of Laboratory Animal Care, conforming to National Institutes of Health guidelines under approved protocols. For timed pregnancies, the day of vaginal plug discovery was considered E0.5. BAC-EGFP transgenic mice and embryos were genotyped by PCR for the EGFP transgene using the following primers: 5′-CGGCGAGCTGCACGCTGCGTCCTC-3′, 5′-CCTACGGCGTGCAGTGCTTCAGC-3′. Tg (Pde1C-Cre) mice and embryos were genotyped by PCR for the Cre transgene using the following primers: 5′-CCGGTGAACGTGCAAAACAGGCTCTA-3′ 5′-CTTCCAGGGCGCGAGTTGATAGC-3′. Homozygous Cre reporter lines used were ROSA26R and ROSA26-GFP (Jackson).
Preparation of Cryostat Sections for Immunohistochemistry and In Situ Hybridization
For embryos, the dam was killed by cervical dislocation and embryos dissected in ice-cold phosphate-buffered saline (PBS). Embryos E13.5 and younger were immediately immersed in fixative (4% paraformaldehyde/PBS). For embryos E14.5 and older, brains were dissected out, then immersion fixed. Brains/embryos were then transferred to 30% sucrose/PBS for cryoprotection overnight at 4 °C. Brains/embryos were embedded in Neg-50 embedding medium (Richard-Allen Scientific, Kalamazoo, MI), and 20-μm sections were cut on a Microm HM500 cryostat, mounted on Permafrost slides (Fisher Scientific, Pittsburgh, PA), and dried for several hours at room temperature.
Immunohistochemistry
For fluorescent staining, cryostat sections were rinsed in PBS to remove mounting medium and blocked for 1 h at room temperature in 5% normal donkey or goat serum/0.02% Triton X-100/PBS. For mouse IgG primary antibodies, slides were then rinsed again in PBS and an additional blocking step with donkey anti-mouse IgG Fab fragments (Jackson ImmunoResearch, West Grove, PA) in PBS for 1 h was carried out. Slides were rinsed again in PBS before adding diluted primary antibodies. Primary antibodies were diluted in blocking buffer with serum and incubated overnight at 4 °C. Slides were rinsed in PBS, incubated with fluorescent-conjugated secondary antibodies diluted in blocking buffer for 1 h at room temperature, then rinsed again in PBS and mounted in Gelmount mounting medium (Biomeda, Foster City, CA). As controls for secondary antibody background and autofluorescence, slides were subjected to this protocol omitting the primary antibody. Images of single optical sections were acquired with a Radiance 2000 confocal laser-scanning microscope (Biorad, Hercules, CA).
Primary antibodies used were: mouse anti-Cajal–Retzius cell marker reelin (RELN) (1:1000, MAB5364, Chemicon, Billerica, MA), rabbit anti-GFP (1:2000, Molecular Probes, Carlsbad, CA), rabbit anti-calretinin (1:2000, Swant, Bellinzona, Switzerland), rabbit anti-hippocalcin (1:2000, Abcam, Cambridge, MA), goat anti-EAAC1 (1:2000, Chemicon), rabbit anti-Ryr1 (1:1000, Chemicon), rabbit anti-serum response factor (SRF) (1:100, Abcam), rat anti-L1 (1:20, a gift from Dr J. Trotter), mouse IgM anti-TAG1 (1:2, gift from Dr Jane Dodd, Columbia University, NY).
Secondary antibodies used were: donkey anti-rabbit IgG, anti-mouse IgM, anti-goat IgG, and anti-rat (IgG) Alexas 488, 555, 594, and 647 (1:500, Molecular Probes), and donkey anti-mouse IgG Cy3 (1:700, Jackson ImmunoResearch).
X-Gal Staining
Paraformaldehyde-fixed cryostat sections were rinsed several times in PBS and permeabilized in a detergent solution (0.01% deoxycholate, 0.02% NP-40, 2 mM MgCl2 in PBS) for 10 min at 4 °C. Sections were developed in staining solution (detergent solution plus 5 mM K3Fe(CN)6, 5 mM K4Fe(CN)6, 5 mM ethylene glycol tetraacetic acid (EGTA), 1 mg/mL X-gal (5-bromo-4-chloro-3-indolyl-β-D-galactoside, Invitrogen, Carlsbad, CA) at 37 °C for several hours or overnight. After staining, sections were rinsed in PBS and mounted in Crystal/Mount (Biomeda).
Dissociation and Purification of EGFP+ Cortical Neurons by Fluorescent Activated Cell Sorting
Pregnant dams were killed by cervical dislocation, and E12 embryos collected in ice-cold Earles Balanced Salt Solution (EBSS) (Sigma-Aldrich, St Louis, MO) with 0.2% glucose. Embryos were genotyped by EGFP fluorescence under a Leica M2FL111 dissection microscope (Leica, Bannockburn, IL) equipped with fluorescence. Brains were removed and cortices dissected away from basal forebrain. During dissection, cortices were kept at 37 °C in EBSS. Cortices were pooled by genotype, the EBSS removed, and activated papain (Worthington Biochemical, Lakewood, NJ) in EBSS, passed through a 20-μm filter, was added. Cortices were incubated in papain solution at 37 °C for approximately 30 min. The tube was inverted several times during this incubation. Digested tissue was pelleted by centrifugation at low speed (500 g for 5 min). The papain solution was removed and the pellet triturated gently with a 5-mL pipet in trituration solution preheated to 37 °C (1 mg/mL ovomucoid trypsin inhibitor/bovine serum albumin [BSA] [Worthington Biochemical], 0.1% DNAse [Worthington Biochemical] in EBSS/0.2% glucose). Two milliliters of ovomucoid/BSA (10 mg/mL) was added to the bottom of the tube to form 2 aqueous layers, and the tube was centrifuged at 500 g for 5 min. The pellet was washed and resuspended in ACSF (125 mM NaCl, 2.5 mM KCl, 1 mM NaH2PO4, 25 mM NaHCO3, 11 mM glucose, 1.25 mM MgCl2, 2.5 mM CaCl2). Immediately prior to sorting, the cell suspension was filtered through a cell-strainer cap (Falcon, Franklin Lakes, NJ).
The cell suspension was presorted by size to remove debris and clumps of cells (Supplemental Fig. 3a). For sorting based on EGFP fluorescence, gates were set by sorting a cell suspension prepared from wild-type littermates as an EGFP-negative baseline (Supplemental Fig. 3b). EGFP+ cells were collected based on these gates (Supplemental Fig. 3c,d) and total RNA was extracted from the purified populations.
Preparation of Total RNA from Fluorescent Activated Cell Sorting-Purified Cells
Total RNA was extracted from EGFP+ purified cells with the RNAqueous Micro kit (Ambion, Austin, TX), with the following modifications to the manufacturer's protocol. All buffers were RNAse free. Cells were pelleted by centrifugation at 3000 g, and lysed by trituration in 200 μL of Lysis Buffer. Fifty microliters of EtOH was added, and the resulting suspension was applied to Micro Filter Columns. Next, an on-column DNAse digestion was carried out (RNAse-free DNAse kit, Qiagen, Valencia, CA) for 15 min at room temperature. The column was washed once in Wash Solution 1, twice in Wash Solution 2/3, and centrifuged at high speed to dry the membrane prior to eluting twice with 5 μL of Elution Solution preheated to 75 °C, yielding 10 μL of total eluate. RNA samples were stored at −80 °C. An Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA) was used to check for total RNA integrity and genomic DNA contamination, and RNA concentration was measured on a NanoDrop-1000 Spectrophotometer (Thermo Scientific, Wilmington, DE).
Affymetrix Genechip Probe Preparation and Hybridization
Amplified biotinylated cRNA was produced from total RNA using the Two-Cycle Labeling Kit with spike-in controls (Affymetrix, Inc., Santa Clara, CA). Briefly, double-stranded cDNA was produced from total RNA using a T7-oligo dT primer for the first strand, and amplified antisense cRNA was produced from the resulting cDNA template by in vitro transcription. A second round of cDNA synthesis from amplified cRNA was performed, and another round of in vitro transcription was used to produce biotinylated cRNA. Labeled cRNA was fragmented before hybridization, and an Agilent 2100 Bioanalyzer was used to ensure proper fragment size.
Hybridization to Mouse Genome 430A 2.0 Microarrays (Affymetrix, Inc.) and scanning was performed by the Rockefeller University Genomics Facility. Raw data files were processed with Affymetrix GCOS software to generate CEL files. Data for all samples were imported together into Genespring 7.3 (Agilent) as CEL files and normalized by Genechip Robust Multiarray Averaging (GC-RMA). For each line, genes were considered expressed in a subpopulation and included in the analysis only if they were flagged as present in 2 out of 3 replicates by the Affymetrix GeneChip Operating Software, and if average normalized signal intensity was above an arbitrary cutoff of 1.
Results
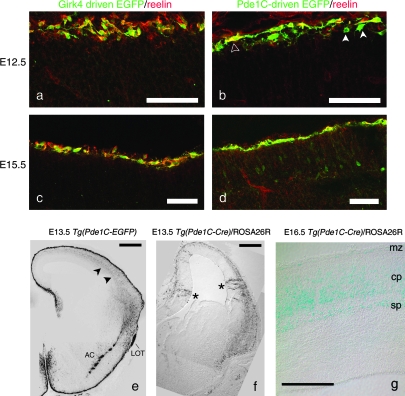
To identify novel genes expressed in preplate neurons that later segregated into the subplate, we screened the GENSAT database (www.gensat.org) for lines that express EGFP in the preplate layer at E12.5, and in the marginal zone (future layer I) or the subplate layer at E15.5. Among the lines we screened, we selected 2 lines for further study: the Tg(Girk4-EGFP) line and the Tg(Pde1C-EGFP) line. To determine whether EGFP-labeled cells in the preplate represented precursors of Cajal–Retzius cells, the principal neurons of the marginal zone, we double-labeled cryostat sections of E12.5 brains from Tg(Girk4-EGFP) and Tg(Pde1C-EGFP) embryos with antibodies against EGFP and RELN. Girk4-driven EGFP expression was restricted to Cajal–Retzius neurons at all of the embryonic stages we examined. Girk4-driven EGFP was expressed in RELN-positive cells at E12.5 (Fig. 1a and Supplemental Fig. 1). Although Girk4-driven EGFP-positive cells coexpressed RELN, a subset of RELN-positive, EGFP-negative cells were observed at E12.5 (Fig. 1a). Expression of Girk4-driven EGFP in RELN-positive cells was maintained in the marginal zone at E15.5 (Fig. 1c). Examination of the postnatal expression pattern in the GENSAT database showed that Girk4-driven EGFP expression was weakly maintained in layer I of postntatal day 7 (P7) cortex. Thus, Girk4-driven EGFP is expressed by Cajal–Retzius neurons during embryonic stages of corticogenesis.
Figure 1.
Pde1C-driven EGFP and Girk4-driven EGFP are markers for future subplate and Cajal–Retzius neurons. Girk4-driven EGFP labels Cajal–Retzius neurons, whereas Pde1C-driven EGFP labels all preplate neurons. (a–d) Immunostaining for EGFP (green) and RELN (red) in cryostat sections of E12.5 (a, b) and E15.5 (c, d) cortex of Tg(Girk4-EGFP) (a, c) and Tg(Pde1C-EGFP) (b, d) embryos. At E12.5, Pde1C-driven EGFP is expressed in both RELN-positive and RELN-negative preplate neurons (b), whereas Girk4-driven EGFP is expressed exclusively in the majority of RELN-positive neurons (a). At E15.5, both EGFP transgenes are expressed only in RELN-positive neurons of the marginal zone (c, d). (e) Immunostaining for EGFP in coronal cryostat section of E13.5 Tg(Pde1C-EGFP) brain. Pde1C-driven EGFP is strongly expressed in the preplate/marginal zone, and more weakly expressed in corticofugal projections (arrowheads). (f) Expression of the Pde1C-driven Cre transgene replicates that of the Pde1C-driven EGFP transgene. X-gal staining in coronal cryostat section of Pde1C-Cre+/−/ROSA26R+/− E13.5 brain. Beta-galactosidase is expressed by cells in the preplate/marginal zone and lateral cortex. Radially oriented beta-gal expressing cells are occasionally observed in the ventricular zone/intermediate zone of the cortical hem and neocortex (asterisks). (g) X-gal staining in coronal cryostat section of Tg(Pde1C-Cre)+/−/ROSA26R+/− E16.5 brain. Beta-gal is expressed in scattered cells in the marginal zone and in the subplate, as well as in cells of the lower cortical plate. Scale bars in a–d = 50 μm, e–f = 400 μm, g = 200 μm. Abbreviations: lateral olfactory tract (LOT), anterior commissure (AC), marginal zone (MZ), cortical plate (CP), subplate (SP).
Phosphodiesterase 1C–driven EGFP is Expressed in RELN-positive Cajal–Retzius Neurons and, Transiently, in Future Subplate Neurons
To examine the colocalization of Pde1C-driven EGFP with RELN in the preplate, we immunostained cryostat sections of E12.5 and E15.5 Tg(Pde1C-EGFP) brains with antibodies against EGFP and RELN (Fig. 1b,d and Supplemental Fig. 1). At E12.5, RELN-positive cells were localized beneath the pia in the most superficial aspect of the neuroepithelium (Fig. 1b, open arrowhead), whereas the RELN-negative, Pde1C-driven EGFP-positive cells were usually located just beneath the RELN-positive population (Fig. 1b, filled arrowheads). RELN-positive cells had the bipolar morphology and horizontal orientation characteristic of Cajal–Retzius neurons, whereas the RELN-negative cells had more rounded cell somas and were not necessarily oriented parallel to the pial surface. By E15.5, the Pde1C-driven EGFP-positive, RELN-negative cells expressed only very low levels of EGFP (Fig. 1d), and the RELN-negative, Pde1C-driven EGFP-positive cells coalesced into a deeper layer of cortex, corresponding to subplate. In postnatal cortex, Pde1C-driven EGFP expression was restricted to RELN-positive cells (data not shown).
Low levels of Pde1C-driven EGFP expression were detectable at E13.5 in early corticofugal pioneer axons extended by subplate neurons, which at this age had not yet exited the cortex (Fig. 1e, arrowheads). The relatively low level of EGFP expressed in preplate/subplate projections and cell bodies after the commencement of preplate splitting suggested that Pde1C-driven EGFP is transiently expressed in future subplate neurons during the preplate stage, and is subsequently downregulated in these cells. In contrast, Pde1C-driven EGFP was expressed at high levels in the lateral olfactory tract and the anterior commissural projections (Fig. 1e). Because the anterior commissure contains projections from both neocortical neurons and the olfactory bulb, we attribute the expression in the anterior commissure to the high levels of Pde1C-driven EGFP expressed in the olfactory bulb (data not shown), as the projections from cortical neurons did not display this level of expression.
Fate Mapping of Pde1c-Expressing Neurons in the Developing Neocortex
To confirm that a subset of Pde1C-driven EGFP-positive preplate neurons were destined for the subplate, we used a Pde1C-driven Cre recombinase BAC transgenic line (Tg[Pde1C-Cre IT150]), provided by the GENSAT Project (Gong et al. 2007), to fate-map Pde1C-positive neurons. Beta-galactosidase expression in the cortex of Tg(Pde1C-Cre IT150)/ROSA26R animals closely matched the expression pattern of Pde1C-driven EGFP in the dorsal and lateral cortex at E13.5, and was therefore selected for further study (Fig. 1e,f). At E13.5, scattered radial columns of beta-galactosidase-positive cells were observed throughout the cortex (Fig. 1f, asterisks), suggesting that the Pde1C-driven transgene was transiently expressed in progenitor neuroblasts in the ventricular zone, and thus that subplate neurons and cortical projection neurons may be derived from common progenitors.
At E16.5, after the cortical plate had formed, beta-galactosidase was detected in some cells in the marginal zone, in the subplate layer, and in the lower cortical plate (Fig. 1g). This suggests that Pde1C is transiently expressed in early, deep layer cortical plate neurons, as well as in Cajal–Retzius and future subplate neurons. Taken together, these data show that Pde1C-driven transgenes are expressed transiently in preplate cells destined for the subplate and marginal zone, as well in some neurons destined for the cortical plate. Pde1C-driven EGFP is strongly expressed in both Cajal–Retzius neurons and future subplate neurons at E12.5, but expression of this marker subsequently decreases in future subplate neurons, while it is maintained in Cajal–Retzius neurons.
Pde1C-driven Transgenes are Expressed in at Least Two Specific Populations of Subplate Neurons
Prior studies provide evidence that subplate neurons pioneer projections to the thalamus (McConnell et al. 1989) as well as the contralateral cortical hemisphere, via the anterior commissure and corpus callosum (Jacobs et al. 2007). All projections from subplate neurons express the axonal protein L1, including those projecting towards the anterior commissure, whereas only projections toward the thalamus express the transient axonal glycoprotein TAG1 (Denaxa et al. 2001). Thus, these projections can be distinguished from one another by the differential expression of TAG1 (Fukuda et al. 1997; Jones et al. 2002; Morante-Oria et al. 2003). L1-positive, TAG1-negative projections destined for the anterior commissure turn slightly medial to the TAG1-positive projections as they exit the lateral cortex (Supplemental Fig. 2, compare arrowheads in panels a and b). To examine whether subpopulations of subplate neurons express Pde1C-driven transgenes, we examined coexpression of EGFP and the axonal proteins L1 or TAG1 in EGFP-positive subplate axons.
To map the colocalization of these axonal markers with Pde1C-driven transgenes, we crossed the Pde1C-driven Cre line to a ROSA26-stop-GFP reporter line (Soriano 1999). At E13.5, GFP expression was detected in projections emanating from future subplate neurons (Supplemental Fig. 2). Double immunostaining for the axonal markers L1 and TAG1 was performed to examine their colocalization with GFP-labeled projections. The majority (>80%) of GFP-labeled projections expressed TAG1 and L1. However, some GFP-labeled projections were TAG1-negative (Supplemental Fig. 2c), projecting slightly medial to the TAG1-positive projections (Supplemental Fig. 2b,c). We conclude that expression of Pde1C transgenes transiently labels a heterogenous population of subplate neurons—some sending TAG1-positive projections towards the thalamus, and others projecting TAG1-negative axons toward the anterior commissure.
Gene Expression Profiling of Future Subplate Neurons
To identify genes expressed in future subplate neurons, we compared gene expression profiles of Fluorescent Activated Cell Sorting (FACS)-purified Pde1C-driven EGFP-positive E12.5 cortical neurons (representing a mixture of both types of preplate neurons, Cajal–Retzius cells and future subplate neurons) with profiles of purified Girk4-driven EGFP-positive E12.5 cortical neurons (representing only Cajal–Retzius cells). We hypothesized that genes upregulated in the Pde1C-driven EGFP-positive population compared with the Girk4-driven EGFP-positive population had a high likelihood of being expressed in future subplate cells.
Purification of EGFP-Positive Preplate Neurons for Microarray Analysis
EGFP-positive neurons were purified by FAC sorting cell suspensions prepared from E12.5 mouse cortices (Supplemental Fig. 3a–d). Total RNA was extracted from purified cells, and labeled cRNA was produced using the Affymetrix Two-Cycle amplification kit and used to probe Affymetrix Mouse Genome 420A2.0 gene arrays. Each litter of E12.5 embryos yielded between 5 and 20 ng of total RNA, and was used for one array hybridization. Three replicates of each line (Tg(Pde1C-EGFP) and Tg(Girk4-EGFP)) were carried out, along with 3 replicates of control unpurified cortical cells.
Analysis of Microarray Gene Expression Data
We estimated the quality and reproducibility of the microarray data using 2 separate methods. First, we calculated correlation coefficients for pairwise comparisons of replicates. The average correlation coefficient for pairwise comparisons was 0.99 (Girk4-driven EGFP-positive cells, unpurified whole cortex) and 0.97 (Pde1C-driven EGFP-positive cells), indicating a high degree of reproducibility between replicate samples. Next, we compared the expression of individual probe sets in replicate samples in scatter plots, and calculated the number of probe sets that increased or decreased 2-fold between replicates for each pairwise comparison (Supplemental Fig. 3e,f). Girk4-driven EGFP-positive cell and whole cortex samples had respectively an average of 2.3% and 4.7% probe sets with greater than 2-fold differences in expression between pairwise replicates. Replicates for Pde1C-driven EGFP-positive cells had an average of 10% probe sets with greater than 2-fold differences in expression. Probe sets with low or unreliable expression values were excluded from our analysis (see Methods for details).
Genes with Enriched Expression in Future Subplate Neurons
Probe sets enriched by 1.5-fold or greater in Pde1C-driven EGFP-positive cells compared with whole cortex (Supplemental Fig. 3g) were analyzed further. Among genes that were upregulated in Pde1C-driven EGFP-positive cells, a subset of genes were identified that were enriched by 1.5-fold in Pde1C-driven EGFP-positive preplate cells compared with Girk4-driven EGFP-positive Cajal–Retzius neurons (Supplemental Fig. 3h). Only probe sets with P values below 0.05 by t-test between samples were included. 229 probe sets met these criteria (Supplemental Fig. 3h, Supplemental Table 1).
As a first step, we used the DAVID bioinformatics software (Hosack et al. 2003) to categorize the gene expression data, identifying 4 prominent categories of genes expressed in subplate neurons: cell adhesion and axonogenesis (Cck, Cckra, Slitrk1, Sema4d, Dscam, Ncam2, Nrcam, 9030425E11Rik; P = 9.3 × 10−8), exocytosis and the formation of coated vesicles (Ica1, Sec24d, Wyn2, Syngr1, Syt4; P = 1.8 × 10−8), regulators of transcription (32 genes, see Supplemental Table 1; P = 3.1 × 10−24), and steroid biosynthesis, metabolism, and signaling (Cy51, Cyp7b1, Tm7sf2, mevalonate (diphospho) decarboxylase; P = 1.1 × 10−4). Analysis of the gene expression data sets with the Ingenuity Pathways Analysis software (Ingenuity Systems, Redwood City, CA), revealed a network of subplate-specific genes regulated by beta-estradiol (estrogen) signaling (Supplemental Fig. 4), which included multiple genes likely to be involved in neurite extension and guidance (Dscam, Sez6, Cdh11, Ccl25, Nell2). Together, the enrichment of genes involved in steroid biosynthesis and signaling in the subplate and the presence of subplate-specific beta-estradiol (estrogen)–regulated gene expression imply a role for estrogen signaling in the development of subplate neurons.
As many of the genes we identified were not placed into categories by these 2 software programs, we curated the dataset using information available in standard NCBI databases (Pubmed, Genbank, etc.) to highlight subsets of genes with important putative functions in CNS development and/or disease, and genes that participate in basic cellular processes, such as transcriptional pathways, protein trafficking, receptors and G-protein pathways, primary effectors and second messenger systems, cytoskeletal and motility systems, growth, survival, and apoptosis pathways and metabolic processes. Among these, we further categorized genes of particular significance to neuronal development and function, including genes involved in cortical neuron specification, axon outgrowth and synapse formation and function. Subplate genes in each of these general categories are listed in Supplemental Table 1. Approximately 20% of the subplate genes identified in this study have not been characterized.
Expression Verification of Future Subplate-Specific Genes
We verified the enrichment and specific expression of genes in preplate/subplate neurons identified by microarray analysis using 2 separate methods. First, the expression of 40 genes chosen randomly from the set of genes enriched in the subplate was checked in preplate/subplate at E14.5, using the Genepaint in situ hybridization database (www.genepaint.org). Future subplate genes were scored blind alongside an equal number of genes randomly selected from the complete array. Fifty-nine percent of genes identified as having enriched expression in future subplate neurons were present in preplate/subplate at E14.5, whereas 28% of randomly selected genes were present in preplate/subplate at this stage (P < 0.05 by chi-square test). The resolution of the in situ images at this early stage did not allow us to determine whether candidate genes were specifically expressed in future subplate neurons.
To confirm that genes identified as having enriched expression in the RELN-negative future subplate neurons were specific to subplate neurons, we performed immunohistochemistry for a selection of candidate marker proteins and RELN. We also examined BAC transgenic mice from GENSAT by costaining tissue sections for expression of the EGFP transgene and RELN. Double immunofluorescence antibody staining in wild-type cortex for the candidate marker proteins hippocalcin (Hpca) and EAAC1 showed that these proteins were specifically expressed in RELN-negative, future subplate neurons at E12.5 (Fig. 2b,c), and were not coexpressed in RELN-positive cells. At this stage, prior to preplate splitting and the formation of the cortical plate, RELN-negative cells were located directly beneath the RELN-positive layer, with little intermingling. Hpca and EAAC1 were localized in the cytoplasm of RELN-negative cells, revealing the more rounded cell bodies characteristic of the future subplate neurons. At E14.5, after preplate splitting, expression of these proteins was maintained in the cell bodies of neurons located in the subplate (Fig. 2d,e). The expression of these markers at E14.5 also revealed the localization pattern of future subplate cells during preplate splitting—some RELN-negative preplate cells remain adjacent to the Cajal–Retzius cell layer at this early stage of cortical plate formation, whereas others descend into the subplate layer. This suggests that the descent of subplate cells to their destination beneath the cortical plate does not occur all at once, but rather occurs progressively during cortical plate formation.
Figure 2.
Future subplate genes are expressed in RELN-negative preplate and subplate neurons. (a) Genes expressed in future subplate neurons categorized by function. (b–h) Double immunofluorescent staining for RELN (red) and marker proteins for future subplate candidate genes (green) in coronal cryostat sections of wild-type E12.5 (b, c) and E14.5 (c–h) brains. Hpca (c, e) and EAAC1 (b, d) are expressed in RELN-negative preplate cells and in subplate cell bodies. At E14.5, SRF (f) and RyR1 (h) are expressed in RELN-positive marginal zone neurons, as well as in intermediate zone fibers originating in the subplate, whereas Sez6-driven EGFP (g) is expressed in subplate and cortical plate neurons, but not in RELN-positive cells. Scale bars = 50 μm.
Although microarray analysis identified genes with enriched expression in subplate neurons, it did not exclusively identify genes whose expression was restricted to the subplate. Some markers examined by antibody staining were expressed in subplate neurons but were not entirely specific to the subplate. At E14.5, SRF and ryanodine receptor 1 (RyR1) were detected on axonal projections located in the intermediate zone, originating from the subplate neurons (Fig. 2f,h, Supplemental Fig. 6a,b). These proteins were also weakly expressed in the RELN-positive marginal zone at E14.5 (Fig. 2f,h, Supplemental Fig. 6a,b), and thus were expressed in both cell types derived from the preplate. In contrast, in the Sez6-driven EGFP BAC transgenic line, Sez6-driven EGFP expression was excluded from RELN-positive cells but was detected in both subplate cells and cortical plate neurons at E14.5 (Fig. 2g, Supplemental Fig. 6c). Expression of this transgene was seen in axonal projections in the intermediate zone, which originate from the subplate. Taken together, these data confirmed that the genes identified by microarray analysis were indeed expressed in preplate neurons destined for the subplate layer.
Discussion
Subplate neurons are among the earliest born neurons of the cerebral cortex, and provide a template for the development of cortical laminar and axonal architectonics. To date, studies on the subplate have focused on its critical role in the establishment of cortical circuitry, because subplate neurons pioneer the first subcortical and contralateral projections in the early cortex (McConnell et al. 1989; Jacobs et al. 2007), guide thalamocortical axons into the cortex (Ghosh et al. 1990) and selectively express genes required to establish and later to finely tune cortical circuits. This study provides putative antigen markers with which to follow the patterning of subplate neurons and their axon projections. More importantly, the gene expression profile reported in this study reveals ongoing biological processes in subplate neurons at the developmental stage when cortical architectonics emerge. Not surprisingly, the largest group of genes, identified by bioinformatics or by manual curation, are those involved in transcriptional processes. Particular subplate genes from general categories, including receptor signaling, G-protein second messenger systems, survival and apoptosis pathways, protein trafficking, and cytoskeletal dynamics and motility, provide insights into pathways important for known subplate functions and for discovering novel subplate processes. Importantly, this study also identified sets of genes involved in CNS disease and in the sexual differentiation of the brain that were enriched in future subplate neurons.
To identify genes expressed in future subplate neurons at the preplate stage, we compared gene expression in purified Pde1C-driven EGFP-positive preplate cells, representing a mixture of both types of preplate neurons, Cajal–Retzius cells and future subplate neurons, to gene expression in Girk4-driven EGFP-positive Cajal–Retzius cells (Fig. 1). We hypothesized that genes upregulated in Pde1C-driven EGFP-positive cells as compared to Girk4-driven EGFP-positive cells would have enriched expression in future subplate neurons. This screen identified 229 microarray features enriched in future subplate neurons (Fig. 2a, Supplemental Table 1). Candidates from this list were selected for expression analysis by immunostaining, and all were found to be expressed in future subplate at E12.5 and in the developing subplate layer at E14.5. The expression of some candidate genes was restricted to subplate neurons at the embryonic time points examined (Fig. 2b–d), whereas others were also expressed in Cajal–Retzius neurons (Fig. 2f,h) or cortical plate neurons (Fig. 2g).
Subplate-Specific Markers
In the present study, we identified 2 genes, Hpca and Eaac1, that are expressed specifically in the future subplate neurons at the preplate stage (Fig. 2b,d), and later in subplate (Fig. 2c,e). Hippocalcin, a calcium-binding protein of the NCS (neuronal calcium sensor) protein family is expressed in retinal photoreceptors and hippocampal pyramidal neurons at later stages of development (Burgoyne and Weiss 2001). The physiological roles of NCS proteins include modulation of neurotransmitter release, control of cyclic nucleotide metabolism, biosynthesis of polyphosphoinositides, regulation of gene expression and regulation of ion channels. EAAC1 is a Na+-dependent high affinity glutamate transporter, which regulates the uptake of glutamate, a key excitatory neurotransmitter. At later stages of development, EAAC1 is expressed in both pre- and postsynaptic elements of cortical pyramidal neurons and of cerebellar Purkinje neurons (Rothstein et al. 1994). These 2 genes provide novel subplate cellular markers and underscore the importance of genes involved in regulating neurotransmitters in developing subplate neurons.
Genes Involved in Synaptic Function
In addition to the subplate-specific genes Eaac1 and Hippa1, other genes involved in synaptic function were enriched in subplated neurons. These included a gamma-aminobutyric acid receptor (GABARα2) and somatostatin receptor (Sstr2) II, synaptotagmin, complexin1, synapsin II, and synaptotagmins 1 and 4 (Supplemental Table 1). Genes encoding other receptors, G-proteins, and transducers and second messenger systems, any of which might influence subplate neuron development and function, were also enriched in subplate neurons.
Genes involved in Cortical Development and CNS Disease
A number of the genes identified in this study have been shown to be important for corticogenesis, including the transcription factors NeuroD2, NeuroD6, and Mef2c, which is thought to upregulate expression of the transcription factor Mash1. We also identified genes associated with CNS disease, including several forms of ataxia (Atcay, A2ba2bp), Down syndrome (Tollip, Dscam), the Fragile-X mental retardation gene, and genes with potential roles in Huntington's Disease and seizures. It will be important to analyze the role of these genes in more detail to understand any putative role of the subplate in corticogenesis or cortical malformation.
Subplate Genes Involved in Axonogenesis and Axon Guidance
Functional classification of the subplate-enriched gene list using the DAVID Bioinformatics Software (Hosack et al. 2003) identified a cluster of 8 genes coding for proteins likely to be involved in neural development and neuritogenesis (Cck, Cckra, Slitrk1, Sema4d, Dscam, Ncam2, Nrcam, 9030425E11Rik, Supplemental Table 1). Several broad categories of genes involved in cell and axon motility were identified, including those which encode adhesion proteins (NCAM2, cadherins, Cdh11, Cdh13, Pcdh8) and axon guidance molecules (Sema7d, Sema7a, EphA3, EphA5, Nelf, Adamts4) (Supplemental Table 1). Other genes important for axonogenesis included the cyctoskeletal proteins (neurofilament proteins and MAP2) and those involved in cytoskeletal dynamics (Rho GTPases, Septin4) as well as genes involved in cell polarity (ligand of Numb-X).
Estrogen Signaling Network Genes
Our analysis of genes expressed in subplate neurons suggests a role for steroid hormone signaling in the development and function of subplate neurons. We identified an enriched cluster of 4 genes involved in cholesterol metabolism or estrogen signaling (Cy51, Cyp7b1, Tm7sf2, mevalonate (diphospho) decarboxylase (Supplemental Table 1). Network analysis using Ingenuity Pathway Analysis (Ingenuity Systems) revealed that a number of the genes with enriched expression in the subplate may be regulated by beta-estradiol (estrogen) signaling (Supplemental Fig. 4). Estrogen is synthesized de novo in brain and influences dendritogenesis in a number of different types of neurons, including hippocampal neurons (Prange-Kiel and Rune 2006) and cerebellar Purkinje neurons (Sasahara et al. 2007). In addition, estrogen receptors are required for normal migration and survival of some cortical plate neurons (Wang et al. 2003). Among the genes regulated by beta-estradiol are some of the candidate axon extension and signaling molecules described above (Dscam, Cdh11, Ccl25), and other genes known to be involved in neuritogenesis (Sez6, Nell2). These findings suggest that steroid hormone signaling may be involved in the development and maturation of subplate neurons, consistent with the important role of estrogen in the maturation of the neocortex (MacLusky et al. 1987; Ivanova and Beyer 2000), and imply a potential role for the preplate in the sexual differentiation of the brain (MacLusky et al. 1987; Hutchison 1997).
Genes Involved in Survival, Growth, and Apoptosis
The current model for subplate function is that neurons pattern the connectivity in cortical circuits during early periods of cortical development, and are gradually eliminated at later developmental stages. Our data sets include a number of apoptosis and anti-apoptosis genes (Casp9, Bid) and genes involved in insulin growth factor pathways (Insl6, Igfbpl1) and an opioid-growth factor like receptor gene (Ogfrl1), suggesting putative pathways involved in subplate neuron survival. These results underscore the critical role of genes involved in subplate neuron survival during early stages of cortical development.
Genes Involved in Transcriptional Processes
The largest group of genes expressed in subplate neurons was those involved in transcriptional processes. We identified 51 transcriptional regulators enriched in future subplate neurons, including several genes previously known to be involved in subplate specification or function (Supplemental Table 1). The transcription factor Sox5 was one of the transcription factors enriched in subplate neurons. Sox5 is of special interest as prior studies demonstrate that Sox5 is required for the proper specification of subplate neurons and early born projection neurons (Lai et al. 2008). Lmo4, which we found to be enriched in subplate neurons, is a calcium-activated transcription factor which, when conditionally deleted in forebrain, results in errors in thalamocortical pathfinding (Kashani et al. 2006), which may be attributable to defects in subplate gene expression (Ghosh et al. 1990). A similar phenotype of mistargeted thalamocortical axons has been observed in the cortex of mice lacking the basic helix–loop–helix transcription factor NeuroD2 (Ince-Dunn et al. 2006), which was also found to be enriched in future subplate. The other transcription factors identified as enriched in future subplate neurons could potentially be involved in the specification of subplate neuron identity, and in the regulation of subplate-specific signaling molecules which are instructive in the patterning of the cortex.
Our observation of scattered radial columns of beta-galactosidase-positive cells throughout the cortex of E13.5 Pde1C-driven Cre mice suggests that subplate neurons and cortical projection neurons may be derived from common progenitors. If this is the case, the stepwise formation of cortical circuits by subplate neurons and later-born cortical neurons may depend upon the activation of a common set of transcription factors and signaling pathways. The activation of a common set of transcription factors and pathways in cells derived from a common progenitor has been observed in both neuronal and non-neuronal development (Noctor et al. 2002; Johnson et al. 2005). The subplate neuron transcription factors and pathways identified in this study may reveal patterns of transcriptional activation that play an important role in cortical circuit formation. Detailed studies on the lineal relationship of subplate neurons and of later-born cortical neurons will be needed to examine this issue.
Genes that Define Subsets of Subplate Neurons
Subplate neurons pioneer axonal projections to multiple targets in the thalamus and contralateral hemisphere (McConnell et al. 1989; De Carlos and O'Leary 1992; Jacobs et al. 2007), raising the question as to whether subsets of subplate neurons have distinct lineages or patterns of gene expression. Prior immunocytochemical studies have identified interneuron markers (calbindin, neuropeptides) that are expressed in subsets of interneurons located in the subplate (Del Rio et al. 2000), and restricted patterns of gene expression in subpopulations of glutamatergic subplate neurons have been reported in the neonatal cortex (Hoerder-Suabedissen et al. 2008). The present study provides a broad set of potential markers for subsets of subplate neurons, based on genes that putatively guide axons or support excitatory versus inhibitory connections. In other work in our laboratory, we recently identified Mig13a/Lrp12 and showed that in Tg(Mig13a-EGFP) mice transgene expression is restricted to only those subplate neurons projecting to the anterior commissure, and is not expressed in those neurons projecting to the thalamus (S. Schneider and M.E. Hatten, manuscript in submission). Comparative analyses of BAC transgenic lines marking subpopulations of subplate neurons will be an invaluable tool in correlating molecular phenotypes with functional phenotypes in the subplate layer. The use of BAC transgenics to purify EGFP-tagged ribosomes from identified populations of CNS neurons offers the promise of more detailed molecular analyses of subplate neurons in specific stages of development or under specific physiological conditions (Doyle et al. 2008).
Supplementary Material
Supplementary material can be found at: http://www.cercor.oxfordjournals.org/.
Funding
National Institutes of Health grant (NINDS R01 NS15429) to M.E.H.
Supplementary Material
Acknowledgments
We gratefully acknowledge the critical advice of Drs S. Schneider, E.-E. Govek, N. Heintz, and C. Bargmann in this study. The Tg(Girk4-EGFP), Tg(Pde1C-EGFP), and Tg(Pde1C-Cre IT150) mice were provided by the GENSAT Project. Drs Joe Doyle and Joe Doughtery provided invaluable assistance with the Affymetrix gene expression data analysis. Conflict of Interest: None declared.
References
- Allendoerfer KL, Shatz CJ. The subplate, a transient neocortical structure: its role in the development of connections between thalamus and cortex. Annu Rev Neurosci. 1994;17:185–218. doi: 10.1146/annurev.ne.17.030194.001153. [DOI] [PubMed] [Google Scholar]
- Burgoyne RD, Weiss JL. The neuronal calcium sensor family of Ca2+-binding proteins. Biochem J. 2001;353:1–12. [PMC free article] [PubMed] [Google Scholar]
- Caviness VS., Jr Neocortical histogenesis in normal and reeler mice: a developmental study based upon [3H]thymidine autoradiography. Brain Res. 1982;256:293–302. doi: 10.1016/0165-3806(82)90141-9. [DOI] [PubMed] [Google Scholar]
- De Carlos JA, O'Leary DD. Growth and targeting of subplate axons and establishment of major cortical pathways. J Neurosci. 1992;12:1194–1211. doi: 10.1523/JNEUROSCI.12-04-01194.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Rio JA, Martinez A, Auladell C, Soriano E. Developmental history of the subplate and developing white matter in the murine neocortex. Neuronal organization and relationship with the main afferent systems at embryonic and perinatal stages. Cereb Cortex. 2000;10:784–801. doi: 10.1093/cercor/10.8.784. [DOI] [PubMed] [Google Scholar]
- Denaxa M, Chan CH, Schachner M, Parnavelas JG, Karagogeos D. The adhesion molecule TAG-1 mediates the migration of cortical interneurons from the ganglionic eminence along the corticofugal fiber system. Development. 2001;128:4635–4644. doi: 10.1242/dev.128.22.4635. [DOI] [PubMed] [Google Scholar]
- Doyle JP, Dougherty JD, Heiman M, Schmidt EF, Stevens TR, Ma G, Bupp S, Shrestha P, Shah RD, Doughty ML, et al. Application of a translational profiling approach for the comparative analysis of CNS cell types. Cell. 2008;135:749–62. doi: 10.1016/j.cell.2008.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda T, Kawano H, Ohyama K, Li HP, Takeda Y, Oohira A, Kawamura K. Immunohistochemical localization of neurocan and L1 in the formation of thalamocortical pathway of developing rats. J Comp Neurol. 1997;382:141–152. [PubMed] [Google Scholar]
- Ghosh A, Antonini A, McConnell SK, Shatz CJ. Requirement for subplate neurons in the formation of thalamocortical connections. Nature. 1990;347:179–181. doi: 10.1038/347179a0. [DOI] [PubMed] [Google Scholar]
- Gong S, Doughty M, Harbaugh CR, Cummins A, Hatten ME, Heintz N, Gerfen CR. Targeting Cre recombinase to specific neuron populations with bacterial artificial chromosome constructs. J Neurosci. 2007;27:9817–9823. doi: 10.1523/JNEUROSCI.2707-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong S, Zheng C, Doughty ML, Losos K, Didkovsky N, Schambra UB, Nowak NJ, Joyner A, Leblanc G, Hatten ME, et al. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature. 2003;425:917–925. doi: 10.1038/nature02033. [DOI] [PubMed] [Google Scholar]
- Hevner RF, Daza RA, Rubenstein JL, Stunnenberg H, Olavarria JF, Englund C. Beyond laminar fate: toward a molecular classification of cortical projection/pyramidal neurons. Dev Neurosci. 2003;25:139–151. doi: 10.1159/000072263. [DOI] [PubMed] [Google Scholar]
- Hevner RF, Shi L, Justice N, Hsueh Y, Sheng M, Smiga S, Bulfone A, Goffinet AM, Campagnoni AT, Rubenstein JL. Tbr1 regulates differentiation of the preplate and layer 6. Neuron. 2001;29:353–366. doi: 10.1016/s0896-6273(01)00211-2. [DOI] [PubMed] [Google Scholar]
- Hoerder-Suabedissen A, Wang WZ, Lee S, Davies KE, Goffinet AM, Rakic S, Parnavelas J, Reim K, Nicolic M, Paulsen O, et al. Novel markers reveal subpopulations of subplate neurons in the murine cerebral cortex. Cereb Cortex. 2008 doi: 10.1093/cercor/bhn195. [DOI] [PubMed] [Google Scholar]
- Hosack DA, Dennis G, Jr., Sherman BT, Lane HC, Lempicki RA. Identifying biological themes within lists of genes with EASE. Genome Biol. 2003;4:R70. doi: 10.1186/gb-2003-4-10-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison JB. Gender-specific steroid metabolism in neural differentiation. Cell Mol Neurobiol. 1997;17:603–626. doi: 10.1023/A:1022581902880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ince-Dunn G, Hall BJ, Hu SC, Ripley B, Huganir RL, Olson JM, Tapscott SJ, Ghosh A. Regulation of thalamocortical patterning and synaptic maturation by NeuroD2. Neuron. 2006;49:683–695. doi: 10.1016/j.neuron.2006.01.031. [DOI] [PubMed] [Google Scholar]
- Ivanova T, Beyer C. Ontogenetic expression and sex differences of aromatase and estrogen receptor-alpha/beta mRNA in the mouse hippocampus. Cell Tissue Res. 2000;300:231–237. doi: 10.1007/s004410000199. [DOI] [PubMed] [Google Scholar]
- Jacobs EC, Campagnoni C, Kampf K, Reyes SD, Kalra V, Handley V, Xie YY, Hong-Hu Y, Spreur V, Fisher RS, et al. Visualization of corticofugal projections during early cortical development in a tau-GFP-transgenic mouse. Eur J Neurosci. 2007;25:17–30. doi: 10.1111/j.1460-9568.2006.05258.x. [DOI] [PubMed] [Google Scholar]
- Johnson K, Shapiro-Shelef M, Tunyaplin C, Calame K. Regulatory events in early and late B-cell differentiation. Mol Immunol. 2005;42:749–761. doi: 10.1016/j.molimm.2004.06.039. [DOI] [PubMed] [Google Scholar]
- Jones L, Lopez-Bendito G, Gruss P, Stoykova A, Molnar Z. Pax6 is required for the normal development of the forebrain axonal connections. Development. 2002;129:5041–5052. doi: 10.1242/dev.129.21.5041. [DOI] [PubMed] [Google Scholar]
- Kashani AH, Qiu Z, Jurata L, Lee SK, Pfaff S, Goebbels S, Nave KA, Ghosh A. Calcium activation of the LMO4 transcription complex and its role in the patterning of thalamocortical connections. J Neurosci. 2006;26:8398–8408. doi: 10.1523/JNEUROSCI.0618-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai T, Jabaudon D, Molyneaux BJ, Azim E, Arlotta P, Menezes JR, Macklis JD. SOX5 controls the sequential generation of distinct corticofugal neuron subtypes. Neuron. 2008;57:232–247. doi: 10.1016/j.neuron.2007.12.023. [DOI] [PubMed] [Google Scholar]
- Landry CF, Pribyl TM, Ellison JA, Givogri MI, Kampf K, Campagnoni CW, Campagnoni AT. Embryonic expression of the myelin basic protein gene: identification of a promoter region that targets transgene expression to pioneer neurons. J Neurosci. 1998;18:7315–7327. doi: 10.1523/JNEUROSCI.18-18-07315.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLusky NJ, Clark AS, Naftolin F, Goldman-Rakic PS. Estrogen formation in the mammalian brain: possible role of aromatase in sexual differentiation of the hippocampus and neocortex. Steroids. 1987;50:459–474. doi: 10.1016/0039-128x(87)90032-8. [DOI] [PubMed] [Google Scholar]
- Marin-Padilla M. Cajal-Retzius cells and the development of the neocortex. Trends Neurosci. 1998;21:64–71. doi: 10.1016/s0166-2236(97)01164-8. [DOI] [PubMed] [Google Scholar]
- McConnell SK, Ghosh A, Shatz CJ. Subplate neurons pioneer the first axon pathway from the cerebral cortex. Science. 1989;245:978–982. doi: 10.1126/science.2475909. [DOI] [PubMed] [Google Scholar]
- Morante-Oria J, Carleton A, Ortino B, Kremer EJ, Fairen A, Lledo PM. Subpallial origin of a population of projecting pioneer neurons during corticogenesis. Proc Natl Acad Sci USA. 2003;100:12468–12473. doi: 10.1073/pnas.1633692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor SC, Flint AC, Weissman TA, Wong WS, Clinton BK, Kriegstein AR. Dividing precursor cells of the embryonic cortical ventricular zone have morphological and molecular characteristics of radial glia. J Neurosci. 2002;22:3161–3173. doi: 10.1523/JNEUROSCI.22-08-03161.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prange-Kiel J, Rune GM. Direct and indirect effects of estrogen on rat hippocampus. Neuroscience. 2006;138:765–772. doi: 10.1016/j.neuroscience.2005.05.061. [DOI] [PubMed] [Google Scholar]
- Rothstein JD, Martin L, Levey AI, Dykes-Hoberg M, Jin L, Wu D, Nash N, Kuncl RW. Localization of neuronal and glial glutamate transporters. Neuron. 1994;13:713–25. doi: 10.1016/0896-6273(94)90038-8. [DOI] [PubMed] [Google Scholar]
- Sasahara K, Shikimi H, Haraguchi S, Sakamoto H, Honda S, Harada N, Tsutsui K. Mode of action and functional significance of estrogen-inducing dendritic growth, spinogenesis, and synaptogenesis in the developing Purkinje cell. J Neurosci. 2007;27:7408–7417. doi: 10.1523/JNEUROSCI.0710-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki K, Miyagi T, Yoshida M, Miyata T, Ogawa M, Aizawa S, Suda Y. Absence of Cajal-Retzius cells and subplate neurons associated with defects of tangential cell migration from ganglionic eminence in Emx1/2 double mutant cerebral cortex. Development. 2002;129:3479–3492. doi: 10.1242/dev.129.14.3479. [DOI] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Wang L, Andersson S, Warner M, Gustafsson JA. Estrogen receptor (ER)beta knockout mice reveal a role for ERbeta in migration of cortical neurons in the developing brain. Proc Natl Acad Sci USA. 2003;100:703–708. doi: 10.1073/pnas.242735799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood JG, Martin S, Price DJ. Evidence that the earliest generated cells of the murine cerebral cortex form a transient population in the subplate and marginal zone. Brain Res. 1992;66:137–140. doi: 10.1016/0165-3806(92)90150-u. [DOI] [PubMed] [Google Scholar]
- Xie Y, Skinner E, Landry C, Handley V, Schonmann V, Jacobs E, Fisher R, Campagnoni A. Influence of the embryonic preplate on the organization of the cerebral cortex: a targeted ablation model. J Neurosci. 2002;22:8981–8991. doi: 10.1523/JNEUROSCI.22-20-08981.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C, Qiu Y, Pereira FA, Crair MC, Tsai SY, Tsai MJ. The nuclear orphan receptor COUP-TFI is required for differentiation of subplate neurons and guidance of thalamocortical axons. Neuron. 1999;24:847–859. doi: 10.1016/s0896-6273(00)81032-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.