Summary
G protein-coupled receptors (GPCRs) mediate responses to a broad range of chemical and environmental signals. In yeast a pheromone-binding GPCR triggers events leading to the fusion of haploid cells. In general, GPCRs function as guanine nucleotide exchange factors (GEFs); upon agonist binding the receptor induces a conformational change in the G protein α subunit, resulting in exchange of GDP for GTP and in signal initiation. Signaling is terminated when GTP is hydrolyzed to GDP [1]. This well-established paradigm has in recent years been revised to include new components that alter the rates of GDP release, GTP binding [2-8], and GTP hydrolysis [9, 10]. Here we report the discovery of a non-receptor GEF, Arr4. Like receptors, Arr4 binds directly to the G protein, accelerates guanine nucleotide exchange, and stabilizes the nucleotide-free state of the α subunit. Moreover, Arr4 promotes G protein-dependent cellular responses including mitogen-activated protein kinase (MAPK) phosphorylation, new gene transcription and mating. In contrast to known GPCRs, however, Arr4 is not a transmembrane receptor, but rather a soluble intracellular protein. Our data suggest that intracellular proteins function in cooperation with mating pheromones to amplify G protein signaling, thereby leading to full pathway activation.
Results
Screen for a novel GEF in yeast
Heterotrimeric G proteins are well known to function as signal transducers, coupling receptors at the cell surface to specific enzymes inside the cell. Recent research has uncovered the existence of non-receptor activators of G protein signaling, including a non-receptor GEF Ric-8a [2-4, 6-8]. It is not known however whether such factors contribute to GPCR signaling. To address this question, we investigated if the yeast mating-response pathway makes use of a non-receptor GEF. Accordingly, we considered 27 proteins reported to bind to the yeast Gα subunit Gpa1. Of the proteins we tested, seven exhibited preferential interaction with GDP-bound Gpa1 compared to the constitutively-active GTP-bound Gpa1Q323L (see Supplementary Data, Table 1). GDP-dependent binding is a common characteristic of GEFs for small GTPases [11], as well as for Ric-8a [3]. Since Gβ subunits and other GDIs also bind preferentially to Gα in the GDP-bound state, we next determined which of the candidate proteins preferentially interact with the Gα mutant Gpa1N388D, previously determined to form a stable but non-productive complex with a known GPCR and GEF Ste2 (binding was observed even in the absence of pheromone) [12]. We anticipated that the mutant G protein might bind preferentially to other proteins having exchange factor activity. Of the seven candidates tested, only Arr4 and Reg1 bound preferentially to Gpa1N388D. Notably, Arr4 is expressed as a dimer, as are many GPCRs including Ste2 [13, 14]. Relative to total expression Arr4 co-purified 8-fold more Gpa1N388D than Gpa1, and 3-fold more Gpa1 than Gpa1Q323L (Fig. 1 A and B). A similar preference was shown previously for binding to Ste2 [12].
Figure 1. Arr4 as a candidate GEF.
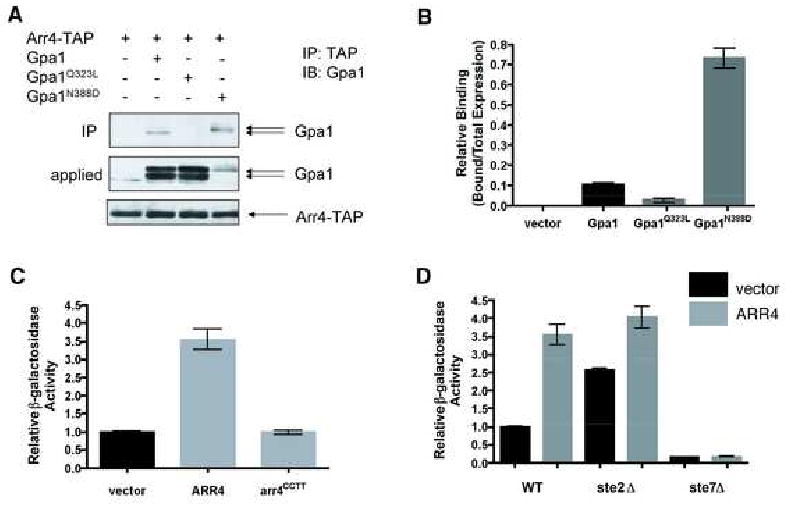
(A) Purification of Arr4-TAP from yeast. Cells expressing Arr4-TAP were transformed with vector (pAD4M) containing either no insert, GPA1, GPA1Q323L or GPA1N388D. TAP fusion protein was purified (IP) using Calmodulin-Sepharose resin, eluted in SDS-PAGE sample buffer, and resolved by immunoblotting (IB) with Gpa1 antibody. Gpa1 typically migrates as a doublet of 52 and 54 kDa, representing the myristoylated and unmyristoylated forms of the protein, respectively [27].
(B) Densitometry of data in panel A expressed as Gpa1 bound relative to total Gpa1 expressed. Data are mean ± SD of 3 separate experiments.
(C) Mating gene transcription-reporter assay: effect of ARR4 over-expression. Cells co-expressing FUS1-lacZ and vector (pAD4M) containing either no insert, ARR4, or dimerization-deficient mutant arr4CCTT were monitored for β-galactosidase activity.
(D) Effect of known mating components on Arr4 activity. Experiment performed as in C, except that receptor (ste2) or MAPK kinase (ste7) deletion strains were used when noted. Results for C and D are the mean ± SEM for 3 individual experiments each performed in triplicate.
Direct, nucleotide-dependent, subtype-selective interaction between Arr4 and Gpa1
We next investigated whether Arr4 over-expression activates G protein signaling in vivo. Over-expression of a GEF, even in the absence of any stimulus, often increases basal activation of the G protein. For example, over-expression of Ric-8a leads to increased basal phospho-ERK production in CHO cells [15]. Similarly, we found that over-expression of ARR4 induced a 3.5-fold increase in the basal transcription of Gpa1-dependent mating genes (FUS1-lacZ, see below) (Fig. 1C). Overexpression of the receptor Ste2 yields a comparable (∼ 3-fold) increase in basal FUS1-lacZ expression (data not shown), while pheromone binding produces an even larger (∼50-fold) increase in activity. This phenotype was dependent on the function of Arr4, as over-expression of a dimerization-deficient mutant Arr4CCTT had no effect [13]. Furthermore, Arr4-stimulated transcription required mating pathway effectors, such as Ste7, but not the pheromone receptor Ste2 (Fig. 1D). Basal activation by Arr4 was equivalent in the presence and absence of receptor; however the baseline activation was higher in the absence of receptor, as noted previously [16]. The mechanism by which Ste2 dampens basal activity is unknown, but may result from sequestration of the G protein heterotrimer and dampening of spontaneous activation in the absence of a pheromone signal. These data suggest that Arr4 functions as a GEF for Gpa1 in vivo.
To determine whether the interaction between Arr4 and Gpa1 is direct or facilitated by another protein, we purified 6xHIS-Gpa1 and GST-Arr4 from E. coli and reconstituted these proteins in the presence of GDP or GDP plus AlF4- (a transition state mimic that induces the activated conformation of Gα). Consistent with our in vivo observations, purified Arr4 co-precipitated more efficiently with the GDP-bound form of Gpa1 than with GDP-AlF4--bound Gpa1 (Fig. 2B, lanes 1-6). Arr4 did not interact with Gpa2, the only other Gα subunit in yeast (Fig. 2B, lanes 7-12).
Figure 2. Arr4 binds directly to Gpa1 and stabilizes the nucleotide-free state of the G protein.
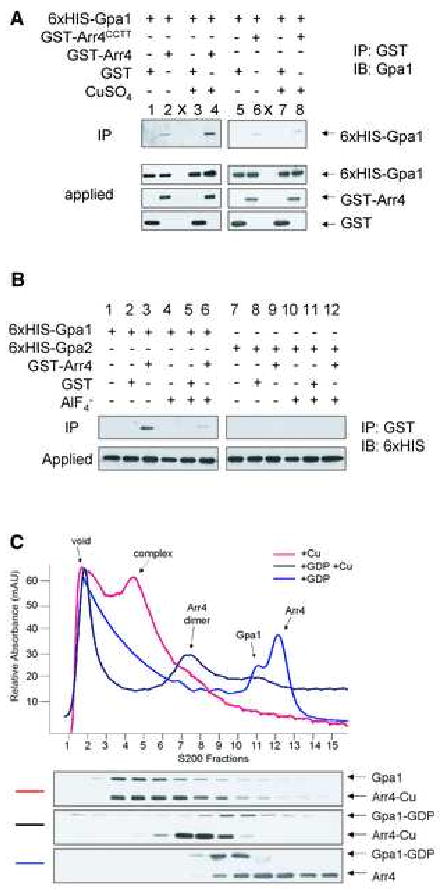
(A) and (B), Direct in vitro binding using recombinant purified components (E. coli). 100 nM of each protein was combined, purified with GST-Sepharose resin (IP), resolved by SDS-PAGE and probed with penta-HIS, Gpa1, or GST antibodies (IB). Note that the same protein preparations were used in the functional assays presented in Fig. 3.
(A) Binding with or without 150 nM CuSO4 (Cu) when noted.
(B) Arr4 binding to Gpa1 versus Gpa2 was performed as in panel A except that CuSO4 was present in all lanes.
(C) Arr4-Gpa1 complex formation using purified components. 4 mg of 6xHIS-Arr4 and 2 mg of 6xHIS-Gpa1 were combined and resolved by steric-exclusion chromatography in the presence or absence of excess GDP and CuSO4, as indicated. Note that the void volume elutes 200 minutes after sample loading. Top panel, A260nm chromatogram. The peak of UV absorbance in the void volume is evidently due to a non-protein buffer component, as determined by Coomassie staining as well as by immunoblotting with penta-His antibodies. Bottom panels, immunoblots using penta-HIS antibody. 20 μl of each 7 ml elution fraction were loaded and resolved by SDS-PAGE. All data are representative of 3 separate experiments.
Arr4 was shown to be a copper-binding protein, and copper induces Arr4 dimer formation [13]. As noted above Arr4 dimerization is required for Gpa1-mediated signaling in vivo (Fig. 1C). Dimerization is likewise required for strong binding to Gα, since Arr4 in the absence of copper bound poorly to Gpa1 in vitro (Fig. 2A). Copper did not stimulate binding between Gpa1 and the dimerization deficient Arr4CCTT mutant (Fig. 2A), consistent with the established evidence that copper binding induces dimerization. These data together show that dimerized Arr4 activates Gpa1-mediated signaling, and interacts with Gpa1 in a direct, subtype-selective, and GDP-dependent manner consistent with known GEFs.
Arr4 stabilizes nucleotide-free Gpa1
GPCRs are known to bind to and stabilize the nucleotide-free form of Gα proteins. To determine if Arr4 can perform this function, we resolved purified Arr4 and Gpa1 by steric-exclusion chromatography. Proteins were detected by absorbance, by immunoblotting, and by protein staining (Fig. 2C, and data not shown). In the presence of copper, Arr4 and Gpa1 co-eluted with a mass corresponding to that predicted for the Arr4-dimer/Gpa1 complex at a 2:1 stoichiometry. This complex was disrupted by the addition of excess GDP, yielding two distinct peaks: one at the predicted mass of Gpa1 and another corresponding to dimerized Arr4. The Arr4 dimer was further dissociated by the removal of copper (Fig. 2C). These results demonstrate that Arr4 dimer binds stably to the nucleotide-free form of Gα.
Arr4 promotes nucleotide exchange on Gpa1 in vitro
In the absence of a GEF, the rate-limiting step in the G protein nucleotide cycle is the release of GDP [17]. A GEF accelerates GTP binding by stabilizing the nucleotide-free state of Gα, thereby promoting GDP release. Using purified components, we found that a 2:1 molar ratio of Arr4 to Gpa1 accelerates the rate of GTP binding, from 0.085 pmol to 0.36 pmol GTPγS·pmol Gpa1-1·min-1 (Fig. 3A). This is comparable to the GEF activity reported for Ric-8a [3, 4] (the GEF activity of the receptor Ste2 has never been quantified). In the absence of copper Arr4 had almost no effect on GTPγS binding to Gpa1 (Fig. 3B). Arr4 alone, with or without copper, could not itself bind GTPγS (Fig. 3A and B).
Figure 3. Arr4 is a GEF for Gpa1.
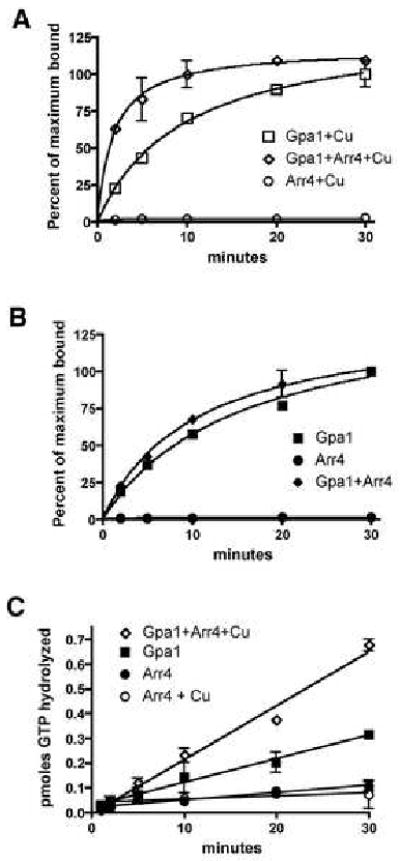
(A) and (B), Single turnover GTP binding assay. Time-course of [35S]GTPγS binding to 100 nM purified 6xHIS-Gpa1 in the presence of 200 nM GST-Arr4. Results are the mean ± SEM of duplicate samples, and are presented as percent of maximum bound (saturated binding occurred between 50-75%).
(A) with 500 nM CuSO4 added.
(B) Same as in A, but no copper added to the reaction.
(C) Steady state GTP hydrolysis assay. Time-course of Pi released in the presence or absence of 6xHIS-Gpa1 (250 nM), GST-Arr4 (500 nM), and copper (500 nM). Results are the mean ± SEM of duplicate samples.
Because GEFs function at the rate-limiting step in the GTPase cycle, accelerating nucleotide exchange should also increase the rate of GTP hydrolysis. As a second measure of Arr4 GEF activity, we showed that Arr4 accelerates the rate of [32P]GTP hydrolysis, from 0.009 to 0.021 pmol Pi·pmol Gpa1-1·min-1 (Fig. 3C). Arr4 itself, with or without copper, could not hydrolyze GTP. We also purified a related human protein (hAsna-I) [18], but detected no GEF activity towards Gpa1, Giα or Gsα (data not shown).
Arr4 promotes pheromone signaling in vivo
To determine the role of Arr4 in GPCR signaling, we used three measures of pathway activity: MAPK activation, gene transcription, and mating. Pheromone is known to stimulate a downstream kinase cascade consisting of Ste11, Ste7, and the partially redundant MAPKs Fus3 and Kss1. To determine if Arr4 modulates this response pathway, we first measured phosphorylation of Fus3 and Kss1 using a phospho-MAPK specific antibody. As shown in Fig. 4A, basal and pheromone-induced MAPK activation was substantially diminished in arr4 mutant cells. We also tested salt stress-dependent activation of another MAPK, Hog1. In this case we found no difference between wild-type cells and mutant cells that lack ARR4 (Fig. 4B).
Figure 4. Arr4 is necessary for maximal transmission of the mating signal.
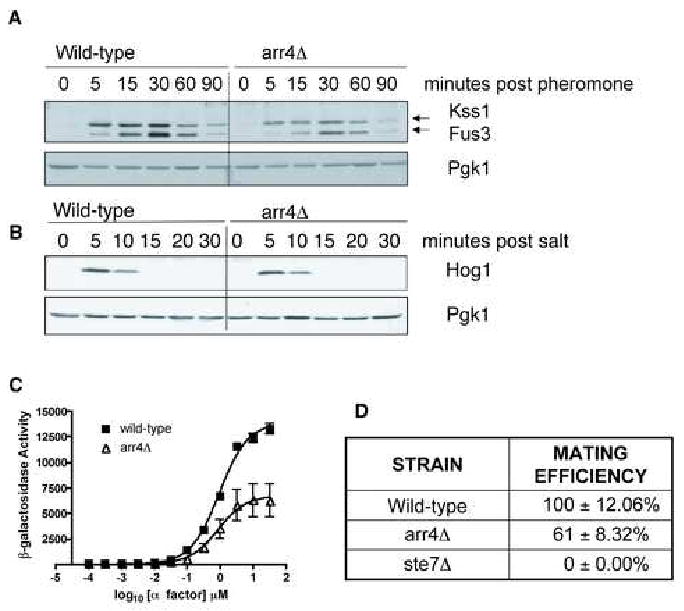
(A) Phospho-mating MAPK time course. Yeast cells were treated with 3 μM α-factor mating pheromone and samples were removed at times indicated. MAPK activation was determined by immunoblotting using a phospho-p44/42 MAPK antibody. Pgk1, loading control.
(B) Phospho-Hog1 MAPK time course. Performed as in (A) except cells were stimulated with 500 mM KCl, and probed with phospho-p38 MAPK antibody.
(C) Transcription-reporter dose-response. Cells expressing a FUS1-lacZ reporter were treated with the indicated concentrations of mating pheromone for 90 mins. Results are the mean ± SEM for 3 individual experiments each performed in triplicate.
(D) Mating efficiency assay. DC17 MATα cells were mixed with BY4741 (wild-type MATa cells), arr4 deletion, or ste7 deletion as a control. Mating was performed by co-incubation of cells on nitrocellulose filters. Maximum mating efficiency of the wild-type cells was approximately 75%. Percent mating efficiency was calculated as number of diploids/total number of MATa cells. Data are mean ± SD of 3 separate experiments.
Activation of the MAPKs Fus3/Kss1 results in selective induction of genes, such as FUS1. To further confirm that Arr4 is a component of the mating-response pathway, we measured transcription of mating-specific genes using the highly selective FUS1-lacZ transcription reporter. Compared to wild-type, deletion of ARR4 caused a ∼ 50% decrease in pheromone-induced transcription (Fig. 4C). Finally, to determine directly if Arr4 contributes to cell fusion, we measured mating efficiency of MATα cells with wild-type MATa cells or MATa cells that lack ARR4. When compared to wild-type, arr4 mutant cells were ∼40% less likely to mate successfully (Fig. 4D). The observed decrease in mating efficiency indicates that Arr4 contributes to mating, but is not essential for this activity. Deletion of other pathway modulators that bind Gpa1, such as the effector protein Vps34 and the RGS protein Sst2, exhibit a comparable degree of mating impairment [19-21]. The observed reduction in mating mirrors the reduction in MAPK and transcription-reporter activities, and further establishes Arr4 as a positive modulator of the mating pathway.
Discussion
For more than 20 years the yeast pheromone-response pathway has served as a model system for G protein signaling [9]. For the last decade, our understanding of the proteins involved in this pathway has remained relatively unchanged. Here, we report the discovery of a novel activator of yeast mating, one operating upstream of the G protein in conjunction with cell surface pheromone receptors. Specifically, we find that Arr4 binds directly to the Gα subunit Gpa1 and induces nucleotide exchange. Arr4 binds as a dimer, since the stoichiometry of the Arr4-Gpa1 complex is 2:1. Binding is selective, since Arr4 co-precipitates with Gpa1 but not Gpa2. Binding is stable, since the complex can be resolved by steric-exclusion chromatography. Binding appears to be ligand-dependent, since the complex is dissociated by removal of copper. Most of these behaviors are characteristic of known GPCRs, including the pheromone receptor Ste2. Finally, using complementary assays of mating pathway activation, we find that Arr4 promotes Fus3 (but not Hog1) MAPK activation, promotes Fus3-mediated gene induction, and enhances mating efficiency. Again, all of these behaviors are characteristic of pheromone receptor activation. Taken together, our results suggest that Arr4 functions as a GEF for Gpa1, and this GEF activity serves to amplify the pheromone-response pathway.
It is worth emphasizing that all of the bioassays used here are highly specific for the mating pathway. Fus3 and the transcription reporter FUS1-lacZ are activated only in mating-competent haploid cells, and only upon stimulation by mating pheromones. Neither response occurs in the absence of a functional G protein. While it is conceivable that Arr4 has additional functions in the cell, Arr4 clearly binds selectively and directly to Gpa1, and loss of this binding has significant consequences for the pheromone response. Moreover, comparable differences were observed for MAP kinase activity, transcription induction, and mating efficiency. Such differences are likely to be especially important in non-ideal (non-laboratory) growth conditions.
Arr4 binds preferentially to the nucleotide-free form of Gpa1, although binding can still be dectected for the GDP-bound form of the protein. Other cellular proteins, such as Gβγ, may further modulate the nucleotide-dependence of binding. Our working model, fully supported by the data, is that Arr4 acts after dissociation of G protein subunits, and thereby sustains the signal. Alternatively, Arr4 could function like Ric-8a to promote GTP binding to Gpa1 directly, in the absence of bound Gβγ. This model seems plausible considering that Gα expression was estimated to be ∼2-fold higher than that of Gβγ [22]. Moreover, we have shown that Arr4 binds stably to Gα in the absence of Gβγ (Fig. 2). Other possibilities are that Arr4 activates the G protein heterotrimer in cooperation with pheromone receptors, or competes with Gβγ for binding to Gα. Testing these more complex models will be challenging, since they require the ability to express and purify functional receptor and Gβγ.
Another open question is how Arr4 is itself regulated, and whether Arr4 is activated by any internal or external stimulus. Arr4 was originally named because of its resemblance to bacterial ARsenicals Resistance proteins. More recently, the ARR4 gene was implicated through hierarchical clustering analysis to be involved in ER/Golgi trafficking, and renamed GET3 [23]. ARR4/GET3 was also shown to function as an extragenic suppressor of npl4, encoding a component in the ubiquitin-proteasome system [24]. Gpa1 is ubiquitinated, but a role for Gpa1 activation in its ubiquitination has not been established [25]. Another possibility is that Arr4 is dynamically regulated by copper. In fact, Arr4 has been reported to localize to the cytoplasm but then translocate to punctate intracellular structures when cells are stressed with heat and/or metal ions [26]. The effect of these stresses on copper homeostasis is not known however. Regardless, Gpa1 is expressed on the cytoplasmic face of the plasma membrane and would therefore be fully accessible to Arr4 in vivo.
Identification of non-receptor exchange factors has been difficult, as no signature sequence or domain exists that accurately predicts activity. Indeed, there is no sequence similarity and no common predicted fold among receptor and non-receptor GEFs. Here we have identified a novel GEF based on its preferential binding to the Gpa1N388D mutant. Our data suggest that analogous mutations in other Gα proteins could be used to identify GEFs in other systems as well. Our genetic studies in yeast reveal a critical role for Arr4 signaling in vivo, and suggest further that non-receptor GEFs may be more common and have a broader physiological function than previously appreciated.
Experimental Procedures
Standard methods for maintenance of yeast and bacteria, manipulation of DNA, purification and detection of proteins, and analysis of G protein guanine nucleotide binding and hydrolysis were used throughout. Full Experimental Procedures and any associated references are available on the online version of the paper.
Supplementary Material
Acknowledgments
We thank D. Siderovski and C. Johnston for the gift of Giα and Gsα and for their guidance with the in vitro GEF assays, J. Snyder for assistance with protein purification, P. Naredi for the gift of hAsna-I, as well as J. Jones, J. Slessareva, C. Zeller, and S. Parnell for helpful comments. This work was supported by NIH grants GM065533 (to H.G.D.), F31-MH081472 (to M.J.L.), and the UNC CMB predoctoral fellowship program.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sprang SR. G protein mechanisms: insights from structural analysis. Annu Rev Biochem. 1997;66:639–678. doi: 10.1146/annurev.biochem.66.1.639. [DOI] [PubMed] [Google Scholar]
- 2.Cismowski MJ, Takesono A, Ma C, Lizano JS, Xie X, Fuernkranz H, Lanier SM, Duzic E. Genetic screens in yeast to identify mammalian nonreceptor modulators of G-protein signaling. Nat Biotechnol. 1999;17:878–883. doi: 10.1038/12867. [DOI] [PubMed] [Google Scholar]
- 3.Tall GG, Krumins AM, Gilman AG. Mammalian Ric-8A (synembryn) is a heterotrimeric Galpha protein guanine nucleotide exchange factor. J Biol Chem. 2003;278:8356–8362. doi: 10.1074/jbc.M211862200. [DOI] [PubMed] [Google Scholar]
- 4.Afshar K, Willard FS, Colombo K, Johnston CA, McCudden CR, Siderovski DP, Gonczy P. RIC-8 is required for GPR-1/2-dependent Galpha function during asymmetric division of C. elegans embryos. Cell. 2004;119:219–230. doi: 10.1016/j.cell.2004.09.026. [DOI] [PubMed] [Google Scholar]
- 5.Willard FS, Kimple RJ, Siderovski DP. Return of the GDI: the GoLoco motif in cell division. Annu Rev Biochem. 2004;73:925–951. doi: 10.1146/annurev.biochem.73.011303.073756. [DOI] [PubMed] [Google Scholar]
- 6.Hess HA, Roper JC, Grill SW, Koelle MR. RGS-7 completes a receptor-independent heterotrimeric G protein cycle to asymmetrically regulate mitotic spindle positioning in C. elegans. Cell. 2004;119:209–218. doi: 10.1016/j.cell.2004.09.025. [DOI] [PubMed] [Google Scholar]
- 7.Du Q, Macara IG. Mammalian Pins is a conformational switch that links NuMA to heterotrimeric G proteins. Cell. 2004;119:503–516. doi: 10.1016/j.cell.2004.10.028. [DOI] [PubMed] [Google Scholar]
- 8.Blumer JB, Cismowski MJ, Sato M, Lanier SM. AGS proteins: receptor-independent activators of G-protein signaling. Trends Pharmacol Sci. 2005;26:470–476. doi: 10.1016/j.tips.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 9.Dohlman HG, Thorner JW. Regulation of G protein-initiated signal transduction in yeast: Paradigms and principles. Annu Rev Biochem. 2001;70:703–754. doi: 10.1146/annurev.biochem.70.1.703. [DOI] [PubMed] [Google Scholar]
- 10.Neubig RR, Siderovski DP. Regulators of G-protein signalling as new central nervous system drug targets. Nat Rev Drug Discov. 2002;1:187–197. doi: 10.1038/nrd747. [DOI] [PubMed] [Google Scholar]
- 11.Rossman KL, Der CJ, Sondek J. GEF means go: turning on RHO GTPases with guanine nucleotide-exchange factors. Nat Rev Mol Cell Biol. 2005;6:167–180. doi: 10.1038/nrm1587. [DOI] [PubMed] [Google Scholar]
- 12.Wu YL, Hooks SB, Harden TK, Dohlman HG. Dominant-negative inhibition of pheromone receptor signaling by a single point mutation in the G protein alpha subunit. J Biol Chem. 2004;279:35287–35297. doi: 10.1074/jbc.M404896200. [DOI] [PubMed] [Google Scholar]
- 13.Metz J, Wachter A, Schmidt B, Bujnicki JM, Schwappach B. The yeast Arr4p ATPase binds the chloride transporter Gef1p when copper is available in the cytosol. J Biol Chem. 2006;281:410–417. doi: 10.1074/jbc.M507481200. [DOI] [PubMed] [Google Scholar]
- 14.Overton MC, Blumer KJ. G-protein-coupled receptors function as oligomers in vivo. Curr Biol. 2000;10:341–344. doi: 10.1016/s0960-9822(00)00386-9. [DOI] [PubMed] [Google Scholar]
- 15.Malik S, Ghosh M, Bonacci TM, Tall GG, Smrcka AV. Ric-8 enhances G protein betagamma-dependent signaling in response to betagamma-binding peptides in intact cells. Mol Pharmacol. 2005;68:129–136. doi: 10.1124/mol.104.010116. [DOI] [PubMed] [Google Scholar]
- 16.Hasson MS, Blinder D, Thorner J, Jenness DD. Mutational activation of the STE5 gene product bypasses the requirement for G protein beta and gamma subunits in the yeast pheromone response pathway. Mol Cell Biol. 1994;14:1054–1065. doi: 10.1128/mcb.14.2.1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ferguson KM, Higashijima T, Smigel MD, Gilman AG. The influence of bound GDP on the kinetics of guanine nucleotide binding to G proteins. J Biol Chem. 1986;261:7393–7399. [PubMed] [Google Scholar]
- 18.Kurdi-Haidar B, Heath D, Aebi S, Howell SB. Biochemical characterization of the human arsenite-stimulated ATPase (hASNA-I) J Biol Chem. 1998;273:22173–22176. doi: 10.1074/jbc.273.35.22173. [DOI] [PubMed] [Google Scholar]
- 19.Chan RK, Otte CA. Physiological characterization of Saccharomyces cerevisiae mutants supersensitive to G1 arrest by a factor and α factor pheromones. Mol Cell Biol. 1982;2:21–29. doi: 10.1128/mcb.2.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chan RK, Otte CA. Isolation and genetic analysis of Saccharomyces cerevisiae mutants supersensitive to G1 arrest by a factor and α factor pheromones. Mol Cell Biol. 1982;2:11–20. doi: 10.1128/mcb.2.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Slessareva JE, Routt SM, Temple B, Bankaitis VA, Dohlman HG. Activation of the phosphatidylinositol 3-kinase Vps34 by a G protein alpha subunit at the endosome. Cell. 2006;126:191–203. doi: 10.1016/j.cell.2006.04.045. [DOI] [PubMed] [Google Scholar]
- 22.Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O'Shea EK, Weissman JS. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 23.Schuldiner M, Collins SR, Thompson NJ, Denic V, Bhamidipati A, Punna T, Ihmels J, Andrews B, Boone C, Greenblatt JF, Weissman JS, Krogan NJ. Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell. 2005;123:507–519. doi: 10.1016/j.cell.2005.08.031. [DOI] [PubMed] [Google Scholar]
- 24.Auld KL, Hitchcock AL, Doherty HK, Frietze S, Huang LS, Silver PA. The conserved ATPase Get3/Arr4 modulates the activity of membrane-associated proteins in Saccharomyces cerevisiae. Genetics. 2006;174:215–227. doi: 10.1534/genetics.106.058362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Madura K, Varshavsky A. Degradation of Gα by the N-end rule pathway. Science. 1994;265:1454–1458. doi: 10.1126/science.8073290. [DOI] [PubMed] [Google Scholar]
- 26.Shen J, Hsu CM, Kang BK, Rosen BP, Bhattacharjee H. The Saccharomyces cerevisiae Arr4p is involved in metal and heat tolerance. Biometals. 2003;16:369–378. doi: 10.1023/a:1022504311669. [DOI] [PubMed] [Google Scholar]
- 27.Stone DE, Cole GM, de Barros Lopes M, Goebl M, Reed SI. N-myristoylation is required for function of the pheromone-responsive G alpha protein of yeast: conditional activation of the pheromone response by a temperature-sensitive N-myristoyl transferase. Genes Dev. 1991;5:1969–1981. doi: 10.1101/gad.5.11.1969. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


