Abstract
Colicin D has long been thought to stop protein synthesis in infected Escherichia coli cells by inactivating ribosomes, just like colicin E3. Here, we show that colicin D specifically cleaves tRNAsArg including four isoaccepting molecules both in vivo and in vitro. The cleavage occurs in vitro between positions 38 and 39 in an anticodon loop with a 2′,3′-cyclic phosphate end, and is inhibited by a specific immunity protein. Consistent with the cleavage of tRNAsArg, the RNA fraction of colicin-treated cells significantly reduced the amino acid-accepting activity only for arginine. Furthermore, we generated a single mutation of histidine in the C-terminal possible catalytic domain, which caused the loss of the killing activity in vivo together with the tRNAArg-cleaving activity both in vivo and in vitro. These findings show that colicin D directly cleaves cytoplasmic tRNAsArg, which leads to impairment of protein synthesis and cell death. Recently, we found that colicin E5 stops protein synthesis by cleaving the anticodons of specific tRNAs for Tyr, His, Asn, and Asp. Despite these apparently similar actions on tRNAs and cells, colicins D and E5 not only exhibit no sequence homology but also have different molecular mechanisms as to both substrate recognition and catalytic reaction.
Colicins are plasmid-encoded proteins that are toxic to Escherichia coli cells that do not have the same plasmid or a cognate Col plasmid (1–5). Most colicins are produced in response to SOS-inducing signals and are secreted into the medium. After binding to cell-surface receptors on sensitive cells, they are translocated across the membrane and then exert their final cytotoxic activities, which are attributable to their C-terminal domains. Two major modes of toxicity are well known; colicins A, B, E1, Ia, Ib, K, and N are ion-channel formers attacking the cytoplasmic membrane, and colicins E2 to E9 are nucleases. In the latter group, E2, E7, E8, and E9 are DNases, and E3 is a special kind of RNase that cleaves 16S rRNA within ribosomes.
Colicins E4 to E6 quickly stop amino acid incorporation in treated cells (6), suggesting impairment of protein synthesis analogous in the established case of E3, which specifically cleaves 16S-RNA at the 49th bond from the 3′ end leading to inactivation of ribosomes (7- 9). This is also the case with colicin D, which Timmis and Hedges have characterized as to the physiological response of treated cells (10, 11), although the actual molecular basis of the cytotoxic effect of colicin D, as well as those of E4 to E6, remained to be elucidated. Colicins E4 and E6 proved to be E3-homologs and showed comparable activity toward ribosomes (ref. 12; GenBank accession number X63621; Y. Gunji, M. Ohno, T.O., H.M., and T.U., unpublished data), but colicin E5 exhibits no similarity to E3 in the C-terminal active domain. We recently showed that colicin E5 comprises a third category of nuclease-type colicins, which does not attack ribosomes but specific tRNAs (13). E5 is a novel RNase that cleaves the anticodons of tRNATyr, tRNAHis, tRNAAsn, and tRNAAsp, which leads to impairment of protein synthesis and cell death. Thus, colicin D might be another “tRNase” candidate because it exhibits no sequence homology to E3 nor to E5.
On the other hand, colicin D shares the FepA receptor and the translocation pathway including TonB with colicin B, which belongs to the channel-former type. Thus, the N-terminal 45% region of colicin D, which is highly homologous to that of colicin B, is thought to function in receptor binding and membrane transfer. In contrast, the remaining C-terminal regions of colicins B and D, which should be responsible for their cytotoxic activities, are entirely different (14, 15). Col plasmids synthesize specific immunity proteins to evade the lethality of their own colicins. The immunity proteins of channel-former colicins, including colicin B, are membrane-bound, and imm genes are expressed independently of col genes whereas the immunity proteins of colicins E2 to E9 bind tightly to cognate colicins forming heterodimer complexes and their imm genes are co-transcribed with col genes. The latter is the case for the colicin D immunity protein (ImmD) and the corresponding gene. The molecular mechanism underlying the cytotoxicity of colicin D is not known, but the above comparison with other colicins led us to the question of whether or not colicin D is some kind of RNase.
Here, we provide an answer to this question. Colicin D cleaves specific tRNAs both in vivo and in vitro. Thus, it is the second member of the tRNase-type colicin family, but, interestingly, so many traits are different between colicins D and E5.
Materials and Methods
Chemicals and Bacterial Strains.
[γ-32P]ATP (111 TBq/mmol; Amersham) was used for 5′-labeling of RNAs with T4 polynucleotide kinase (Toyobo, Osaka), and [5′-32P]cytidine-3′,5′-bisphosphate (pCp) (111 TBq/mmol; Amersham) for 3′-labeling with T4 RNA ligase (Pharmacia). Nineteen kinds of [14C] or [35S] amino acids, excluding asparagine, a radiolabeled compound not being available, were purchased from Amersham or NEN. RNaseT1, RNaseT2, and RNaseU2 were purchased from Sigma. RNasePhyM, RNaseCL3, NucleaseP1, and E. coli A19 alkaline phosphatase were obtained from Pharmacia, Boehringer Mannheim, Yamasa Shoyu (Choshi, Japan), and Takara Shuzo (Kyoto), respectively. Streptavidin-agarose was from BRL.
Cells were aerobically grown in LB medium (16) at 37°C. A streptomycin-resistant derivative of E. coli K12 W3110 was used as the colicin-sensitive strain, from which a spontaneous colicin D-resistant mutant, DR1, was isolated. DR1 was also resistant to colicin B and sensitive to colicins Ia and Ib, suggesting that the mutation is in the gene for the colicin B/D receptor fepA.
Preparation of Colicin D and ImmD.
A log phase culture of E. coli K12 RR1 [ColD-CA23] in 1 liter of medium was incubated for 3 h after the addition of 0.4 mg/l mitomycin C. The cells were harvested, were resuspended in 30 ml of 20 mM potassium phosphate (pH 6.8) (buffer A) and 0.5 mM phenylmethanesulfonyl fluoride, were sonicated, and then were centrifuged at 100,000 × g for 3 h. The supernatant was applied to a DEAE-TOYOPEARL 650S (Tosoh, Tokyo) column and was eluted with a KCl gradient in buffer A. The colicin fraction was applied to a Mono-S (Pharmacia) column, was eluted with a KCl gradient in 10 mM sodium acetate buffer (pH 5.0), and then was dialyzed against buffer A. The purified colicin, as a complex with ImmD, was incubated in buffer A containing 8 M urea for 2 h at room temperature. The unfolded colicin D and ImmD proteins were separated on a Superose 12HR 10/30 (Pharmacia) or Bio-gel P30 (Bio-Rad) column, and then were refolded by stepwise dialysis against 50 mM Hepes⋅KOH (pH 7.8). Quantification of the proteins was carried out with a Protein Assay Kit (Pierce).
Preparation and Aminoacylation of RNAs from Colicin-Treated Cells.
W3110 or DR1 cells grown in 100 ml medium to A660 = 0.5 were incubated with or without 22.5 μg of partially purified colicin D (DEAE fraction) for 55 min and then were quickly harvested. RNAs were prepared from the cells by the guanidinium thiocyanate method (17). For aminoacylation, 0.06 A260 units of total RNA was incubated for 15 min at 37°C in a 20-μl reaction mixture comprising 100 mM Hepes⋅KOH (pH 7.8), 15 mM MgCl2, 20 mM KCl, 1 mM DTT, 1 mM ATP, 2 μl of the S100 fraction prepared from E. coli K12 A19, and 10–50 μM 1 of 19 kinds of [14C] amino acids (2.0–16.8 GBq/mmol) or [35S] cysteine (800 GBq/mmol). A 17-μl aliquot of each reaction mixture was withdrawn, was spotted onto Whatman 3MM paper, and then was washed with ice-cold 5% trichloroacetic acid for 15 min three times, followed by washing with cold ethanol. The radioactivity incorporated into the acid-insoluble fraction was measured with a liquid scintillation counter.
Northern Blot Hybridization.
Total RNA (2.0 μg) isolated as above was electrophoresed on a 10% polyacrylamide gel containing 7 M urea and then was transferred to a Hybond-N+ membrane (Amersham) according to the manufacturer's instructions. Hybridization was carried out overnight at 55°C in a buffer comprising 900 mM NaCl, 90 mM Tris⋅HCl (pH 7.5), 6 mM EDTA, and 0.3% (wt/vol) SDS. The membrane was washed in 6 × standard saline citrate (SSC) (16) at room temperature for 15 min and then in 3× SSC for a total of 60 min at 55°C. The DNA sequences used as specific probes were 5′-CCTCCGACCGCTCGG-3′ for tRNAArgICG, 5′-CCTGAGACCTCTGCC-3′ for tRNAArgCCG, 5′-CCTGCGGCCCACGAC-3′ for tRNAArgU*CU (U*: 5-methylaminomethyluridine), and 5′-AACCTGCAATTAGCCC-3′ for tRNAArgCCU (18). The DNA probes for control RNAs, tRNATyr, tRNAHis, tRNAAsn tRNAAsp, tRNA2Gln, tRNALys, tRNAGlu, and 16S-rRNA were described previously (13). The DNA probes for tRNATrp, tRNACys, tRNA3Ser, tRNA2Gly, and tRNA3Gly were designed to cover the corresponding tRNA regions of the above tRNAsArg.
In Vitro Cleavage Analysis of tRNAs.
To determine the cleavage sites in vitro, tRNAArgICG, tRNAArgU*CU, and tRNAArgCCU were purified from an unfractionated tRNA mixture by the solid-phase hybridization method (19, 20). The nucleotide sequences of the 3′-biotinylated oligonucleotides used in this tRNA isolation were 5′-AACCTCCGACCGCTCGGTTCGTAGCCG-3′ for tRNAArgICG, 5′-CCTGCGGCCCACGACTTAGAAGGTC-3′ for tRNAArgU*CU, and 5′-CCTGCAATTAGCCCTTAGGAGGGGCT-3′ for tRNAArgCCU. The isolated tRNAsArg were purified by PAGE, and their nucleotide sequences were confirmed by Donis-Keller's method (21). Each purified tRNA was 5′- or 3′-32P-labeled and then was subjected to an in vitro cleavage reaction in such a way that about 20,000 cpm was contained in the reaction mixture. The extents of cleavage were determined with an imaging analyzer BAS 1000 (Fuji). Enzymatic and alkaline cleavage sequence ladders of 32P-labeled tRNAsArg were used as references to determine the cleavage sites.
Mutagenesis of the Colicin D Gene.
To manipulate plasmid ColD-CA23, the SalI cartridge of the pUC-4K kanamycin resistance gene (22) was flush-ended and then inserted into the unique PvuII site in the mob region of ColD-CA23, giving rise to plasmid ColD-Km. Each histidine codon (CAT) in the colD gene corresponding to His536, His545, His611, and His637 in the C-terminal domain of colicin D was mutated to a tyrosine codon (TAT) using a Quick Change Mutagenesis Kit (Stratagene). The oligonucleotides used for the mutagenesis were 5′-GATACAGGTAATTATCAACCCGTTCCG-3′, 5′-GTTACACCAGTGTATACAGGAACGGAAG-3′, 5′-GATAAAAAATATAAATATGCTGGTGATTTTG-3′, and 5′-GCTATTGAGGAGTATTTATCGGATAAGG-3′ for the His536, His545, His611, and His637 mutants, respectively. The DNA sequences of the mutant plasmids were confirmed. The plasmids were introduced into W3110, and then mutant colicins were prepared, as in the case of the wild type. No differences were observed in the chromatographic and electrophoretic patterns between the mutant and wild-type colicins. For in vivo and in vitro tRNA cleavage experiments, partially purified DEAE fractions of colicin D mutants and the purified mutant colicins devoid of ImmD, respectively, were used.
Results
Colicin D-Treated Cells Produce a Pair of tRNAArgICG Fragments.
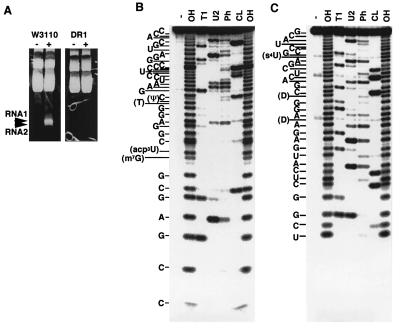
Based on the assumption that colicin D is some kind of a nuclease, we first looked for specific nucleic acid molecules that might arise on the treatment of cells with colicin D. E. coli K12 W3110 and its colicin-resistant derivative, DR1, were challenged with colicin D during the logarithmic growth phase, and total RNA was isolated from each cell sample. On electrophoretic analysis of the RNA, two close bands, RNA1 and RNA2, corresponding to about 30–40 nucleotides were observed, only for the colicin treated colicin-sensitive cells (Fig. 1A).
Figure 1.
RNA fragments that appeared in response to the action of colicin D. (A) RNAs were prepared from W3110 or DR1 cells grown in 100 ml of medium and then incubated with or without partially purified colicin D. The RNAs (20 μg) were separated on a 10% polyacrylamide gel containing 7 M urea in TBE buffer (16) and then were stained with ethidium bromide. RNA1 and RNA2, which appeared specifically with colicin-treated W3110, are indicated by arrowheads. Direct sequencing according to Donis-Keller (21) showed that 5′-labeled RNA1 corresponded to the 3′-fragment (positions 39–76) of tRNAArgICG (B) and that 3′-labeled RNA2 corresponded to the 5′-fragment (positions 1–34) of tRNAArgICG (C). -, OH, T1, U2, Ph, and CL indicate samples without digestion, and digested with alkali, and RNasesT1, U2, PhyM, and CL3, respectively.
RNA1 and RNA2 were extracted and purified from a preparative gel for further sequence analysis. Because no information was available on their 3′ and 5′ end forms, we first treated the RNAs with alkaline phosphatase before 5′-end labeling with T4 polynucleotide kinase and [γ-32P]ATP or 3′-end labeling with T4 RNA ligase and [5′-32P]pCp. The sequencing of the 5′- labeled RNA1 showed that it corresponded to the 3′- half fragment of tRNAArgICG, i.e., positions 39–76 (Fig. 1B). Consistent with this, RNA2 corresponded to the 5′-fragment of the tRNAArgICG, i.e., positions 1 to 34 (Fig. 1C). Both the 5′-end of RNA1 (3′-fragment of tRNAArgICG) and the 3′-end of RNA2 (5′-fragment of tRNAArgICG) proved susceptible to 32Plabeling without dephosphorylation by alkaline phosphatase, indicating that each end consists of a hydroxyl group. In conclusion, colicin D-treated cells produce a pair of specific fragments of tRNAArgICG, from positions 1–34 (RNA2) and 39–76 (RNA1). Both molecules contain new ends lacking a phosphate group.
tRNAsArg Are Cleaved Through the Action of Colicin D in Vivo.
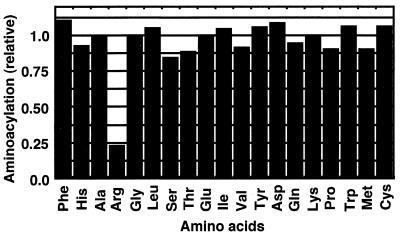
To determine whether the activity of cytoplasmic tRNAsArg is in fact impaired through the action of colicin D and whether tRNA species other than tRNAArgICG are affected, the aminoacylation of tRNAs was examined for each amino acid. Total RNA was prepared from a pair of growing W3110 cultures and was incubated with and without colicin D. The amino acid-accepting activities of these two RNA preparations were compared. Fig. 2 shows that only arginine-acceptance was drastically reduced for the RNA from the cells treated with colicin D, with the level decreasing to about 25% of that in the cells without treatment. This indicates that tRNAArg, among all of the tRNAs, was specifically inactivated in vivo through the action of colicin D. This result is consistent with the appearance of fragments of the major tRNAArg in response to colicin D (Fig. 1).
Figure 2.
Aminoacylation activities of the tRNAs in colicin D-treated cells. The incorporation of each amino acid into the RNA fraction prepared from colicin-treated cells using the S100 fraction of E. coli K12 A19 is indicated relative to that without colicin treatment. The data represent the averages of two independent experiments.
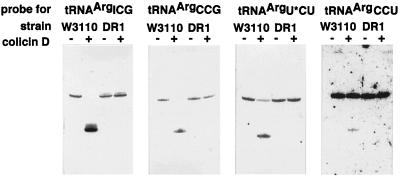
The results of Fig. 2 suggest that tRNA species other than those for arginine are not sensitive to colicin D, but does not definitely predict whether minor isoaccepting tRNAsArg are as sensitive as the major one. Therefore, several species of tRNAs prepared from colicin-treated cells were analyzed by Northern blot hybridization. Oligo DNA probes were designed to discriminate tRNAArgICG, tRNAArgCCG, tRNAArgU*CU, and tRNAArgCCU. As shown in Fig. 3, not only tRNAArgICG but also tRNAArgCCG, tRNAArgU*CU, and tRNAArgCCU proved to be cleaved in vivo through the action of colicin D. The cleavage of these tRNAs was not observed without colicin treatment nor for RNA prepared from a colicin-treated colicin-resistant strain, DR1. All other tRNAs examined were not cleaved by colicin D. These include tRNATyr, tRNAHis, tRNAAsn, and tRNAAsp, which are cleaved by colicin E5 (13), in addition to tRNA2Gln, tRNALys, and tRNAGlu. This is also the case for tRNACys, tRNATrp, tRNA3Ser, tRNA2Gly, and tRNA3Gly, which share C35 at the anticodon second position with tRNAsArg. Furthermore, 16S-rRNA was never cleaved by colicin D in vivo (data not shown).
Figure 3.
Northern blot hybridization of tRNAs prepared from W3110 or DR1 after colicin D treatment using four DNA probes specific to tRNAsArg.
Colicin D Cleaves tRNAsArg Between Positions 38 and 39 in Vitro.
The above findings do not exclude the possibility that the cleavage of tRNAsArg is caused indirectly through some cellular mechanism in response to the colicin D treatment. The in vitro action of purified colicin D on E. coli total tRNA was thus analyzed by Northern blot hybridization with DNA probes specific to individual tRNAs used in the in vivo experiment. The colicin D was found to directly cleave tRNAArgICG and tRNAArgCCG, although the extent of cleavage by colicin D in vitro may vary with the tRNAArg species. These changes were inhibited by the addition of ImmD, implying that this immunity protein is the inhibitor of the tRNA-cleaving activity of colicin D (Fig. 4). At the same time, this inhibition excludes the possibility of contamination of the reaction mixture by unknown nucleases. No other tRNAs were cleaved by colicin D in vitro, which is consistent with the in vivo results (data not shown).
Figure 4.
In vitro cleavage of tRNAsArg by colicin D. The purified colicin D (2–50 nM) was preincubated with or without 100–500 nM ImmD for 15 min at 37°C in 10 mM Hepes⋅KOH (pH 7.8) and 1 mM DTT. E. coli tRNA mixture (derived from MRE 600; Sigma) was then added to the reaction mixture to 5.0 A260 units/ml, followed by incubation for 10 min at 37°C. The RNAs were separated by electrophoresis on a 10% polyacrylamide gel containing 7 M urea, and then were analyzed by Northern blot hybridization using DNA probes specific to tRNAArgICG (A) and tRNAArgCCG (B). The arrows indicate cleaved 3′ fragments of the tRNAsArg. The weaker signal for the uncleaved tRNAArgCCG band compared with that for the cleaved one is possibly attributable to different efficiencies of hybridization and/or transfer to the membrane.
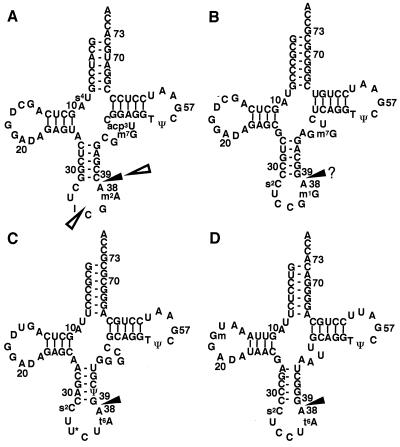
To exactly map the cleavage sites in tRNAsArg, three tRNAArg species, tRNAArgICG, tRNAArgU*CU, and tRNAArgCCU, were purified. We could not purify another isoacceptor, tRNAArgCCG, possibly because of its low content in the total RNA. As shown in Fig. 5, the cleavage sites in tRNAsArg examined were unique between nucleotide positions 38 and 39 (Fig. 6). Because tRNAArgICG was cleaved in vivo into two discontinuous fragments, 1–34 (RNA2) and 39–76 (RNA1) (Figs. 1 B and C, and 6A), four nucleotide residues corresponding to 35–38 were missing from the in vivo products compared with the in vitro ones. Probably in a cell, tRNAArgICG is first cleaved by colicin D between positions 38 and 39, and subsequently the resulting 5′-fragment corresponding to positions 1–38 may be digested to one comprising positions 1–34 by some other nuclease(s).
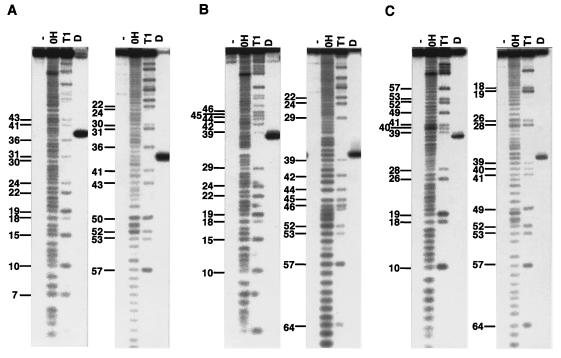
Figure 5.
Cleavage sites of tRNAArgICG (A), tRNAArgU*CU (B), and tRNAArgCCU (C) with purified colicin D. To exactly map the cleavage sites, both 5′-32P-labeled (left panels of A, B, and C) and 3′-32P-labeled (right panels of A, B, and C) tRNAsArg were subjected to the in vitro cleavage reaction with the purified colicin D. -, OH, T1, and D indicate samples without digestion, and digested with alkali, RNaseT1, and colicin D, respectively. The numbering system conforms to the proposal of Sprinzl et al. (18) (see Fig. 6).
Figure 6.
The clover-leaf structures of E. coli tRNAArgICG (A), tRNAArgCCG (B), tRNAArgU*CU (C), and tRNAArgCCU (D). The cleavage sites in vivo and in vitro are shown by open and solid triangles, respectively. For the cleavage sites in tRNAArgICG, two sites were identified in vivo but only one in vitro (see Results). The cleavage site of tRNAArgCCG, which is indicated by a solid triangle with the “?,” was deduced from the other tRNAArg cleavage sites. The numbering system conforms to the proposal of Sprinzl et al. (18).
The 5′ termini of the 3′-half fragments of tRNAsArg were found to have a hydroxyl group because these fragments were 5′-32P-labeled without prior dephosphorylation with alkaline phosphatase, as in the case of that obtained on in vivo colicin treatment. In contrast, the 3′ termini of the 5′-half fragments of tRNAsArg were deduced to have a 2′,3′-cyclic phosphate group because the fragments could not be 3′-labeled with T4 RNA ligase and [5′-32P]pCp even after alkaline phosphatase treatment. Alkaline phosphatase treatment made the 5′-half fragments of tRNAsArg prone to 3′-labeling only after acid treatment, which cleaves 2′,3′-cyclic phosphate (data not shown). Thus, the mode of cleavage of tRNAsArg by colicin D is typical of most usual ribonucleases with the 5′-hydroxyl and 2′,3′-cyclic phosphate ends being retained.
A Mutation Possibly Involved in the Catalytic Reaction.
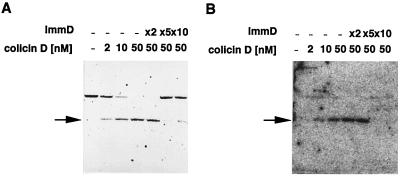
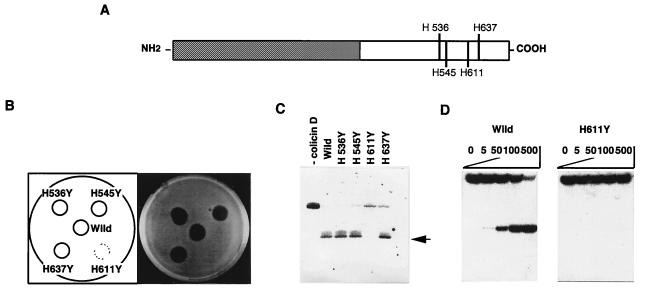
To elucidate the correlation between the ribonuclease activity of colicin D and its cytotoxicity, the putative catalytic residues in colicin D were mutated. Because all ribonucleases reported to date, except for colicin E5, have at least one histidine as a catalytic residue (23, 24), each of the four histidine residues located in the C-terminal domain of colicin D was changed to tyrosine (Fig. 7A). The four mutant colicins were stably produced in response to mitomycin C, but only mutant H611Y completely lost the lethal activity toward E. coli (Fig. 7B), suggesting the involvement of residue H611 in the cytotoxic action of colicin D. Northern blot analysis of tRNAArgICG from cells treated with the colicin mutants confirmed this suggestion. No cleavage of tRNAArgICG was observed in vivo with the mutant colicin H611Y (Fig. 7C). In contrast, with the other mutant colicins, H536Y, H545Y, and H637Y, cleavage of tRNAArgICG was observed to almost the same extent as the wild-type colicin D. In addition, the H611Y mutation of colicin D has abolished most of the in vitro RNase activity toward the purified and 5′-labeled tRNAArgICG, the level being less than 0.1% of that observed for the wild-type colicin D (Fig. 7D). These results show that H611 is essential for both the nuclease activity and the cytotoxicity of colicin D.
Figure 7.
Effects of replacement of a histidine with a tyrosine residue in the C-terminal region of colicin D (A) on its cytotoxicity (B), and nuclease activity in vivo (C) and in vitro (D). (B) One microgram of an extract (S100 fraction) prepared from cells producing each mutant colicin was spotted onto a W3110 layer on an agar plate, followed by incubation overnight at 37°C. (C) Cleavage of the intracellular tRNAArgICG with various colicin D mutants, as observed on Northern blot hybridization with the probe for tRNAArgICG. Arrows indicate the cleaved 3′ fragment of tRNAArgICG. (D) Cleavage of the purified and 5′-32P-labeled tRNAArgICG by the purified wild-type (Left) and H611Y (Right) colicin D.
Discussion
We have revealed that colicin D kills E. coli cells by means of its specific ribonuclease activity toward tRNAsArg. Direct sequencing of the RNA fragments, which appeared in colicin D-treated cells (Fig. 1 B and C), and aminoacylation assaying (Fig. 2) and Northern hybridization (Fig. 3) of the tRNA fraction of colicin D-treated cells all consistently showed that intracellular tRNAsArg were specifically cleaved through the action of colicin D. The possibility that the changes in these tRNAs in vivo were indirectly caused by cell death or by an unknown nuclease activated somehow by colicin D was excluded by in vitro experiments involving purified colicin D and its immunity protein (Fig. 4). At least three of the four tRNAsArg examined (tRNAArgICG, tRNAArgU*CU and tRNAArgCCU) were found to be cleaved between nucleotide positions 38 and 39 in vitro (Fig. 5). We confirmed that the purified colicin D significantly reduced the amino acid incorporation activity in vitro of the E. coli S-30 fraction with MS2 RNA as the template, as previously demonstrated for colicin E5 (ref. 13; data not shown).
Furthermore, a single mutation of H611 in the C-terminal domain of colicin D resulted in the complete loss of its lethal activity (Fig. 7B) as well as loss of its ribonuclease activity, both in vivo and in vitro (Fig. 7 C and D), indicating that the ribonuclease activity of colicin D is directly correlated with its lethal action. Thus, the most likely scenario is that the cleavage of tRNAsArg by colicin D results in the exhaustion or a shortage of the cytoplasmic pool of these tRNAArg species, which impairs protein synthesis and finally causes cell death. An alternative or additional possibility remains that the cleavage of a specific isoacceptor of the four species is critically responsible for the cell death. dnaY is a mutant allele of argU encoding tRNAArgU*CU and has been reported to cause a temperature-sensitive defect in replication (25, 26). Thus, colicin D might act as an inhibitor of replication by cleaving tRNAArgU*CU.
Until quite recently, based only on the phenotypic analogy to colicin E3, both colicins E5 and D were believed to stop protein synthesis by nucleolytically inactivating ribosomes. We showed that the target of colicin E5 is not ribosomes but the anticodons of tRNAs for Tyr, His, Asn, and Asp (13). In this paper, we demonstrated that the target of colicin D is the anticodon loops of tRNAs for Arg. Here we propose a new category of “cytotoxic tRNases” comprising highly toxic colicins impairing protein synthesis, although this term was first used by Saxena et al. (27) for an RNase A-type enzyme, angiogenin. Angiogenin was claimed to specifically cleave tRNAs. But, neither a specific cleavage site nor a specific tRNA species as a substrate has been reported (28), which is in clear contrast to colicins D and E5.
Considering these apparent similar actions of colicins E5 and D, however, it is a great surprise to find so many different molecular traits underlying the activities of these two colicins. First, no significant homology in amino acid sequence was found between them, even in their C-terminal catalytic or cytotoxic domains. Second, and more importantly, their catalytic mechanisms are entirely different. Colicin E5 is unique as the first ribonuclease lacking histidine residues in its catalytic domain because histidine is indispensable as a general acid or acid-base catalyst in all ribonucleases examined so far. However, in colicin D, we have identified a crucial histidine, H611, which is probably involved in its catalytic reaction. Although the structure of colicin D does not resemble other ribonucleases, a very local similarity can be seen around H611 between colicin D and E. coli RNase I.
Third, the modes of substrate recognition of colicins E5 and D seem to be different. Besides the different groups of tRNAs targeted as their substrates, the relative cleavage sites are different within anticodon loops; colicin E5 cleaves between positions 34 and 35, and colicin D between positions 38 and 39. Furthermore, when protein-free rRNAs were used as substrates, colicin E5 degraded the RNAs to some extent, but colicin D showed little degradation, if any (data not shown). The common feature in the substrate tRNAs of colicin E5 is a local RNA sequence around the anticodons of the tRNAs concerned. We recently showed that the dinucleotide at positions 34 and 35 is the major determinant of the tRNA recognition by E5-CRD (T.O., T.U., and H.M., unpublished work). In contrast, the most conspicuous feature of the substrate recognition of colicin D is that only tRNAsArg and all isoaccepting tRNAsArg are susceptible, just as in the case of the cognate tRNA recognition by arginyl-tRNA synthetase (ArgRS). The main identity elements needed for the recognition of tRNAsArg by ArgRS were elucidated to be A20, C35, and the discriminator (29–31). Detailed analyses are now in progress to determine whether colicin D uses a similar recognition mechanism as ArgRS. In contrast to colicin E5 as “an RNA restriction enzyme,” colicin D seems to recognize several features shared by the tRNA molecules concerned in a more specific manner.
The differences between colicins E5 and D in their molecular structures, catalytic mechanisms, and substrate recognition suggest that they convergently acquired very close functions during evolution. In this context, the anticodon nuclease PrrC is another interesting cytotoxic tRNase, although it is not a physiological toxin. PrrC is produced by a clinical E. coli strain in response to phage T4 infection and specifically cleaves intracellular tRNALys between positions 33 and 34 to interfere with propagation of the infected phage (32–34). Despite apparently similar activities toward tRNAs, again significant homology has not been found between PrrC and colicin E5 or D.
If the goal of these tRNases is just cytotoxicity, it seems curious that they all have distinct target specificities toward unique cleavage sites of different tRNAs. Nonspecific ribonuclease activity should be enough or even more efficient to kill a cell. What is the advantage of having specific tRNAs as targets? This question is as yet unanswered. Considering that as many as three nonhomologous ribonucleases evolved convergently, other cytotoxic tRNases with different specificities may well be discovered in the future. Thus, it is intriguing to imagine an unknown world of cytotoxic tRNases, which individually target specific tRNAs, possibly, for the sake of some cell-cell interaction, communication or competition.
Acknowledgments
This work was supported by a Grant-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Science, Sports and Culture, Japan.
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.140213797.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.140213797
References
- 1.Konisky J. Annu Rev Microbiol. 1982;36:125–144. doi: 10.1146/annurev.mi.36.100182.001013. [DOI] [PubMed] [Google Scholar]
- 2.Pugsley A. Microbiol Sci. 1984;1:168–175. [PubMed] [Google Scholar]
- 3.Pugsley A. Microbiol Sci. 1984;1:203–205. [PubMed] [Google Scholar]
- 4.James R, Kleanthous C, Moore G R. Microbiology. 1996;142:1569–1580. doi: 10.1099/13500872-142-7-1569. [DOI] [PubMed] [Google Scholar]
- 5.Cramer W A, Lindeberg M, Taylor R. Nat Struct Biol. 1999;6:295–297. doi: 10.1038/7520. [DOI] [PubMed] [Google Scholar]
- 6.Mock M, Pugsley A P. J Bacteriol. 1982;150:1069–1076. doi: 10.1128/jb.150.3.1069-1076.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bowman C M, Sidikaro J, Nomura M. Nat New Biol. 1971;234:133–137. doi: 10.1038/newbio234133a0. [DOI] [PubMed] [Google Scholar]
- 8.Bowman C M, Dahlberg J E, Ikemura T, Konisky J, Nomura M. Proc Natl Acad Sci USA. 1971;68:964–968. doi: 10.1073/pnas.68.5.964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dahlberg A E, Dahlberg J E. Proc Natl Acad Sci USA. 1975;72:2940–2944. doi: 10.1073/pnas.72.8.2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Timmis K. J Bacteriol. 1972;109:12–20. doi: 10.1128/jb.109.1.12-20.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Timmis K, Hedges A J. Biochem Biophys Acta. 1972;262:200–207. doi: 10.1016/0005-2787(72)90233-x. [DOI] [PubMed] [Google Scholar]
- 12.Akutsu A, Masaki H, Ohta T. J Bacteriol. 1989;171:6430–6436. doi: 10.1128/jb.171.12.6430-6436.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ogawa T, Tomita K, Ueda T, Watanabe K, Uozumi T, Masaki H. Science. 1999;283:2097–2100. doi: 10.1126/science.283.5410.2097. [DOI] [PubMed] [Google Scholar]
- 14.Schramm E, Mende J, Braun V, Kamp R M. J Bacteriol. 1987;169:3350–3357. doi: 10.1128/jb.169.7.3350-3357.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roos U, Harkness R E, Braun V. Mol Microbiol. 1989;3:891–902. doi: 10.1111/j.1365-2958.1989.tb00238.x. [DOI] [PubMed] [Google Scholar]
- 16.Maniatis T, Fritsch E F, Sambrook J. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1982. [Google Scholar]
- 17.Chomczynski P, Sacchi N. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 18.Sprinzl M, Horn C, Brown M, Ioudovitch A, Steinberg S. Nucleic Acids Res. 1998;26:148–153. doi: 10.1093/nar/26.1.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsurui H, Kumazawa Y, Sanokawa R, Watanabe Y, Kuroda T, Wada A, Watanabe K, Shirai T. Anal Biochem. 1994;221:166–172. doi: 10.1006/abio.1994.1393. [DOI] [PubMed] [Google Scholar]
- 20.Wakita K, Watanabe Y, Yokogawa T, Kumazawa Y, Nakamura S, Ueda T, Watanabe K, Nishikawa K. Nucleic Acids Res. 1994;22:345–353. doi: 10.1093/nar/22.3.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Donis-Keller H. Nucleic Acids Res. 1980;8:3133–3142. doi: 10.1093/nar/8.14.3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vieira J, Messing J. Gene. 1982;19:259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- 23.Takahashi K. J Biochem. 1970;67:833–839. doi: 10.1093/oxfordjournals.jbchem.a129315. [DOI] [PubMed] [Google Scholar]
- 24.Gerlt J A. In: Nucleases. Linn S M, Lloyd R S, Roberts R J, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1993. pp. 1–34. [Google Scholar]
- 25.Mullin D A, Garcia G M, Walker J R. Cell. 1984;37:669–674. doi: 10.1016/0092-8674(84)90399-4. [DOI] [PubMed] [Google Scholar]
- 26.Garcia G M, Mar P K, Mullin D A, Walker J R, Prather N E. Cell. 1986;45:453–459. doi: 10.1016/0092-8674(86)90331-4. [DOI] [PubMed] [Google Scholar]
- 27.Saxena S K, Rybak S M, Davey R T, Jr, Youle R Y, Ackerman E J. J Biol Chem. 1992;267:21982–21986. [PubMed] [Google Scholar]
- 28.Leonidas D D, Shapiro R, Allen S C, Subbarao G V, Veluraja K, Acharya K R. J Mol Biol. 1999;285:1209–1233. doi: 10.1006/jmbi.1998.2378. [DOI] [PubMed] [Google Scholar]
- 29.Atilgan T, Nicholas H B, McClain W H. Nucleic Acids Res. 1986;14:375–380. doi: 10.1093/nar/14.1.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chakraburtty K. Nucleic Acids Res. 1975;2:1793–1804. doi: 10.1093/nar/2.10.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McClain W H, Foss K, Jenkins R A, Schneider J. Proc Natl Acad Sci USA. 1990;87:9260–9264. doi: 10.1073/pnas.87.23.9260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Amitsur M, Levitz R, Kaufmann G. EMBO J. 1987;6:2499–2503. doi: 10.1002/j.1460-2075.1987.tb02532.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meidler R, Morad I, Amitsur M, Inokuchi H, Kaufmann G. J Mol Biol. 1999;287:499–510. doi: 10.1006/jmbi.1999.2634. [DOI] [PubMed] [Google Scholar]
- 34.Kaufmann G. Trends Biochem Sci. 2000;25:70–74. doi: 10.1016/s0968-0004(99)01525-x. [DOI] [PubMed] [Google Scholar]