Abstract
Eukaryotic DNA replication requires the coordinated activity of the multi-subunit DNA polymerases: Pol α, Pol δ and Pol ɛ. The conserved catalytic and regulatory B subunits associate in a constitutive heterodimer that represents the functional core of all three replicative polymerases. Here, we combine X-ray crystallography and electron microscopy (EM) to describe subunit interaction and 3D architecture of heterodimeric yeast Pol α. The crystal structure of the C-terminal domain (CTD) of the catalytic subunit bound to the B subunit illustrates a conserved mechanism of accessory factor recruitment by replicative polymerases. The EM reconstructions of Pol α reveal a bilobal shape with separate catalytic and regulatory modules. Docking of the B–CTD complex in the EM reconstruction shows that the B subunit is tethered to the polymerase domain through a structured but flexible linker. Our combined findings provide a structural template for the common functional architecture of the three major replicative DNA polymerases.
Keywords: DNA polymerase α, DNA replication, electron microscopy, X-ray crystallography
Introduction
Three polymerases—Pol α, Pol δ and Pol ɛ—cooperate to synthesise the bulk of novel DNA during replication in the eukaryotic cell. According to current models, the primase subunit of the Pol α/primase complex initiates synthesis by oligomerising short RNA primers on both leading and lagging strands. These primers are initially extended by Pol α and subsequently transferred to Pol δ and Pol ɛ for processive synthesis on the lagging and leading strand, respectively (Stillman, 2008). Pol α, Pol δ and Pol ɛ are large, multi-subunit enzymes and their size has so far complicated their structural analysis: the lack of high-resolution information severely restricts our mechanistic understanding of their central role in eukaryotic replication.
All three polymerases share unifying features of their subunit organisation that reveal their evolutionary relationship (Figure 1A). The catalytic subunits of Pol α, Pol δ and Pol ɛ are phylogenetically related and conserved from yeast to humans (Makiniemi et al, 1999; Stillman, 2008; Weill and Reynaud, 2008). Of their different cohorts of accessory subunits, only the so-called B subunit is present in all three polymerase assemblies and—similarly to the catalytic subunit—is clearly conserved in eukaryotic organisms. Interestingly, an orthologue of the B subunit has also been found in archaeal organisms as the single accessory polypeptide of a replicative polymerase.
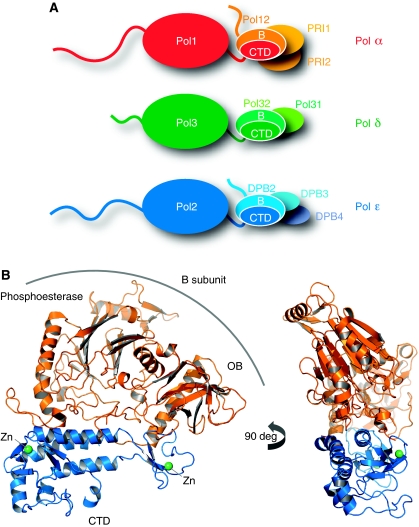
Figure 1.
Crystal structure of the yeast Pol α CTD–B subunit complex. (A) The eukaryotic DNA polymerases α, δ and ɛ are multi-subunit enzymes. The drawing shows the subunit organisation of yeast Pol α, Pol δ and Pol ɛ. A heterodimeric complex of catalytic subunit and accessory B subunit forms their common functional core. The interaction is mediated by the conserved C-terminal domain of the catalytic subunit. The extended intervening sequence between polymerase domain and CTD in the catalytic subunit of Pol ɛ is not drawn to scale. (B) Ribbon diagram of the crystal structure of the yeast Pol α CTD–B subunit complex, showing two orientations of the complex related by a 90 ° rotation. The CTD is drawn in blue and the B subunit in orange. The position of the phosphoesterase and OB domains in the B subunit are indicated. The zinc atoms are shown as green spheres.
Reflecting their high degree of conservation, the catalytic and B subunits are the only indispensable polymerase components. The sequence conservation of the catalytic subunit extends past the polymerase fold to a cysteine-rich C-terminal domain (CTD) that is necessary for DNA replication and cell viability (Navas et al, 1995; D'Urso and Nurse, 1997; Kesti et al, 1999; Feng and D'Urso, 2001). A large body of experimental evidence has highlighted the functional importance of the CTD interaction with the B subunit (Morrison et al, 1990; Giot et al, 1997; Dua et al, 1998, 1999; Mizuno et al, 1999; Sanchez Garcia et al, 2004). Thus, a heterodimer of the catalytic and B subunits represents the conserved functional core of the three replicative polymerases.
Here, we provide a comprehensive structural description of heterodimeric yeast Pol α by a combination of X-ray crystallography and electron microscopy approaches. We reconstitute biochemically and determine the X-ray crystal structure of a complex between the CTD of Pol α and its B subunit. Furthermore, we show by EM analysis that Pol α forms an elongated shape with distinct lobes occupied by the catalytic and B subunits. Docking of the CTD–B complex within the 3D reconstruction of Pol α allows the accurate determination of the spacial relationship between catalytic and B subunits. Our combined findings provide a structural foundation on which to build our growing mechanistic understanding of the DNA polymerases that replicate the genome of eukaryotic organisms.
Results
Architecture of the CTD–B subunit complex
We have biochemically reconstituted, crystallised and determined the 2.5 Å X-ray crystal structure of a complex between amino acids 1263–1468 (natural C-end; CTD) of Pol1, the catalytic subunit of Saccharomyces cerevisiae DNA polymerase α, and amino acids 246–705 (natural C-end) of Pol12, its B subunit (see Supplementary Figure 1 for complex purification; Supplementary Table 1 for crystallographic analysis). The N-terminal region of the B subunit was excluded from our analysis as it is not evolutionarily conserved and superfluous for the interaction with Pol1.
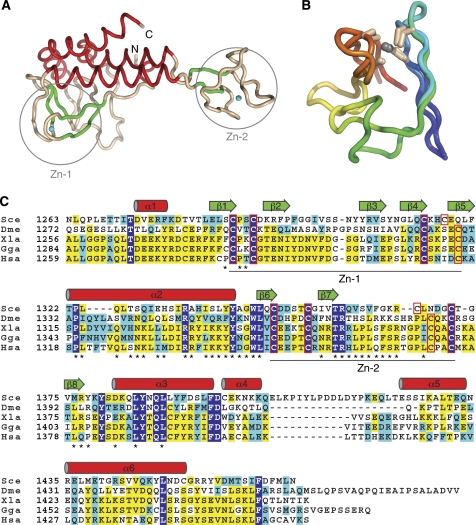
The association of CTD with the B subunit results in a compact, slab-like structure of approximate triangular shape and overall dimensions: 80 Å (height), 70 Å (width) and 25 Å (depth) (Figure 1B). The structure reveals that the CTD of yeast Pol α adopts an elongated, bilobal shape reminiscent of an asymmetrically proportioned saddle (Figures 1B and 2A). Each lobe contains a zinc-binding module: the lobe with the four N-terminal cysteine ligands (Zn-1) is larger and includes additional secondary structure elements as well as irregular coil structure; the lobe with the four C-terminal cysteine ligands is smaller and formed entirely by the zinc-binding module (Zn-2) (Figure 2A). The two lobes are connected by a three-helix bundle that represents the central portion or ‘backbone' of the saddle-shaped CTD. The two zinc-binding motifs bear a clear structural relationship to each other; in both cases, metal-binding results from the ‘handshake' interaction of two β ribbons, each providing a pair of cysteine ligands for the tetrahedral coordination of the zinc atom (Figure 2B). The mode of zinc binding observed in the yeast Pol α CTD resembles most closely the iron-binding motif found in rubredoxin (PDB id 1RDV) (Misaki et al, 1999). A published structural analysis of a Zn-2 peptide of human Pol α differs markedly from the conformation of the Zn-2 region that we observe in the crystal of the CTD–B subunit complex (Evanics et al, 2003). The discrepancy might be because of the lack of structural context of the earlier study. The structure of the Pol α CTD does not match any fold in the protein databank and it clearly represents a specialised fold that evolved for the specific recognition of the B subunit.
Figure 2.
The yeast Pol α CTD. (A) Tube diagram of the CTD, coloured according to secondary structure (red: α helices; green: β strand; wheat: irregular coil). The zinc atoms are drawn as cyan spheres. The position of the two zinc-binding modules, Zn-1 and Zn-2, are indicated by circles. (B) Superposition of the Zn-1 and Zn-2 modules of CTD. The side chains of the cysteine ligands are drawn as sticks and the polypeptide chains are coloured blue (N-terminus) to red (C-terminus). (C) Multiple sequence alignment of the CTD region of Pol α from distantly related eukaryotic species (Sce, Saccharomyces cerevisiae; Dme, Drosophila melanogaster; Xla, Xenopus laevis; Gga, Gallus gallus; Hsa, Homo sapiens). Completely conserved residues are shown in white in blue background, identical residues in yellow and conserved residues in cyan. The elements of secondary structure as observed in the crystal structure of the complex are drawn above the alignment (α helices as cylinders and β strands as arrows). The cysteine ligands that coordinate the zinc atoms are boxed in red, and the extent of Zn-1 and Zn-2 modules is marked under the alignment. Residues at the interface with the B subunit are indicated by an asterisk.
The B subunit of replicative polymerases is organised as an evolutionarily conserved, folded C-terminal region fused to a relatively unstructured amino-terminal tail, which is phosphorylated in a cell-cycle-dependent fashion (Foiani et al, 1995; Kuroda and Ueda, 1999). Our crystallographic analysis reveals that the tertiary structure of the B subunit derives from the intimate association of an N-terminal oligonucleotide/oligosaccharide (OB) domain with a C-terminal phosphoesterase domain (Figure 1B). The structure of the B subunit confirms earlier bioinformatic predictions and more recent structural analysis, as a calcineurin-like phosphoesterase that has evolutionarily converted into a role as a catalytically inactive scaffold deputed to the assembly of multi-subunit complexes (Aravind and Koonin, 1998; Koonin et al, 2000; Baranovskiy et al, 2008).
High-resolution view of the CTD–B interface
The structure of the complex reveals that the association of the B subunit with the CTD is not limited to the two conserved zinc-binding modules but involves the upper, flat side of the CTD saddle in its entirety (Figures 1B and 3A). Thus, an important functional role of the CTD is to provide a structural platform for extensive interactions with both OB and phosphoesterase domains of the B subunit. Interestingly, the modular composition of the B subunit is mirrored in its mode of association with the CTD, as the OB fold makes exclusive contact with the Zn-2 module, whereas the phosphoesterase domain interacts with the larger, N-terminal lobe of the CTD saddle. The presence of two juxtaposed but largely distinct CTD–B interfaces might reflect independent events in the evolutionary process that led to a stable association between catalytic and B subunits of Pol α.
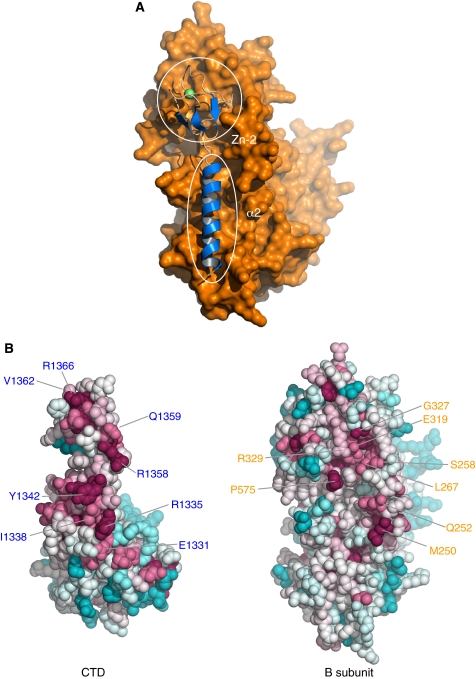
Figure 3.
The interface of the CTD–B subunit complex. (A) Overall view highlighting the involvement of CTD helix α2 and the Zn-2 module in the interaction with the B subunit. The structural elements of the CTD are depicted as blue ribbons, the B subunit as a molecular surface in orange. (B) Evolutionary conservation of surface amino acids mapped on the Pol α CTD and B subunit structures in space-fill representation, calculated with ConSurf (Landau et al, 2005) from an alignment of 50 Pol α CTD and 66 B subunit sequences. Amino-acid conservation is indicated by a transition in colour hues, from magenta (most conserved) to cyan (most variable). The position of some of the strongly conserved CTD and B subunit residues at the interface is indicated.
A defining feature of the CTD–B subunit interaction is the very large size of its interface: burying 4500 Å2 of total surface area, it is at the upper end of the known size distribution for heterodimeric interfaces and clearly defines the CTD–B subunit interaction as a constitutive association. The structure shows that the majority of the interactions with the B subunit are mediated by helix α2 and Zn-2 module of the CTD (Figures 2C and 3A). These are cradled within a long, shallow groove formed by flexible loops emanating from the core secondary structure elements of the phosphoesterase and OB domains of the B subunit. The chemical nature of the interactions at the interface is varied and includes networks of direct and water-mediated buried polar contacts, as well as extensive contacts between hydrophobic residues (Figure 4A and B). The remarkable plasticity of the CTD–B interface is underlined by the large proportion of polar contacts (16 hydrogen bonds out of 29) that are formed between CTD side chains and backbone atoms of the B subunit. As predicted by the crystallographic analysis, the Pol α CTD and B residues identified as important for the interaction display a higher degree of evolutionary conservation relative to the rest of their solvent-exposed regions (Figure 3B).
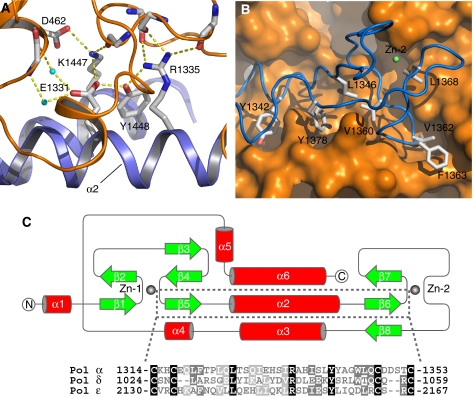
Figure 4.
Details of subunit interactions at the CTD–B interface. (A) Close-up views of hydrophilic interactions at the CTD α2–B subunit interface. Side chains are drawn as sticks, water molecules as cyan spheres and hydrogen bonds as dashed yellow lines. (B) Hydrophobic interactions at the CTD Zn-2–B subunit interface. CTD side chains are represented as in panel A. The molecular surface of the B subunit is shown in orange. (C) Topology diagram of the CTD structure. The α helices are shown as red cylinders and the β strands as green arrows. A sequence alignment for CTD helix α2 of yeast Pol α, Pol δ and Pol ɛ is shown below the diagram; identical residues are highlighted in black and conserved residues in grey.
Biological implications of the CTD–B subunit structure
The organisation of the C-terminal region as a tandem repeat of zinc-binding modules is a shared feature of the catalytic subunit of Pol α, Pol δ and Pol ɛ. Thus, the crystal structure of the yeast Pol α CTD–B complex represents a valid structural template for understanding the interaction between catalytic and B subunits in all three polymerases. The structure highlights helix α2 in the central region of the CTD as an essential structural element in the recognition of the B subunit. The importance of helix α2 is reflected in the conservation of residues that are critical for the interaction, such as R1335 and Y1342, in yeast Pol δ and Pol ɛ (Figure 4C). The structure of the yeast Pol α CTD–B subunit complex further allows the rationalisation of a number of experimental observations concerning the role of the CTD and its interaction with the B subunit. In particular, the structure provides an explanation for the observations that, in addition to mutations affecting the conserved cysteine ligands, alterations in the sequence between the two zinc-binding modules or C-terminal to Zn-2 have a profound negative impact on CTD function (Budd and Campbell, 1993; Navas et al, 1995; Dua et al, 1998, 1999). Thus, a 10aa-long deletion spanning residues 2143–2152 in the CTD of Pol ɛ, which does not support cell growth (Dua et al, 1998), map to helix α2 that is essential for interaction with the B subunit in our structure. Premature truncations at E2192 and Q2196 of Pol ɛ, which confer cells a temperature-sensitive phenotype, compromise the stability of the CTD fold by removing a C-terminal segment spanning helices α3–α6 (Navas et al, 1995).
3D reconstruction of heterodimeric yeast Pol α
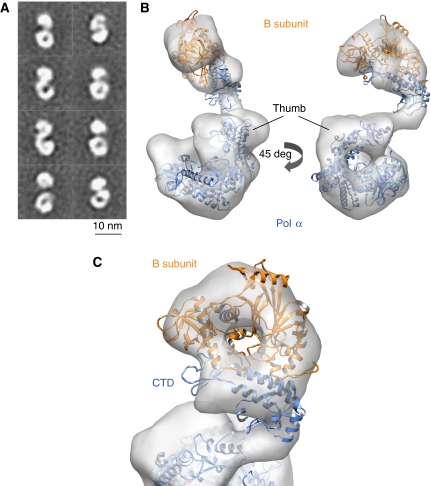
To gain a functional insight into the orientation of the CTD–B complex relative to the catalytic domain of Pol α, we purified and analysed the heterodimeric yeast Pol α by EM. The EM images show a prevalence of bilobal molecules with densities of different shape and size at each end of their longitudinal axis (Figure 5A). These views represent different projections of Pol α after rotating along its longitudinal axis, combined with multiple tilt angles on the support film (Supplementary Figure 2). The 3D reconstruction based on 12 913 side-view images of the Pol α at 23 Å resolution reveals clearly distinct structural modules: a smaller, flatter region corresponding to the CTD–B complex and a larger, globular domain that agrees with the known structural features of related archaeal DNA polymerases (Firbank et al, 2008) (Figure 5B and C). The two lobes do not contact each other but are connected by a structured ‘arm' that keeps the flat shape of the CTD–B complex suspended over the ‘palm' of the polymerase fold (Figure 5A and B).
Figure 5.
3D reconstruction of heterodimeric yeast Pol α using electron microscopy. (A) Reference-free 2D averages obtained from images of the heterodimeric Pol α reveal the presence of two distinct regions in the structure. (B) Ribbon models for the crystal structures of the CTD–B complex and the catalytic domain of the archaeal polymerase from T. gorgonarius (PDB id 2VWJ) are fitted to the EM reconstruction. Two rotated views of the EM reconstruction are shown, represented as white transparent density. The Pol α CTD and the catalytic domain of the archaeal polymerase are shown in blue and the B subunit in orange. (C) Close-up view of the EM reconstruction of Pol α, demonstrating the fitting of the CTD–B complex within the smaller lobe of the EM reconstruction.
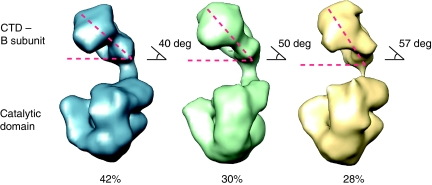
Inspection of the EM images further revealed the presence of limited conformational heterogeneity in the Pol α–B complex. Refinement of the data set by maximum-likelihood methods yielded three solutions, one major class comprising 42% of the molecules—selected for representation in Figure 5—and two other classes representing 30 and 28% of the data set. Comparison of the three solutions shows that the CTD–B complex can sample a range of positions relative to the catalytic domain of Pol α that are related by a moderate degree of rocking motion (Figure 6). Detection of such movement suggests that conformational changes mediated by the ‘arm' region connecting the CTD to the catalytic domain of Pol α could be part of the functional mechanism of the polymerase.
Figure 6.
Conformational flexibility of Pol α. The EM reconstructions highlight the variability in the respective position of the lobes containing catalytic and B subunits. The structural heterogeneity is illustrated by a representative view of the three reconstructions obtained. The plasticity of the ‘arm' region suggests that controlled changes in the relative position of the two subunits might be relevant to the function of Pol α. The percentage values reported under the EM reconstructions reflect the proportion of particles assigned to the three 3D solutions and represents an estimate of the prevalence of each conformation in the data set.
Discussion
In this paper, we have analysed the 3D architecture of heterodimeric yeast Pol α by X-ray crystallography and electron microscopy. Our findings provide the first accurate determination of the interaction mechanism and spatial relationship between catalytic and accessory B subunits. The conservation in sequence and architecture of the functional cores of Pol α, Pol δ and Pol ɛ implies that features of our 3D model of yeast Pol α will be of general applicability to the three major replicative polymerases.
Our structural analysis defines the C-terminal region of the replicative Pols α, δ and ɛ as an independently folded protein domain, separate from the polymerase fold, in agreement with the biological evidence that points to the existence of an independent role of the CTD in DNA replication, separate from the catalytic function (Kesti et al, 1999). The size and nature of the interface in the complex indicates that the CTD is a specific molecular tether for the constitutive engagement of the B subunit with the polymerase subunit.
The structure further reveals that the two zinc-binding modules, although structurally related, fulfil different roles in the interaction with the B subunit. Thus, Zn-2 is directly and extensively implicated in binding to the B subunit, whereas Zn-1 is only marginally involved in the interaction and protrudes instead into solvent from the side of the CTD, where it might be poised for interactions with the rest of the replication machinery. This structural evidence provides an explanation for the observation that Zn-2 of Pol δ is necessary and sufficient for interaction with the B subunit in yeast two-hybrid experiments (Sanchez Garcia et al, 2004). The relevance of the Pol α CTD structure might extend to other polymerases—such as REV3, the catalytic subunit of the translesion polymerase ζ and archaeal DNA polymerases—for which the existence of zinc-binding motifs at their C-terminus has been predicted, based on the presence of a conserved pattern of cysteine residues (Cann et al, 1998; Shen et al, 2004; Gan et al, 2008).
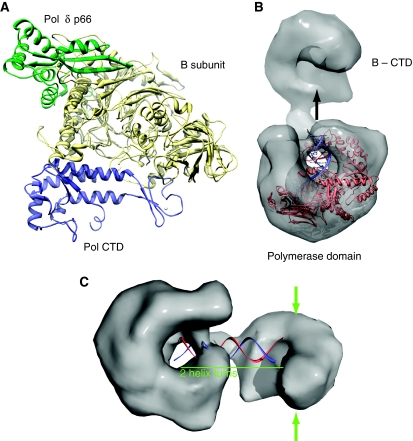
The clear sequence similarity between the heterodimeric cores of Pol α and Pol δ implies that the functional insight derived from our analysis of Pol α's 3D architecture is likely to be applicable to both lagging strand polymerases. In its role as a hub of protein–protein interactions that mediate and regulate DNA synthesis, the B subunit participates in multiple simultaneous interactions with different polymerase subunits. Superposition of the available B subunit structures bound to Pol α CTD (this study) and Pol δ p66 (PDB id 3E0J) shows how this can be achieved in the case of Pol δ and provides a first glimpse of the multi-subunit polymerase architecture that contains the B subunit as its central scaffold (Figure 7A).
Figure 7.
Functional interactions of the heterodimeric Pol α. (A) Superposition of B subunits of the CTD–B complex of yeast Pol α and the p50-p66 complex (PDB id 3E0J) of human Pol δ. The superposition illustrates how the B subunit of replicative polymerases can act as a structural scaffold by engaging in multiple, concomitant protein–protein interactions. The protein chains are drawn as ribbons, coloured pale yellow for the B subunits, blue for the Pol α CTD and green for Pol δ p66. (B) The position of the DNA molecule bound to the T. gorgonarius polymerase (PDB id 2VWJ) fitted to the EM reconstruction suggests a possible trajectory for the nascent DNA emerging from the active site of Pol α. (C) Proposed model for the interaction of Pol α with DNA. An idealised B-form DNA molecule spanning two helical turns has been modelled within Pol α, to span the distance between active site and the smaller lobe containing the B subunit. The green arrows highlight possible sites on the surface of the smaller lobe that might be involved in the interaction with the primase subunits.
Placing the co-crystal structure of the archaeal DNA polymerase from Thermococcus gorgonarius (PDB id 2VWJ) within the EM structure of Pol α indicates that the nascent DNA helix emerging from the active site of Pol α would grow in the direction of the CTD–B complex (Figure 7B) (Firbank et al, 2008). In our fitting of the CTD–B complex, the active site of the phosphoesterase domain of the B subunit faces the DNA and is available to interact with it. Although the eukaryotic B subunit is known to be no longer catalytically active, our reconstruction suggests that it might still interact with the emerging double-stranded nucleic acid. This observation prompted us to measure and to detect a micromolar affinity of the CTD–B complex for double-stranded DNA (Supplementary Figure 3). The interaction of the CTD–B with DNA does not alter the enzymatic properties of the catalytic domain of Pol α (SK and LP, unpublished observation), but it might be important for the proper orientation of the B subunit relative to the DNA template. Our structural characterisation of the heterodimeric yeast Pol α as a catalytic engine tethered by a flexible ‘arm' to its accessory subunit poised to interact with DNA downstream of the growing 3′-end of the nascent strand is in agreement with a recent low-resolution 3D model of yeast Pol ɛ (Asturias et al, 2006).
Finally, experimental evidence shows that the CTD–B complex is necessary and sufficient for recruitment to Pol α of the heterodimeric primase, the specialised polymerase that initiates nucleic acid synthesis in DNA replication (Mizuno et al, 1999) (SK and LP, unpublished observation). We note that in the EM reconstruction the flanks of the CTD–B complex are easily accessible for interactions with the primase (Figure 7C). In the light of the elongated, bilobal shape of Pol α's 3D architecture described here, association of the primase with the lobe occupied by the CTD–B raises the question of how the catalytic activities of RNA primer synthesis and extension can be coordinated over the considerable distance separating the two catalytic subunits in the Pol α/primase complex. A conformational rearrangement bringing the two lobes of the complex in closer proximity would represent a plausible structural basis for the mechanism of transfer of the RNA primer between active sites. The observed flexibility in the linker that connects catalytic and B subunits of Pol α provides indirect evidence in favour of this hypothesis. Further structural investigation will be necessary to improve our understanding of the mechanism of initiation of DNA synthesis in eukaryotic cells.
Materials and methods
Crystallisation and X-ray structure determination of the CTD–B complex
Residues 1263–1468 (C-terminus) of S. cerevisiae DNA polymerase α and 246–705 (C-terminus) of the B subunit were cloned into an RSF1-Duet expression vector (Novagen), to produce the B subunit fused to an N-terminal six-histidine tag and Pol α CTD as an untagged protein. The protein complex was overexpressed in Escherichia coli strain BL21(DE3) Rosetta2 at 18°C by the addition of 1 mM IPTG and purified using Co-NTA and heparin sepharose chromatography followed by Tev cleavage and size exclusion chromatography. The purified protein was concentrated to 5 mg/ml in 20 mM Tris·Cl pH 8.0, 200 mM KCl, flash frozen in liquid nitrogen and stored at −80°C.
Crystals were grown by vapour diffusion in hanging drops at 18°C, by mixing the protein sample at 5 mg/ml with a reservoir solution containing 0.1 M MES pH 6.0, 1.26 M ammonium sulfate, in a 2:1 volume ratio. For best crystallisation results 3 mM TCEP was added to the protein sample immediately before hanging drop set-up. The crystals belong to space group P21 (a=85.9 Å, b=143.0 Å, c=175.6 Å and β=102.2 deg), with four copies of the complex in the asymmetric unit (asu).
The crystal structure of the CTD–B complex was determined using phase information derived from anomalous scattering data collected at the Se K-edge, obtained from selenomethionine-containing crystals at beamline ID29 of the European Synchrotron Radiation Facility (ESRF) in Grenoble, France. An initial, partial model for the CTD–B complex was obtained using PHENIX (Zwart et al, 2008), completed manually in Coot (Emsley and Cowtan, 2004) and refined in REFMAC5 (Murshudov et al, 1997) using NCS restraints and one TLS parameter per protein chain in the asu, to R/Rfree values of 19.1/21.8 at 2.5 Å resolution. The final crystallographic model contains 2464 residues, 8 zinc atoms, 65 sulphate ions and 571 waters. 97.3% of all residues are in favoured regions of the Ramachandran plot, 99.8% in allowed regions and 0.2% in the outlier regions. The conformation of solvent-exposed regions of the B subunit spanning residues 306–310 of chains A, C, E, G and residues 674–688 of chain E, as well as residues 1411–1420 of chains D, F and H of the CTD must be considered tentative as the electron density of these regions is weak. Residues 581–605 in all chains of the B subunit are presumably disordered as no interpretable electron density was detected; they were therefore omitted from the final model.
The four copies of the CTD–B complex in the asu are arranged as a pair of dimers. Within each dimer, the two copies of the CTD–B complex are related by non-crystallographic two-fold symmetry originating from the same mode of interaction. In particular, the N-terminal tail of each CTD chain in the asu interacts with the second copy of the B subunit of the non-crystallographic dimer, by addition of a short parallel strand to one of the β sheets of its phosphoesterase domain. Although the path of the polypeptide chain is clearly visible in the density map, the quality of the density is not sufficient for unambiguous tracing of the chain, except for the interacting β strand that was modelled as a tri-alanine peptide. The repeated involvement of the same surface areas in the non-crystallographic packing between copies of the CTD–B complex suggests that some features of the association might be physiologically relevant.
Electron microscopy and 3D reconstruction of heterodimeric Pol α
Residues 349–1468 (C-terminus) of the catalytic subunit and 246–705 (C-terminus) of the B subunit of Pol α were cloned to produce recombinant versions of the two proteins, fused to N-terminal strepII- and six-histidine tags, respectively. The heterodimeric Pol α was overexpressed in Sf9 cells using the MultiBac system (Berger et al, 2004). Cells were harvested 4 days post-infection and the complex was purified using streptactin and heparin sepharose chromatography and flash frozen in liquid nitrogen in 100 mM Tris·Cl pH 8.0, 500 mM NaCl. A diluted solution of Pol α was adsorbed onto glow-discharged carbon-coated grids, stained with 2% uranyl formate and visualised in a JEOL 1230 transmission electron microscope operating at 100 kV. Micrographs were recorded at a magnification of 50 000 under low-dose conditions and digitised using a Minolta Dimage Scan Multi Pro scanner at a final pixel size of 3.8 Å. The contrast transfer function of the microscope for each micrograph was estimated using CTFFIND3 (Mindell and Grigorieff, 2003) and corrected using Bsoft (Heymann, 2001); 12 913 side-view images of the complex were manually extracted using XMIPP (Sorzano et al, 2004) and 2D reference-free classification of the data was performed using EMAN (Ludtke et al, 1999) and 2D maximum-likelihood methods (Scheres et al, 2005). These 2D averages were used to build an initial template using common lines, which was then refined using EMAN.
To investigate the presence of possible heterogeneity in Pol α, the average model obtained after refinement with EMAN (Ludtke et al, 1999) was used as the initial template for 3D maximum-likelihood (ML3D) classification (Scheres et al, 2007), to split the images into more homogeneous subsets of the data. It has been shown earlier that ML3D can resolve several protein conformations without earlier knowledge of the source of heterogeneity (Scheres et al, 2007). Three initial template volumes for ML3D classification were prepared by splitting the data into three random subsets. After convergence of the ML3D refinement, the images were sorted in three output volumes resembling the average structure, but showing some conformational differences. The three reconstructions contained, respectively, 42% (conformation 1), 30% (conformation 2) and 28% (conformation 3) of the 12 913 initial particles. The resolution of each structure, calculated using an FSC cut-off of 0.5, was 22.9, 24.5 and 25.5 Å, respectively. Conformation 1 was chosen for further analysis as it was derived from the largest portion of the data set and was obtained at the highest resolution. It was also used for representation in Figures 5 and 7. The final volumes and fitting were visualised using UCSF Chimera (Goddard et al, 2007).
Fitting of crystallographic models into the EM density was performed automatically using ADP_EM (Garzon et al, 2007) and testing both possible hands of the reconstruction. The CTD–B complex was confronted with the complete EM map in an exhaustive 6D search. All top solutions placed the flat shape of the CTD–B crystal structure within the smaller lobe of the EM reconstruction. The top solution (cross-correlation coefficient >0.77) located the CTD closer to the linker region of density in the reconstruction, in agreement with the requirement for continuity in the polypeptide chain of Pol α. The crystal structure of the T. gorgonarius polymerase bound to DNA (PDB id 2VWJ) was used as a template for the catalytic domain of Pol α and fitted similarly to the CTD–B complex. The top solution placed its C-terminus in the proximity of the CTD region (cross-correlation coefficient >0.7).
The crystallographic coordinates and structure factors of the CTD–B complex have been deposited in the Protein Databank under accession code 3FLO. The EM map of the heterodimeric yeast Pol α has been deposited in the 3D EM database under accession code EMD-1618 (http://www.ebi.ac.uk/pdbe/emdb/).
Supplementary Material
Supplementary Material
Review Process File
Acknowledgments
We would like to thank Joseph Maman for performing the SPR experiments. This work was funded by a Wellcome Trust senior research fellowship in basic biomedical science to LP and by funding of the Spanish Ministry of Science (SAF2008-00451), the Autonomous Region of Madrid (CAM S-BIO-0214-2006) and the ‘Red Temática Investigación Cooperativa en Cáncer (RTICC)' from the ‘Instituto de Salud Carlos III' (RD06/0020/1001) to OL. OL is additionally supported by the Human Frontiers Science Program (RGP39/2008). SK is funded by a Gates Cambridge Trust scholarship and the Medical Research Council.
Footnotes
The authors declare that they have no conflict of interest.
References
- Aravind L, Koonin EV (1998) Phosphoesterase domains associated with DNA polymerases of diverse origins. Nucleic Acids Res 26: 3746–3752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asturias FJ, Cheung IK, Sabouri N, Chilkova O, Wepplo D, Johansson E (2006) Structure of Saccharomyces cerevisiae DNA polymerase epsilon by cryo-electron microscopy. Nat Struct Mol Biol 13: 35–43 [DOI] [PubMed] [Google Scholar]
- Baranovskiy AG, Babayeva ND, Liston VG, Rogozin IB, Koonin EV, Pavlov YI, Vassylyev DG, Tahirov TH (2008) X-ray structure of the complex of regulatory subunits of human DNA polymerase delta. Cell Cycle 7: 3026–3036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger I, Fitzgerald DJ, Richmond TJ (2004) Baculovirus expression system for heterologous multiprotein complexes. Nat Biotechnol 22: 1583–1587 [DOI] [PubMed] [Google Scholar]
- Budd ME, Campbell JL (1993) DNA polymerases delta and epsilon are required for chromosomal replication in Saccharomyces cerevisiae. Mol Cell Biol 13: 496–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cann IKO, Komori K, Toh H, Kanai S, Ishino Y (1998) A heterodimeric DNA polymerase: evidence that members of Euryarchaeota possess a distinct DNA polymerase. Proceedings of the National Academy of Sciences of the United States of America 95: 14250–14255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Urso G, Nurse P (1997) Schizosaccharomyces pombe cdc20+ encodes DNA polymerase ɛ and is required for chromosomal replication but not for the S phase checkpoint. Proceedings of the National Academy of Sciences of the United States of America 94: 12491–12496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dua R, Levy DL, Campbell JL (1998) Role of the putative zinc finger domain of Saccharomyces cerevisiae DNA polymerase epsilon in DNA replication and the S/M checkpoint pathway. J Biol Chem 273: 30046–30055 [DOI] [PubMed] [Google Scholar]
- Dua R, Levy DL, Campbell JL (1999) Analysis of the essential functions of the C-terminal protein/protein interaction domain of Saccharomyces cerevisiae pol epsilon and its unexpected ability to support growth in the absence of the DNA polymerase domain. J Biol Chem 274: 22283–22288 [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K (2004) Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Evanics F, Maurmann L, Yang WW, Bose RN (2003) Nuclear magnetic resonance structures of the zinc finger domain of human DNA polymerase alpha. Biochim Biophys Acta 1651: 163–171 [DOI] [PubMed] [Google Scholar]
- Feng W, D'Urso G (2001) Schizosaccharomyces pombe cells lacking the amino-terminal catalytic domains of DNA polymerase epsilon are viable but require the DNA damage checkpoint control. Mol Cell Biol 21: 4495–4504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firbank SJ, Wardle J, Heslop P, Lewis RJ, Connolly BA (2008) Uracil recognition in archaeal DNA polymerases captured by X-ray crystallography. J Mol Biol 381: 529–539 [DOI] [PubMed] [Google Scholar]
- Foiani M, Liberi G, Lucchini G, Plevani P (1995) Cell cycle-dependent phosphorylation and dephosphorylation of the yeast DNA polymerase alpha-primase B subunit. Mol Cell Biol 15: 883–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan GN, Wittschieben JP, Wittschieben BO, Wood RD (2008) DNA polymerase zeta in higher eukaryotes. Cell Res 18: 174–183 [DOI] [PubMed] [Google Scholar]
- Garzon JI, Kovacs J, Abagyan R, Chacon P (2007) ADP_EM: fast exhaustive multi-resolution docking for high-throughput coverage. Bioinformatics 23: 427–433 [DOI] [PubMed] [Google Scholar]
- Giot L, Chanet R, Simon M, Facca C, Faye G (1997) Involvement of the yeast DNA polymerase delta in DNA Repair in vivo. Genetics 146: 1239–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard TD, Huang CC, Ferrin TE (2007) Visualizing density maps with UCSF Chimera. J Struct Biol 157: 281–287 [DOI] [PubMed] [Google Scholar]
- Heymann JB (2001) Bsoft: image and molecular processing in electron microscopy. J Struct Biol 133: 156–169 [DOI] [PubMed] [Google Scholar]
- Kesti T, Flick K, Keränen S, Syväoja JE, Wittenberg C (1999) DNA polymerase epsilon catalytic domains are dispensable for DNA replication, DNA repair, and cell viability. Mol Cell 3: 679–685 [DOI] [PubMed] [Google Scholar]
- Koonin EV, Wolf YI, Aravind L (2000) Protein fold recognition using sequence profiles and its application in structural genomics. Adv Protein Chem 54: 245–275 [DOI] [PubMed] [Google Scholar]
- Kuroda K, Ueda R (1999) Phosphorylation and dephosphorylation of the B subunit of DNA polymerase alpha-primase complex in the early embryogenesis of Drosophila. Biochem Biophys Res Commun 254: 372–377 [DOI] [PubMed] [Google Scholar]
- Landau M, Mayrose I, Rosenberg Y, Glaser F, Martz E, Pupko T, Ben-Tal N (2005) ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res 33 (Web Server issue): W299–W302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludtke SJ, Baldwin PR, Chiu W (1999) EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol 128: 82–97 [DOI] [PubMed] [Google Scholar]
- Makiniemi M, Pospiech H, Kilpelainen S, Jokela M, Vihinen M, Syvaoja JE (1999) A novel family of DNA-polymerase-associated B subunits. Trends Biochem Sci 24: 14–16 [DOI] [PubMed] [Google Scholar]
- Mindell JA, Grigorieff N (2003) Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol 142: 334–347 [DOI] [PubMed] [Google Scholar]
- Misaki S, Morimoto Y, Ogata M, Yagi T, Higuchi Y, Yasuoka N (1999) Structure determination of rubredoxin from Desulfovibrio vulgaris Miyazaki F in two crystal forms. Acta Crystallogr D Biol Crystallogr 55(Pt 2): 408–413 [DOI] [PubMed] [Google Scholar]
- Mizuno T, Yamagishi K, Miyazawa H, Hanaoka F (1999) Molecular architecture of the mouse DNA polymerase alpha-primase complex. Mol Cell Biol 19: 7886–7896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison A, Araki H, Clark AB, Hamatake RK, Sugino A (1990) A third essential DNA polymerase in S. cerevisiae. Cell 62: 1143. [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr 53(Pt 3): 240–255 [DOI] [PubMed] [Google Scholar]
- Navas TA, Zhou Z, Elledge SJ (1995) DNA polymerase epsilon links the DNA replication machinery to the S phase checkpoint. Cell 80: 29–39 [DOI] [PubMed] [Google Scholar]
- Sanchez Garcia J, Ciufo LF, Yang X, Kearsey SE, MacNeill SA (2004) The C-terminal zinc finger of the catalytic subunit of DNA polymerase delta is responsible for direct interaction with the B-subunit. Nucleic Acids Res 32: 3005–3016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres SH, Gao H, Valle M, Herman GT, Eggermont PP, Frank J, Carazo JM (2007) Disentangling conformational states of macromolecules in 3D-EM through likelihood optimization. Nat Methods 4: 27–29 [DOI] [PubMed] [Google Scholar]
- Scheres SH, Valle M, Nunez R, Sorzano CO, Marabini R, Herman GT, Carazo JM (2005) Maximum-likelihood multi-reference refinement for electron microscopy images. J Mol Biol 348: 139–149 [DOI] [PubMed] [Google Scholar]
- Shen Y, Tang XF, Matsui E, Matsui I (2004) Subunit interaction and regulation of activity through terminal domains of the family D DNA polymerase from Pyrococcus horikoshii. Biochem Soc Trans 32(Pt 2): 245–249 [DOI] [PubMed] [Google Scholar]
- Sorzano CO, Marabini R, Velazquez-Muriel J, Bilbao-Castro JR, Scheres SH, Carazo JM, Pascual-Montano A (2004) XMIPP: a new generation of an open-source image processing package for electron microscopy. J Struct Biol 148: 194–204 [DOI] [PubMed] [Google Scholar]
- Stillman B (2008) DNA polymerases at the replication fork in eukaryotes. Mol Cell 30: 259–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weill J-C, Reynaud C-A (2008) DNA polymerases in adaptive immunity. Nat Rev Immunol 8: 302–312 [DOI] [PubMed] [Google Scholar]
- Zwart PH, Afonine PV, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, McKee E, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Storoni LC, Terwilliger TC, Adams PD (2008) Automated structure solution with the PHENIX suite. Methods Mol Biol 426: 419–435 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material
Review Process File