Abstract
Retrovirus integrase (IN) integrates the viral linear DNA genome (~10 kb) into a host chromosome, a step which is essential for viral replication. Integration occurs via a nucleoprotein complex, termed the preintegration complex (PIC). This article focuses on the reconstitution of synaptic complexes from purified components whose molecular properties mirror those of the PIC, including the efficient concerted integration of two ends of linear viral DNA into target DNA. The methods described herein permit the biochemical and biophysical analyses of concerted integration. The methods enable: 1) the study of interactions between purified recombinant IN and its viral DNA substrates at the molecular level; 2) the identification and characterization of nucleoprotein complexes involved in the human immunodeficiency virus type-1 (HIV-1) concerted integration pathway; 3) the determination of the multimeric state of IN within these complexes; 4) dissection of the interaction between HIV-1 IN and cellular proteins such as lens epithelium-derived growth factor (LEDGF/p75); 5) the examination of HIV-1 Class II and strand transfer inhibitor resistant IN mutants; and 6) the mechanisms associated with strand transfer inhibitors directed against HIV-1 IN that have clinical relevance in the treatment of HIV-1/AIDS.
Keywords: retrovirus concerted integration, integrase, preintegration complex, reconstituted system, synaptic complex
1. Introduction
Significant progress has augmented our understanding of the molecular processes involved in human immunodeficiency virus type-1 (HIV-1), gamma, and alpha retrovirus integration in vivo [1]. Integrase (IN) catalyzes two independent reactions in different cellular compartments [2, 3]. Within the cytosolic preintegration complex (PIC), IN removes the terminal dinucleotides from the U5 and U3 blunt-ends synthesized during reverse transcription, exposing an active 3′ OH group adjacent to the conserved CA motif. Upon transport of the PIC into the nucleus, IN mediates the concerted insertion of the 3′ OH recessed DNA ends into a chromosome by a cutting and joining reaction known as strand transfer. With the emergence of resistant HIV-1 strains against highly active antiretroviral therapy (HAART), IN is an attractive alternative and new therapeutic target. Strand transfer inhibitors directed against IN effectively prevent HIV-1 integration at low nM concentrations in vivo and prevent the development of HIV-1/AIDS in infected individuals [4–6].
We and others have developed a reconstituted system to investigate concerted integration that uses purified recombinant HIV-1 IN [7–13], alpha retrovirus IN [14–17], or IN purified from avian myeloblastosis virus (AMV) particles [17–19]. IN efficiently mediates the concerted insertion of relatively large size DNA substrates (0.5 to 4 kb) harboring U3 or U5 viral end sequences into a supercoiled plasmid as target DNA. The methods described in this article are useful in investigating the retrovirus concerted integration pathway in vitro. These methods have provided insights into: (i) the biochemical mechanisms of HIV-1 integration and identification of novel nucleoprotein intermediates in the concerted integration pathway; (ii) the mechanisms by which strand transfer inhibitors block the catalytic activity of HIV-1 IN; and (iii) the role of cellular cofactors in the HIV-1 integration process. Emphasis will be placed on biochemical and biophysical studies that help to understand the mechanistic basis of PIC function in HIV-1 infected cells and the mechanisms through which strand transfer inhibitors block this process.
2. Description of Methods
2.1. Overview of the concerted integration assay
Efficient reconstitution of concerted retroviral DNA integration in vitro requires three macromolecular reaction components (IN, viral DNA substrate, and target DNA), all present in optimized solution conditions for IN catalysis. High quality purified IN appears to be a prerequisite for effective assembly of the HIV-1 synaptic complex (SC). Within the SC, IN non-covalently juxtaposes the two viral DNA ends (Fig. 1a, 1b) [12, 20]. The SC is a transient intermediate in the concerted integration pathway (Fig. 1b) [12, 20]. We purify bacterially expressed HIV-1 IN at relatively low protein concentrations at pH 6.8, a feature which seems to minimize severe aggregation of IN in solution [9]. The final concentration of purified IN is ~2.5 to 3 μM. The supercoiled target as well as supercoiled donor DNA plasmids that contain viral long terminal repeat (LTR) sequences, are purified by velocity sedimentation on sucrose gradients to remove small-sized DNA contaminants. Linear viral DNA substrates (~0.5 to 4 kb) are obtained by restriction digestion of LTR-containing donor DNA plasmids, followed by one of several conventional purification techniques (see section 3.2.1 below). Using the above purified components, the reconstitution system led to the identification of the transient SC [12] and strand transfer complex (STC) [11] in the HIV-1 concerted integration pathway (Fig. 1a and 1b).
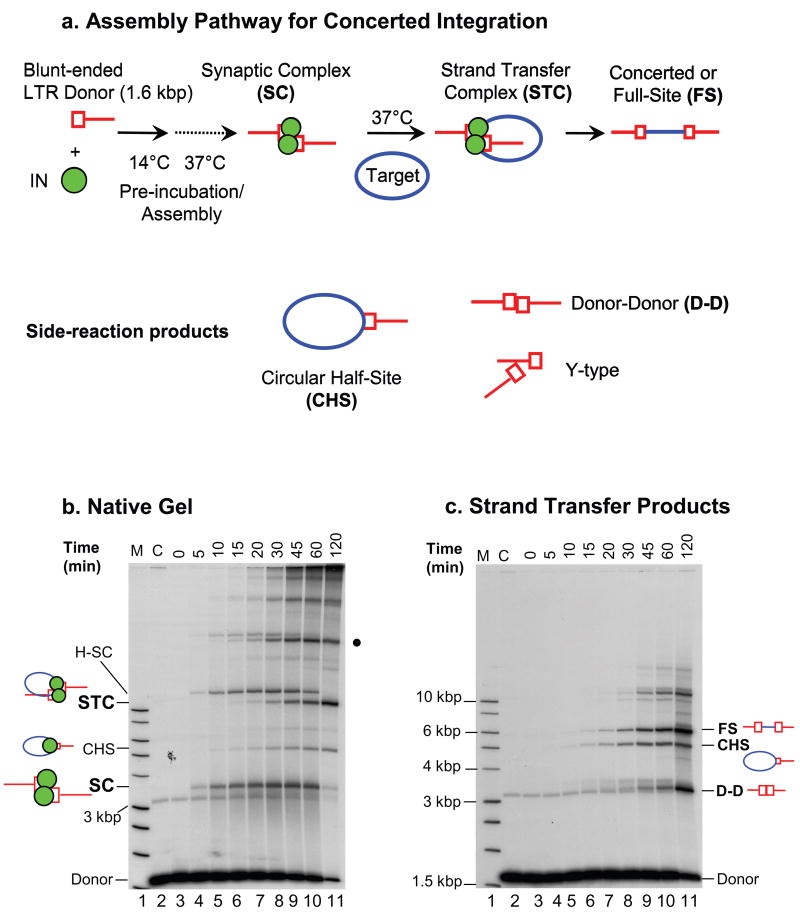
Fig. 1.
Concerted retroviral DNA integration. (a) Schematic for HIV-1 SC and STC assembly, which results in concerted or full-site (FS) integration product formation. The reaction, depicted with a 1.6 kb blunt-ended HIV-1 U5-containing substrate, is applicable to a variety of linear viral DNA substrates (see Fig. 2 for additional HIV-1 examples). Side-reaction products (CHS, D-D, and Y-type) are also formed in the reaction mixture. (b) A typical time course experiment revealing the differential kinetics of SC and STC formation. IN (20 nM) was assembled with the 1.6 kb U5 blunt-ended donor DNA substrate (0.5 nM) at 14°C, and further incubated with supercoiled target DNA at 37°C for various times. The transient SC, which is identified here on a 0.7% native agarose gel, appears at ~5 min (lane 4), reaches near maximum between 30 min (lane 8) and 45 min (lane 9), and nearly disappears by 120 min (lane 11). The H-SC, presumably formed by SC multimers, follows similar kinetics. Upon binding target DNA, the SC is converted to the STC, with maximum quantities of STC detectable at ~120 min. The various IN-DNA complexes are marked on the left. Lane 1 contains a 32P-labeled 1 kb ladder (marked M) and lane 2 is without IN (marked C). (c) Strand transfer products obtained after deproteinization of the same samples shown in panel b. The quantities of FS product parallel the quantities of STC. Other labeling is the same as in panel b.
Assembly of the HIV-1 SC and subsequent concerted integration into target DNA is depicted in Fig. 1a. A pre-assembly step where IN and the linear blunt-ended viral DNA substrate are incubated together at 14°C for 15 min, followed by incubation at 37°C, results in the assembly of the transient SC (Fig. 1b). As revealed by native agarose gel electrophoresis, maximum accumulation of the SC occurs at ~30–45 min (Fig. 1b, lanes 8 and 9), which is followed by its subsequent disappearance by ~120 min [12, 20]. IN processes the viral 3′ OH ends gradually, between 5 and 60 min during the incubation at 37°C [12] (data not shown). Simultaneously, the accumulating SC associates with supercoiled target DNA, resulting in the concerted insertion of the two 3′ OH recessed ends, producing the STC [11, 12]. The STC accumulates gradually and its formation is essentially saturated by 120 min (Fig. 1b, lane 11). Higher-order forms of the SC (H-SC) appear on the native gel and may be due to looping of DNA by IN [21, 22]. The STC also forms a higher-order complex on native gels (Fig. 1b, dark circle on right). The SC and H-SC are also formed in the absence of target (data not shown).
The STC consists of the concerted or full-site (FS) integration product [11] in association with IN. Using HIV-1 IN mutant W235F, it has been shown that a tetrameric form of IN is present in the SC when it is produced in the presence of a strand transfer inhibitor [11]. The FS integration product, along with other side-reaction products, including the circular half-site (CHS) and donor-donor (D-D) products, are visualized by deproteinization of the samples prior to agarose gel electophoresis [7–11] (Fig. 1c). Following isolation of the concerted integration product, the size of the host-site duplication that results from DNA repair of the staggered insertion of the viral DNA ends into target DNA can be determined. Individual recombinant DNA molecules are genetically selected in bacteria, followed by DNA sequencing [9, 13, 23]. The methods and experimental approaches used to investigate HIV-1 concerted integration in vitro are outlined below.
2.2. HIV-1 IN expression
BL21(DE3)(Stratagene) cells are transformed with expression vector pET11a-HIV-1 IN (pNY clone) [8], and the transformants are selected on 1.5% agar plates containing Terrific Broth (TB) and carbenicillin (50 μg/ml). A single colony is grown to an absorbance at 600 nm (A600) of ~0.5 in 100 ml of TB containing 50 μg/ml carbenicillin. Sterile glycerol (50% solution) is added to the culture to a final concentration of 10%. Ten ml aliquots are stored at −70°C. Newly transformed BL21(DE3) cells are made as needed.
We inoculate 350 ml of TB containing carbenicillin with 10 ml of the thawed, transformed BL21(DE3) bacterial stock. The cells are grown at 37°C (250 rpm) until A600 reaches ~0.5.
Fifty ml of the culture is transferred to 450 ml of prewarmed TB with carbenicillin in each of six baffled flasks (2 liters). The bacteria are grown to A600 0.8 to 0.9 at 250 rpm. An aliquot (1.5 ml) of the culture is removed as the uninduced sample for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis. The sample is centrifuged and the pellet stored on ice.
The culture is induced by adding isopropyl-β-D-thiogalactopyranoside to the final concentration of 0.4 mM, and incubation for 3 h at 37°C with shaking. As above, an aliquot is removed from each flask to determine IN expression levels.
The cells are harvested by centrifugation at 5000 rpm (Beckman JA-10 rotor) for 15 min at 4°C. The supernatant is removed and the wet weight of the bacterial pellet is determined. The wet cell weight is generally ~2.5 g. The pellets can be stored at −70°C for at least four months.
2.3. Purification of HIV-1 IN
2.3.1. Cell lysis and IN extraction
The following protocol is used for extraction and purification of IN from bacterial cells.
The bacterial pellet is placed on ice for ~10 to 15 min. The pellet is resuspended in four volumes (~8 ml) of ice-cold low-salt lysis buffer (50 mM HEPES, 1 mM EDTA, 100 mM NaCl, 3 mM dithiothreitrol (DTT), 0.1 mM 4-(2-aminoethyl) benzenesulfonyl fluoride hydrochloride (AEBSF), pH 6.8) in a shaker on ice for 10 min. The sample is kept on ice for an additional 30 min. DTT and AEBSF are added to the buffer just before use. The pH of the ice-cold buffer is adjusted at this temperature with NaOH.
The suspension is subjected to sonication with a small probe (Sonifer Cell Disruptor W-350) at 70% duty cycle setting for a total of 2 min, using 30 sec bursts followed by 30 sec of cooling. The samples are maintained on ice throughout the procedure. Over-sonication of the pellet results in low IN yields. The bacterial DNA must remain intact because (i) IN binds to the DNA in the pellet after centrifugation and (ii) the DNA is avoided in the purified IN fractions upon column chromatography.
The lysate suspension is centrifuged at 30,000 rpm (~110,000g) in an Beckman SW41-Ti rotor for 30 min at 4°C. It is important to use this g force to pellet IN, while the majority of the bacterial contaminants remain in the supernatant. The supernatant (1st lysate) is removed, and 20 μl is saved for SDS-PAGE analysis to confirm that IN has partitioned to the pellet.
The pellet is resuspended in 8 ml of the same low-salt lysis buffer with two plastic spatula to break up the pellet and major aggregates. The pellet is further resuspended by sonication at 70% duty cycle for 30 sec (only once). If necessary, use the plastic spatula to resuspend the aggregates further. The samples are centrifuged at 30,000 rpm for 15 min at 4°C. Again, pour off the supernatant (2nd lysate) and save an aliquot (20 μl) for SDS-PAGE.
Suspend the pellet in 8 ml of ice-cold low-salt lysis buffer with the use of two plastic spatula to disperse the aggregates. Spin the sample at 30,000 rpm for 15 min at 4°C. Pour off the supernatant (1st wash) and save an aliquot (20 μl) for SDS-PAGE.
Repeat the wash step as described in step five (2nd wash).
Homogenize the pellet (2 ml/g initial wet weight of bacteria) with high-salt extraction buffer (low-salt lysis buffer supplemented with 1 M NaCl) to extract IN from the DNA. Incubate the sample on ice for 45 min with frequent mixing, using two plastic spatula as needed to disperse the aggregates.
Centrifuge the 1M NaCl extract at 30,000 rpm for 15 min at 4°C, and transfer the supernatant that contains the majority of IN to a new tube. The transferred supernatant is re-spun at 30,000 rpm for 15 min at 4°C to pellet any small particulate contaminants from the 1 M NaCl extract. An aliquot of the high salt extract is removed to check the purity of IN. Save the supernatant in a new tube, which can be stored at −70°C for up to at least four months with no loss of IN activities upon purification. We initially quick-freeze the samples on a dry ice-ethanol bath. Check all of the samples on SDS-PAGE. Usually, 10 μl of each sample with an equal volume of SDS denaturing buffer is sufficient for evaluation of purity. We routinely obtain preparations where IN represents ~60% of the proteins observed in the 1 M NaCl extract fraction.
2.3.2. SP-Sepharose column chromatography
The following purification protocol has been optimized with a Bio-Rad Biologic Work Station using a GE Healthcare-HiTrap SP HP-5ml cation exchange column.
Buffer A and Buffer B are adjusted to pH 6.8 with NaOH when the solutions are ice-cold. The composition of Buffer A is 50 mM HEPES, 1 mM EDTA, 10 mM MgSO4, and 3 mM DTT. Buffer B is the same as Buffer A except it contains 1 M NaCl. All of the following steps are performed on ice or at 4°C.
The 1 M NaCl extract containing IN is thawed on ice and filtered through a 0.45 μm filter.
The extract is diluted slowly to 80 mM NaCl with Buffer A with intermittent mixing. A calibration curve using various NaCl concentrations for measuring conductivity is essential.
The diluted high-salt extract is immediately loaded at 3 ml/min onto a pre-equilibrated SP-Sepharose column with Buffer A. Wash the column in two steps at 2 ml/min; first with 2 column volumes (CVs) of a 5–20% linear gradient of Buffer B; second by isocratic flow with 6 CVs of 80% Buffer A–20% Buffer B.
IN is eluted with 10 CVs of a linear gradient (Buffer B, 20%–100%) at 1 ml/min. Fractions (0.75 ml) are collected. The peak fractions of IN elute between 340 mM to 360 mM NaCl as judged by conductivity and UV absorbance.
Save an aliquot (20 μl) for SDS-PAGE from each of the IN containing fractions (as identified on the chromatogram), and freeze the fractions using a dry ice-ethanol bath. Store the samples at −70°C. The emphasis should be on purity of IN rather than quantity. Fractions having the maximum amounts of IN, with the least amounts of protein and DNA contaminants as judged by SDS-PAGE and A280/260, respectively, are pooled. We pool material derived from two separate SP-Sepharose IN preparations and apply this to a Heparin-Sepharose column to increase the concentration and purity of IN.
2.3.3. Heparin-Sepharose column chromatography (HiTrap Heparin HP, 5 ml)
Buffer A and Buffer B are the same as described above, except both buffers contain 10% glycerol and Buffer B contains 1.3 M NaCl. Prepare the Heparin column as described by the manufacturer. Equilibrate the column with Buffer A (5 CVs) at 2 ml/min.
Dilute the pooled SP-Sepharose fractions to 200 mM NaCl with Buffer A (usually a ~1:1 dilution). Dilute the sample slowly as described previously. Immediately load the sample onto the column at 2 ml/min. Wash the column with 2 CVs using a linear 5–15% gradient of Buffer B, followed by an isocratic wash with 85% Buffer A–15% Buffer B at 2 ml/min for 15 min.
Elute IN from the column with a linear gradient of 15%–100% of Buffer B at 1 ml/min for 15 min (3 CVs), collecting 0.5 ml fractions. Wash the column with 100% Buffer B at 1 ml/min for 10 min. Store the fractions at −70°C after flash-freezing.
Analyze the peak IN-containing fractions (10 μl) by SDS-PAGE. The peak fraction of IN elutes at ~550 mM NaCl. As before, pool the fractions for purity to maximize the amount of IN with the least contaminants. Generally, IN at this step is >95% pure as judged by SDS-PAGE.
The concentration of IN is evaluated by A280 and/or analytical SDS-PAGE. The gel is stained with Coomassie Fluor Orange (Invitrogen) with a protein standard for comparison [9, 12]. The concentration of IN obtained using the molar extension coefficient of 50,460 M−1 cm−1 at 280 nm is very similar to the above staining method. We also perform a scan from 220 to 380 nm to check for purity and aggregation. The 280/260 ratio is usually ~1.7, indicating a negligible amount of DNA contamination. Wavelength scans reveal a distinct peak at 295 nm, characteristic of tryptophan absorbance. The concentrations of IN are generally ~3 μM, and possess minimal light scattering (A360 < 0.01), suggesting nominal aggregation of the IN protein.
The above purification protocol for wild type HIV-1 IN (pNY and HXB2-IIIB strains) has been successfully used to purify IN mutant proteins N155H, N155S, and S153Y that are resistant to different strand transfer inhibitors [20] as well as Class II IN mutant proteins Q168A, V165A, and R166A that impart pleiotropic replication defects during HIV-1 infection.
3. Concerted integration assays using large-size DNA substrates
3.1. Development of direct physical assays probing concerted integration
Early attempts to reconsitute assay mixtures employing recombinant HIV-1 IN and ~ 20 bp LTR oligodeoxynucleotides yielded concerted integration products at a very low level (<1%) [24]. We developed a direct physical assay using relatively large size linear DNA fragments (~0.5 to 4 kb), possessing one or two LTR ends, with AMV IN purified from virions or recombinant alpha retrovirus IN. The concerted integration products are readily identifiable on agarose gels, and yielded a significant increase (~20% to 30%) in product formation [17, 18, 23, 25, 26]. With these size DNA fragments, we were able to significantly decrease the concentration of viral DNA ends (0.2 nM to 1 nM) relative to the ends in the oligodeoxynucleotide-based assays, and, subsequently, the concentration of IN could also be lowered into the low nM (10 nM to 40 nM) range. Below, we describe our optimized conditions for concerted integration using recombinant HIV-1 IN.
3.2. HIV-1 concerted integration assay
3.2.1. Preparation of DNA substrates
We have used a variety of DNA substrates, which are derived by restriction digestion of plasmids. A linear 4.7 kb substrate containing the natural HIV-1 U5 and U3 DNA blunt-ends is produced by digesting plasmid pU3U5 [22], which contains a unique ScaI site at the U3-U5 LTR circle junction (Fig. 2a). After dephosphorylation, the linearized plasmid is 5′ end-labeled on the U3 plus and U5 minus strands using [γ-32P] ATP and T4 polynucleotide kinase. Subsequent digestion with NcoI liberates 32P-labeled 1.6 kb U5 and 2.4 kb U3 blunt-ended substrates (Fig. 2a). The specific activities are generally 1,000 cpm (Cerenkov) per ng of single-end labeled substrate DNA. The fragments are isolated from agarose gels and extracted using the Qiaquick Gel Extraction Kit (Qiagen, Valencia, CA) or electroelution.
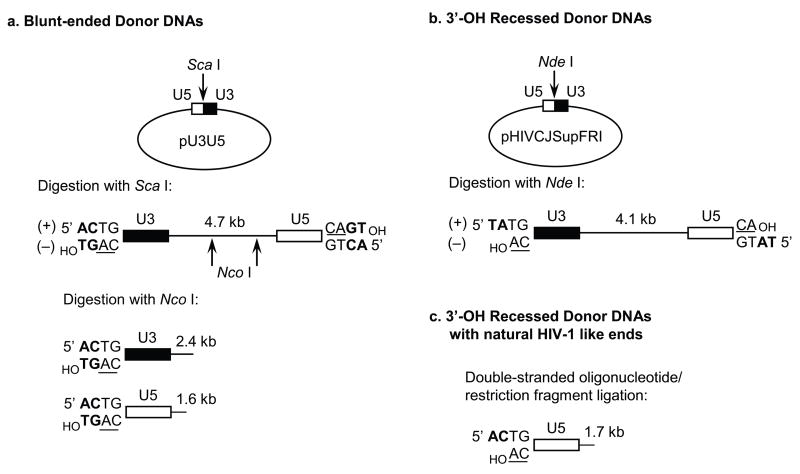
Fig. 2.
Generation of HIV-1 LTR donor DNA substrates. (a) Plasmid pU3U5 harbors a unique ScaI restriction site at the natural junction of abutted HIV-1 U3 and U5 blunt-ended sequences. ScaI digestion yields a linearized plasmid that contains 175 bp of the viral U3 region at one end and all 85 bp of U5 at the other [22]. The conserved CA dinucleotide is underlined. (b) Plasmid pHIVCJSupFRI harbors a unique NdeI site at the engineered U5/U3 circle junction. Enzyme cleavage yields a pre-processed linearized substrate that differs from the bona fide viral sequence at the terminal two base 5′ extensions (5′-TA instead of 5′-AC). (c) Alternatively, a 1.7 kb preprocessed U5 end substrate containing the natural viral sequence is generated by the forced ligation of an annealed 91/93 bp oligonucleotide onto a 1.6 kb linear fragment derived from pUC19.
A 1.7 kb U5 DNA substrate possessing the pre-processed 3′ OH end is constructed by ligation of an annealed 91/93 bp double stranded oligonucleotide possessing HIV-1 U5 terminal sequences to a 1.6 kb linear pUC19-derived fragment (Fig. 2c) [12, 13, 20]. The ligated fragments are separated from unligated DNA by agarose gel electrophoresis and purified by the Qiaquick gel extraction kit. The pre-processed substrate is then labeled using T4 polynucleotide kinase. Alternatively, a linear 4.1 kb substrate containing HIV-1 U3 and U5 termini is produced by digesting plasmid pHIVCJSupFRI with NdeI, which yields a pre-processed substrate containing 5′-TA at the LTR termini as compared to the natural 5′-AC overhangs (Fig. 2c) [9, 13]. As described above, the linearized plasmid is labeled using T4 polynucleotide kinase. Subsequent digestion by different restriction enzymes allows the isolation of various smaller-sized, single LTR end labeled substrates [7, 8, 27].
The concerted insertion of two LTR DNA ends by IN into supercoiled DNA (Fig. 1a) is a biomolecular (two molecules of donor DNA) reaction [23]. The substrate mixture can contain a viral DNA substrate possessing only a U5 end, a U3 end, or a combination of both ends (Fig. 2). The U3 substrate supports ~30 to 40% of the level of IN strand transfer activity observed with the U5 substrate [10, 13]. The concerted insertion of the U3 and U5 ends within the PIC into target DNA, either in vitro or in vivo, is a unimolecular (insertion of one viral DNA molecule) reaction [3, 4, 28, 29]. However, pre-cleavage of HIV-1 DNA in purified PICs by a restriction enzyme that recognizes a single internal site allows the biomolecular reaction to occur, suggesting the two DNA ends are properly juxtaposed by IN [2]. A unimolecular reaction using a viral DNA substrate that contains two active LTR ends on a single molecule (~300 bp) is possible in the presence of an added DNA bending protein, although this reaction [30–32] is significantly less efficient than the bimolecular reaction. A small size DNA molecule in-solution has a more favorable cyclization probability (j factor) than a larger size molecule [23].
3.2.2. Solution chemistry for HIV-1 concerted integration
The concerted integration assay using radioactively labeled DNAs is described here. IN is pre-assembled with the linear 1.6 kb U5 blunt-ended substrate at 14°C for 15 min in the presence of 20 mM HEPES (pH 7.0), 5 mM DTT, 10 mM MgCl2, 25 μM ZnCl2, 100 mM NaCl, and 10% polyethylene glycol (6000 daltons). Reaction volumes are generally 100 μl but can be readily scaled-up for preparative analysis. The IN concentration is usually between 15 to 60 nM, and the 32P-labeled LTR DNA substrate concentration is 0.5 nM to 3 nM. Concerted integration activity is initiated by addition of supercoiled target DNA (1.5 nM to 3.0 nM; typically 2.86 kb pGEM-3) followed by incubation for 1 to 2 h at 37°C. The reactions are stopped by addition of EDTA, SDS, and proteinase K to the final concentrations of 25 mM, 0.5%, and 1 mg/ml, respectively. Samples are incubated for 1 h at 37°C, and subjected to electrophoresis on 0.7% agarose gels in TBE buffer (89 mM Tris, 89 mM borate, 2 mM EDTA, pH 8.3) at 100 volts for 16 h (Fig. 1c). After drying, the gels are exposed to a PhosphorImager screen. The different DNA products can be quantified using a Storm 860 system (Amersham Biosciences) or Typhoon (GE Healthcare).
3.2.3. Evaluation of concerted integration products
It is essential to establish that the reaction products possess the correct host-site duplication indicative of concerted two-ended integration. The host-site duplications for HIV-1, alpha, and gamma retroviruses are 5 bp, 6 bp, and 4 bp, respectively [1]. There are multiple methods to determine the host-site duplication sizes, for example via using supercoiled plasmid [23] or linear bacteriophage λ as target DNAs in vitro [33]. These early selection methods for isolating recombinants employed the presence of the bacterial SupF gene in the viral DNA substrate, and subsequent outgrowth of integration reaction mixtures in E. coli strain CA244 that possesses amber mutations in the lacZ gene. In all cases, individual recombinants are genetically selected and the host-site duplication between the U5 and U3 viral DNA ends determined by DNA sequencing [10, 13, 34, 35].
Here we describe a recent method for determining the size of the HIV-1 host site duplication in reaction products derived from blunt-ended viral DNA substrates (Fig. 2a). After incubation of the reaction mixture at 37°C, the deproteinized and phenol extracted strand transfer products are digested with a restriction enzyme having a unique site close to each LTR end. The restriction enzyme should be unique to each U3 and U5 donor DNA and absent in the target DNA. Alternatively, two different enzymes that are unique to each LTR end, but still absent in the target DNA, can be used. The two enzymes used with linearized pU3U5, BglII in U5 and BamHI in U3, have compatible ends for ligation. To accomplish genetic selection, supercoiled pBSK2-ΔZeo carrying the zeocin resistance gene is used as target DNA [36]. The digested DNA products are subjected to electrophoresis on agarose gels, and the linear DNA species that corresponds to concerted integration is isolated. The DNAs are self-ligated, transformed into E. coli strain DH5α, and transformants are selected using zeocin. The LTR-target DNA junctions are sequenced using U5- (5′-TGACCTGCCAGATCTGCTAACTAGGGAACCCAC 3′) or U3-specific (5′-CGCCCGCCACTAGTGGATCCAAAGAATTCTATC 3′) primers. Generally, the fidelity of the proper-sized host-site duplications varies from 60 to 80% of the sequenced recombinants, with the majority of the remaining clones containing repetitive small size deletions (~17–20 bp and ~28–32 bp) [8–10, 31] and one to two bp shorter or longer duplications than normal. Other minor species containing alterations of the LTR ends or host target site are also observed.
4. Applications of the concerted integration assay
4.1. Native agarose gel electrophoresis for SC formation
We have used native agarose gel electrophoresis to study the assembly and molecular properties of the SC and STC produced by HIV-1 IN (Fig. 1b) [12, 13, 20]. IN juxtaposes the two LTR ends in a non-covalent manner, as defined by two-dimensional gel electrophoresis of the SC [12]. The two-dimensional gel technique was originally developed by the Craigie lab to investigate the STC [11]. Several native agarose gel electrophoresis options are available including assembly of the SC in the presence or absence of target DNA, variations of IN and viral DNA substrate concentrations, defining IN subunit compositions by protein-protein crosslinking, investigation of strand transfer inhibitors, interactions of cellular co-factors with the SC, and in-gel fluorescence resonance energy transfer (FRET) analysis.
The assay conditions to form the SC are described above in section 3.2.2. In general, aliquots (~40 μl) taken at different times for native agarose analysis are stopped by adding EDTA to 25 mM (Fig. 1b). Samples are stored on ice prior to electrophoresis, or immediately applied to gels for separation. Larger volumes can be analyzed using preparative gel combs for scaled-up analysis of these assays. The nucleoprotein complexes are subjected to electrophoresis on 0.7% native agarose gels in TBE buffer at 4°C for 16 h and 100 volts. The gels should be pre-cooled prior to application of the samples. After electrophoresis, the gels can be dried and subjected to PhosphorImager analysis if 32P-labeled DNA is used and/or stained with SYBR Gold (Invitrogen) and analysis by ChemiDoc XRS (BioRad) QuantityOne software. In addition, the DNA products associated with the SC and STC can be eluted, deproteinized, and subjected to further agarose gel electrophoresis to verify their structures. For preparative analysis, the SC and STC are readily identifiable upon staining with SYBR Gold using unlabeled viral DNA substrates (3 nM).
We have successfully investigated the interactions of HIV-1 IN with the LTR ends within the SC and STC using native agarose gel electrophoresis (manuscript in preparation). We have probed these assembled complexes by mild DNaseI treatment of reaction mixtures at 14°C, prior to electrophoresis and their isolation from native agarose gels. Denaturing gel electrophoresis of the isolated DNAs from these complexes is used to define the terminal LTR sequences protected by IN. The in-solution DNaseI protection assay was used to characterize the interaction of alpha retrovirus IN with its respective LTR substrates [16, 19]. We have determined the HIV-1 IN subunit composition of these complexes by protein-protein cross-linking of the complexes prior to native gel electrophoresis. The cross-linked subunits are eluted from the isolated complexes, separated by SDS-PAGE, and probed with rabbit antisera directed against peptides located at different regions of HIV-1 IN. Lastly, we have attached Cy3 and Cy5 fluorophores to the 5′-end of a 1.5 kb blunt-ended U5 DNA substrate (Fig. 3) to investigate the interactions of the DNA ends in the SC and STC by FRET.
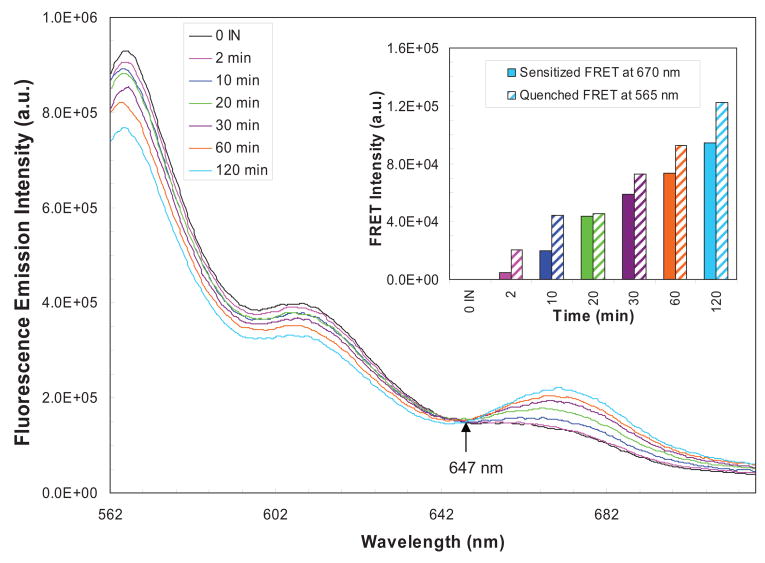
Fig. 3.
In-solution FRET analysis of HIV-1 nucleoprotein complexes. The spectra of quenched emission of donor fluorophore (Cy3) and sensitized emission of acceptor fluorophore (Cy5) as a result of energy transfer is displayed. An equimolar (1.5 nM) mixture of U5-Cy3 and U5-Cy5 DNA substrates were titrated with 60 nM IN. After each timepoint (2, 10, 20, 30, 60 and 120 min; magenta, blue, green, violet, orange and sky blue, respectively) at 37°C, the samples were adjusted to 500 mM NaCl and placed on ice. The spectra of each sample were obtained between 560 nm to 720 nm in a temperature regulated cell holder at 14°C. The black line emission spectrum was obtained in the absence of IN. The excitation wavelength was 550 nm while the excitation and emission monochromator slit width were 5 nm each. All spectra are instrument and buffer corrected. Quenched FRET at 565 nm (upward diagonal) and sensitized FRET intensities at 670 nm (solid) were determined by subtracting corresponding emission intensity from the sample with no IN and plotted with time (inset).
4.2. Defining the biochemical mechanisms of strand transfer inhibitors
The strand transfer inhibitors directed against HIV-1 IN block virus replication in vivo [4] and prevent HIV-1 replication in humans. Raltegravir (Isentress), a hydroxypyrimidinone carboxamide strand transfer inhibitor, has received FDA approval for treatment of HIV-1/AIDS [6]. We have determined that strand transfer inhibitors (L-870,810, L-870,812, and L-841,411) are effective for inhibiting concerted integration at low nM concentrations (IC50 of 55, 102, and 110, respectively) using the 1.6 kb blunt-ended U5 [12, 20] or 2.4 kb U3 DNA substrate (data not shown). In contrast, 3′ OH recessed substrates require significantly higher concentration of inhibitors (IC50 > 1000 nM) for effective inhibition [12]. These results suggest that inhibitor binding occurs prior to or during 3′ OH processing in the transient SC. Only one viral blunt DNA end juxtaposed with a 3′ OH recessed end within the SC is sufficient for low nM inhibition [20]. We suggest that the IN-DNA complex is “trapped” by the strand transfer inhibitors via a transient intermediate within the cytoplasmic PIC [12].
4.2.1. Assay conditions to determine IC50 values for inhibition of concerted integration
The naphthyridine carboxamide inhibitors L-870,810 and L-870,812 and the diketo-acid-2 inhibitor L-841,411 have been described [5, 37]. Stocks (10 mM) of each inhibitor are made in 100% dimethyl sulfoxide (DMSO) and stored in small aliquots at −70°C. Aliquots of the stock solution are discarded after single use. Multiple working solutions at different inhibitor concentrations in DMSO are prepared upon usage. The normal concerted integration assay conditions described in section 3.2.2 are employed, and two order-of-addition approaches are used to investigate the mechanism of inhibitor action. After assembly of the IN-DNA complexes at 14°C for 15 min, target DNA is added followed immediately (~20 sec) by inhibitors. The samples are immediately incubated for 2 h at 37°C, deproteinized, and the quantities of FS, CHS, and D-D product formation are determined by PhosphorImager analysis of dried gels. In the second approach, the inhibitors are added after pre-assembly of the IN-DNA complexes at 14°C and further incubated at this temperature for up to 20 min, prior to addition of target DNA and incubation at 37°C [20]. We observed no differences in the IC50 values obtained with either approach using blunt-ended or 3′ OH recessed DNA substrates [20].
4.2.2. Effects of protein concentrations on observed IC50 values
Using the 1.6 kb U5 blunt-ended DNA substrate, a four-fold variation in IN concentration (10 to 40 nM) yielded an ~2-fold difference in IC50 values [12]. The data suggest that higher numbers of active IN-DNA complexes in the concerted integration reaction mixture requires a higher concentration of inhibitor for effective inhibition.
4.3. LEDGF/p75 affects HIV-1 integration
Numerous laboratories have clearly demonstrated that the cellular protein lens epithelium-derived growth factor (LEDGF/p75) significantly influences the ability of the HIV-1 PIC to preferentially select active transcription units as integration sites on host chromosomes, recently reviewed by investigators active in the field [38, 39]. In HIV-1 and equine infectious anemia virus infected cells, extensive knockdown of LEDGF/p75 decreases viral DNA integration into chromosomes ~90% and results in the redistribution of integration sites from transcription units to CpG islands and other regions [40, 41]. However, LEDGF/p75 is not absolutely essential for HIV-1 integration as demonstrated by the use of Ledgf-null mouse cells where again, residual integration sites were redistributed from the norm [41, 42]. Retroviral integration sites reveal a weak consensus DNA sequence centered around the 4–6 bp target site duplication [43, 44]. The HIV-1 consensus site was maintained in LEDGF/p75 knockout cells, consistent with the notion that IN-target DNA interactions important for strand transfer activity are independent from the host mechanisms that help guide PICs to specific genomic areas for integration [41].
4.3.1. LEDGF/p75 modulates HIV-1 concerted integration
Methods to purify recombinant LEDGF/p75 and several of its independently expressed structural domains have been described by others [45, 46]. Using standard assay conditions (section 3.2.2), we found that LEDGF/p75 modulates the ability of HIV-1 IN to promote concerted integration in vitro [13]. Under normal concerted integration assay conditions, LEDGF/p75 modestly stimulates (two- to three-fold) concerted integration at low molar ratios to IN (<1). Concerted integration is inhibited if the molar ratio of LEDGF/p75 to IN is >1, most likely due to disruption of essential IN-IN interactions for the formation of the SC prior to its association with the target DNA to produce the STC. The order in which LEDGF/p75 is added to the reaction mixture significantly affects the outcomes. Addition of LEDGF/p75 at low concentrations prior to or during the pre-assembly step promotes concerted integration, while high concentrations of LEDGF/p75 disrupt this reaction [13]. There is no effect of LEDGF/p75 if added late in the reaction [13, 47]. These order-of-addition experiments should be explored if other cellular co-factors involved in integration are identified in vivo.
4.4. In solution FRET analyses
FRET is based on a relatively long range (10 – 100Å) dipole-dipole energy transfer between nonradiative donor and acceptor fluorophore molecules. This phenomenon results in a decrease in fluorescence emission intensity of a donor fluorophore, and an increase in the acceptor fluorophore emission intensity while exciting the donor fluorophore. The energy transfer efficiency (E) depends on the inverse-sixth power of the distance between the fluorophores, and in doing so provides distance information for labeled biomolecules in a range which is not readily available using other biophysical techniques. An increase or decrease in E between fluorophores generally results from conformational change(s) between the tagged-biomolecules. Here we will discuss our in vitro approaches starting from the design and synthesis of the Cy3 (donor) and Cy5 (acceptor) fluorophore labeled viral DNAs for in solution [48] and in-gel-FRET methods and analysis of concerted integration.
4.4.1. Design and synthesis of Cy3 and Cy5 fluorophore-labeled LTR DNA substrates
We investigated the assembly properties of alpha retroviral IN synaptic complexes at 14°C using in solution FRET analysis [48]. The 5′ end labeled Cy3 and Cy5 LTR oligodeoxynucleotides (91/93 bp, dual HPLC purified) are purchased from Integrated DNA Technologies, Coralville, IA. The fluorophores are attached at the 5′ end of the DNA (U3 plus or U5 minus) (Fig. 2a). We positively correlated significant fluorescence signal changes with concerted integration activity at 37°C and DNaseI protection of ~20 bp of the LTR end by IN. FRET was not observed with IN mutants lacking catalytic activity or using fluorophore-labeled non-LTR substrates. Fluoroscence measurements are performed on a Fluoromax-3 spectrofluorometer with a termperature-regulated cell holder (Jobin Yvon, Inc., Edison, NJ). The FRET methods utilized for alpha retroviral IN generally apply to the study of HIV-1 IN as outlined below.
4.4.2. Design of Cy3 and Cy5 labeled 1.5 kb DNA substrates
We have designed and synthesized Cy3 (donor) and Cy5 (acceptor) fluorophore labeled 1.5 kb U5 DNA substrates. As described above, each fluorophore is attached at the 5′-end of the U5 minus strand. The HPLC/PAGE purified 5′-end fluorescently labeled primers and unlabeled plus-strand primers are from Integrated DNA Technologies Inc. and Operon Technologies, respectively. The fluorescently labeled 1.5 kb DNA containing a single U5 end is prepared by PCR using Cy3 or Cy5 labeled and unlabeled primers and pU3U5 template DNA [22]. PCR cycling parameters includes initial denaturation at 94°C for 2 min, followed by 30 cycles of denaturation at 94°C for 1 min, annealing at 58°C for 30 sec, and extension at 72°C for 4 min, followed by a final extension at 72°C for 10 min. The DNA product containing Cy3 or Cy5 labeled ends is purified on 0.7% agarose gels. The dye labeling efficiency (DLE) is given by:
where ADNA and εDNA are the absorbance and molar extinction coefficient of the purified DNA at 260 nm, respectively. ADye is the absorbance of the same fluorophore labeled DNA solution at 550 nm or 650 nm (for Cy3 or Cy5, respectively). εDye is the molar extinction coefficient of the dye (150,000 M−1cm−1 at 550 nm for Cy3 and 250,000 M−1cm−1 at 650 nm for Cy5). The DLE for the purified fluorophore labeled DNAs are between 0.75 to 0.82.
4.4.3. In solution FRET analysis of HIV-1 nucleoprotein complexes
We describe a method here which demonstrates that HIV-1 nucleoprotein complexes formed in solution are capable of both sensitized and quenched FRET (Fig. 3, see inset) as previously observed with the SC formed with alpha retrovirus IN [48]. The 1.5 kb Cy3 and Cy5 U5 substrates, at 1.5 nM each, were pre-assembled with IN (60 nM) at 14°C followed by the addition of supercoiled DNA (section 3.2.2.) and incubation at 37°C. At different time points, the reactions were stopped by adding EDTA to the final concentration of 25 mM, prior to storage on ice. The samples were adjusted to 0.5 M NaCl to measure the emission spectra (at 14°C) in the range of 560 to 720 nm while exciting the donor fluorophore at 550 nm with 5 nm bands pass for excitation and emission (Fig. 3). A clear isosbestic point exists at 647±1 nm in the emission spectra of donor and acceptor fluorophores, strongly suggesting energy transfer between the 1.5 kb donor and acceptor DNA substrates (Fig. 3). The energy transfer likely results from both the SC and STC, since both complexes are stable to 0.5 M NaCl treatment, which eliminates IN non-specific interactions with DNA [11, 13] (data not shown). Concerted integration by IN is directly correlated with the FRET signal. No FRET signal is observed with Cy3 and Cy5 labeled non-specific DNAs. This system should be expandable for in-gel FRET analysis of the SC and STC on native agarose gels, for example to determine the distance between the LTR ends in these complexes, and within SCs treated with clinically relevant strand transfer inhibitors.
5. Summary
Investigating the biochemical and biophysical mechanisms associated with concerted integration catalyzed by the HIV-1 PIC are limited by the complexities associated with the isolation and the study of this complex from infected cells. Here we emphasize the utility of the reconstituted system to understand the basic mechanisms of concerted integration, to dissect the mechanisms of IN strand transfer inhibitors, and to investigate the potential roles of cellular cofactors that influence the selection of target sites in vivo. The reconstituted system can only partially address the many unknown facets of concerted integration. Still, further development of this system may provide useful tools to understand the next generation of IN inhibitors, as well as the interaction of IN with cellular cofactors and small molecule inhibitors directed against these protein-protein interactions [49, 50].
Acknowledgments
This work was supported by grants from the National Cancer Institute (CA-16312) and the National Institutes of Allergy and Infectious Diseases (AI31334).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Craigie R. In: Mobile DNA II. Craig NL, Craigie R, Gellert M, Lambowitz AM, editors. ASM Press; Washington, DC: 2002. pp. 613–630. [Google Scholar]
- 2.Miller MD, Farnet CM, Bushman FD. J Virol. 1997;71:5382–5390. doi: 10.1128/jvi.71.7.5382-5390.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen H, Engelman A. Mol Cell Biol. 2001;21:6758–6767. doi: 10.1128/MCB.21.20.6758-6767.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hazuda DJ, Felock P, Witmer M, Wolfe A, Stillmock K, Grobler JA, Espeseth A, Gabryelski L, Schleif W, Blau C, Miller MD. Science. 2000;287:646–650. doi: 10.1126/science.287.5453.646. [DOI] [PubMed] [Google Scholar]
- 5.Hazuda DJ, Anthony NJ, Gomez RP, Jolly SM, Wai JS, Zhuang L, Fisher TE, Embrey M, Guare JP, Jr, Egbertson MS, Vacca JP, Huff JR, Felock PJ, Witmer MV, Stillmock KA, Danovich R, Grobler J, Miller MD, Espeseth AS, Jin L, Chen IW, Lin JH, Kassahun K, Ellis JD, Wong BK, Xu W, Pearson PG, Schleif WA, Cortese R, Emini E, Summa V, Holloway MK, Young SD. Proc Natl Acad Sci U S A. 2004;101:11233–11238. doi: 10.1073/pnas.0402357101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Evering TH, Markowitz M. Drugs Today (Barc) 2007;43:865–877. doi: 10.1358/dot.2007.43.12.1146063. [DOI] [PubMed] [Google Scholar]
- 7.Goodarzi G, Im GJ, Brackmann K, Grandgenett D. J Virol. 1995;69:6090–6097. doi: 10.1128/jvi.69.10.6090-6097.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sinha S, Pursley MH, Grandgenett DP. J Virol. 2002;76:3105–3113. doi: 10.1128/JVI.76.7.3105-3113.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sinha S, Grandgenett D. J Virol. 2005;79:8208–8216. doi: 10.1128/JVI.79.13.8208-8216.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li M, Craigie R. J Biol Chem. 2005;280:29334–29339. doi: 10.1074/jbc.M505367200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li M, Mizuuchi M, Burke TR, Jr, Craigie R. EMBO J. 2006;25:1295–1304. doi: 10.1038/sj.emboj.7601005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pandey KK, Bera S, Zahm J, Vora A, Stillmock K, Hazuda D, Grandgenett DP. J Virol. 2007;81:12189–12199. doi: 10.1128/JVI.02863-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pandey KK, Sinha S, Grandgenett DP. J Virol. 2007;81:3969–3979. doi: 10.1128/JVI.02322-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McCord M, Stahl SJ, Mueser TC, Hyde CC, Vora AC, Grandgenett DP. Protein Expr Purif. 1998;14:167–177. doi: 10.1006/prep.1998.0954. [DOI] [PubMed] [Google Scholar]
- 15.Hindmarsh P, Johnson M, Reeves R, Leis J. J Virol. 2001;75:1132–1141. doi: 10.1128/JVI.75.3.1132-1141.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chiu R, Grandgenett DP. J Virol. 2003;77:6482–6492. doi: 10.1128/JVI.77.11.6482-6492.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vora AC, Grandgenett DP. J Virol. 1995;69:7483–7488. doi: 10.1128/jvi.69.12.7483-7488.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vora AC, Chiu R, McCord M, Goodarzi G, Stahl SJ, Mueser TC, Hyde CC, Grandgenett DP. J Biol Chem. 1997;272:23938–23945. doi: 10.1074/jbc.272.38.23938. [DOI] [PubMed] [Google Scholar]
- 19.Vora A, Grandgenett DP. J Virol. 2001;75:3556–3567. doi: 10.1128/JVI.75.8.3556-3567.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zahm JA, Bera S, Pandey KK, Vora A, Stillmock K, Hazuda D, Grandgenett DP. Antimicrob Agents Chemother. 2008;52:3358–3368. doi: 10.1128/AAC.00271-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grandgenett DP, Inman RB, Vora AC, Fitzgerald ML. J Virol. 1993;67:2628–2636. doi: 10.1128/jvi.67.5.2628-2636.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cherepanov P, Surratt D, Toelen J, Pluymers W, Griffith J, De Clercq E, Debyser Z. Nucleic Acids Res. 1999;27:2202–2210. doi: 10.1093/nar/27.10.2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vora AC, McCord M, Fitzgerald ML, Inman RB, Grandgenett DP. Nucleic Acids Res. 1994;22:4454–4461. doi: 10.1093/nar/22.21.4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bushman FD, Craigie R. Proc Natl Acad Sci U S A. 1991;88:1339–1343. doi: 10.1073/pnas.88.4.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vora A, Bera S, Grandgenett D. J Biol Chem. 2004;279:18670–18678. doi: 10.1074/jbc.M314270200. [DOI] [PubMed] [Google Scholar]
- 26.Goodarzi G, Chiu R, Brackmann K, Kohn K, Pommier Y, Grandgenett DP. Virology. 1997;231:210–217. doi: 10.1006/viro.1997.8558. [DOI] [PubMed] [Google Scholar]
- 27.Goodarzi G, Pursley M, Felock P, Witmer M, Hazuda D, Brackmann K, Grandgenett D. J Virol. 1999;73:8104–8111. doi: 10.1128/jvi.73.10.8104-8111.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen H, Wei SQ, Engelman A. J Biol Chem. 1999;274:17358–17364. doi: 10.1074/jbc.274.24.17358. [DOI] [PubMed] [Google Scholar]
- 29.Farnet CM, Haseltine WA. Proc Natl Acad Sci U S A. 1990;87:4164–4168. doi: 10.1073/pnas.87.11.4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moreau K, Torne-Celer C, Faure C, Verdier G, Ronfort C. Virology. 2000;278:133–136. doi: 10.1006/viro.2000.0641. [DOI] [PubMed] [Google Scholar]
- 31.Brin E, Leis J. J Biol Chem. 2002;277:18357–18364. doi: 10.1074/jbc.M201354200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hindmarsh P, Ridky T, Reeves R, Andrake M, Skalka AM, Leis J. J Virol. 1999;73:2994–3003. doi: 10.1128/jvi.73.4.2994-3003.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fitzgerald ML, Grandgenett DP. J Virol. 1994;68:4314–4321. doi: 10.1128/jvi.68.7.4314-4321.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aiyar A, Hindmarsh P, Skalka AM, Leis J. J Virol. 1996;70:3571–3580. doi: 10.1128/jvi.70.6.3571-3580.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brin E, Leis J. J Biol Chem. 2002;277:10938–10948. doi: 10.1074/jbc.M108116200. [DOI] [PubMed] [Google Scholar]
- 36.Moreau K, Faure C, Violot S, Verdier G, Ronfort C. Eur J Biochem. 2003;270:4426–4438. doi: 10.1046/j.1432-1033.2003.03833.x. [DOI] [PubMed] [Google Scholar]
- 37.Hazuda DJ, Young SD, Guare JP, Anthony NJ, Gomez RP, Wai JS, Vacca JP, Handt L, Motzel SL, Klein HJ, Dornadula G, Danovich RM, Witmer MV, Wilson KA, Tussey L, Schleif WA, Gabryelski LS, Jin L, Miller MD, Casimiro DR, Emini EA, Shiver JW. Science. 2004;305:528–532. doi: 10.1126/science.1098632. [DOI] [PubMed] [Google Scholar]
- 38.Engelman A, Cherepanov P. PLoS Pathog. 2008;4:e1000046. doi: 10.1371/journal.ppat.1000046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Poeschla EM. Cell Mol Life Sci. 2008;65:1403–1424. doi: 10.1007/s00018-008-7540-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Llano M, Saenz DT, Meehan A, Wongthida P, Peretz M, Walker WH, Teo W, Poeschla EM. Science. 2006;314:461–464. doi: 10.1126/science.1132319. [DOI] [PubMed] [Google Scholar]
- 41.Marshall HM, Ronen K, Berry C, Llano M, Sutherland H, Saenz D, Bickmore W, Poeschla E, Bushman FD. PLoS ONE. 2007;2:e1340. doi: 10.1371/journal.pone.0001340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shun MC, Raghavendra NK, Vandegraaff N, Daigle JE, Hughes S, Kellam P, Cherepanov P, Engelman A. Genes Dev. 2007;21:1767–1778. doi: 10.1101/gad.1565107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Holman AG, Coffin JM. Proc Natl Acad Sci U S A. 2005;102:6103–6107. doi: 10.1073/pnas.0501646102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu X, Li Y, Crise B, Burgess SM, Munroe DJ. J Virol. 2005;79:5211–5214. doi: 10.1128/JVI.79.8.5211-5214.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cherepanov P, Maertens G, Proost P, Devreese B, Van Beeumen J, Engelborghs Y, De Clercq E, Debyser Z. J Biol Chem. 2003;278:372–381. doi: 10.1074/jbc.M209278200. [DOI] [PubMed] [Google Scholar]
- 46.Cherepanov P, Sun ZY, Rahman S, Maertens G, Wagner G, Engelman A. Nat Struct Mol Biol. 2005;12:526–532. doi: 10.1038/nsmb937. [DOI] [PubMed] [Google Scholar]
- 47.Raghavendra NK, Engelman A. Virology. 2007;360:1–5. doi: 10.1016/j.virol.2006.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bera S, Vora AC, Chiu R, Heyduk T, Grandgenett DP. Biochemistry. 2005;44:15106–15114. doi: 10.1021/bi0508340. [DOI] [PubMed] [Google Scholar]
- 49.Al-Mawsawi LQ, Christ F, Dayam R, Debyser Z, Neamati N. FEBS Lett. 2008;582:1425–1430. doi: 10.1016/j.febslet.2008.02.076. [DOI] [PubMed] [Google Scholar]
- 50.Hayouka Z, Rosenbluh J, Levin A, Loya S, Lebendiker M, Veprintsev D, Kotler M, Hizi A, Loyter A, Friedler A. Proc Natl Acad Sci U S A. 2007;104:8316–8321. doi: 10.1073/pnas.0700781104. [DOI] [PMC free article] [PubMed] [Google Scholar]