Abstract
The impact of genome length on replication and genome stability was assessed for guinea pig cytomegalovirus (GPCMV), a member of the Herpesviridae. The 233-kb genome could be decreased by 15.1 kb without discernable impact on viral replication efficiency in vitro. Viruses with genomes under-length by up to 31 kb replicated with decreased efficiencies but this appeared to arise from the loss of augmenting viral genes rather than decreased genome length. Two deletions that were non-lethal on their own were lethal when combined, suggesting that the resulting 40.1 kb under-length genome fell below a minimum packageable size. Genomes over-length by 8.8 kb gave rise to spontaneous deletions just to the right of the major immediate early locus, the same region that undergoes deletions during fibroblast passage of human and rhesus cytomegaloviruses. These results suggest that genome integrity should be confirmed for herpesvirus mutants in which genome length is increased even modestly.
Keywords: herpesvirus, genome length, genome maturation
Introduction
Herpesviruses have large (130 - 235 kb) linear double-stranded DNA genomes that replicate via concatemeric intermediates consisting of head-to-tail linked genomes (Bataille and Epstein, 1994; Ben-Porat, 1983; Jacob, Morse, and Roizman, 1979; Martinez et al., 1996; McVoy and Adler, 1994; Roizman and Sears, 1996; Severini et al., 1994; Zhang, Efstathiou, and Simmons, 1994). The concatemers are packaged into preformed capsids and cleaved at precise locations to release unit length genomes within the capsids (Brown, McVoy, and Homa, 2002). These sequence-specific cleavage events are governed by nearby cis-acting DNA sequences; however, even when the necessary cis sequences are present DNA cleavage will not occur unless a sufficient amount of DNA has entered the capsid (Bloss and Sugden, 1994; Deiss and Frenkel, 1986; McVoy et al., 1998; McVoy et al., 2000; McVoy and Ramnarain, 2000; Roizman and Sears, 1996; Wang, Nixon, and McVoy, 2008). This “head-full” constraint appears to impose a lower limit on genome length. Conversely, an upper limit must be imposed, if not by the packaging machinery then ultimately by the interior volume of the capsid. Although examining the lengths of preferentially packaged defective genomes has allowed an estimation of the size range for efficient DNA packaging for herpes simplex virus type 1 (Vlazny, Kwong, and Frenkel, 1982), human cytomegalovirus (HCMV) (Borst and Messerle, 2003), and Epstein-Barr virus (Bloss and Sugden, 1994; Kempkes et al., 1995), the impact of genome length on viral replication has not been methodically studied by engineering viruses with under-length or over-length genomes.
In the process of constructing a bacterial artificial chromosome (BAC) clone of the guinea pig cytomegalovirus (GPCMV) genome, BAC clones with substantial deletions on one or both sides of the 8.8-kb BAC origin insertion (BAC ori) were invariably recovered, suggesting that GPCMV is highly intolerant of this large an increase in genome length (Cui et al., 2008). In this report the importance of genome length was investigated further using BACs predicted to reconstitute viruses with genomes ranging from 48.9 kb under-length to 8.8 kb over-length. Surprisingly, while genomes under-length by up to 31 kb were tolerated, genomes over-length by 8.8 kb quickly gave rise to compensatory deletions that occurred predominantly in a region just to the right of the GPCMV major immediate early locus (Fig. 1), a region noted for spontaneous deletions and rearrangements in both HCMV and rhesus cytomegalovirus (RhCMV) (Oxford et al., 2008; Prichard et al., 2001).
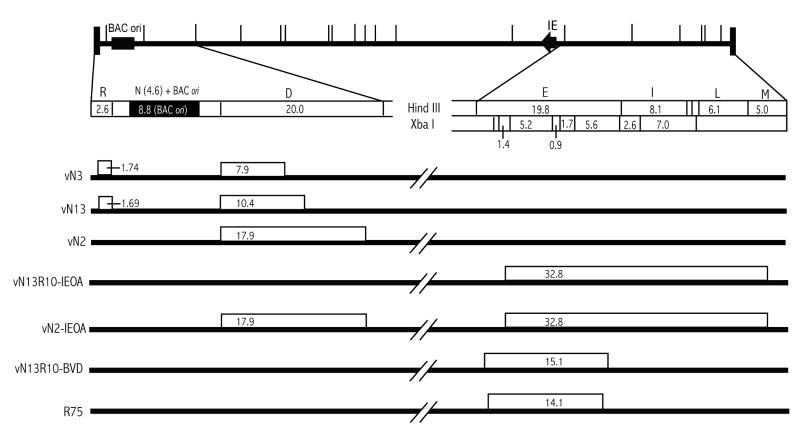
Fig. 1. Viral deletion mutants.
A HindIII map of the BAC ori-containing GPCMV genome (top) shows the locations of the BAC ori in HindIII N (black box) and the major immediate early (IE) gene locus (black arrow). The left end from HindIII R to D and the right end from HindIII E to M are expanded below. Sequence-based HindIII and XbaI restriction maps indicate the sizes of predicted fragments in kb. Thick lines below represent the genomes of mutant viruses. Open boxes represent deletions and numbers within indicate the sizes of deletions in kb.
Results
Insertion of an 8.8-kb BAC origin gives rise to spontaneous deletions flanking the insertion
In recently reported work we undertook to clone the GPCMV strain 22122 genome as an infectious BAC (Cui et al., 2008). The cloning strategy involved inserting an 8.8-kb BAC ori cassette into the HindIII N region of the GPCMV genome (Fig. 1) by homologous recombination, then deriving BAC clones of this genome by transformation of viral DNA into E. coli. While the majority of BAC clones were infectious (able to reconstitute replication competent virus upon transfection of BAC DNA into guinea pig cells), restriction analysis of 13 initial clones revealed that all lacked a 2.3-kb EcoRI fragment that lies within HindIII D just to the right of HindIII N (Cui et al., 2008). An additional 59 clones were screened using a PCR reaction specific to sequences within the 2.3-kb EcoRI fragment and all but one were negative; the one positive clone proved to be non-infectious and lacked large regions from elsewhere in the viral genome (Cui et al., 2008). Three of the initial 13 clones were further characterized by restriction mapping and sequencing to establish the exact breakpoints of their deletions. All three BACs had substantial deletions within HindIII D. In each case the leftward break points coincided precisely with the HindIII site defining the boundary between HindIII N and D, whereas the rightward break points differed among BACs and resulted in deletions that varied in size (7.9 kb in pN3, 10.4 kb in pN13, and 17.9 kb in pN2 (Fig. 1). The BACs with the smaller deletions in HindIII D (pN3 and pN13) were found to have additional small deletions of 1.69- and 1.74-kb in HindIII R, which lies to the left of HindIII N. Again, the rightward break points coincided precisely with the HindIII site that defines the boundary between HindIII R and N, while the leftward break points differed slightly (Fig. 1). These deletions were not unique to this particular attempt to recombine the BAC ori into the viral genome as similar deletions occurred upon two additional attempts (not shown).
That the deletional break points proximal to HindIII N coincided precisely with the ends of the linear HindIII N fragment (modified to contain the BAC ori, see Fig. 1) that was used for recombination suggested that the deletions arose during and not subsequent to the recombination process. That all infectious BAC clones that were recovered had deletions associated with the HindIII D region suggests that genomes lacking compensatory deletions were strongly selected against, presumably at the time of genome packaging.
Viruses derived from BACs with deletions have modest growth defects that are not associated with genome length
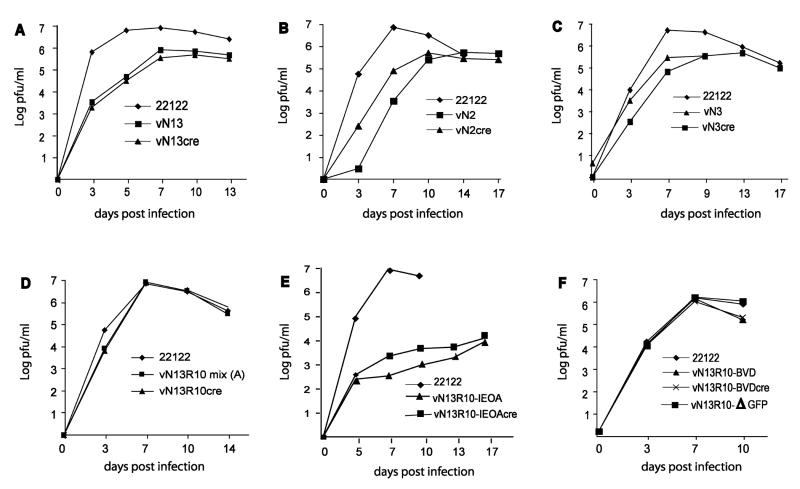
To investigate the impact of these deletions on viral replication, BACs were transfected into guinea pig lung fibroblast (GLF) cells and infectious viruses were recovered. Moreover, because the BAC ori is flanked by LoxP sites and can be excised by cotransfection of BAC DNA with a plasmid that expresses cre recombinase, pCre, it was possible to reconstitute from each BAC virus pairs that were predicted to differ in genome length by 8.8 kb due to retention or excision of the BAC ori. Consequently, although the viruses vN2 and vN2cre were both derived from BAC pN2 and are therefore missing 17.9 kb of viral sequences, the genome of virus vN2 is predicted to be 9.1 kb under-length because it retains the 8.8-kb BAC ori (-17.9 + 8.8 = -9.1), while that of vN2cre should be 17.9 kb under-length because the BAC ori was excised. Similarly, the genomes of vN13cre and vN13, derived from BAC pN13, were predicted to be 12.1 kb (-10.4 -1.69 = -12.1) and 3.3 kb (-10.4 -1.69 + 8.8 = -3.3) under-length, while those of vN3cre and vN3, derived from BAC pN3, were predicted to be 9.6 (-7.9 -1.74 = -9.6) and 0.84 (-7.9 -1.74 + 8.8 = -0.84) kb under-length respectively. All six BAC-derived viruses replicated with modest growth impairments relative to the parental strain 22122 virus from which the BACs were derived. Peak titers were generally one log below that of the parental virus and times to reach peak titers were delayed by several days (Fig. 2A-C). Genome length, however, did not appear to be a factor in these growth defects as retention of the BAC ori, which in the case of vN3 essentially restores normal genome length, did not restore wild type replication kinetics or levels (Fig. 2A-C). A more likely explanation for these replication deficiencies is the loss of specific augmenting genes that lie within the deleted regions of HindIII D that are common to all three mutants.
Fig. 2. Growth properties of BAC-derived deletion mutants.
GPL cells were infected with the indicated viruses at MOIs of 0.01 PFU/cell and viral titers in the culture supernatants were determined on the days post infection indicated.
A 32.8-kb deletion near the right end of the genome causes a severe growth defect and is lethal when combined with the N2 deletion
Both deletions in the pN13 BAC were repaired to generate BAC pN13R10 such that upon excision of BAC ori, pN13R10 reconstitutes a virus, vN13R10cre, that contains a complete and authentic GPCMV genome and replicates with fully wild type kinetics ((Cui et al., 2008) and Fig. 2D). Allelic exchange was used to introduce a 32.8-kb deletion of viral sequences spanning HindIII E - M and designated IEOA into either pN2 or pN13R10 and the resulting BACs were designated pN2-IEOA and pN13R10-IEOA, respectively (Fig. 1). Two candidate clones representing pN13R10-IEOA and nine clones representing BAC pN2-IEOA were confirmed by restriction pattern analysis to have the correct predicted patterns (details of restriction pattern analyses are described in Materials and Methods and examples are shown in Fig. 3A). As the deleted sequence was replaced by a 1.8-kb kanamycin-resistance/lacZα (kanr/lacZ) marker gene cassette, the net decrease in genome length was predicted to be 31 kb. The two pN13R10-IEOA clones were both infectious and reconstituted viruses that grew extremely poorly, reaching peak titers of only 104 pfu/ml. Again, retention or excision of the BAC ori did not significantly impact replication kinetics or efficiency (Fig. 2E). Surprisingly, all nine of the pN2-IEOA clones were non-infectious, whether or not pCre was included to excise the BAC ori. That the N2 and IEOA mutations were not lethal in separate viruses but were lethal when combined within the same BAC-cloned genome suggests that the 48.9 (-17.9 −32.8 +1.8 = -48.9) or 40.1 (-17.9 -32.8 +1.8 + 8.8 = -40.1) kb under-length genomes of vN2-IEOAcre or vN2-IEOA (respectively) may have fallen below the minimum necessary for viral replication.
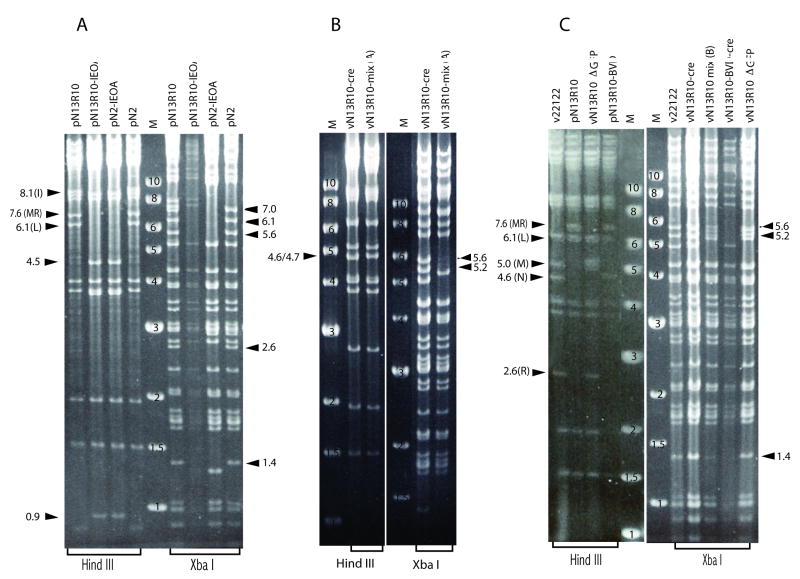
Fig. 3. Restriction pattern analyses of BACs and viral DNAs.
(A) BACs with IEOA deletions were compared to their respective parental BACs by restriction with HindIII or XbaI, separation on agarose gels, and ethidium bromide staining. (B) Virion DNA from vN13R10-mix(A) was similarly compared to vN13R10cre (reconstituted from BAC pN13R10 with BAC ori excision). (C) BAC DNAs pN13R10 and pN13R10-BVD were similarly compared to virion DNAs from the indicated viruses. Numbers on 1 kb ladder markers (M) indicate their sizes in kb. Arrows indicate the positions and sizes (in kb) of relevant restriction fragments. Letters in parentheses indicate letter designations for restriction fragments that are referred to in the text.
An 8.8-kb over-length genome undergoes spontaneous deletions within a “deletional hot spot”
Thorough restriction pattern analyses indicate that the GPCMV genome cloned in BAC pN13R10 is full length (Cui et al., 2008). Thus, when pN13R10 was transfected into cells without pCre, retention of the BAC ori was predicted to make the genome over-length by 8.8 kb. Transfection of pN13R10 DNA without pCre gave rise to a virus stock designated vN13R10-mix(A) that was heterogeneous. Individual foci of infected cells could be identified by their characteristic viral cytopathic effect (CPE) and while most of these foci contained GFP-positive cells, occasional GFP-negative foci were also present (not shown). Growth curve analysis of vN13R10-mix(A), however, detected no evidence of a growth defect when compared to either the parental virus or to vN13R10cre (Fig. 2D).
Restriction of virion DNA extracted from vN13R10-mix(A) revealed that 5.2- and 5.6-kb XbaI fragments were greatly under-represented (Fig. 3B). Based on their sizes these fragments were identified as lying within the HindIII E region of the GPCMV genome (Fig. 1). Due to the large size of HindIII E (19.8 kb) it was not possible to determine if HindIII E was altered, however, vN13R10-mix(A) DNA also contained a novel 4.7-kb HindIII fragment (Fig. 3B). The mixture of viruses in vN13R10-mix(A) was subjected to limiting-dilution in 96-well plates. At a dilution sufficient to achieve predominantly singular viral infections per well 10% of virus-positive wells were GFP-negative, suggesting that some viruses had undergone spontaneous deletions that impacted gfp, which lies within the BAC ori. Restriction analyses and sequencing of one GFP-negative clone, designated vN13R10-ΔGFP, revealed an 8.4-kb deletion that removed gfp, most of the BAC ori, and a small amount of adjacent HindIII N sequence. The deletion, however, had no impact on viral replication (Fig. 2F).
Similar analyses were performed on a GFP+ virus clone designated vN13R10-BVD. This virus contained the novel 4.7-kb HindIII fragment (not shown) and completely lacked the 5.2- and 5.6-kb XbaI fragments (Fig. 3C). A BAC clone of the vN13R10-BVD genome designated pN13R10-BVD was obtained by transformation of vN13R10-BVD DNA into E. coli. Restriction analysis again confirmed the absence of the 5.2- and 5.6-kb XbaI fragments (data not shown), while the 4.7-kb HindIII fragment could be readily identified as a novel fragment because the closely migrating 4.6-kb HindIII N fragment was enlarged to 13.4 kb by retention of the BAC ori (Fig. 3C). Sequencing of pN13R10-BVD revealed a 15.1-kb deletion that removed the 5.2- and 5.6-kb XbaI fragments and reduced the size of HindIII E from 19.8 kb to 4.7 kb. (Fig. 1). Transfection of pN13R10-BVD into cells with or without pCre was used to derive viruses in which the BAC ori was either retained (vN13R10-BVD) or excised (vN13R10-BVDcre). Growth curves revealed that both viruses replicate with efficiencies and kinetics similar to wild-type (Fig. 2F). Thus, neither the 15.1-kb deletion in HindIII E nor the presence or absence of the BAC ori had an impact on viral replication. This result further confirms that the replication defects of viruses derived from pN2, pN3, and pN13 are caused by the absence of specific augmenting genes, and not decreased genome length, as vN13R10-BVDcre has a similarly undersized genome yet replicates normally.
The near absence of 5.2- and 5.6-kb XbaI fragments from vN13R10-mix(A) DNA suggested that the majority of viruses in the mix had undergone spontaneous deletions within HindIII E. To determine if this is a consistent phenomenon, pN13R10 BAC DNA was again transfected into cells to derive a second virus mixture designated vN13R10-mix(B). Limiting dilution of vN13R10-mix(B) revealed a somewhat higher proportion of GFP-negative viruses (38%), and while the 5.2- and 5.6-kb XbaI fragments were again under-represented, this was less profound than in the previous experiment (Fig. 3C). This can be explained by the higher proportion of GFP-negative viruses, assuming they, like vN13R10-ΔGFP (Fig. 3C), lack deletions in HindIII E and therefore contribute 5.2- and 5.6-kb XbaI fragments to the mixture. These results indicate that on two independent occasions, a preponderance of spontaneous deletions occurred within HindIII E.
Discussion
Infectious BAC clones of herpesvirus genomes provide powerful tools for mutagenesis of viral sequences as genetic changes can be created in E. coli irrespective of their impact on virus replication. In most cases BAC clones have proven to be extremely stable when propagated or manipulated in E. coli and the reconstitution of viruses from BAC clones has generally proven to be highly reproducible and not prone to spontaneous deletions or rearrangements. However, while elaborate manipulations to viral genomes can now be engineered in BACs, the impact that large-scale engineered deletions, insertions, or other rearrangements might have on viral replication efficiency or genome stability has often been overlooked. The flexibility of the herpesvirus genome packaging machinery in terms of genome size or cleavage site spacing has not to date been carefully examined. Experience has been largely anecdotal, with various reports noting that certain viruses appear to tolerate marker gene insertions of a few kb, or in some cases relatively large deletions, without a significant impact on replication efficiency; however, the integrity of the resulting mutant viral genomes has not always been closely examined.
Our analyses of BAC-derived GPCMV mutants demonstrates that GPCMV can tolerate loss of up to 15.1 kb of non-essential sequences without any impact on replication efficiency. Mutants with larger deletions were viable but had modest (N2) to severe (IEOA) growth defects, but this appears to be unrelated to genome length. Similar results have been recently reported for murine cytomegalovirus (MCMV), which tolerated a 26-kb decrease in genome length without loss of replication efficiency (Cicin-Sain et al., 2007). However, our failure to reconstitute infectious virus from a BAC that combined the 17.9-kb N2 deletion with the 32.8-kb IEOA deletion suggests that there is a limit to which the spacing between packaging signals can be reduced; while decreasing this spacing by 17.9 (vN2cre), 22.2(vN13R10-IEOA), or 31 kb (vN13R10-IEOAcre) was tolerated, decreasing it by 40.1 (pN2-IEOA) or 48.9 (pN2-IEOAcre) kb was not. Also consistent with this hypothesis, concatemers of an MCMV genome containing an engineered ectopic cleavage site are not cleaved at adjacent sites, which are spaced 45 or 191 kb apart, but rather are cleaved at alternate sites that are spaced the normal 236-kb genome length apart (Wang, Nixon, and McVoy, 2008). It should be noted, however, that the available data do not rule out an alternative explanation that the two deleted regions each contain a gene that can compensate for the other, such that only combining the two deletions results in a lethal phenotype.
In contrast, GPCMV is relatively intolerant of over-length genomes. Although small 2-3 kb marker gene insertions have been tolerated by GPCMV (Abbate et al., 2001) and many other herpesviruses, insertion of the 8.8-kb BAC origin cassette apparently exceeded the packaging machinery's upper limit. During recombinant virus construction this resulted in selection of deletion mutants that arose concomitant with recombination between plasmid and viral DNA. However, when an 8.8 kb over-length BAC-cloned genome was transfected into guinea pig cells, spontaneous deletions in the HindIII E region were predominantly selected. Even the normal GPCMV genome appears to be unstable in this region as a 1.6-kb deletion (Fig. 1) in the region corresponding to the left-hand break point of the BVD deletion was recently found in 17% of genomes in the ATCC stock of GPCMV strain 22122 (Nozawa et al., 2008). Why this particular region is favored for deletion is not known. Other similarly-sized tracts of sequence elsewhere in the genome would seem equally suitable for deletion. For example, viruses with a 6-kb deletion from the right half of HindIII D (Lacayo and McVoy, unpublished data) or an 8.2-kb deletion spanning HindIII L and M (Reeves et al., manuscript in preparation) replicate with fully wild-type growth properties in vitro. Moreover, examination of the nucleotide sequence of this region or the specific break points of the deletions did not reveal evidence for repeats or partially homologous sequences that could have recombined.
Interestingly, a GPCMV mutant named R-75 was derived after serial passage of strain 22122 in the presence of the cleavage/packaging inhibitor 2-bromo-5,6-dichloro-1-β-D-riborfuranosyl benzimidazole riboside (BDCRB). While the BDCRB resistance of this virus may be associated with an amino acid change in the GP89 terminase subunit gene, the genome of R-75 has two other very striking features – extensive reiterations at both ends of up to 10 copies of the 1-kb terminal repeat (which are normally present as either one or no copies), and a 14.1-kb deletion within HindIII E that is very similar but not identical to the BVD deletion (Fig. 1) (Sauer et al., manuscript in preparation). We hypothesize that during passage of virus in the presence of BDCRB amplification of the terminal repeats may have occurred, perhaps conferring resistance by increasing the number of cleavage sites available for terminase recognition. As additional terminal repeats accumulated a compensatory deletion in HindIII E occurred to prevent the genome from becoming over-length.
The importance of maintaining an optimal genome length may help to explain why many herpesviruses have evolved and retained highly complex genome structures that are characterized by large direct repeats, highly reiterated small direct repeats, or large inverted repeats (Roizman, 1993). Maintaining such repeats may facilitate viral adaptation by providing the genome with plasticity. If acquisition of a new gene is advantageous to the virus it would not be necessary to delete sequences elsewhere in the genome, but rather, genome length could be maintained by modulating either the size or the number of repeats. Conversely, loss of certain genes or sequences might be advantageous and the genome could readily increase in length by increasing the size or number of its repeats. Indeed, this appears to have occurred on several independent occasions during serial passage of HCMV in cultured fibroblasts; several fibroblast-adapted HCMV laboratory strains have deleted up to 15.5 kb of sequence from the UL/b′ region of the genome, yet genome length has remained essentially unchanged due to compensatory increases in the sizes of the inverted repeats (Prichard et al., 2001). Analogous deletions have apparently also occurred upon fibroblast passage of rhesus cytomegalovirus (RhCMV) (Oxford et al., 2008). Curiously, deletions in HCMV and RhCMV laboratory strains occurred in roughly the same location as the deletional hot spot in GPCMV, just to the right of the major immediate early gene locus. While these deletions are speculated to confer a replication advantage for HCMV and RhCMV in fibroblasts, our data further suggest that this region is inherently prone to deletions. In all three viruses the proximity of the major immediate early promoter to the deleted regions suggests a possible mechanism by which strong transcriptional activity may promote DNA instability.
In practical terms, our results suggest that genome length should be an important consideration when designing mutations. While substantial amounts of sequence can be deleted without impacting virus replication, a lower limit may be reached. Conversely, genomes exceeding a critical length are strongly selected against and this gives rise to spontaneous deletions within the first rounds of viral replication. These observations may have implications for the use of recombinant HCMVs as gene therapy vectors or as recombinant live attenuated vaccines expressing heterologous antigens since insertion of heterologous sequences may cause compensatory deletions (Rizvanov et al., 2003). As such mutants may exhibit no alterations in replication efficiency, care should be taken when an over-length genome is anticipated to examine mutant virus genomes for deletions after reconstitution from BACs.
Materials and Methods
Virus and cell culture
GPCMV strain 22122 (ATTC VR682) and BAC-derived viruses were propagated in guinea pig lung fibroblast (GLF) cells (ATCC CCL 158) in minimum essential medium (Invitrogen, Carlsbad) supplemented with 10% fetal calf serum (FCS, HyClone, Logan,), 10,000 IU/L penicillin, 10 mg/L streptomycin (Invitrogen).
DNA preparation
Virion DNA was prepared from culture supernatants of virus-infected cells when the cells reached full CPE. Virions were pelleted from supernatants at 25,000 rpm for 30 min. and viral DNA was isolated as previously described (McVoy, Nixon, and Adler, 1997). Mini- and Midi-prep BAC DNAs were prepared as described previously (Cui et al., 2008).
Virus reconstitution from BAC DNA and isolation of GFP-negative viruses
Midi-prep BAC DNA (0.2-0.4 μg) was co-transfected with 0.2 μg pCre plasmid DNA (constructed by Wolfram Brune and kindly provided by Gabriele Hahn) into subconfluent GLF cells in 6-well plates using 20 μl Effectene (Qiagen, Valencia). Cells were incubated at 37°C for 10-14 days until extensive CPE was observed. Typically, cre-mediated excision of BAC ori was ∼50%- efficient, giving rise to both GFP+ and GFP-negative CPE+ cells. BAC ori-excised (GFP-negative) viruses were isolated by limiting-dilution in 96-well plates containing confluent GLF cells and selection of CPE+ wells that exhibited no detectable GFP expression. Viruses predicted to retain the BAC ori were reconstituted by transfection of BAC DNAs without pCre.
Viral growth curves
Confluent GLF cells in 75 cm2 flasks were infected with carefully matched viral inocula at an MOI of 0.01 PFU/cell. The cultures were washed 3 h post infection to remove unattached virus. Samples of culture media were titered every 3 or 4 days for up to 17 days. Viral titers were determined using a 96-well method described elsewhere (Cui et al., 2008).
Construction IEOA deletion mutants
The IEOA deletions were introduced to BACs by allelic exchange using the vector IE-kanr/lacZ-OA-pGS284. To construct this vector a 799-bp region from within HindIII E was PCR amplified using primers IE-f (5′-GGA GCT CCG TTA CGG TTG CTG ACG AAC TGG-3′) and IE-r (5′-GCG GCC GCT TTT GCG GCC GCG AAA TGG ACG CTA CGG ATA CGA G-3′) and designated IE. A 929-bp region within HindIII O was then amplified using primers OA-f (5′-GCG GCC GCA AAA GCG GCC GCG TAA CGG GTA GAT GGA ACT GG-3′) and OA-r (5′-AAG ATC TGG AGG TGG AAA TCG GGG TTG AG-3′) and designated OA. Both products were amplified from 0.5 μg GPCMV strain 22122 virion DNA using the Bio-X-ACT-Short PCR kit (Bioline, Boston) according to the manufacturer's instructions. IE and OA PCR products were each T/A cloned into vector pCR-XL-TOPO (Invitrogen) to make plasmids pCRXL-IE and pCRXL-OA, respectively. Non-viral sequences within the above described primers (underlined) generated 5′ SacI and 3′ NotI sites flanking IE and 5′ NotI and 3′ BglII sites flanking OA. The IE fragment was excised from pCRXL-IE by SacI/NotI double digestion and OA was excised from pCRXL-OA by NotI/BglII double digestion. A kanr/lacZ marker gene cassette was then excised from pYD-Tn1721 (Yu et al., 2002) (kindly provided by Dong Yu) as a 1.8-kb NotI fragment. The IE, OA, and kanr/lacZ fragments were annealed and ligated into SacI/BglII-digested plasmid pGS284ΔNotI (Cui et al., 2008), a modified version of the allelic exchange vector pGS284 (Smith and Enquist, 1999) (kindly provided by Greg Smith). Blue colonies that grew on plates containing ampicillin, kanamycin, and X-gal were characterized by restriction pattern analysis and sequencing to confirm the correct order and orientation of each fragment (SacI-IE-NotI-kanr/lacZ-NotI-OA-BglII) and correct insertion into the vector. The resulting clone was designated IE-kanr/lacZ-OA-pGS284.
Allelic exchange was conducted essentially as described previously (Cui et al., 2008). Briefly, E. coli strain GS500 cells (kindly provided by Greg Smith) (Smith and Enquist, 1999) containing either pN2 or pN13R10 were mated with S17λpir cells containing IE-kanr/lacZ-OA-pGS284. Clones containing cointegrates were selected on plates containing 20 μg/ml chloramphenicol, 10 μg/ml kanamycin, 20 μg/ml ampicillin. Colonies containing BACs that had lost sacB by resolution and left the kanr/lacZ cassette in place of 32.8 kb of viral sequence were then negatively selected on plates containing 10 μg/ml kanamycin, 20 μg/ml chloramphenicol, 5% sucrose, 100 μg/ml X-gal, and 40 μg/ml IPTG. BACs derived from blue colonies were compared to pN13R10 and pN2 by digestion with HindIII or XbaI. Based on the assembled genomic sequence, BACs with the IEOA deletion were predicted to lack HindIII E (19.8 kb), I (8.1 kb), L (6.1 kb), and MR (7.6 kb) and to lack XbaI fragments of 0.9, 1.4, 1.7, 2.6, 5.2, 5.6 and 7.0 kb (Fig. 1), while insertion of the kanr/lacZ cassette was predicted to create novel 4.5-kb and 0.9-kb HindIII and 1.3-kb XbaI fragments. Examples of restriction patterns from representative BAC clones pN13R10-IEOA and pN2-IEOA are shown in Fig. 3. The predicted fragment changes were observed with the following exceptions. The 18.1-kb HindIII E fragment and 0.9-kb XbaI fragments could not be resolved from other fragments. The 5.2-kb XbaI fragment was missing from all BACs but instead a 6.1-kb fragment was present in pN13R10 and pN2 but absent from IEOA and BVD deletion mutants. However, the 6.1-kb fragment was lost and the 5.2-kb fragment regained upon reconstitution of vN13R10cre viral DNA from pN13R10 (Fig. 3), suggesting that the XbaI site that lies between the 5.2- and the adjacent 0.9-kb fragment is blocked by Dam methylase in E. coli-derived BAC DNA (the site has the appropriate sequence for Dam methylation). BACs containing IEOA deletions were sequenced across the breakpoints between IE and kanr/lacZ and between kanr/lacZ and OA.
Sequencing
Sanger dideoxy sequencing was performed by the Nucleic Acids Research Facilities of Virginia Commonwealth University using either midi-prep BAC or virion DNA templates. Deletions were identified relative to the complete sequence of the GPCMV genome, GenBank accession #FJ355434 (Schleiss et al., 2008). Table 1 (supplemental data) gives exact nucleotide positions for all deletions, as numbered in #FJ355434.
Supplementary Material
Acknowledgments
The authors are grateful to Gabriele Hahn and Wolfram Brune for providing pCre, Dong Yu for providing pYD-Tn1721, and Greg Smith for providing pGS284 and E. coli strain GS500. This work was supported by a grant R01HD044864 from the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abbate J, Lacayo JC, Prichard M, Pari G, McVoy MA. Bifunctional protein conferring enhanced green fluorescence and puromycin resistance. Biotechniques. 2001;31(2):336–40. doi: 10.2144/01312st05. [DOI] [PubMed] [Google Scholar]
- Bataille D, Epstein A. Herpes simplex virus replicative concatemers contain L components in inverted orientation. Virology. 1994;203(2):384–8. doi: 10.1006/viro.1994.1498. [DOI] [PubMed] [Google Scholar]
- Ben-Porat T. Replication of herpesvirus DNA. In: Roizman B, editor. The herpesviruses. Plenum Press; New York, N.Y.: 1983. pp. 81–106. [Google Scholar]
- Bloss TA, Sugden B. Optimal lengths for DNAs encapsidated by Epstein-Barr virus. J Virol. 1994;68(12):8217–22. doi: 10.1128/jvi.68.12.8217-8222.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borst EM, Messerle M. Construction of a cytomegalovirus-based amplicon: a vector with a unique transfer capacity. Hum Gene Ther. 2003;14(10):959–70. doi: 10.1089/104303403766682223. [DOI] [PubMed] [Google Scholar]
- Brown JC, McVoy MA, Homa FL. Packaging DNA into Herpesvirus Capsids. In: Bogner E, Holzenburg A, editors. Structure-Function Relationships of Human Pathogenic Viruses. Kluwer Academic/Plenum Publishers; London: 2002. [Google Scholar]
- Cicin-Sain L, Bubic I, Schnee M, Ruzsics Z, Mohr C, Jonjic S, Koszinowski UH. Targeted deletion of regions rich in immune-evasive genes from the cytomegalovirus genome as a novel vaccine strategy. J Virol. 2007;81(24):13825–34. doi: 10.1128/JVI.01911-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X, McGregor A, Schleiss MR, McVoy MA. Cloning the complete guinea pig cytomegalovirus genome as an infectious bacterial artificial chromosome with excisable origin of replication. J Virol Methods. 2008;149(2):231–9. doi: 10.1016/j.jviromet.2008.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deiss LP, Frenkel N. Herpes simplex virus amplicon: cleavage of concatemeric DNA is linked to packaging and involves amplification of the terminally reiterated a sequence. J Virol. 1986;57(3):933–41. doi: 10.1128/jvi.57.3.933-941.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob RJ, Morse LS, Roizman B. Anatomy of herpes simplex virus DNA. XII. Accumulation of head-to-tail concatemers in nuclei of infected cells and their role in the generation of the four isomeric arrangements of viral DNA. J Virol. 1979;29(2):448–57. doi: 10.1128/jvi.29.2.448-457.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempkes B, Pich D, Zeidler R, Sugden B, Hammerschmidt W. Immortalization of human B lymphocytes by a plasmid containing 71 kilobase pairs of Epstein-Barr virus DNA. J Virol. 1995;69(1):231–8. doi: 10.1128/jvi.69.1.231-238.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez R, Sarisky RT, Weber PC, Weller SK. Herpes simplex virus type 1 alkaline nuclease is required for efficient processing of viral DNA replication intermediates. J Virol. 1996;70(4):2075–85. doi: 10.1128/jvi.70.4.2075-2085.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVoy MA, Adler SP. Human cytomegalovirus DNA replicates after early circularization by concatemer formation, and inversion occurs within the concatemer. J Virol. 1994;68(2):1040–51. doi: 10.1128/jvi.68.2.1040-1051.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVoy MA, Nixon DE, Adler SP. Circularization and cleavage of guinea pig cytomegalovirus genomes. J Virol. 1997;71(6):4209–17. doi: 10.1128/jvi.71.6.4209-4217.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVoy MA, Nixon DE, Adler SP, Mocarski ES. Sequences within the herpesvirus-conserved pac1 and pac2 motifs are required for cleavage and packaging of the murine cytomegalovirus genome. J Virol. 1998;72(1):48–56. doi: 10.1128/jvi.72.1.48-56.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVoy MA, Nixon DE, Hur JK, Adler SP. The ends on herpesvirus DNA replicative concatemers contain pac2 cis cleavage/packaging elements and their formation is controlled by terminal cis sequences. J Virol. 2000;74(3):1587–92. doi: 10.1128/jvi.74.3.1587-1592.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVoy MA, Ramnarain D. Machinery to support genome segment inversion exists in a herpesvirus which does not naturally contain invertible elements. J Virol. 2000;74(10):4882–7. doi: 10.1128/jvi.74.10.4882-4887.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozawa N, Yamamoto Y, Fukui Y, Katano H, Tsutsui Y, Sato Y, Yamada S, Inami Y, Nakamura K, Yokoi M, Kurane I, Inoue N. Identification of a 1.6 kb genome locus of guinea pig cytomegalovirus required for efficient viral growth in animals but not in cell culture. Virology. 2008;379(1):45–54. doi: 10.1016/j.virol.2008.06.018. [DOI] [PubMed] [Google Scholar]
- Oxford KL, Eberhardt MK, Yang KW, Strelow L, Kelly S, Zhou SS, Barry PA. Protein coding content of the ULb′ region of wild-type rhesus cytomegalovirus. Virology. 2008;373(1):181–8. doi: 10.1016/j.virol.2007.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prichard MN, Penfold ME, Duke GM, Spaete RR, Kemble GW. A review of genetic differences between limited and extensively passaged human cytomegalovirus strains. Rev Med Virol. 2001;11(3):191–200. doi: 10.1002/rmv.315. [DOI] [PubMed] [Google Scholar]
- Rizvanov AA, van Geelen AG, Morzunov S, Otteson EW, Bohlman C, Pari GS, St Jeor SC. Generation of a recombinant cytomegalovirus for expression of a hantavirus glycoprotein. J Virol. 2003;77(22):12203–10. doi: 10.1128/JVI.77.22.12203-12210.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roizman B. The herpesviridae, a brief introduction. In: Roizman B, Whitley RJ, Lopez C, editors. The human herpesviruses. Raven Press; New York: 1993. pp. 1–9. [Google Scholar]
- Roizman B, Sears AE. Herpes simplex viruses and their replication. In: Fields BN, Knipe DM, Howley PM, Chanock RM, Melnick JL, Monath TP, Roizman B, editors. Fundamental Virology. Raven Press; New York: 1996. pp. 1048–1066. [Google Scholar]
- Schleiss MR, McGregor A, Choi KY, Date SV, Cui X, McVoy MA. Analysis of the Nucleotide Sequence of a BAC-Derived Clone of the Guinea Pig Cytomegalovirus (GPCMV) Genome. Virology Journal. 2008;5:139. doi: 10.1186/1743-422X-5-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severini A, Morgan AR, Tovell DR, Tyrrell DL. Study of the structure of replicative intermediates of HSV-1 DNA by pulsed-field gel electrophoresis. Virology. 1994;200(2):428–35. doi: 10.1006/viro.1994.1206. [DOI] [PubMed] [Google Scholar]
- Smith GA, Enquist LW. Construction and transposon mutagenesis in Escherichia coli of a full- length infectious clone of pseudorabies virus, an alphaherpesvirus. J Virol. 1999;73(8):6405–14. doi: 10.1128/jvi.73.8.6405-6414.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlazny DA, Kwong A, Frenkel N. Site-specific cleavage/packaging of herpes simplex virus DNA and the selective maturation of nucleocapsids containing full-length viral DNA. Proc Natl Acad Sci U S A. 1982;79(5):1423–7. doi: 10.1073/pnas.79.5.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JB, Nixon DE, McVoy MA. Definition of the minimal cis-acting sequences necessary for genome maturation of the herpesvirus murine cytomegalovirus. J Virol. 2008;82(5):2394–404. doi: 10.1128/JVI.00063-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Smith GA, Enquist LW, Shenk T. Construction of a self-excisable bacterial artificial chromosome containing the human cytomegalovirus genome and mutagenesis of the diploid TRL/IRL13 gene. J Virol. 2002;76(5):2316–28. doi: 10.1128/jvi.76.5.2316-2328.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Efstathiou S, Simmons A. Identification of novel herpes simplex virus replicative intermediates by field inversion gel electrophoresis: implications for viral DNA amplification strategies. Virology. 1994;202(2):530–9. doi: 10.1006/viro.1994.1375. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.