Abstract
Cleavage of proteins inserted into the plasma membrane (shedding) is an essential process controlling many biological functions including cell signaling, cell adhesion and migration as well as proliferation and differentiation. ADAM surface metalloproteases have been shown to play an essential role in these processes. Gene inactivation during embryonic development have provided evidence of the central role of ADAM proteins in nematodes, flies, frogs, birds and mammals. The relative contribution of four subfamilies of ADAM proteins to developmental processes is the focus of this review.
Keywords: ADAM, Disintegrin, Metalloproteases, Cell Migration, Cell Adhesion, embryo
1. Introduction
ADAM proteins are single-pass transmembrane metalloproteases that contain a disintegrin domain. They were initially identified as sperm-specific proteins with similarity to snake venom metalloproteases [1-7]. In snakes the disintegrin domain binds with high affinity to the platelet integrins, inhibiting platelet aggregation, while the metalloprotease domain cleaves components of the endothelial basement membrane inducing blood vessel leakage and hemorrhage. During the past 12 years, ADAMs have been identified in a wide variety of animal species including nematodes, sea urchin, fruit flies, frogs, birds and mammals. Detailed analyses of the expression of ADAMs have shown that far from being restricted to spermatozoa, these proteins are expressed in all cell types. While our understanding of ADAM protein function has improved tremendously, it is still a growing field with many questions left unanswered. Many excellent reviews have looked at the various aspects of ADAM biology [8-18].
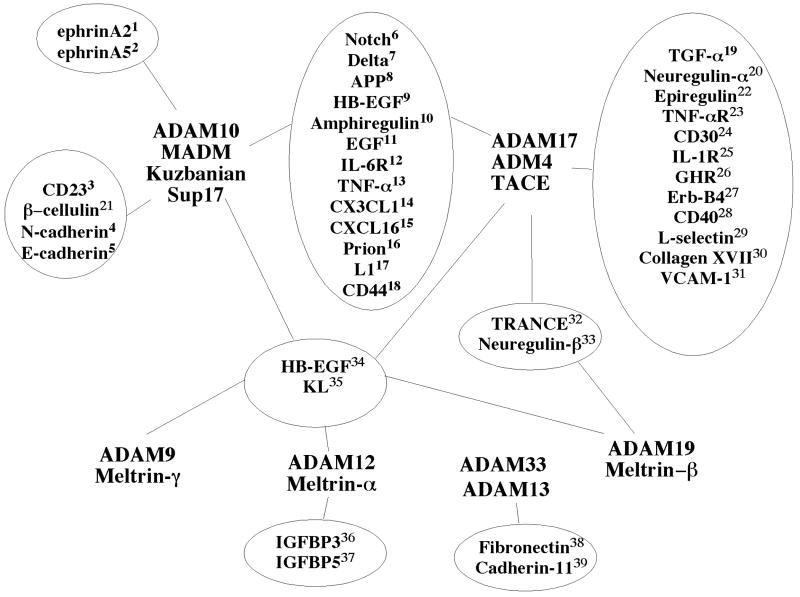
The basic ADAM structure is represented in Figure 1. All ADAMs contain a prodomain, a metalloprotease domain, a disintegrin and cysteine-rich domain. Most ADAMs also contain an EGF repeat, a transmembrane domain and a cytoplasmic domain. The pro-domain is thought to be essential for protein folding and to prevent the metalloprotease activity [19]. The metalloprotease domain is present even in ADAMs that do not have proteolytic activity. The activity of the metalloprotease domain can be predicted by the presence or absence of the consensus catalytic active site (HEXGHXXGXXHD). The substrates for ADAM metalloproteases include, cell adhesion molecules, proteins of the extracellular matrix, growth factors and cytokines, as well as receptors and ligands of signaling molecules (Figure 2).
Figure 1. Schematic diagram of ADAM protein organization and processing.

(I) ADAMs are single-pass transmembrane proteins composed of a pro-domain (P) a metalloprotease domain (M), a disintegrin domain (D) a cysteine-rich domain (C) an EGF repeat (E) and a cytoplasmic domain (Cy). (II) The pro-domain of ADAMs is removed by furin cleavage either during the transit through the trans golgie network or at the cell surface producing the metalloprotease active form. (III) In some cases, a second cleavage occurs removing the metalloprotease domain and potentially “unmasking” the disintegrin domain to promote integrin binding [63, 73]. (IV) Additional shedding has been reported for both ADAM13 and ADAM33 whereby the metalloprotease domain is shed with the disintegrin and cysteine-rich domains [75, 80]. (V) A similar soluble active metalloprotease can be obtained by alternative splicing as shown for ADAM9s and ADAM12s [67, 100]. (VI) Alternative splicing can also generate a proteolytically inactive ADAM, anchored in the membrane as shown for ADAM19 [101].
Figure 2. ADAM proteolytic substrates.
Diagram of selected ADAMs and substrates. The potential overlap between different ADAMs is presented. The number of potential substrates and the overlap makes the analysis of phenotype very difficult. 1 [55], 2 [23], 3 [102], 4 [49], 5 [50], 6 [26], 7 [35], 8 [103], 9 [104], 10 [105], 11 [71], 12 [106], 13 [107], 14 [108, 109], 15 [110, 111], 16 [112], 17 [78], 18 [113, 114], 19 [115], 20 [116], 21 [51], 22 [71], 23 [115], 24 [117], 25 [118], 26 [119], 27 [120], 28 [121], 29 [115, 122], 30 [123], 31 [124], 32 [125-127], 33 [83, 128, 129], 34 [83, 130-132], 35 [127, 133], 36 [134, 135], 37 [134], 38 [27], 39 McCusker et al., submitted.
The study of ADAM proteins has shown that many cell surface proteins are indeed cleaved from the cell surface in a process called shedding [20]. This can either activate (Notch or EGF ligands) or inactivate the cleaved proteins (ephrin, cadherins). The disintegrin and cysteine-rich domain of ADAMs have been shown to interact with cell adhesion molecules including the receptors of the extracellular matrix, integrins [21], as well as proteoglycans (e.g. syndecans) [22]. Studies of ephrin cleavage have shown that the cysteine-rich domain also interacts with the substrate and could be responsible for ADAM specificity [23]. The cytoplasmic domain of ADAMs is the most variable part of the molecule and, in some ADAMs, controls the function and the subcellular localization.
In this review we will focus on ADAM function during embryo development using data generated by the powerful genetics in nematodes, fly and mouse, together with results from gain-of- and loss-of-function experiments both in the bird and in frog embryos. Given the large number of potential substrates for each ADAM (Figure 2), we will only discuss those with direct links to the observed phenotypes. Since many reviews have already addressed the role of ADAMs during fertilization, we will focus on later aspects of embryonic development. We will describe the functional evidence by subsets of ADAMs across all species to try to find common trends. The subgroup of ADAMs discussed will be the following: (1) the two main sheddases; ADAM10 and 17, (2) the meltrins; ADAM9, 12, 13/33, and 19, and (3) the ADAMs that have a non-functional metalloprotease domain; ADAM11, 22, 23, and Unc-71. While these groups may appear at first as an artificial distribution, they in fact represent the different ADAMs found in the most primitive organisms such as the nematode, C. elegans, in which at least one member of each subgroup is found. Because of space limitations, we will not discuss the role of secreted ADAM-TS despite their essential function during embryogenesis reviewed in [24, 25].
2. How to study ADAM function in embryos
The study of ADAM metalloproteases during embryo development provides some specific challenges that may be addressed differently depending on the model chosen. Clearly, the first and most efficient approach has been the loss-of-function experiments. These have been achieved using genetics to inactivate selected genes in the fruit fly, the nematode and the mouse. While this is an extremely powerful approach, it sometimes prevents the study of tissue-specific phenotypes if the overall effect on embryo development is too severe. Loss-of-function studies have also been performed in other vertebrates, such as frog and chick embryos, using Morpholino antisense oligonucleotides that prevent translation of specific mRNA(s) (Figure 3). These can either be injected (in frogs) or electroporated (in chickens). The last approach is the use of Dominant Negative (DN) constructs whose protein product can interfere with the function of an ADAM. This can be achieved by using an ADAM lacking the metalloprotease domain or having a single point mutation in the active proteolytic site, as was shown for ADAM10 and ADAM13 [26, 27]. The DN approach was efficiently used for ADAM10 in Drosophila to selectively target different tissues and different stages of development (see below). The study of mosaic embryos in Drosophila whereby cells lacking ADAMs are placed in embryos possessing the wild type protein, have contributed immensely to our understanding of the ADAM function. In addition, at least one study in mice has tested the ability of wild type ADAM19 to rescue heart defects by expressing the protein in different cell types (described below [28]). This type of study, together with conditional knock outs, will be essential to understanding the function of ADAMs in vertebrate embryos.
Figure 3. How to study ADAM function during development.
Example of the strategy employed in Xenopus to test the function and potential substrate of ADAM proteases during development. The ADAM protein translation is inhibited by injection of a Morpolino oligonucleotide into the fertilized egg (A). At the blastula stage, mRNA encoding the wild type protein or various mutant lacking selected domains can be injected in a cell that will give rise to a specific tissue, for example the cranial neural crest cells (B). A lineage tracer is co-injected to compare the behavior of these cells to the rest of the embryo missing the ADAM protein. If an appropriate substrate is identified (C), the putative cleavage product (EC) can be produced and injected to test its ability to rescue the loss of ADAM phenotype. Phenotypical analysis can be performed at the tailbud stage (D) using the lineage tracer and tissue specific marker to determine the position and differentiation of the injected cells progeny. Similar techniques can be used in genetic models using tissue specific promoter rather than targeted injection.
While this is the first step of any functional study, in the case of ADAM metalloproteases, the identification of the substrates must follow and is, in most cases, the most challenging part. This is due to the lack of specific cleavage sequences, the large overlap in substrate specificity (Figure 2), and either the absence of phenotype (for most ADAMs) or the multitude of phenotypes (as for ADAM10). This has been addressed very differently in vertebrates and invertebrates. In Drosophila and C.elegans, genetic complementation has been used to identify target genes and has proven very effective at identifying the various components of the Notch pathway affected by ADAM10. Support for other substrates has not yet been clearly identified using this approach. On the other hand, studies using mammalian cell culture have identified a large number of substrates for each ADAM, defined the overlap and, in some cases the cleavage site, without a direct link to the physiology. By comparing the ability of cells isolated either from wild-type or KO mice to shed the various substrates, the physiological role of each cleavage can be established and, in some cases, linked to the phenotype. In frogs, similar approaches have been undertaken using candidate genes from cell-based assays. In order to confirm the importance of each substrate, rescue experiments introducing the artificially cleaved product of the putative substrate have been initiated (Figure 3). Clearly, the combination of all approaches will be necessary to identify the function and substrates of each ADAM during embryo development.
3. ADAM10 and ADAM17 are the main sheddases
ADAM10 and ADAM17 are the best-characterized ADAMs thus far and appear to have the most important function during embryo development. Mutation of either ADAM10, or ADAM17 produces dramatic phenotypes during early embryogenesis both in vertebrates and invertebrates (see below). Surprisingly, both Drosophila and the frog, Xenopus tropicalis, have an additional duplication of ADAM10. In Drosophila Kuzbanian-like (Kul) function appears critical for a subset of Notch signaling activity. The Notch/Delta signaling pathway is one of the most important during early development and is used primarily when cells are faced with a binary decision: one cell can become or not become a specific cell type. The best example of this has been shown during Drosophila neurogenesis when a uniform population of cells needs to decide whether to become neural or epidermal cells. The first cell that expresses an excess of Notch (compared to its neighbors) will become neural and signal to all the cells that are in direct contact that they can't become neural. These cells will up regulate Delta and down regulate Notch, amplifying the initial difference and stabilizing the cell fate decision [29-31].
3.1. ADAM10 role in early cell fate decision
ADAM10 loss-of-function mutation was first identified in Drosophila as a mutant with neurogenic phenotype [32]. In these flies, the neural tissue was abnormally large at the expense of the epidermis. Using analyses of mosaic embryos, in which a subset of cells are mutant in a wild-type background, Rooke and colleagues further showed that Kuzbanian was required in two separate events. First, in ectodermal cells to receive the signal that prevents them from adopting the neural fate (lateral inhibition) and second, in cells to generate a signal to promote neuronal fate. They went on to clone the Kuzbanian gene and found that it was homologous to a previously known metalloprotease cloned in bovine and known to cleave the myelin basic protein [33]. This metalloprotease is now known as ADAM10. The same year, Fambrough and colleagues found that in zygotic Kuzbanian-null mutants, axon extension was severely impaired. This was not simply due to the earlier defect in neurogenesis, as some neurons were not affected by the mutation [34]. To be able to turn off the function of ADAM10 in embryos, Pan and colleagues produced a “putative” DN form of the protein by removing the metalloprotease domain [26]. Using this technique, they were able to induce the production of the ADAM10 mutant in selected tissues using tissue-specific promoters and at specific times during embryogenesis using the heat shock promoter. The mutant ADAM10 produced phenotypes in the retina and the wing that were reminiscent of the Notch loss-of-function mutation. The “putative dominant negative” could be rescued by overexpression of the wild-type protein, arguing that the phenotype was specific to ADAM10 function. By expressing the DN in frog embryos, they also showed that the number of neurons was increased, suggesting that in vertebrates, the role of ADAM10 is conserved.
While our understanding of the function of ADAM10 has greatly improved since these early studies, the mechanism by which the proteolytically inactive ADAM10 protein may act as a dominant negative remains unclear. The simplest and most likely explanation is that the mutant binds to the substrate and competes with the active protein. Given the large overlap of substrates between the various ADAMs, it is likely that the dominant negative form of ADAM10 may also interfere with the function of related ADAMs expressed in the same cells and this should be kept in mind during the analysis of the phenotypes.
By specifically expressing the DN ADAM10 in neurons, Pan and colleagues were able to reproduce the axon extension phenotype obtained with the zygotic null mutant, further arguing that the mutant acted as a DN. Using the Drosophila S2 cells, as well as embryos expressing the DN and embryos with the null mutation, they showed that ADAM10 cleaved the extracellular domain of Notch and that in embryos lacking ADAM10 function the Notch receptor was not cleaved. The cleavage of Notch is essential for its activation and, therefore, loss of ADAM10 results in inactive Notch signaling. Interestingly, another study showed that in S2 cells the processing of Notch was not compromised by the ADAM10 DN, but that ADAM10 could process Delta and release a soluble fragment that retained the ability to bind to the Notch receptor, suggesting that this could promote signaling in the absence of cell contact [35]. They showed both that the DN ADAM10 and the loss of ADAM10 resulted in the absence of cleaved Delta protein in embryos. Cleavage of both Notch and Delta could explain how ADAM10 is required in the signal-receiving cells (Notch) and the signal-generating cells (Delta) for proper neurogenesis [32]. In fact, another study showed that a Kuzbanian-like protein (also related to ADAM10) was expressed in Drosophila embryos and appeared to process Delta in the wing imaginal disc [36]. In this study, they showed that cells that receive the Notch signal maintain a high level of Notch on their surface while reducing the level of Delta. In the absence of Kuzbanian-like, the level of Delta increases interfering with proper Notch signaling.
Thus, in Drosophila the receiving cell expresses both Kuzbanian and Kuzbanian-like, two proteins closely related to ADAM10. Kuzbanian processes Notch to activate the receptor while Kuzbanian-like cleaves Delta to prevent its signaling to neighboring cells. While this has not been demonstrated, it is likely that DN Kuzbanian may interfere with both functions, while the loss of one of the genes may or may not be compensated by the other. Until a clear understanding of the control of ADAM cleavage is achieved, we can only speculate about the function of individual ADAMs in developmental models. Evidence for the importance of ADAM10 in the cleavage of Notch and Delta in Drosophila have been confirmed by many studies in flies, nematodes and mice, making it the best-characterized signaling pathway involving an ADAM [26, 35-48]. In C. elegans, several studies have confirmed the role of ADAM10 in Notch signaling and have also shown that ADAM17 can compensate, at least partially, for the loss of ADAM10 in cells that expresses both [39, 45]. Surprisingly, while both ADAM10/Sup17 and ADAM17/ADM-4 genetically interact with Notch/Lin12 signaling, the absence of one or both of these genes does not produce the Notch phenotype, suggesting that, in the nematode, other proteases may also cleave Notch, or that Notch may also signal in a cleavage-independent manner.
Given the pleiotropic nature of Notch signaling during development, loss of ADAM10 function is likely to affect most developmental processes. While this was a strength for the initial discovery of ADAM10 function, it also makes it very difficult to identify other functional substrates in the absence of proper Notch signaling.
3.2 Role of ADAM10 in organ morphogenesis
In Drosophila, ADAM10 is also required for heart morphogenesis. Loss of zygotic ADAM10 function results in an increase in cardioblasts and a decrease in pericardial cells [46]. This phenotype can be phenocopied by expressing the DN in dorsal mesoderm cells prior to the specification of cardioblast precursors, but not if the DN is expressed in the cardioblast, suggesting a role in the early cell specification. At later stages, cardioblast migration was not affected by ADAM10 loss-of-function but the morphology of the cells in the heart was.
3.3 ADAM10: a master controller of epithelium to mesenchyme transition (EMT)
The mouse model has provided answers and functional evidence about other embryologically-relevant substrates of ADAM10. Mice lacking ADAM10 die at embryonic day 9.5 with severe defects in the central nervous system (CNS) and both the dorsal (somitic) and ventral (cardiovascular) mesoderm [41]. Using in situ hybridization with Hes-5, a gene induced by Notch signaling, they further showed that Notch signaling was decreased in ADAM10 KO embryos. In addition, Delta-1 protein expression was found to be increased in these embryos, suggesting that ADAM10 in mouse embryos functions similarly to Kuzbanian to cleave both Notch (to activate) and Delta (to reduce).
In addition to the Notch phenotype, one of the most striking features of these embryos was their small size. In the epithelium, E-cadherin and N-cadherin proteins accumulated to abnormally high levels [49, 50]. These proteins were found to be cleaved by ADAM10 to promote epithelium to mesenchyme transition (EMT) essential for both embryonic development and wound healing. In addition, the accumulation of Cadherins at the cell membrane sequestered β-catenin, decreasing its translocation to the nucleus and subsequent signaling. Using cell lines from the KO, ADAM10 was also shown to be a major contributor of EGF and β-cellulin shedding [51]. Defects in the processing of these growth factors are likely to contribute to the overall phenotype of the ADAM10 KO, but direct proof of this has not been shown. Other cell adhesion molecules, such as the protocadherin, PCDHγ, are cleaved by ADAM10 and could contribute to the neural phenotype by perturbing the “adhesive code” used during neuronal wiring [52]. While this has not been demonstrated, it is likely that ADAM10 cleavage of cell adhesion molecules, like Cadherins and protocadherins, also occurs in Drosophila and may be responsible for some of the phenotypes observed in the Kuzbanian-mutant embryos. In particular, phenotypes involving cell migration and neurite outgrowth could be due to defects in cell adhesion molecules or ephrin signaling (see 3.4).
In the chicken, ADAM10 is widely-expressed and most abundant in epithelial structures. Knock down of ADAM10 with an antisense Morpholino oligonucleotide induces a thickening of the epithelium together with increased expression of N-Cam protein [53]. At the moment it is unclear if this phenotype is due to defects in the processing of cell adhesion molecules (N- and E-cadherin) or Notch or Delta. In addition, in quail, the metalloprotease inhibitor, GI254023X, which blocks ADAM10, was shown to prevent N-cadherin cleavage during neural crest emigration. This resulted in a reduction of β-catenin signaling and the inhibition of neural crest delamination from the neural tube [54]. These results provide additional evidence that ADAM10 cleavage of Cadherins may play an essential role in the developmental EMT in vertebrates. It would be interesting to see if this is also the case in invertebrates.
3.4 ADAM10 switches ephrins off in neurons
One more family of substrates for ADAM10 discovered in the mouse model is the ephrin. Two ephrin ligands, ephrin-A2 and -A5, have been shown to be cleaved by ADAM10. ADAM10 and ephrin-A2 are co-expressed on axons. When axons contact a cell expressing the Eph-A3 receptor, ephrin-A2 is cleaved by ADAM10, inducing a signaling cascade that promotes axon retraction. In the absence of ADAM10 the repulsion signaling cascade still occurs, but the axon remains attached to the repulsing cells as the ephrin/Eph interaction is not released [55]. In the case of ephrin-A5, ADAM10 is constitutively associated with the Eph-A3 receptor. When clustered ephrin-A5 is engaged with the Eph-A3 receptor, ADAM10 associates in trans via its cysteine-rich domain and cleaves the ephrin-A5 ligand, once again resulting in the repulsion and release of the axon [23]. While this has not been demonstrated in embryos, it is tempting to speculate that the loss of cleavage of ephrins by ADAM10 is, at least in part, responsible for axon extension defects both in mice and Drosophila lacking the ADAM10 protein.
3.5 ADAM17 is the master controller of EGF signaling
While ADAM17 is one of the most-studied ADAMs in mammals, there is only one report addressing the function of this protein in non-mammalian species. In C.elegans, ADAM17/ADM-4 null mutants are viable. They show a low-penetrance phenotype related to abnormal germ cell division. This phenotype is exacerbated in embryos lacking both ADAM10 and ADAM17, further demonstrating the functional redundancy of these two proteins [45]. At this point, it is unclear which of the ADAM17 substrates are involved in C.elegans development.
Mice lacking ADAM17 die perinataly and suffer from multiple defects in most organs [56]. The defects appear to occur from problems arising in the epithelial layer of each organ. Phenotypes include abnormal development of the skin, the eye, digestive tract, lungs and heart. Cells isolated from the KO mice have significantly less shedding of TNF-α, TGF-α, and the Erb-B4 receptor [56, 57]. Mice lacking ADAM17 have enlarged hearts due both to increased cardiomyocyte proliferation and a lack of proper morphogenesis. Isolated hearts have a significant reduction in EGF signaling. It has been proposed that the function of ADAM17 in the heart is to regulate cell proliferation and cell organization during morphogenesis [57]. In the lung, lack of ADAM17 results in reduced organ size due to decreased cell proliferation of epithelial cells and a decrease in branching morphogenesis [58-60]. Interestingly, the ADAM17 protein is found in the plasma membrane of epithelial cells at their contact with the mesenchymal layer. Organ culture confirmed that branching morphogenesis was severely reduced in KO lungs and this could be rescued by adding exogenous TNF-α or EGF, suggesting that ADAM17 function during lung morphogenesis is as a sheddase mediating EGF signaling. This is also supported by the fact that the ADAM17 knock outs phenotypes are very similar to that of EGF receptor knock outs [59, 60] as well as the triple knock outs for EGF, amphiregulin and TGF-α [61]. While in cell culture, ADAM17 can cleave many substrates (Figure 2), although the relative importance of each substrate during embryogenesis has not been established. For example, while ADAM17 can cleave Notch in vitro [48] and is essential for Notch processing in bone-marrow cells [62], none of the phenotypes observed in the mouse KO suggest that ADAM17 is essential for Notch function during early embryogenesis. Conditional knock out and rescue experiments expressing either the wild-type protein or the “shed” ligands in a tissue-specific manner should help further elucidate the relative contribution of ADAM17 substrates during embryogenesis.
4. The mesenchymal ADAMs, (Meltrins: ADAM9, 12, 13/33, 19)
The meltrins were originally cloned in an effort to identify proteins involved in cell fusion. ADAM12 (meltrin-α) and ADAM19 (meltrin-β) were found to be expressed in muscle precursor cells. In addition, ADAM12 was also expressed prior to myoblast fusion in the form of three main polypeptides. Based on their sizes, these polypeptides likely represent the precursor (including pro-domain), the mature protease (lacking prodomain) and a shorter form most likely lacking both the pro- and metalloprotease domains. Reduction of ADAM12 in C2C12 cells (a muscle cell line), using antisense approaches reduced cell fusion in vitro. In contrast, overexpression of the short form lacking both pro- and metalloprotease domains increased cell fusion. Surprisingly, overexpression of the full length ADAM12 decreased fusion suggesting that only the truncated form was competent to promote cell fusion and that an excess of the pro-form may compete with endogenous short ADAM12 binding to elements of the cellular machinery [63]. These key experiments promoted the notion that the disintegrin domain of ADAMs became accessible as ligands for cell adhesion molecules following the removal of the metalloprotease domain similar to what was already known for the fertilins (ADAM1 and 2) at the surface of spermatozoids [5, 64, 65]. However, ADAM12 deletion in mice shows no obvious defect in myoblast fusion, demonstrating that ADAM12 is not absolutely essential for muscle development in vivo. While the ADAM12 knockout did not have the expected muscle phenotype, mice lacking ADAM12 showed approximately 30% perinatal mortality. About one third of the embryos lacking ADAM12 show some reduction of the interscapular brown adipose tissue, and in the neck and interscapular muscles. Cells isolated from the knock out mice showed reduced shedding of HB-EGF in response to phorbol ester stimulation [66]. While the ADAM12 loss-of-function experiments are relatively uninformative, gain-of-function experiments have produced some interesting results. In particular, study of ADAM12 proteins expression during myoblast development has revealed the presence of a secreted form of ADAM12 lacking the transmembrane and cytoplasmic domains. When human rabdocarcinoma cells were transfected with the secreted ADAM12 and introduced into nude mice, the tumors formed by the human cells were invaded by mouse myoblasts, suggesting that the secreted ADAM12 protein could recruit muscle precursor cells [67]. Similarly overexpression of ADAM12 in muscle improves muscle regeneration following injury and alleviates some of the symptoms of mdx mice (dystrophyn deficient) [68], suggesting a role for this protein in muscle regeneration. However, loss-of-function experiments show that mice lacking ADAM12 possess normal regeneration capacity [66]. Overexpression of ADAM12 in mice induces ectopic adipocytes resulting in increased weight [69]. Together with the knock out data, these data suggest that ADAM12 may participate in adipogenesis.
4.1 Are meltrins useless or replaceable?
The lack of an obvious phenotype in the ADAM12 knock out is one of the most frustrating characteristics of the meltrin subfamily of ADAMs. For example, there are two meltrins in Drosophila, D-meltrin and Mmd (Mind meld), but to date there are no phenotypes reported for either genes. In C-elegans, ADM-2 is the closest relative of the meltrins, and as with Drosophila there are no phenotypes associated with the loss of ADM-2 function during embryogenesis. In mice, loss of ADAM9, which is ubiquitously expressed, has no phenotype [70]. Furthermore, mice lacking ADAM9, 12 and 15 are viable and fertile [71]. Taken together these results suggest that either these genes have no function during embryogenesis or that other metalloproteases (ADAMs or others) can substitute for the absence of these proteins. It is hard to believe that genes expressed at significant levels in various tissues during embryogenesis do not have a function and, therefore, the second hypothesis seems more likely.
In the frog, three meltrins are expressed during early embryogenesis. ADAM9 is present maternally and in all cells [72], whereas ADAM13 and 19 are both expressed zygotically in very similar patterns, in the somitic mesoderm, the neural tube and cranial neural crest cells (CNC). In addition, ADAM19 is expressed in the notochord as early as gastrulation while ADAM13 appears to be absent from this structure ([73]; Neuner et al., submitted). Experiments using the proteolytically inactive form of ADAM13, as a DN, have shown that ADAM13 function is essential for the migration of the CNC [27]. Loss of the ADAM13 protein using a Morpholino AS approach has been reported to decrease CNC induction and migration in Xenopus [74], but we have not been able to confirm these results. In our hands, the ADAM13 MO efficiently eliminates the endogenous protein and interferes with gastrulation movements but has only marginal effect on CNC induction and migration (Cousin et al., in preparation, McCusker et al., submitted). In contrast, elimination both of ADAM13 and 19 in the CNC completely inhibits CNC migration, while ADAM19 MO by itself has only marginal effects on migration but does decrease CNC induction (Neuner et al., submitted, Cousin et al., in preparation, McCusker et al., submitted). These results confirm the potential compensation of meltrin ADAMs during early embryo development.
4.2 How does ADAM13 control CNC migration?
We have shown that ADAM13 can bind and cleave fibronectin in vitro, suggesting that it may interfere with cell-ECM interactions [27, 75]. In addition, ADAM13 and ADAM9 cleave Cadherin-11, a cell adhesion molecule expressed in Xenopus CNC and known in mammals as a mesenchymal Cadherin. Loss of ADAM9 or ADAM13 results in an accumulation of Cadherin-11 in the embryo (McCusker et al., submitted). Overexpression of Cadherin-11 prevents CNC migration [76] and this can be rescued by the overexpression of ADAM13. Similarly, expression of the cleaved, extracellular domain of Cadherin-11 can rescue migration in embryos overexpressing Cadherin-11, as well as those lacking ADAM9, 13 and 19 (McCusker et al., submitted). This suggests that one of ADAM13 main functions in CNC is to cleave Cadherin-11, in order to release its extracellular domain and promote cell migration. This is reminiscent of ADAM10 cleavage of another adhesion molecule, L1. The extracellular domain of L1 binds to the αvβ3 integrin and promotes cell migration [77, 78]. Additional work will be needed to understand how the Cadherin-11 extracellular domain promotes cell migration. Another interesting aspect of Cadherin-11 cleavage by ADAM13 is that it occurs some distance from the plasma membrane, between the EC3 and EC4 domains, while cleavage of N- and E-cadherin by ADAM10 is near the membrane. This is significant because cleavage of N- and E-cadherin by ADAM10 is immediately followed by a second cleavage by a gamma-secretase, releasing its cytoplasmic domain and β-catenin, which immediately translocates to the nucleus and activates downstream genes including c-myc and cyclin-D1 [49, 50]. In contrast, cleavage of Cadherin-11 by ADAM13 does not induce β-catenin signaling, suggesting that it can control cell adhesion and migration without modifying the signaling capacity of Cadherin-11 (McCusker et al., submitted). In Xenopus tropicalis, ADAM13 may also function in cleaving ephrin-B ligands to interrupt signaling. Negative control of ephrin-B signaling appears to be essential for head patterning and neural crest cell induction (DeSimone, personal communication). In mice, the knock out of ADAM33, the most likely homologue of ADAM13, shows no phenotype. There are no observable defects and the adults are fertile [79]. As with ADAM13, the extracellular domain of ADAM33 can be shed from the cell surface [75], and for ADAM33 the soluble form can induce angiogenesis in vitro and ex vivo (organ culture) [80]. Given our results with ADAM13, it would be interesting to produce multiple ADAM knock outs, including ADAM9, 12, 19 and 33, to investigate their role in development and migration of neural crest cells as well as the specification and fusion of myoblasts.
4.3. The exception, ADAM19
The only member of the ADAM family that shows a clear phenotype following knock out in mice is ADAM19. Mice lacking ADAM19 die soon after birth. Careful examination of ADAM19 null embryos shows severe defects in heart morphogenesis, including a ventricular septal defects and abnormal valve formation [81, 82]. In addition, Kurohara and colleagues found defects in muscle due to decreased thickness of the myofibers, and some abnormal positioning of the thick preganglionic neuron bundle in the adrenal medula. Zhou and colleagues also showed that many of the capillaries were abnormal and eventually ruptured. Using β–galactosidase as a cell lineage tracer they showed that the cardiac neural crest of ADAM19 null embryos still migrated to the proper heart field. These observations were followed by another elegant study using the full “tool kit” of mammalian genetics [28]. In this report they studied, in detail, the contribution of cardiac neural crest cells and endothelial cells to heart formation in wild-type and knock out mice. By reintroducing the ADAM19 protein specifically in cardiac neural crest or endothelial cells, they showed that ADAM19 was required in the cardiac neural crest to promote normal heart morphogenesis. They further showed that cardiac neural crest cells lacking ADAM19 were capable of migrating into the proper heart region but were unable to participate in the formation of the ventricular septum. While ADAM19 can cleave many substrates, including EGF ligands, the physiological substrate responsible for the observed phenotype is not known. As another testimony to redundancy, ADAM9-ADAM19 double knock outs have a more severe heart phenotype, including defects in the mitral valve [83]. Interestingly, in frogs, ADAM19 knock down also induces phenotypes. In particular, expression of transcript for the general mesoderm marker Brachyury is decreased in the notochord, while other dorsal mesoderm markers (Goosecoid and Chordin) are increased. At later stages of development, some tissue-specific markers are also decreased, including markers of neurons (N-tubulin), neural crest (Slug, Twist and ADAM11) and muscle (Myosin light chain). Together with a reduction in these markers, the organization of muscle precursors in the somites is perturbed, while the formation of the somite itself is not (Neuner et al., submitted). This suggests that, as shown in the mouse, frog ADAM19 possesses unique functions that are not compensated for by other related ADAMs. The analysis of multiple meltrin knock downs in frogs should help elucidate the overall redundancy and physiological substrates involved in early embryogenesis (Figure 3).
5. Non proteolytic ADAMs
These ADAMs have all the characteristics of the ADAM family including the metalloprotease domain, but the consensus active site sequence is missing, suggesting that they are not proteolytically active. Because of this particularity, the use of prototypical ADAM DN constructs was unavailable, and progress regarding their function has been slow. Fortunately, genetics has brought some invaluable information both in C.elegans and in mouse. The original work done on fertilin function during fertilization suggested that these non-proteolytic ADAMs functioned as ligands for the integrin family of receptors. While it is clear that most isolated disintegrin domains tested so far can promote cell adhesion mediated via selected integrins, the physiological role of these interactions remain controversial. Overexpression studies using full-length ADAM proteins have shown that they can regulate the activity of integrins, expressed in the same cells, with some selectivity, but the direct proof of this functional interaction in physiological conditions is lacking.
5.1 The nematode Unc-71/ADM1 controls cell migration
In C.elegans, Unc-71/ADM1/ADAM14 was identified during a search for potential fusion proteins (as proposed for ADAM function in myoblast and sperm/egg fusion). The protein was found in syncytial organs, sperm and sheath cells of sensory organs [84]. Mutation in the Unc-71 gene induces defects in axon guidance and axon fasciculation [85, 86], as well as in the migration of hermaphrodite sex myoblast [87]. Detailed analyses of multiple alleles of Unc-71, show that most of the mutations are in sequences coding for the disintegrin and cysteine-rich domains of the protein. Subtle mutations in the disintegrin loop (D504 to Q) drastically affect protein function, suggesting that this region of the protein is important either for folding or for interaction with other proteins. Surprisingly, while the metalloprotease domain is likely inactive, rescue experiments demonstrate that this domain is essential for the proper function of the protein. Further experiments are needed to test whether this domain is required for the proper folding and export of the protein or for its function. Other rescue experiments showed that Unc-71 protein acts in a non cell-autonomously fashion in sexual myoblast migration, meaning that the protein does not rescue migration if it is expressed directly in the sexual myoblasts, but rather, needs to be expressed in cells along the pathway [85]. In addition, the presence of the transmembrane and cytoplasmic domains of the protein is required for the rescue, suggesting that it helps guide cell migration by providing a permissive cue on the cell surface. This is also true for the function of Unc-71 in type-D motor axon guidance [85].
5.2 Non-proteolytic ADAMs in vertebrates: The “smart” ADAMs
In the mouse, non-proteolytically active ADAMs are expressed both in the central nervous system (CNS) and in the testis. We will not discuss the function related to fertilization as these have been described in detail elsewhere [11, 88-91]. ADAM2, ADAM11, ADAM22 and ADAM23 are expressed in the CNS and have been knocked out with significant consequences.
Loss of ADAM2 in the CNS perturbs neuroblast migration along the rostral migratory stream to the olfactory bulb resulting in a smaller olfactory bulb. Both in vitro and ex vivo (in organ culture) experiments have shown that the migration of the neuroblasts was decreased. This was associated with an apparent decrease in cell/cell interaction. The defects in cell migration could be reproduced by addition of a peptide corresponding to the disintegrin loop of ADAM2 in the organ culture experiments, suggesting that the disintegrin domain is essential to promote or guide cell migration [92].
ADAM11 is expressed in the CNS of mice and both the CNS and cranial neural crest cells of frogs [72, 93]. Mice lacking ADAM11 are viable and fertile with no major histological defects but show behavior defects related to learning, motor coordination and response to pain [94, 95]. The cellular basis for these defects is unknown.
Loss of ADAM22 and ADAM23 results in death shortly after birth (1 to 3 weeks) due to tremors and ataxia [96, 97]. For ADAM22, detailed analysis of the brain of the homozygotes revealed a normal organization with the exception of a deficit in myelination of the peripheral sciatic and trigeminal nerves while the myelination of other nerves in the spinal chord and the brain was not affected. Myelination of peripheral nerves depends on the function of Schwann cells. These cells were more abundant in the mutant embryos suggesting that they did not stop dividing and failed to differentiate to produce myelin. In support of this hypothesis, ADAM22 gene is not expressed in highly proliferative gliomas (cancer), and artificial expression of the protein in these cells can inhibit proliferation. Inhibition of cell proliferation could also be achieved by incubating cells with a bacterially expressed ADAM22 disintegrin domain. In addition, overexpression of integrin linked kinase (ILK) in glioma cells could prevent the inhibition of proliferation suggesting that the disintegrin domain was reducing cell proliferation by binding to an integrin at the cell surface [98]. Finally, ADAM22 was also found to be present in the post-synaptic membrane where it could act as a receptor for LGI1, a secreted protein involved in epilepsy. A mutated form of the LGI1 protein, found in autosomal dominant partial epilepsy with auditory feature, failed to bind to ADAM22 [99]. It is tempting to speculate that the phenotypes associated with the loss of ADAM22 in mice may be related to its role as a receptor for LGI1.
In summary, non-proteolytic ADAMs are involved in nervous system formation both in vertebrates and invertebrates. Their functions are likely to involve the regulation of cell adhesion to promote axon guidance, neuroblast migration and Schwann cell differentiation. The identification of ligands for non-proteolytic ADAMs will be essential to understanding the mechanisms by which these ADAMs perform their function.
6. Conclusions
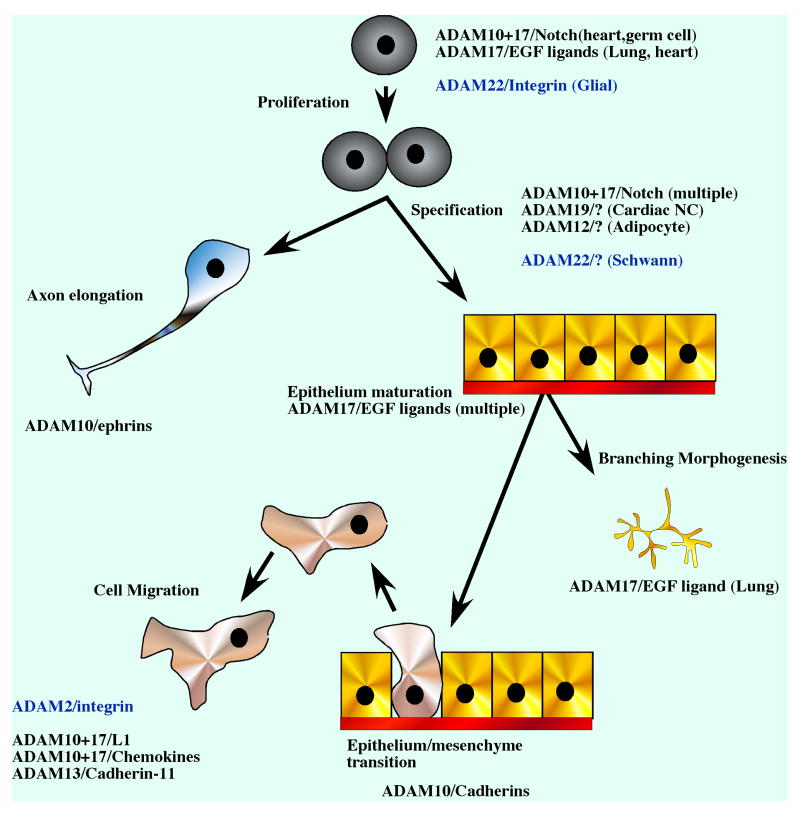
ADAMs are essential proteins involved at every step of embryo development. They control cell proliferation, cell specification, cell migration, axon elongation and organ morphogenesis (Figure 4). They are so essential that built-in redundancy and functional overlap is the rule, with only a few exceptions. Attributing “the” functional substrate for each ADAM in each cellular context will be the next great challenge.
Figure 4. ADAM function in Development.
ADAM proteins are involved at every step of embryogenesis. They can control cell proliferation, cell specification and differentiation as well as various aspects of morphogenesis. ADAMs that have been implicated with each step are indicated together with the substrate if it is known. In parenthesis the cell type or tissue in which the function was shown is indicated. Non-proteolytic ADAMs are in blue.
Acknowledgments
This work was supported by support from NIH (DE016289) to Dr. Alfandari. Thanks to Erin Kerdavid Drs. Lisa Minter and Alban Gaultier for critical reading of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Blobel CP, Wolfsberg TG, Turck CW, Myles DG, Primakoff P, White JM. A potential fusion peptide and an integrin ligand domain in a protein active in sperm-egg fusion. Nature. 1992;356:248–252. doi: 10.1038/356248a0. [DOI] [PubMed] [Google Scholar]
- 2.Perry AC, Jones R, Barker PJ, Hall L. A mammalian epididymal protein with remarkable sequence similarity to snake venom haemorrhagic peptides. Biochem J. 1992;286(Pt 3):671–675. doi: 10.1042/bj2860671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wolfsberg TG, Bazan JF, Blobel CP, Myles DG, Primakoff P, White JM. The precursor region of a protein active in sperm-egg fusion contains a metalloprotease and a disintegrin domain: structural, functional, and evolutionary implications. Proc Natl Acad Sci U S A. 1993;90:10783–10787. doi: 10.1073/pnas.90.22.10783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Muga A, Neugebauer W, Hirama T, Surewicz WK. Membrane interaction and conformational properties of the putative fusion peptide of PH-30, a protein active in sperm-egg fusion. Biochemistry. 1994;33:4444–4448. doi: 10.1021/bi00181a002. [DOI] [PubMed] [Google Scholar]
- 5.Myles DG, Kimmel LH, Blobel CP, White JM, Primakoff P. Identification of a binding site in the disintegrin domain of fertilin required for sperm-egg fusion. Proc Natl Acad Sci U S A. 1994;91:4195–4198. doi: 10.1073/pnas.91.10.4195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weskamp G, Blobel CP. A family of cellular proteins related to snake venom disintegrins. Proc Natl Acad Sci U S A. 1994;91:2748–2751. doi: 10.1073/pnas.91.7.2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wolfsberg TG, Primakoff P, Myles DG, White JM. ADAM, a novel family of membrane proteins containing A Disintegrin And Metalloprotease domain: multipotential functions in cell-cell and cell-matrix interactions. J Cell Biol. 1995;131:275–278. doi: 10.1083/jcb.131.2.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Murphy G, Murthy A, Khokha R. Clipping, shedding and RIPping keep immunity on cue. Trends Immunol. 2008;29:75–82. doi: 10.1016/j.it.2007.10.009. [DOI] [PubMed] [Google Scholar]
- 9.Kveiborg M, Albrechtsen R, Couchman JR, Wewer UM. Cellular roles of ADAM12 in health and disease. Int J Biochem Cell Biol. 2008;40:1685–1702. doi: 10.1016/j.biocel.2008.01.025. [DOI] [PubMed] [Google Scholar]
- 10.Deuss M, Reiss K, Hartmann D. Part-time alpha-secretases: the functional biology of ADAM 9, 10 and 17. Curr Alzheimer Res. 2008;5:187–201. doi: 10.2174/156720508783954686. [DOI] [PubMed] [Google Scholar]
- 11.Okabe M, Cummins JM. Mechanisms of sperm-egg interactions emerging from gene-manipulated animals. Cell Mol Life Sci. 2007;64:1945–1958. doi: 10.1007/s00018-007-7037-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mochizuki S, Okada Y. ADAMs in cancer cell proliferation and progression. Cancer Sci. 2007;98:621–628. doi: 10.1111/j.1349-7006.2007.00434.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang P, Baker KA, Hagg T. The ADAMs family: coordinators of nervous system development, plasticity and repair. Prog Neurobiol. 2006;79:73–94. doi: 10.1016/j.pneurobio.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 14.Reiss K, Ludwig A, Saftig P. Breaking up the tie: disintegrin-like metalloproteinases as regulators of cell migration in inflammation and invasion. Pharmacol Ther. 2006;111:985–1006. doi: 10.1016/j.pharmthera.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 15.Ohtsu H, Dempsey PJ, Eguchi S. ADAMs as mediators of EGF receptor transactivation by G protein-coupled receptors. Am J Physiol Cell Physiol. 2006;291:C1–10. doi: 10.1152/ajpcell.00620.2005. [DOI] [PubMed] [Google Scholar]
- 16.Arribas J, Bech-Serra JJ, Santiago-Josefat B. ADAMs, cell migration and cancer. Cancer Metastasis Rev. 2006;25:57–68. doi: 10.1007/s10555-006-7889-6. [DOI] [PubMed] [Google Scholar]
- 17.Huovila AP, Turner AJ, Pelto-Huikko M, Karkkainen I, Ortiz RM. Shedding light on ADAM metalloproteinases. Trends Biochem Sci. 2005;30:413–422. doi: 10.1016/j.tibs.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 18.Blobel CP. ADAMs: key components in EGFR signalling and development. Nat Rev Mol Cell Biol. 2005;6:32–43. doi: 10.1038/nrm1548. [DOI] [PubMed] [Google Scholar]
- 19.Loechel F, Overgaard MT, Oxvig C, Albrechtsen R, Wewer UM. Regulation of human ADAM 12 protease by the prodomain. Evidence for a functional cysteine switch. J Biol Chem. 1999;274:13427–13433. doi: 10.1074/jbc.274.19.13427. [DOI] [PubMed] [Google Scholar]
- 20.Blobel CP. Functional and biochemical characterization of ADAMs and their predicted role in protein ectodomain shedding. Inflamm Res. 2002;51:83–84. doi: 10.1007/BF02684007. [DOI] [PubMed] [Google Scholar]
- 21.White JM. ADAMs: modulators of cell-cell and cell-matrix interactions. Curr Opin Cell Biol. 2003;15:598–606. doi: 10.1016/j.ceb.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 22.Thodeti CK, Albrechtsen R, Grauslund M, Asmar M, Larsson C, Takada Y, Mercurio AM, Couchman JR, Wewer UM. ADAM12/syndecan-4 signaling promotes beta 1 integrin-dependent cell spreading through protein kinase Calpha and RhoA. J Biol Chem. 2003;278:9576–9584. doi: 10.1074/jbc.M208937200. [DOI] [PubMed] [Google Scholar]
- 23.Janes PW, Saha N, Barton WA, Kolev MV, Wimmer-Kleikamp SH, Nievergall E, Blobel CP, Himanen JP, Lackmann M, Nikolov DB. Adam meets Eph: an ADAM substrate recognition module acts as a molecular switch for ephrin cleavage in trans. Cell. 2005;123:291–304. doi: 10.1016/j.cell.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 24.Porter S, Clark IM, Kevorkian L, Edwards DR. The ADAMTS metalloproteinases. Biochem J. 2005;386:15–27. doi: 10.1042/BJ20040424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Flannery CR. MMPs and ADAMTSs: functional studies. Front Biosci. 2006;11:544–569. doi: 10.2741/1818. [DOI] [PubMed] [Google Scholar]
- 26.Pan D, Rubin GM. Kuzbanian controls proteolytic processing of Notch and mediates lateral inhibition during Drosophila and vertebrate neurogenesis. Cell. 1997;90:271–280. doi: 10.1016/s0092-8674(00)80335-9. [DOI] [PubMed] [Google Scholar]
- 27.Alfandari D, Cousin H, Gaultier A, Smith K, White JM, Darribere T, DeSimone DW. Xenopus ADAM 13 is a metalloprotease required for cranial neural crest-cell migration. Curr Biol. 2001;11:918–930. doi: 10.1016/s0960-9822(01)00263-9. [DOI] [PubMed] [Google Scholar]
- 28.Komatsu K, Wakatsuki S, Yamada S, Yamamura K, Miyazaki J, Sehara-Fujisawa A. Meltrin beta expressed in cardiac neural crest cells is required for ventricular septum formation of the heart. Dev Biol. 2007;303:82–92. doi: 10.1016/j.ydbio.2006.10.037. [DOI] [PubMed] [Google Scholar]
- 29.Watt FM, Estrach S, Ambler CA. Epidermal Notch signalling: differentiation, cancer and adhesion. Curr Opin Cell Biol. 2008;20:171–179. doi: 10.1016/j.ceb.2008.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fiuza UM, Arias AM. Cell and molecular biology of Notch. J Endocrinol. 2007;194:459–474. doi: 10.1677/JOE-07-0242. [DOI] [PubMed] [Google Scholar]
- 31.Osborne BA, Minter LM. Notch signalling during peripheral T-cell activation and differentiation. Nat Rev Immunol. 2007;7:64–75. doi: 10.1038/nri1998. [DOI] [PubMed] [Google Scholar]
- 32.Rooke J, Pan D, Xu T, Rubin GM. KUZ, a conserved metalloprotease-disintegrin protein with two roles in Drosophila neurogenesis. Science. 1996;273:1227–1231. doi: 10.1126/science.273.5279.1227. [DOI] [PubMed] [Google Scholar]
- 33.Chantry A, Gregson NA, Glynn P. A novel metalloproteinase associated with brain myelin membranes. Isolation and characterization. J Biol Chem. 1989;264:21603–21607. [PubMed] [Google Scholar]
- 34.Fambrough D, Pan D, Rubin GM, Goodman CS. The cell surface metalloprotease/disintegrin Kuzbanian is required for axonal extension in Drosophila. Proc Natl Acad Sci U S A. 1996;93:13233–13238. doi: 10.1073/pnas.93.23.13233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qi H, Rand MD, Wu X, Sestan N, Wang W, Rakic P, Xu T, Artavanis-Tsakonas S. Processing of the notch ligand delta by the metalloprotease Kuzbanian. Science. 1999;283:91–94. doi: 10.1126/science.283.5398.91. [DOI] [PubMed] [Google Scholar]
- 36.Sapir A, Assa-Kunik E, Tsruya R, Schejter E, Shilo BZ. Unidirectional Notch signaling depends on continuous cleavage of Delta. Development. 2005;132:123–132. doi: 10.1242/dev.01546. [DOI] [PubMed] [Google Scholar]
- 37.Nye JS. Developmental signaling: notch signals Kuz it's cleaved. Curr Biol. 1997;7:R716–720. doi: 10.1016/s0960-9822(06)00364-2. [DOI] [PubMed] [Google Scholar]
- 38.Sotillos S, Roch F, Campuzano S. The metalloprotease-disintegrin Kuzbanian participates in Notch activation during growth and patterning of Drosophila imaginal discs. Development. 1997;124:4769–4779. doi: 10.1242/dev.124.23.4769. [DOI] [PubMed] [Google Scholar]
- 39.Wen C, Metzstein MM, Greenwald I. SUP-17, a Caenorhabditis elegans ADAM protein related to Drosophila KUZBANIAN, and its role in LIN12/NOTCH signalling. Development. 1997;124:4759–4767. doi: 10.1242/dev.124.23.4759. [DOI] [PubMed] [Google Scholar]
- 40.Jarriault S, Le Bail O, Hirsinger E, Pourquie O, Logeat F, Strong CF, Brou C, Seidah NG, Isra l A. Delta-1 activation of notch-1 signaling results in HES-1 transactivation. Mol Cell Biol. 1998;18:7423–7431. doi: 10.1128/mcb.18.12.7423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hartmann D, de Strooper B, Serneels L, Craessaerts K, Herreman A, Annaert W, Umans L, Lubke T, Lena Illert A, von Figura K, et al. The disintegrin/metalloprotease ADAM 10 is essential for Notch signalling but not for alpha-secretase activity in fibroblasts. Hum Mol Genet. 2002;11:2615–2624. doi: 10.1093/hmg/11.21.2615. [DOI] [PubMed] [Google Scholar]
- 42.Klein T. kuzbanian is required cell autonomously during Notch signalling in the Drosophila wing. Dev Genes Evol. 2002;212:251–255. doi: 10.1007/s00427-002-0233-4. [DOI] [PubMed] [Google Scholar]
- 43.Lieber T, Kidd S, Young MW. kuzbanian-mediated cleavage of Drosophila Notch. Genes Dev. 2002;16:209–221. doi: 10.1101/gad.942302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Six E, Ndiaye D, Laabi Y, Brou C, Gupta-Rossi N, Israel A, Logeat F. The Notch ligand Delta1 is sequentially cleaved by an ADAM protease and gamma-secretase. Proc Natl Acad Sci U S A. 2003;100:7638–7643. doi: 10.1073/pnas.1230693100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jarriault S, Greenwald I. Evidence for functional redundancy between C. elegans ADAM proteins SUP-17/Kuzbanian and ADM-4/TACE. Dev Biol. 2005;287:1–10. doi: 10.1016/j.ydbio.2005.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Albrecht S, Wang S, Holz A, Bergter A, Paululat A. The ADAM metalloprotease Kuzbanian is crucial for proper heart formation in Drosophila melanogaster. Mech Dev. 2006;123:372–387. doi: 10.1016/j.mod.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 47.Delwig A, Bland C, Beem-Miller M, Kimberly P, Rand MD. Endocytosis-independent mechanisms of Delta ligand proteolysis. Exp Cell Res. 2006;312:1345–1360. doi: 10.1016/j.yexcr.2005.12.037. [DOI] [PubMed] [Google Scholar]
- 48.Delwig A, Rand MD. Kuz and TACE can activate Notch independent of ligand. Cell Mol Life Sci. 2008 doi: 10.1007/s00018-008-8127-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reiss K, Maretzky T, Ludwig A, Tousseyn T, de Strooper B, Hartmann D, Saftig P. ADAM10 cleavage of N-cadherin and regulation of cell-cell adhesion and beta-catenin nuclear signalling. EMBO J. 2005;24:742–752. doi: 10.1038/sj.emboj.7600548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maretzky T, Reiss K, Ludwig A, Buchholz J, Scholz F, Proksch E, de Strooper B, Hartmann D, Saftig P. ADAM10 mediates E-cadherin shedding and regulates epithelial cell-cell adhesion, migration, and beta-catenin translocation. Proc Natl Acad Sci U S A. 2005;102:9182–9187. doi: 10.1073/pnas.0500918102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sanderson MP, Erickson SN, Gough PJ, Garton KJ, Wille PT, Raines EW, Dunbar AJ, Dempsey PJ. ADAM10 mediates ectodomain shedding of the betacellulin precursor activated by p-aminophenylmercuric acetate and extracellular calcium influx. J Biol Chem. 2005;280:1826–1837. doi: 10.1074/jbc.M408804200. [DOI] [PubMed] [Google Scholar]
- 52.Reiss K, Maretzky T, Haas IG, Schulte M, Ludwig A, Frank M, Saftig P. Regulated ADAM10-dependent ectodomain shedding of gamma-protocadherin C3 modulates cell-cell adhesion. J Biol Chem. 2006;281:21735–21744. doi: 10.1074/jbc.M602663200. [DOI] [PubMed] [Google Scholar]
- 53.Hall RJ, Erickson CA. ADAM 10: an active metalloprotease expressed during avian epithelial morphogenesis. Dev Biol. 2003;256:146–159. doi: 10.1016/s0012-1606(02)00133-1. [DOI] [PubMed] [Google Scholar]
- 54.Shoval I, Ludwig A, Kalcheim C. Antagonistic roles of full-length N-cadherin and its soluble BMP cleavage product in neural crest delamination. Development. 2007;134:491–501. doi: 10.1242/dev.02742. [DOI] [PubMed] [Google Scholar]
- 55.Hattori M, Osterfield M, Flanagan JG. Regulated cleavage of a contact-mediated axon repellent. Science. 2000;289:1360–1365. doi: 10.1126/science.289.5483.1360. [DOI] [PubMed] [Google Scholar]
- 56.Black RA, Rauch CT, Kozlosky CJ, Peschon JJ, Slack JL, Wolfson MF, Castner BJ, Stocking KL, Reddy P, Srinivasan S, et al. A metalloproteinase disintegrin that releases tumour-necrosis factor-alpha from cells. Nature. 1997;385:729–733. doi: 10.1038/385729a0. [DOI] [PubMed] [Google Scholar]
- 57.Shi W, Chen H, Sun J, Buckley S, Zhao J, Anderson KD, Williams RG, Warburton D. TACE is required for fetal murine cardiac development and modeling. Dev Biol. 2003;261:371–380. doi: 10.1016/s0012-1606(03)00315-4. [DOI] [PubMed] [Google Scholar]
- 58.Zhao J, Chen H, Wang YL, Warburton D. Abrogation of tumor necrosis factor-alpha converting enzyme inhibits embryonic lung morphogenesis in culture. Int J Dev Biol. 2001;45:623–631. [PubMed] [Google Scholar]
- 59.Sibilia M, Wagner EF. Strain-dependent epithelial defects in mice lacking the EGF receptor. Science. 1995;269:234–238. doi: 10.1126/science.7618085. [DOI] [PubMed] [Google Scholar]
- 60.Threadgill DW, Dlugosz AA, Hansen LA, Tennenbaum T, Lichti U, Yee D, LaMantia C, Mourton T, Herrup K, Harris RC, et al. Targeted disruption of mouse EGF receptor: effect of genetic background on mutant phenotype. Science. 1995;269:230–234. doi: 10.1126/science.7618084. [DOI] [PubMed] [Google Scholar]
- 61.Luetteke NC, Qiu TH, Fenton SE, Troyer KL, Riedel RF, Chang A, Lee DC. Targeted inactivation of the EGF and amphiregulin genes reveals distinct roles for EGF receptor ligands in mouse mammary gland development. Development. 1999;126:2739–2750. doi: 10.1242/dev.126.12.2739. [DOI] [PubMed] [Google Scholar]
- 62.Brou C, Logeat F, Gupta N, Bessia C, LeBail O, Doedens JR, Cumano A, Roux P, Black RA, Israel A. A novel proteolytic cleavage involved in Notch signaling: the role of the disintegrin-metalloprotease TACE. Mol Cell. 2000;5:207–216. doi: 10.1016/s1097-2765(00)80417-7. [DOI] [PubMed] [Google Scholar]
- 63.Yagami-Hiromasa T, Sato T, Kurisaki T, Kamijo K, Nabeshima Y, Fujisawa-Sehara A. A metalloprotease-disintegrin participating in myoblast fusion. Nature. 1995;377:652–656. doi: 10.1038/377652a0. [DOI] [PubMed] [Google Scholar]
- 64.Almeida EA, Huovila AP, Sutherland AE, Stephens LE, Calarco PG, Shaw LM, Mercurio AM, Sonnenberg A, Primakoff P, Myles DG, et al. Mouse egg integrin alpha 6 beta 1 functions as a sperm receptor. Cell. 1995;81:1095–1104. doi: 10.1016/s0092-8674(05)80014-5. [DOI] [PubMed] [Google Scholar]
- 65.Blobel CP. Functional processing of fertilin: evidence for a critical role of proteolysis in sperm maturation and activation. Rev Reprod. 2000;5:75–83. doi: 10.1530/ror.0.0050075. [DOI] [PubMed] [Google Scholar]
- 66.Kurisaki T, Masuda A, Sudo K, Sakagami J, Higashiyama S, Matsuda Y, Nagabukuro A, Tsuji A, Nabeshima Y, Asano M, et al. Phenotypic analysis of Meltrin alpha (ADAM12)-deficient mice: involvement of Meltrin alpha in adipogenesis and myogenesis. Mol Cell Biol. 2003;23:55–61. doi: 10.1128/MCB.23.1.55-61.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gilpin BJ, Loechel F, Mattei MG, Engvall E, Albrechtsen R, Wewer UM. A novel, secreted form of human ADAM 12 (meltrin alpha) provokes myogenesis in vivo. J Biol Chem. 1998;273:157–166. doi: 10.1074/jbc.273.1.157. [DOI] [PubMed] [Google Scholar]
- 68.Kronqvist P, Kawaguchi N, Albrechtsen R, Xu X, Schroder HD, Moghadaszadeh B, Nielsen FC, Frohlich C, Engvall E, Wewer UM. ADAM12 alleviates the skeletal muscle pathology in mdx dystrophic mice. Am J Pathol. 2002;161:1535–1540. doi: 10.1016/S0002-9440(10)64431-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kawaguchi N, Xu X, Tajima R, Kronqvist P, Sundberg C, Loechel F, Albrechtsen R, Wewer UM. ADAM 12 protease induces adipogenesis in transgenic mice. Am J Pathol. 2002;160:1895–1903. doi: 10.1016/S0002-9440(10)61136-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Weskamp G, Cai H, Brodie TA, Higashyama S, Manova K, Ludwig T, Blobel CP. Mice lacking the metalloprotease-disintegrin MDC9 (ADAM9) have no evident major abnormalities during development or adult life. Mol Cell Biol. 2002;22:1537–1544. doi: 10.1128/mcb.22.5.1537-1544.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sahin U, Weskamp G, Kelly K, Zhou HM, Higashiyama S, Peschon J, Hartmann D, Saftig P, Blobel CP. Distinct roles for ADAM10 and ADAM17 in ectodomain shedding of six EGFR ligands. J Cell Biol. 2004;164:769–779. doi: 10.1083/jcb.200307137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cai H, Kratzschmar J, Alfandari D, Hunnicutt G, Blobel CP. Neural crest-specific and general expression of distinct metalloprotease-disintegrins in early Xenopus laevis development. Dev Biol. 1998;204:508–524. doi: 10.1006/dbio.1998.9017. [DOI] [PubMed] [Google Scholar]
- 73.Alfandari D, Wolfsberg TG, White JM, DeSimone DW. ADAM 13: a novel ADAM expressed in somitic mesoderm and neural crest cells during Xenopus laevis development. Dev Biol. 1997;182:314–330. doi: 10.1006/dbio.1996.8458. [DOI] [PubMed] [Google Scholar]
- 74.Kee Y, Hwang BJ, Sternberg PW, Bronner-Fraser M. Evolutionary conservation of cell migration genes: from nematode neurons to vertebrate neural crest. Genes Dev. 2007;21:391–396. doi: 10.1101/gad.1509307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gaultier A, Cousin H, Darribere T, Alfandari D. ADAM13 disintegrin and cysteine-rich domains bind to the second heparin-binding domain of fibronectin. J Biol Chem. 2002;277:23336–23344. doi: 10.1074/jbc.M201792200. [DOI] [PubMed] [Google Scholar]
- 76.Borchers A, David R, Wedlich D. Xenopus cadherin-11 restrains cranial neural crest migration and influences neural crest specification. Development. 2001;128:3049–3060. doi: 10.1242/dev.128.16.3049. [DOI] [PubMed] [Google Scholar]
- 77.Mechtersheimer S, Gutwein P, Agmon-Levin N, Stoeck A, Oleszewski M, Riedle S, Postina R, Fahrenholz F, Fogel M, Lemmon V, et al. Ectodomain shedding of L1 adhesion molecule promotes cell migration by autocrine binding to integrins. J Cell Biol. 2001;155:661–673. doi: 10.1083/jcb.200101099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Maretzky T, Schulte M, Ludwig A, Rose-John S, Blobel C, Hartmann D, Altevogt P, Saftig P, Reiss K. L1 is sequentially processed by two differently activated metalloproteases and presenilin/gamma-secretase and regulates neural cell adhesion, cell migration, and neurite outgrowth. Mol Cell Biol. 2005;25:9040–9053. doi: 10.1128/MCB.25.20.9040-9053.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chen C, Huang X, Sheppard D. ADAM33 is not essential for growth and development and does not modulate allergic asthma in mice. Mol Cell Biol. 2006;26:6950–6956. doi: 10.1128/MCB.00646-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Puxeddu I, Pang YY, Harvey A, Haitchi HM, Nicholas B, Yoshisue H, Ribatti D, Clough G, Powell RM, Murphy G, et al. The soluble form of a disintegrin and metalloprotease 33 promotes angiogenesis: implications for airway remodeling in asthma. J Allergy Clin Immunol. 2008;121:1400–1406. e1401–1404. doi: 10.1016/j.jaci.2008.03.003. 1406. [DOI] [PubMed] [Google Scholar]
- 81.Kurohara K, Komatsu K, Kurisaki T, Masuda A, Irie N, Asano M, Sudo K, Nabeshima Y, Iwakura Y, Sehara-Fujisawa A. Essential roles of Meltrin beta (ADAM19) in heart development. Dev Biol. 2004;267:14–28. doi: 10.1016/j.ydbio.2003.10.021. [DOI] [PubMed] [Google Scholar]
- 82.Zhou HM, Weskamp G, Chesneau V, Sahin U, Vortkamp A, Horiuchi K, Chiusaroli R, Hahn R, Wilkes D, Fisher P, et al. Essential role for ADAM19 in cardiovascular morphogenesis. Mol Cell Biol. 2004;24:96–104. doi: 10.1128/MCB.24.1.96-104.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Horiuchi K, Zhou HM, Kelly K, Manova K, Blobel CP. Evaluation of the contributions of ADAMs 9, 12, 15, 17, and 19 to heart development and ectodomain shedding of neuregulins beta1 and beta2. Dev Biol. 2005;283:459–471. doi: 10.1016/j.ydbio.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 84.Podbilewicz B. ADM-1, a protein with metalloprotease- and disintegrin-like domains, is expressed in syncytial organs, sperm, and sheath cells of sensory organs in Caenorhabditis elegans. Mol Biol Cell. 1996;7:1877–1893. doi: 10.1091/mbc.7.12.1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Huang X, Huang P, Robinson MK, Stern MJ, Jin Y. UNC-71, a disintegrin and metalloprotease (ADAM) protein, regulates motor axon guidance and sex myoblast migration in C. elegans. Development. 2003;130:3147–3161. doi: 10.1242/dev.00518. [DOI] [PubMed] [Google Scholar]
- 86.Siddiqui SS, Culotti JG. Examination of neurons in wild type and mutants of Caenorhabditis elegans using antibodies to horseradish peroxidase. J Neurogenet. 1991;7:193–211. doi: 10.3109/01677069109167433. [DOI] [PubMed] [Google Scholar]
- 87.Chen EB, Branda CS, Stern MJ. Genetic enhancers of sem-5 define components of the gonad-independent guidance mechanism controlling sex myoblast migration in Caenorhabditis elegans hermaphrodites. Dev Biol. 1997;182:88–100. doi: 10.1006/dbio.1996.8473. [DOI] [PubMed] [Google Scholar]
- 88.Vjugina U, Evans JP. New insights into the molecular basis of mammalian sperm-egg membrane interactions. Front Biosci. 2008;13:462–476. doi: 10.2741/2693. [DOI] [PubMed] [Google Scholar]
- 89.Rubinstein E, Ziyyat A, Wolf JP, Le Naour F, Boucheix C. The molecular players of sperm-egg fusion in mammals. Semin Cell Dev Biol. 2006;17:254–263. doi: 10.1016/j.semcdb.2006.02.012. [DOI] [PubMed] [Google Scholar]
- 90.Talbot P, Shur BD, Myles DG. Cell adhesion and fertilization: steps in oocyte transport, sperm-zona pellucida interactions, and sperm-egg fusion. Biol Reprod. 2003;68:1–9. doi: 10.1095/biolreprod.102.007856. [DOI] [PubMed] [Google Scholar]
- 91.Miller DJ, Shi X, Burkin H. Molecular basis of mammalian gamete binding. Recent Prog Horm Res. 2002;57:37–73. doi: 10.1210/rp.57.1.37. [DOI] [PubMed] [Google Scholar]
- 92.Murase S, Cho C, White JM, Horwitz AF. ADAM2 promotes migration of neuroblasts in the rostral migratory stream to the olfactory bulb. Eur J Neurosci. 2008;27:1585–1595. doi: 10.1111/j.1460-9568.2008.06119.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Rybnikova E, Karkkainen I, Pelto-Huikko M, Huovila AP. Developmental regulation and neuronal expression of the cellular disintegrin ADAM11 gene in mouse nervous system. Neuroscience. 2002;112:921–934. doi: 10.1016/s0306-4522(02)00124-0. [DOI] [PubMed] [Google Scholar]
- 94.Takahashi E, Sagane K, Oki T, Yamazaki K, Nagasu T, Kuromitsu J. Deficits in spatial learning and motor coordination in ADAM11-deficient mice. BMC Neurosci. 2006;7:19. doi: 10.1186/1471-2202-7-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Takahashi E, Sagane K, Nagasu T, Kuromitsu J. Altered nociceptive response in ADAM11-deficient mice. Brain Res. 2006;1097:39–42. doi: 10.1016/j.brainres.2006.04.043. [DOI] [PubMed] [Google Scholar]
- 96.Leighton PA, Mitchell KJ, Goodrich LV, Lu X, Pinson K, Scherz P, Skarnes WC, Tessier-Lavigne M. Defining brain wiring patterns and mechanisms through gene trapping in mice. Nature. 2001;410:174–179. doi: 10.1038/35065539. [DOI] [PubMed] [Google Scholar]
- 97.Sagane K, Hayakawa K, Kai J, Hirohashi T, Takahashi E, Miyamoto N, Ino M, Oki T, Yamazaki K, Nagasu T. Ataxia and peripheral nerve hypomyelination in ADAM22-deficient mice. BMC Neurosci. 2005;6:33. doi: 10.1186/1471-2202-6-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.D'Abaco GM, Ng K, Paradiso L, Godde NJ, Kaye A, Novak U. ADAM22, expressed in normal brain but not in high-grade gliomas, inhibits cellular proliferation via the disintegrin domain. Neurosurgery. 2006;58:179–186. doi: 10.1227/01.neu.0000192363.84287.8b. discussion 179-186. [DOI] [PubMed] [Google Scholar]
- 99.Fukata Y, Adesnik H, Iwanaga T, Bredt DS, Nicoll RA, Fukata M. Epilepsy-related ligand/receptor complex LGI1 and ADAM22 regulate synaptic transmission. Science. 2006;313:1792–1795. doi: 10.1126/science.1129947. [DOI] [PubMed] [Google Scholar]
- 100.Hotoda N, Koike H, Sasagawa N, Ishiura S. A secreted form of human ADAM9 has an alpha-secretase activity for APP. Biochem Biophys Res Commun. 2002;293:800–805. doi: 10.1016/S0006-291X(02)00302-9. [DOI] [PubMed] [Google Scholar]
- 101.Kurisaki T, Wakatsuki S, Sehara-Fujisawa A. Meltrin beta mini, a new ADAM19 isoform lacking metalloprotease and disintegrin domains, induces morphological changes in neuronal cells. FEBS Lett. 2002;532:419–422. doi: 10.1016/s0014-5793(02)03732-8. [DOI] [PubMed] [Google Scholar]
- 102.Weskamp G, Ford JW, Sturgill J, Martin S, Docherty AJ, Swendeman S, Broadway N, Hartmann D, Saftig P, Umland S, et al. ADAM10 is a principal ‘sheddase’ of the low-affinity immunoglobulin E receptor CD23. Nat Immunol. 2006;7:1293–1298. doi: 10.1038/ni1399. [DOI] [PubMed] [Google Scholar]
- 103.Asai M, Hattori C, Szabo B, Sasagawa N, Maruyama K, Tanuma S, Ishiura S. Putative function of ADAM9, ADAM10, and ADAM17 as APP alpha-secretase. Biochem Biophys Res Commun. 2003;301:231–235. doi: 10.1016/s0006-291x(02)02999-6. [DOI] [PubMed] [Google Scholar]
- 104.Lemjabbar H, Basbaum C. Platelet-activating factor receptor and ADAM10 mediate responses to Staphylococcus aureus in epithelial cells. Nat Med. 2002;8:41–46. doi: 10.1038/nm0102-41. [DOI] [PubMed] [Google Scholar]
- 105.Horiuchi K, Le Gall S, Schulte M, Yamaguchi T, Reiss K, Murphy G, Toyama Y, Hartmann D, Saftig P, Blobel CP. Substrate selectivity of epidermal growth factor-receptor ligand sheddases and their regulation by phorbol esters and calcium influx. Mol Biol Cell. 2007;18:176–188. doi: 10.1091/mbc.E06-01-0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Matthews V, Schuster B, Schutze S, Bussmeyer I, Ludwig A, Hundhausen C, Sadowski T, Saftig P, Hartmann D, Kallen KJ, et al. Cellular cholesterol depletion triggers shedding of the human interleukin-6 receptor by ADAM10 and ADAM17 (TACE) J Biol Chem. 2003;278:38829–38839. doi: 10.1074/jbc.M210584200. [DOI] [PubMed] [Google Scholar]
- 107.Lunn CA, Fan X, Dalie B, Miller K, Zavodny PJ, Narula SK, Lundell D. Purification of ADAM 10 from bovine spleen as a TNFalpha convertase. FEBS Lett. 1997;400:333–335. doi: 10.1016/s0014-5793(96)01410-x. [DOI] [PubMed] [Google Scholar]
- 108.Garton KJ, Gough PJ, Blobel CP, Murphy G, Greaves DR, Dempsey PJ, Raines EW. Tumor necrosis factor-alpha-converting enzyme (ADAM17) mediates the cleavage and shedding of fractalkine (CX3CL1) J Biol Chem. 2001;276:37993–38001. doi: 10.1074/jbc.M106434200. [DOI] [PubMed] [Google Scholar]
- 109.Hundhausen C, Misztela D, Berkhout TA, Broadway N, Saftig P, Reiss K, Hartmann D, Fahrenholz F, Postina R, Matthews V, et al. The disintegrin-like metalloproteinase ADAM10 is involved in constitutive cleavage of CX3CL1 (fractalkine) and regulates CX3CL1-mediated cell-cell adhesion. Blood. 2003;102:1186–1195. doi: 10.1182/blood-2002-12-3775. [DOI] [PubMed] [Google Scholar]
- 110.Abel S, Hundhausen C, Mentlein R, Schulte A, Berkhout TA, Broadway N, Hartmann D, Sedlacek R, Dietrich S, Muetze B, et al. The transmembrane CXC-chemokine ligand 16 is induced by IFN-gamma and TNFalpha and shed by the activity of the disintegrin-like metalloproteinase ADAM10. J Immunol. 2004;172:6362–6372. doi: 10.4049/jimmunol.172.10.6362. [DOI] [PubMed] [Google Scholar]
- 111.Gough PJ, Garton KJ, Wille PT, Rychlewski M, Dempsey PJ, Raines EW. A disintegrin and metalloproteinase 10-mediated cleavage and shedding regulates the cell surface expression of CXC chemokine ligand 16. J Immunol. 2004;172:3678–3685. doi: 10.4049/jimmunol.172.6.3678. [DOI] [PubMed] [Google Scholar]
- 112.Vincent B, Paitel E, Saftig P, Frobert Y, Hartmann D, De Strooper B, Grassi J, Lopez-Perez E, Checler F. The disintegrins ADAM10 and TACE contribute to the constitutive and phorbol ester-regulated normal cleavage of the cellular prion protein. J Biol Chem. 2001;276:37743–37746. doi: 10.1074/jbc.M105677200. [DOI] [PubMed] [Google Scholar]
- 113.Takamune Y, Ikebe T, Nagano O, Nakayama H, Ota K, Obayashi T, Saya H, Shinohara M. ADAM-17 associated with CD44 cleavage and metastasis in oral squamous cell carcinoma. Virchows Arch. 2007;450:169–177. doi: 10.1007/s00428-006-0350-y. [DOI] [PubMed] [Google Scholar]
- 114.Murai T, Miyauchi T, Yanagida T, Sako Y. Epidermal growth factor-regulated activation of Rac GTPase enhances CD44 cleavage by metalloproteinase disintegrin ADAM10. Biochem J. 2006;395:65–71. doi: 10.1042/BJ20050582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Peschon JJ, Slack JL, Reddy P, Stocking KL, Sunnarborg SW, Lee DC, Russell WE, Castner BJ, Johnson RS, Fitzner JN, et al. An essential role for ectodomain shedding in mammalian development. Science. 1998;282:1281–1284. doi: 10.1126/science.282.5392.1281. [DOI] [PubMed] [Google Scholar]
- 116.Montero JC, Yuste L, Diaz-Rodriguez E, Esparis-Ogando A, Pandiella A. Differential shedding of transmembrane neuregulin isoforms by the tumor necrosis factor-alpha-converting enzyme. Mol Cell Neurosci. 2000;16:631–648. doi: 10.1006/mcne.2000.0896. [DOI] [PubMed] [Google Scholar]
- 117.Hansen HP, Dietrich S, Kisseleva T, Mokros T, Mentlein R, Lange HH, Murphy G, Lemke H. CD30 shedding from Karpas 299 lymphoma cells is mediated by TNF-alpha-converting enzyme. J Immunol. 2000;165:6703–6709. doi: 10.4049/jimmunol.165.12.6703. [DOI] [PubMed] [Google Scholar]
- 118.Reddy P, Slack JL, Davis R, Cerretti DP, Kozlosky CJ, Blanton RA, Shows D, Peschon JJ, Black RA. Functional analysis of the domain structure of tumor necrosis factor-alpha converting enzyme. J Biol Chem. 2000;275:14608–14614. doi: 10.1074/jbc.275.19.14608. [DOI] [PubMed] [Google Scholar]
- 119.Wang X, He K, Gerhart M, Huang Y, Jiang J, Paxton RJ, Yang S, Lu C, Menon RK, Black RA, et al. Metalloprotease-mediated GH receptor proteolysis and GHBP shedding. Determination of extracellular domain stem region cleavage site. J Biol Chem. 2002;277:50510–50519. doi: 10.1074/jbc.M208738200. [DOI] [PubMed] [Google Scholar]
- 120.Mohan MJ, Seaton T, Mitchell J, Howe A, Blackburn K, Burkhart W, Moyer M, Patel I, Waitt GM, Becherer JD, et al. The tumor necrosis factor-alpha converting enzyme (TACE): a unique metalloproteinase with highly defined substrate selectivity. Biochemistry. 2002;41:9462–9469. doi: 10.1021/bi0260132. [DOI] [PubMed] [Google Scholar]
- 121.Contin C, Pitard V, Itai T, Nagata S, Moreau JF, Dechanet-Merville J. Membrane-anchored CD40 is processed by the tumor necrosis factor-alpha-converting enzyme. Implications for CD40 signaling. J Biol Chem. 2003;278:32801–32809. doi: 10.1074/jbc.M209993200. [DOI] [PubMed] [Google Scholar]
- 122.Condon TP, Flournoy S, Sawyer GJ, Baker BF, Kishimoto TK, Bennett CF. ADAM17 but not ADAM10 mediates tumor necrosis factor-alpha and L-selectin shedding from leukocyte membranes. Antisense Nucleic Acid Drug Dev. 2001;11:107–116. doi: 10.1089/108729001750171353. [DOI] [PubMed] [Google Scholar]
- 123.Franzke CW, Tasanen K, Schacke H, Zhou Z, Tryggvason K, Mauch C, Zigrino P, Sunnarborg S, Lee DC, Fahrenholz F, et al. Transmembrane collagen XVII, an epithelial adhesion protein, is shed from the cell surface by ADAMs. EMBO J. 2002;21:5026–5035. doi: 10.1093/emboj/cdf532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Garton KJ, Gough PJ, Philalay J, Wille PT, Blobel CP, Whitehead RH, Dempsey PJ, Raines EW. Stimulated shedding of vascular cell adhesion molecule 1 (VCAM-1) is mediated by tumor necrosis factor-alpha-converting enzyme (ADAM 17) J Biol Chem. 2003;278:37459–37464. doi: 10.1074/jbc.M305877200. [DOI] [PubMed] [Google Scholar]
- 125.Lum L, Wong BR, Josien R, Becherer JD, Erdjument-Bromage H, Schlondorff J, Tempst P, Choi Y, Blobel CP. Evidence for a role of a tumor necrosis factor-alpha (TNF-alpha)-converting enzyme-like protease in shedding of TRANCE, a TNF family member involved in osteoclastogenesis and dendritic cell survival. J Biol Chem. 1999;274:13613–13618. doi: 10.1074/jbc.274.19.13613. [DOI] [PubMed] [Google Scholar]
- 126.Schlondorff J, Lum L, Blobel CP. Biochemical and pharmacological criteria define two shedding activities for TRANCE/OPGL that are distinct from the tumor necrosis factor alpha convertase. J Biol Chem. 2001;276:14665–14674. doi: 10.1074/jbc.M010741200. [DOI] [PubMed] [Google Scholar]
- 127.Chesneau V, Becherer JD, Zheng Y, Erdjument-Bromage H, Tempst P, Blobel CP. Catalytic properties of ADAM19. J Biol Chem. 2003;278:22331–22340. doi: 10.1074/jbc.M302781200. [DOI] [PubMed] [Google Scholar]
- 128.Shirakabe K, Wakatsuki S, Kurisaki T, Fujisawa-Sehara A. Roles of Meltrin beta /ADAM19 in the processing of neuregulin. J Biol Chem. 2001;276:9352–9358. doi: 10.1074/jbc.M007913200. [DOI] [PubMed] [Google Scholar]
- 129.Yokozeki T, Wakatsuki S, Hatsuzawa K, Black RA, Wada I, Sehara-Fujisawa A. Meltrin beta (ADAM19) mediates ectodomain shedding of Neuregulin beta1 in the Golgi apparatus: fluorescence correlation spectroscopic observation of the dynamics of ectodomain shedding in living cells. Genes Cells. 2007;12:329–343. doi: 10.1111/j.1365-2443.2007.01060.x. [DOI] [PubMed] [Google Scholar]
- 130.Izumi Y, Hirata M, Hasuwa H, Iwamoto R, Umata T, Miyado K, Tamai Y, Kurisaki T, Sehara-Fujisawa A, Ohno S, et al. A metalloprotease-disintegrin, MDC9/meltrin-gamma/ADAM9 and PKCdelta are involved in TPA-induced ectodomain shedding of membrane-anchored heparin-binding EGF-like growth factor. EMBO J. 1998;17:7260–7272. doi: 10.1093/emboj/17.24.7260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Asakura M, Kitakaze M, Takashima S, Liao Y, Ishikura F, Yoshinaka T, Ohmoto H, Node K, Yoshino K, Ishiguro H, et al. Cardiac hypertrophy is inhibited by antagonism of ADAM12 processing of HB-EGF: metalloproteinase inhibitors as a new therapy. Nat Med. 2002;8:35–40. doi: 10.1038/nm0102-35. [DOI] [PubMed] [Google Scholar]
- 132.Higashiyama S, Nanba D. ADAM-mediated ectodomain shedding of HB-EGF in receptor cross-talk. Biochim Biophys Acta. 2005;1751:110–117. doi: 10.1016/j.bbapap.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 133.Kawaguchi N, Horiuchi K, Becherer JD, Toyama Y, Besmer P, Blobel CP. Different ADAMs have distinct influences on Kit ligand processing: phorbol-ester-stimulated ectodomain shedding of Kitl1 by ADAM17 is reduced by ADAM19. J Cell Sci. 2007;120:943–952. doi: 10.1242/jcs.03403. [DOI] [PubMed] [Google Scholar]
- 134.Shi Z, Xu W, Loechel F, Wewer UM, Murphy LJ. ADAM 12, a disintegrin metalloprotease, interacts with insulin-like growth factor-binding protein-3. J Biol Chem. 2000;275:18574–18580. doi: 10.1074/jbc.M002172200. [DOI] [PubMed] [Google Scholar]
- 135.Loechel F, Fox JW, Murphy G, Albrechtsen R, Wewer UM. ADAM 12-S cleaves IGFBP-3 and IGFBP-5 and is inhibited by TIMP-3. Biochem Biophys Res Commun. 2000;278:511–515. doi: 10.1006/bbrc.2000.3835. [DOI] [PubMed] [Google Scholar]