Abstract
Both fetal ventral mesencephalic (VM) and embryonic stem (ES) cell-derived dopamine neurons have been used successfully to correct behavioral responses in animal models of Parkinson’s disease. However, grafts derived from fetal VM cells or from ES cells contain multiple cell types, and the majority of these cells are not dopamine neurons. Isolation of ES cell-derived dopamine neurons and subsequent transplantation would both elucidate the capacity of these neurons to provide functional input and also further explore an efficient and safer use of ES cells for the treatment of Parkinson’s disease. Toward this goal, we used a Pitx3-enhanced green fluorescent protein (Pitx3-eGFP) knock-in mouse blastocyst-derived embryonic stem (mES) cell line and fluorescence-activated cell sorting (FACS) to select and purify midbrain dopamine neurons. Initially, the dopaminergic marker profile of intact Pitx3-eGFP mES cultures was evaluated after differentiation in vitro. eGFP expression overlapped closely with that of Pitx3, Nurr1, Engrailed-1, Lmx1a, tyrosine hydroxylase (TH), l-aromatic amino acid decarboxylase (AADC), and vesicular monoamine transporter 2 (VMAT2), demonstrating that these cells were of a midbrain dopamine neuron character. Furthermore, postmitotic Pitx3-eGFP+ dopamine neurons, which constituted 2%–5% of all live cells in the culture after dissociation, could be highly enriched to >90% purity by FACS, and these isolated neurons were viable, extended neurites, and maintained a dopaminergic profile in vitro. Transplantation to 6-hydroxydopamine-lesioned rats showed that an enriched dopaminergic population could survive and restore both amphetamine- and apomorphine-induced functions, and the grafts contained large numbers of midbrain dopamine neurons, which innervated the host striatum.
Keywords: Pitx3, Transplantation, Parkinson’s disease, Stage-specific embryonic antigen-1, Mouse embryonic stem cells
INTRODUCTION
Midbrain dopamine neurons, the cells lost during the course of Parkinson’s disease, can be generated from embryonic stem cells either in vitro or in vivo after transplantation [1– 6]. Protocols used to generate midbrain dopamine neurons from embryonic stem (ES) cells in vitro use morphogens aimed at creating an environment similar to that surrounding the developing isthmus [2, 3, 5]. As would be expected, the majority of the cells in differentiating ES cell cultures are not of a dopaminergic phenotype [7, 2], since the cultures are not completely synchronized in either a temporal or a spatial manner. Serotonergic neurons [2], GABAergic neurons [8], and cells that have proliferative capacity [6, 9–11] are also present in culture. Grafts derived from primary ventral mesencephalic cells, usually containing 5%–10% midbrain dopamine neurons, elicit a well-documented effect on Parkinsonian symptoms [12–17]. Similarly, transplantation of in vitro-differentiated ES cell cultures generates grafts containing a portion of midbrain dopamine neurons, which can alleviate symptoms in animal models of Parkinson’s disease [1–6]. However, until analysis of grafts containing mainly midbrain dopamine neurons is done, deducing the functional input provided by each different cell type and elucidating their functional interdependence will remain difficult.
Furthermore, the capacity of ES cells to develop into all cellular types creates the risk of tumor and/or teratoma formation after transplantation. Therefore, transplantation of ES cell-derived dopamine neuron preparations could vastly benefit from an exclusive selection of midbrain dopamine neurons alone from the culture prior to transplantation. Markers used for selection of a specific cellular subtype derived from ES cells need to show a very limited spatial distribution during development or otherwise be used in combination with another marker so that together they encompass a restricted region.
Within the central nervous system, the homeodomain transcription factor Pitx3 is selectively expressed in midbrain dopamine neurons of both the substantia nigra compacta (SNc) and the ventral tegmental area (VTA) [18]. Expression of Pitx3 is initiated at embryonic day (E) 11.5, at the time when tyrosine hydroxylase (TH) expression is induced, and is maintained to adulthood [18]. Pitx3 deficiency results in a selective developmental failure and loss of SNc dopamine neurons and their projections to the dorsal caudate putamen only [19 –22]. The specific dependence on Pitx3 in SNc dopamine neurons appears to be due to the selective expression of its downstream target Raldh1 and the resultant production of retinoic acid there [23].
On the basis of the developmentally restricted expression pattern of Pitx3 and the previous evidence that ES cell-derived TH neurons can indeed be viably isolated from ES cell cultures [11, 24, 25], we deemed Pitx3 a good candidate for the selection of dopamine neurons from ES cell cultures. Toward this goal, first, our aim was to carefully characterize the Pitx3-enhanced green fluorescent protein (Pitx3-eGFP) knock-in (KI) mES cells in vitro prior to isolation by fluorescence-activated cell sorting (FACS) to determine the specificity of the eGFP expression in differentiating ES cell cultures. Second, we wanted to enrich cells for Pitx3-eGFP expression using FACS and to culture these mature midbrain dopamine neurons in vitro. Finally, our aim was to transplant the midbrain dopamine neuron-enriched fraction to 6-hydroxydopamine-lesioned rats and subsequently analyze their functional integration in this model of Parkinson’s disease.
METHODS
Mouse ES Cell Propagation
The mouse blastocyst-derived embryonic stem (mES) cell line Pitx3-eGFP (a gift from Dr. M. Li) [26] was propagated on mitomycin C-treated (10 µg/ml medium; Sigma-Aldrich, St. Louis, http://www.sigmaaldrich.com) PMEFs (StemCell Technologies, Vancouver, BC, Canada, http://www.stemcell.com) in modified Dulbecco’s modified Eagle’s medium (DMEM; Invitrogen, Carlsbad, CA, http://www.invitrogen.com), supplemented with 2 mM l-glutamine (Invitrogen), 1 mM β-mercaptoethanol, 1× nonessential amino acids (NEAA; Invitrogen), 10% fetal bovine serum (FBS; Sigma-Aldrich), 100 U/ml penicillin, 100 µg/ml streptomycin (Invitrogen), and 2,000 U/ml human recombinant leukemia inhibitory factor (R&D Systems Inc., Minneapolis, http://www.rndsystems.com). In this ES cell line, part of intron 1 and exons 2, 3, and 4 were replaced with eGFP [26]. The nuclear translocation signal in the Pitx3 gene lies within the deleted sequence [27], and eGFP is therefore not localized to the nucleus.
In Vitro Differentiation of Pitx3-eGFP mES Cells
Pitx3-eGFP mES cells were differentiated into dopamine neurons using a previously described protocol [2], with some modifications. In brief, mES cells were dissociated using 0.05% trypsin and purified by attachment to tissue culture dishes for 1 hour. Embryoid bodies (EBs) were allowed 3 days to form after plating of mES cells in bacterial dishes in DMEM containing defined 10% FBS (Sigma-Aldrich), 2 mM l-glutamine (Invitrogen), 1× NEAA (Invitrogen), 10 mM HEPES (Invitrogen), 1 mM β-mercaptoethanol, 100 U/ml penicillin, and 100 µg/ml streptomycin (Invitrogen) (EB medium). EBs were allowed 1 day to attach to tissue culture dishes, and neuronal precursors were then selected for by incubation in DMEM F-12 medium containing 50 µg/ml apotransferrin (Sigma-Aldrich), 5 µg/ml insulin (Sigma-Aldrich), 30 nM sodium selenite (Sigma-Aldrich), 250 ng/ml fibronectin (Sigma-Aldrich), 100 U/ml penicillin, and 100 µg/ml streptomycin (Invitrogen) for 9–10 days. Cells were subsequently dissociated by 0.05% trypsin, and neuronal precursors were expanded and patterned for 4 days after plating onto fibronectin-/polyornithine-coated plates at a density of 75,000 cells per cm2 in DMEM F-12 medium containing 100 µg/ml apotransferrin, 5 µg/ml insulin, 30 nM sodium selenite, 20 nM progesterone, 100 nM putrescine, 100 U/ml penicillin, 100 µg/ml streptomycin, 1 µg/ml laminin, 10 ng/ml basic fibroblast growth factor (R&D Systems), 500 ng/ml Shh (R&D Systems), and 100 ng/ml Fgf8 (R&D Systems) (N3 medium). The cells were subsequently differentiated in N3 medium containing 200 µM ascorbic acid (N3AA medium) for 3–14 days (stage 5). Cells used for immunofluorescent staining were fixed in 4% formaldehyde for 30 minutes and rinsed with phosphate-buffered saline (PBS). For the initial evaluation of an optimal time point for FACS, cells were harvested after 7–10 days in stage 5 (stages 5:7–5:10 of differentiation) using 0.05% trypsin/EDTA, with 5:8 and 5:9 determined to be the optimal time points for this procedure. Cells to be further analyzed in vitro after FACS for eGFP expression were replated onto primary rat astrocytes (Lonza, Walkersville, MD, http://www.lonza.com) in N3AA medium supplemented with 10% FBS, 10 ng/ml glial-derived neurotrophic factor (GDNF; Sigma-Aldrich), and 20 ng/ml brain-derived neurotrophic factor (BDNF; Peprotech, Rocky Hill, NJ, http://www.peprotech.com).
FACS
Cells were harvested at stages 5:7–5:10 using 0.05% trypsin/EDTA (Invitrogen), gently dissociated into a single-cell suspension, and resuspended in phenol-free Hanks’ balanced saline solution (HBSS; Invitrogen) containing 20 mM d-glucose (Sigma-Aldrich), penicillin-streptomycin, and 2% FBS. Samples were filtered through cell strainer caps (35 µm mesh; BD Biosciences, San Jose, CA, http://www.bdbiosciences.com) and then either analyzed and sorted immediately for eGFP expression or subjected to surface marker staining as follows: primary mouse IgM anti-stage-specific embryonic antigen-1 (anti-SSEA-1) antibody (0.4 µg/ml; Developmental Studies Hybridoma Bank, Iowa City, IA, http://www.uiowa.edu/~dshbwww), rat anti-CD56 (NCAM; 1–2 µg/ml; BD Pharmingen, San Diego, http://www.bdbiosciences.com), or mouse IgM anti-PSA-NCAM (1:500; Millipore, http://www.millipore.com) was added for 20 minutes, and cells were then washed, incubated with Alexa Fluor 647 fluorescent secondary antibodies, and subsequently washed with the HBSS as described above. FACS was performed using a FACSAria cell sorter and FACSDiva software (BD Biosciences). The population of interest, excluding debris and dead cells, was identified by forward and side scatter gating. Using a 488-nm laser for excitation, eGFP+ was determined according to fluorescence intensity in the eGFP channel (490-nm long-pass [LP], 510/20-nm band-pass [BP] filters), and cells labeled with Alexa Fluor 647 secondary antibodies were detected in the far-red channel (655-nm LP, 655–735-nm BP filters). Naïve E14TG2a mES cells (the parental mES cell line to Pitx3-eGFP KI cells) at the same stage of differentiation were used as the eGFP-negative control. eGFP-fluorescent cells were sorted using a purity mask excluding doublets and conflicts. SSEA-1+, CD56+, and PSA-NCAM+ events were determined according to fluorescence in the far-red channel compared with negative controls lacking the primary and/or secondary antibodies. All FACS experiments were repeated at least three times, and the purity of all sorted eGFP+ fractions was determined by reanalysis using FACS. A 100 µm nozzle, sheath pressure of 20–25 psi, and an acquisition rate of 1,000 –2,000 events per second were used as the standard conditions [11, 25]. Further flow cytometric analysis was performed using FlowJo software (Tree Star, Ashland, OR, http://www.treestar.com).
Animal Procedures
All animal procedures were performed in accordance with National Institute of Health guidelines and were approved by the Animal Care and Use Committee at McLean Hospital, Harvard Medical School. Animals were housed according to standard conditions, with access to food and water ad libitum and with a dark/light cycle of 12 hours.
Transplantation into 6-Hydroxydopamine-Lesioned Rats and Analysis of Drug-Induced Rotational Behavior
Pitx3-eGFP mES cells, sorted for eGFP expression and analyzed for viability, were resuspended in N3AA medium containing 20 ng/ml BDNF and 10 ng/ml GDNF or containing 20 ng/ml BDNF, 10 ng/ml GDNF, and 20 µM Boc-Asp(OMe) fluoromethyl ketone (BAF; Sigma-Aldrich) at a density of 10,000–15,000 cells per microliter. Female Sprague-Dawley rats with unilateral 6-hydroxy-dopamine lesions were obtained from Charles River Laboratories (Wilmington, MA, http://www.criver.com). The severity of the lesions was measured prior to transplantation by rotational behavior in response to amphetamine (4 mg/kg i.p.) and apomorphine (0.05 mg/kg). Rats were grafted into the lesioned striatum with 4–6 µl of cell suspension devoid of BAF (n = 11) or containing BAF (n = 13) or medium only (n = 5) into one tract with two deposits (coordinates from bregma: anterior-posterior 0.0, lateral −0.3, ventral −0.55 and −0.45). Immunosuppression, anesthesia, transplantation, and analgesia were performed as previously described [4]. Amphetamine-induced rotational behavior was measured again at 5, 7, and 9 weeks post-transplantation, and apomorphine-induced rotations were measured at 8 weeks post-transplantation. The animals were sacrificed 9 weeks post-transplantation. Anesthesia was performed by administration of an i.p. overdose of pentobarbital (150 mg/kg), and animals were perfused intracardially with 0.1% heparinized saline followed by 4% paraformaldehyde. Brains were removed, postfixed in 4% paraformaldehyde, equilibrated in 20% sucrose, and sectioned on a freezing microtome in 40-µm coronal slices.
Histological and Stereological Procedures
For immunofluorescent staining, cells on coverslips and tissue sections were rinsed with PBS and incubated with blocking buffer (PBS, 10% normal donkey serum or normal goat serum, 0.1% Triton X-100) for 1 hour. Coverslips/sections were then incubated overnight at 4°C with primary antibodies in blocking buffer. The following primary antibodies were used: rabbit anti-green fluorescent protein (anti-GFP; 1:1,000; Molecular Probes, Eugene, OR, http://probes.invitrogen.com), sheep anti-TH (1:1,000), and rabbit anti-vesicular monoamine transporter 2 (anti-VMAT2; 1:1,000; Pel-Freez, Rogers, AK, http://www.invitrogen.com), rabbit anti-Lmx1a (1:1,000; a kind gift from Dr MS German), sheep anti-l-aromatic amino acid decarboxylase (anti-AADC; 1:200), and mouse anti-nestin (1:100; Millipore), rabbit anti-Pitx3 (1:250; Invitrogen), rabbit anti-glial fibrillary acidic protein (1:500; Dako, Carpinteria, CA, http://www.dako.com), serotonin (5-HT; 1:1,000; Diasorin, Stillwater, MN, http://www.diasorin.com/en/), rabbit anti-Nurr1 (E-20; 1:300; Santa Cruz Biotechnology Inc., Santa Cruz, CA, http://www.scbt.com), mouse anti-engrailed 1 (clone 4G11; 1:40), mouse anti-SSEA-1 (1 µg/ml; Developmental Studies Hybridoma Bank), rabbit anti-Ki67 (1:2,000; Novocastra Ltd., Newcastle upon Tyne, U.K., http://www.novocastra.co.uk), and rabbit anti-Raldh1a1 (1:250; Calbiochem, San Diego, http://www.emdbiosciences.com). The coverslips/tissue sections were subsequently incubated in fluorescent-labeled Alexa Fluor secondary antibodies for 1 hour at room temperature. After rinsing in PBS, Hoechst 33342 (4 µg/ml) was used for counterstaining, and coverslips/tissues sections were mounted onto slides in Mowiol 4–88 (Calbiochem). For light microscopy, biotinylated secondary antibody (1:300; Vector Laboratories, Burlingame, CA, http://www.vectorlabs.com) was used, followed by incubation in streptavidin-biotin complex (Vector Laboratories) for 1 hour and visualization by incubation in 3,3′-diaminobenzidine (Vector Laboratories). Control experiments were performed by omission of primary antibodies and using different combinations of secondary antibodies. Confocal analysis was performed using a Zeiss LSM510/Meta Station (Carl Zeiss, Thornwood, NY, http://www.zeiss.com). For identification of signal colocalization within a cell, optical thickness was kept to a minimum, and orthogonal reconstructions were obtained. Stereology was performed using Stereo Investigator image-capture equipment and software (MicroBrightField Inc., Williston, VT, http://www.mbfbioscience.com) and a Zeiss Axioplan I fluorescent microscope. Graft volumes were calculated using the Cavalieri estimator probe. The number of TH neurons present in Pitx3-eGFP-grafted 6-hydroxydopamine-lesioned rats was calculated using the Optical fractionator probe of Stereo Investigator, on the basis of randomly selected sampling sites from one in six sections. Mounted section thickness was measured in one in two sampling sites for accurate estimation of the number of TH neurons within the whole grafts. The extent of overlap between eGFP and Pitx3, engrailed-1 (En), Nurr1, Lmx1a, TH, and AADC in intact cultures and after FACS was determined by random sampling using Stereo Investigator. At least three coverslips were counted for each immunostaining and feeder layer.
Statistical Analysis
Amphetamine- and apomorphine-induced rotational scores for Pitx3-eGFP mES cell-grafted and vehicle-grafted animals were analyzed using analysis of variance. InStat3 software (GraphPad Software, La Jolla, CA, http://www.graphpad.com) was used for statistical analyses.
RESULTS
Pitx3-eGFP KI mES Cells Showed a Transcription Factor and Enzymatic Profile Characteristic of Midbrain Dopamine Neurons
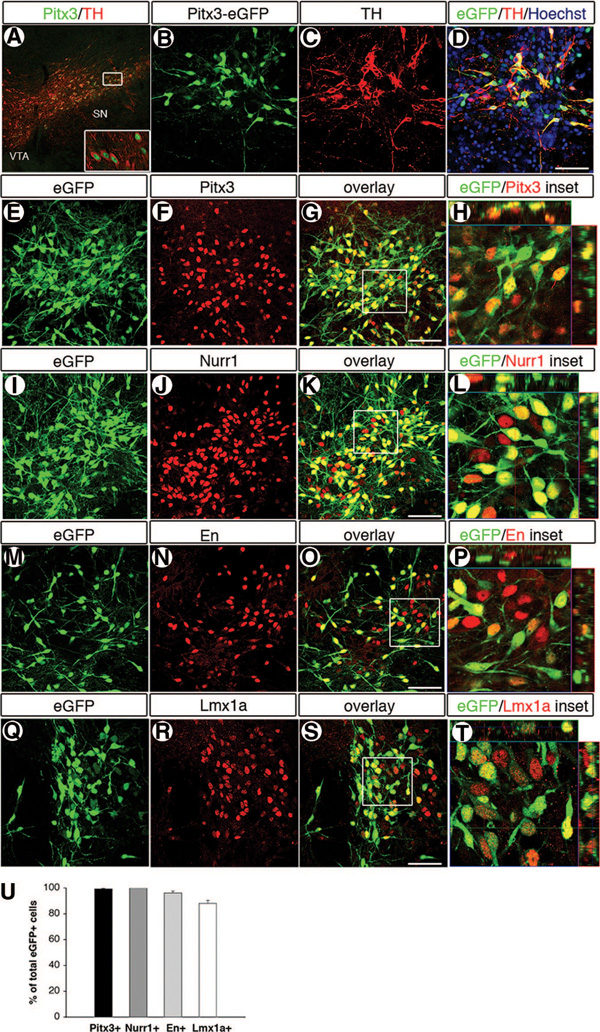
We differentiated Pitx3-eGFP KI mES cells according to the five-stage protocol [2]. To evaluate the phenotype of the eGFP+ cells in culture, the overlapping expression of transcription factors and enzymes characteristic of midbrain dopamine neurons was analyzed at stage 5:8 of differentiation. At this stage, there were many eGFP+/TH+ cells in culture, but the majority of cells were nondopaminergic cells, as visualized by Hoechst staining (and their lack of eGFP and TH expression) (Fig. 1B–1D). Initially, the coexpression of Pitx3 and eGFP (99.4% ± 0.3%) (n = 3 coverslips; 5,033 cells counted), was verified (Fig. 1E–1H, 1U). The specificity of the Pitx3 antibody was confirmed by the localized nuclear staining of TH+ neurons in the VTA and the substantia nigra in the naïve mouse brain (Fig. 1A). Furthermore, 100% of eGFP+ cells were also Nurr1+ (n = 3 coverslips; 2,057 cells counted) (Fig. 1I–1L, 1U). As expected on the basis of the broader expression domain of Nurr1 [27–29] than Pitx3 [18] within the central nervous system, there were Nurr1+ cells present in culture (34.3% ± 4.3%) (n = 3 coverslips) that were negative for Pitx3 expression (Fig. 1I–1L). Of the Pitx3-eGFP+ cells, 96.2% ± 1.6% were En+ (n = 3 coverslips; 1,387 cells counted) (Fig. 1M–1P, 1U) and 88.1% ± 2.3% were Lmx1a+ (n = 3 coverslips, 1,591 cells counted) (Fig. 1Q–1T, 1U). The lower level of coexpression between Lmx1a and eGFP compared with Nurr1 and Engrailed made us consider the possibility that Lmx1a might have a temporally more restricted expression than Nurr1 and Engrailed within dopamine neurons. Careful analysis of intact Pitx3-eGFP cultures revealed that Lmx1a was expressed early, as would be expected, on the basis of its expression within cells of more immature morphology within the neural tube-like (rosette) structures in the culture, which did not yet express TH or Pitx3 (supplemental online Fig. 1A–1D). Confocal imaging also revealed that there were eGFP+TH+ cells of mature neuronal morphology that were lacking Lmx1a expression (supplemental online Fig. 1A–1D). Since Lmx1a is required for the differentiation of dopamine neurons [8], it would be expected that these Lmx1a− dopamine neurons initially expressed Lmx1a and subsequently downregulated the expression once these particular dopamine neurons were formed.
Figure 1.
Analysis of the transcription factor profile of Pitx3-eGFP knock-in (KI) mouse blastocyst-derived embryonic stem (mES) cells during in vitro differentiation showed that eGFP+ cells were of a midbrain dopamine neuron phenotype. Pitx3 is expressed in dopamine neurons of the VTA and the SN, as visualized by staining of the naïve mouse brain (A). Pitx3 is also expressed in mES cell culture during in vitro differentiation, as visualized by eGFP expression in Pitx3-eGFP KI mES cells (B, D), and this expression overlapped with TH immunoreactivity (C). Hoechst staining showed that the majority of the cells in the mES cell culture were not midbrain dopamine neurons (D). eGFP expression in mES cells, driven by the Pitx3 allele, showed high overlapping expression with Pitx3 (99.4%) (E–H and U) ([H] is an orthogonal view of [G]), Nurr1 (100%) (I–L and U) ([L] is an orthogonal view of [K]), En (96.2%) (M–P and U) ([P] is an orthogonal view of [O]), and Lmx1a (88.1%) (Q–T) ([T] is an orthogonal view of [S]). All analyses were performed on cells in stage 5:8 of the differentiation protocol. Scale bars = 50 µm (bars in [D, G, K, O, S] applies to [B, C, E, F, I, J, M, N, Q, R]). Abbreviations: eGFP, enhanced green fluorescent protein; En, engrailed; SN, substantia nigra; TH, tyrosine hydroxylase; VTA, ventral tegmental area.
Most eGFP+ cells also expressed TH (78.5% ± 0.6%) (n = 3 coverslips; 1,404 cells counted) (Fig. 2A–2C, 2M). A previous analysis of these Pitx3-eGFP KI mES cells, using a PA6 feeder-based system for in vitro differentiation of ES cells, showed a slightly higher overlap between TH and eGFP expression at the end of the protocol [26]. The somewhat lower overlap in our system might be due to a relatively earlier time point of differentiation being analyzed. However, a difference in the ratio of ventrolateral versus dorsomedial mesencephalic cells being generated in the two systems could also explain this slight discrepancy. It has been described that ventrolateral mesencephalic cells express Pitx3 prior to TH, whereas the dorsomedial cells express TH before Pitx3 in the developing mouse [30]. Confocal analysis of the overlapping expression patterns of Lmx1a, TH, and Pitx3 showed the presence of midbrain dopamine neurons (Lmx1a+) that expressed either TH first or Pitx3 first, indicating the presence of both ventrolateral and dorsomedial mesencephalic cells in the culture (supplemental online Fig. 1). Importantly, the majority of the TH+ cells in the culture were of a midbrain dopamine neuron type, with only 14.3% ± 1.7% of the TH+ cells lacking Pitx3-eGFP expression. Furthermore, most eGFP+ cells coexpressed AADC (94.7% ± 0.8%) (n = 3 coverslips; 961 cells counted) (Fig. 2D–2F, 2M). The earlier developmental expression of AADC than of TH [22, 31] might explain the better overlap in expression between AADC and Pitx3 compared with TH and Pitx3. Confocal analysis showed that there were AADC+ neurons in the culture that did not show overlapping expression with eGFP. On the basis of the earlier expression of AADC than of Pitx3, some of these AADC+ cells could be developing into dopamine neurons. However, previous analysis of the neuronal subtypes in this culture system has shown that 10% of all neurons generated are serotonergic [2], rendering it likely that some of the AADC+/eGFP+ cells were or were to become serotonergic neurons. 5-HT+ neurons, which were never eGFP+, were indeed present within the cultures (Fig. 2J–2L). eGFP+ cells showed high overlapping expression with VMAT2. There were also VMAT2+ cells in the culture that were not eGFP+, indicating the presence of other monoaminergic neurons, such as serotonergic neurons (Fig. 2G–2I).
Figure 2.
Analysis of the dopamine-synthesizing enzyme profile of Pitx3-eGFP knock-in embryonic stem cells during in vitro differentiation showed that eGFP+ cells were of a midbrain dopamine neuron phenotype. Confocal analysis showed that most eGFP+ cells expressed TH (78.5%) (A–C, M), AADC (94.7%) (D–F, M), and VMAT2 (G–I). eGFP+ cells did not show overlapping expression with serotonin (J–L). All analyses were performed on cells in stage 5:8 of the differentiation protocol. Scale bars = 50 µm (bars in [C, F, I, L] also applies to [A, B, D, E, G, H, J, K]). Abbreviations: 5-HT, serotonin; AADC, l-aromatic amino acid decarboxylase; eGFP, enhanced green fluorescent protein; TH, tyrosine hydroxylase; VMAT2, vesicular monoamine transporter 2.
Postmitotic Pitx3-eGFP+ Neurons Could Be Viably Enriched by FACS with Maintenance of a Midbrain Dopamine Neuron Profile
Intact Pitx3-eGFP KI mES cell cultures at stages 5:8 –5:9 of differentiation showed neuronal morphology, and there were clusters that exhibited a strong expression of eGFP (Fig. 3A, 3B). In our initial experiments we made an effort to increase the percentage of Pitx3-eGFP+ cells in the culture by optimizing the plating density of the EBs and the neuronal precursors in stage 4. Dissociation of the culture at 5:8 –5:9 and FACS for Pitx3-eGFP expression showed that the number of eGFP+ cells could be increased from 0.2%–0.5% (data not shown) to 2%–5% (n > 15) (Fig. 3C). We also analyzed the percentage of Pitx3-eGFP+ cells using differentiation protocols based on PA6 [3] or MS5 [5] feeders, but in our hands, the percentage of Pitx3-GFP+ cells in these culture was always too low to be usable for FACS (<0.2%) (data not shown). Dissociation of the five-stage differentiated culture at stages 5:8 –5:9 consistently resulted in a loss of ~50% of all cells, as evaluated by trypan blue exclusion (data not shown). If we assume that the trypsinization affects all cell types in the culture equally, then 2%–5% of all cells were midbrain dopamine neurons at the time of dissociation also. It has not been previously evaluated how many cells of all cells in the culture become midbrain dopamine neurons, only how many neurons are dopamine neurons, since the three-dimensional nature of the culture does not enable an accurate estimation of all cells in intact cultures. After a single enrichment by FACS, the Pitx3-eGFP+ cells constituted up to 80% of the cell population (Fig. 3D). Further characterization of the dissociated Pitx3-eGFP KI mES cells showed that eGFP expression colocalized with the neuronal markers CD56 (NCAM) and PSA-NCAM but not with SSEA-1, a marker of immature or transformed ES cells (Fig. 3E). Analysis of SSEA-1 expression in dissociated culture at stage 5:8 showed that approximately 3% of the cells prior to FACS were SSEA-1+ (Fig. 3F, left panel). FACS for eGFP expression and reanalysis of the eGFP-enriched fraction showed that the number of SSEA-1+ cells had been reduced 10-fold to ~0.3% of all cells (Fig. 3F, right panel). As previously reported, the purified dopamine neurons required coculturing with primary astrocytes to survive in vitro post-FACS [11, 32]. Analysis of sorted cells 1 day in vitro post-FACS showed that the Pitx3-eGFP+ cells had extended long processes and that Pitx3-eGFP expression remained colocalized with the midbrain dopamine transcription factors Engrailed (86.8% ± 6.6%) (n = 3 coverslips; 428 cells counted) (Fig. 4A–4D, 4L), Pitx3 (99.6% ± 0.4%) (n = 3 coverslips; 555 cells counted) (Fig. 4E–4H, 4L), and Nurr1 (97.4% ± 1.5%) (n = 4 coverslips; 222 cells counted (Fig. 4I–4K, 4L). The coexpression of eGFP and TH was almost complete (99.9% ± 0.1%) (n = 3 coverslips; 295 cells counted) (Fig. 4A–4D, 4L). Colocalization of eGFP and AADC expression remained high after FACS (91.4%) (n = 3 coverslips; 378 cells counted) (Fig. 4E–4H, 4L). To further enrich the fraction of Pitx3-eGFP+ neurons, we subjected already sorted cells, enriched to 75% ± 5.1%, to a second round of FACS for eGFP expression. This double-sorting procedure resulted in an enrichment of Pitx3-eGFP expression to 95.4% ± 2.7% of midbrain dopamine neurons that could be viably cultured post-sorting (supplemental online Fig. 2).
Figure 3.
Fluorescence-activated cell sorting (FACS) for eGFP expression enriched for a neuronal population with expression of midbrain dopamine neuron markers. Intact Pitx3-eGFP knock-in mouse blastocyst-derived embryonic stem cell cultures at stage 5:8 showed neuronal morphology and expression of eGFP (A, B). Dissociation and FACS of these cells showed that a small proportion of the cells in the culture were eGFP+ after dissociation (C), and these cells could be enriched to constitute approximately 80% of the cell population (D). FACS analysis of dissociated cultures at 5:8 stained for CD56, PSA-NCAM, and SSEA-1 showed that eGFP+ cells colocalize with the neuronal surface markers but not with SSEA-1 (E). The left panel of (E) depicts cells that were stained with secondary antibody only. Further analysis showed that approximately 3% the cells in the culture were SSEA-1+ (n = 3) ([F], left panel), but the eGFP-enriched fraction contained less than 0.3% SSEA-1+ cells (n = 3) ([F], right panel). Abbreviations: eGFP, enhanced green fluorescent protein; SSEA-1, stage-specific embryonic antigen-1.
Figure 4.
Pitx3-eGFP expression colocalized with neuronal surface markers, and enrichment for eGFP+ cells removed the majority of undifferentiated cells from the culture. Replating of the eGFP-enriched fraction onto primary astrocytes and analysis 1 day in vitro post-FACS showed that the cells had extended processes and that eGFP expression was highly overlapping with En (A–D, L), TH (A–D, L), Pitx3 (E–H, L), AADC (E–H, L), and Nurr1 (I–L). Scale bars = 50 µm (bars in [D, H, K] also apply to [A–C, E–G, I, J]). Abbreviations: AADC, l-aromatic amino acid decarboxylase; eGFP, enhanced green fluorescent protein; En, engrailed; TH, tyrosine hydroxylase.
The Pitx3-eGFP-Enriched Cell Population Generated Functional Grafts That Could Reverse Drug-Induced Behavior in Parkinsonian Rats
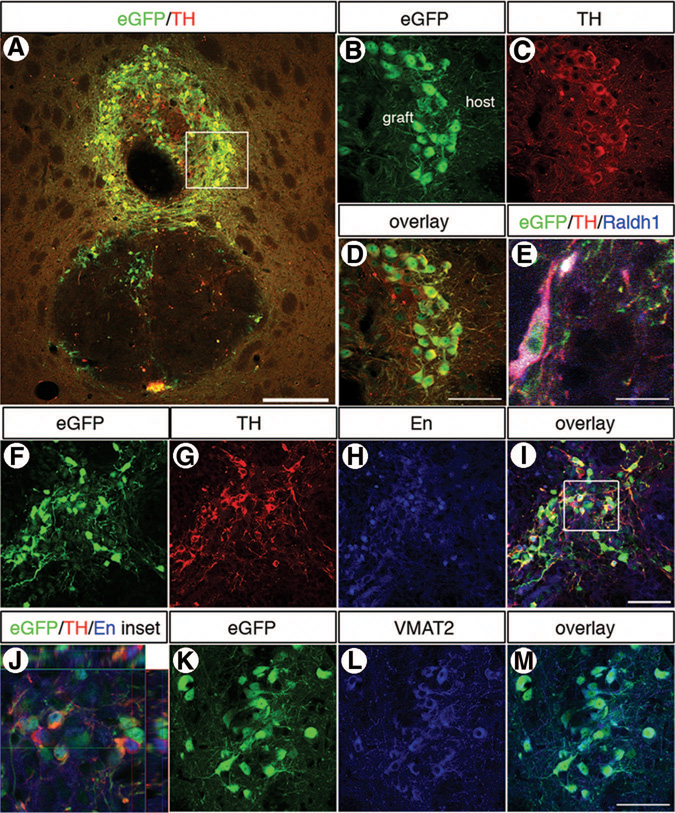
Analysis of amphetamine-induced behavior of 6-hydroxydopamine-lesioned rats showed that several animals transplanted with a cell population enriched for Pitx3-eGFP expression decreased their clockwise rotations dramatically (Fig. 5A) and also initiated counterclockwise rotations (Fig. 5A), indicating significant restoration of the 6-hydroxydopamine lesion. Analysis of the apomorphine turning behavior confirmed that the grafted cells provided functional input (Fig. 5B). All graft analysis was done at 9 weeks post-transplantation. Staining for TH showed that animals displaying improvement in the behavioral assays had grafts containing large numbers of dopamine neurons, 350–7,000 TH+ cells (Fig. 6A–6H). These grafted dopamine neurons extended neurites into the deinnervated host striatum (Fig. 6B–6H). As previously described for fetal dopamine neurons [33], the addition of the pan-caspase inhibitor BAF to the cell suspension prior to engraftment appeared to have significantly enhanced the survival of grafted Pitx3-eGFP neurons, with 30% of all animals displaying grafts containing dopamine neurons with subsequent induced functional recovery. In the group grafted with Pitx3-eGFP cell suspension without BAF, only 9% of the animals had surviving grafts. Confocal analysis of Pitx3-eGFP-enriched grafts showed that the transplants contained large numbers of Pitx3-eGFP+ cells (Fig. 7A). These Pitx3-eGFP+ cells had neuronal morphology and showed a high overlapping expression with TH (Fig. 7A–7D), En (Fig. 7F–7J), and VMAT2 (Fig. 7K–7M), consistent with midbrain dopamine neurons. Some Pitx3-eGFP+ cells expressed Raldh1a1 (Fig. 7E), consistent with previous reports [23, 34, 35]. Some grafts also contained eGFP− cells (Fig. 7A). A portion of these eGFP− cells were Ki67+ cells, and some were nestin+ (supplemental online Fig. 4). The lack of overlap in expression between eGFP and markers of proliferation both in vitro (Fig. 3E, 3F; supplemental online Fig. 3) and in vivo (supplemental online Fig. 4) strongly indicates that the proliferative cells found in the grafts were of non-eGFP origin. Further analysis of the grafts showed that there were also serotonergic neurons present. Consistent with the in vitro data, there was no overlap in expression between eGFP and 5-HT (data not shown).
Figure 5.

A Pitx3-enhanced green fluorescent protein (eGFP)-enriched cell population could reverse amphetamine- and apomorphine-induced rotational behavior upon engraftment into 6-hydroxydopamine-lesioned rats. 6-Hydroxydopamine-lesioned rats were analyzed for amphetamine-induced (4 mg/kg) rotational behavior (90 minutes) pre txp and 5, 7, and 9 w post of 40,000–80,000 Pitx3-eGFP+ cells. Several cell-grafted animals decreased their cw rotations dramatically, whereas the vehicle-grafted animals did not (n = 5) (A). Many animals that showed a decreased in cw rotations also displayed ccw rotations, indicating a complete reversal of the effect of the 6-hydroxydopamie lesion (A). None of the vehicle-grafted animals ever showed any ccw rotations (n = 5) (A). Rotational behavior (30 minutes) in response to apomorphine administration (0.05 mg/kg) was analyzed prior to transplantation and 8 w post. Vehicle-grafted animals did not show a significant decrease in the number of rotations post-transplantation, but several Pitx3-eGFP-grafted animals did (B). Abbreviations: cw-ccw, clockwise-counterclockwise; post, post-transplantation; pre txp, pretransplantation; w, weeks.
Figure 6.
Grafted TH neurons innervated the host striatum of Pitx3-enhanced green fluorescent protein-transplanted animals with behavioral improvement. TH-DAB staining 9 weeks post-transplantation showed that the 6-hydroxydopamine-lesioned animals with improvement in the behavioral assays had grafts containing embryonic stem cell-derived TH+ neurons, which innervated the host striatum. A low-power microphotograph of a grafted animal (rat 19) showed the presence of TH neurons within the host striatum (A), and higher-power photographs confirmed that the grafted TH neurons grew into the deinnervated striatum of this animal ([B] is an enlargement of the boxed area in [A] and [C] is an enlargement of the boxed area in [B]). Analysis of additional animals (D–H) displaying behavioral recovery showed that these animals also contained grafted TH neurons, which innervated the hosts. Scale bars = 200 µm (B) and 100 µm (D, F and H). Abbreviations: DAB, 3,3′-diaminobenzidine; TH, tyrosine hydroxylase.
Figure 7.
Pitx3-eGFP-enriched grafts contained midbrain dopamine neurons. Confocal analysis of Pitx3-eGFP-enriched grafts, 9 weeks post-transplantation, showed that most grafts were composed of both eGFP+ and eGFP− cells (rat 19 is shown here) (A). The eGFP+ cells had neuronal morphology and showed a high overlapping expression with TH ([B–D], which are enlargements of the boxed area in [A]), En ([F–J]; [J] is an orthogonal view of the boxed area in [I]) and VMAT2 (K–M). Some Pitx3-eGFP+ cells expressed Raldh1a1 (E). Scale bars = 100 µm (A), and 50 µm (bars in [D, I, M] also apply to [B, C, F–H, K, L], and 10 µm (E). Abbreviations: eGFP, enhanced green fluorescent protein; En, engrailed; TH, tyrosine hydroxylase; VMAT2, vesicular monoamine transporter 2.
DISCUSSION
Fetal ventral mesencephalic dopamine neurons can be viably enriched from embryos by FACS using either dye labeling or fluorescence based on TH expression [32, 36–38]. Furthermore, primary dopamine neurons are also able to survive dissociation after in vitro culture prior to transplantation [39]. Isolation of dopamine neurons derived from ES cells has proven to be more complicated, since the differentiating cultures are not completely restricted in either a temporal or a spatial manner. TH is expressed in multiple cell types during development [35]. Furthermore, in the peripheral nervous system, TH+ cells with proliferative capacity have been identified [40, 41]. Selection of ES-derived cells based on TH expression, therefore, does not isolate catecholaminergic neurons alone but rather cells of many distinct phenotypes [11].
Within the central nervous system the homeodomain transcription factor Pitx3 is constitutively and selectively expressed in midbrain dopamine neurons. Pitx3 is also transiently expressed in the lens of the eye and in skeletal muscle [22, 26, 42, 43]. On the basis of its restricted expression pattern and the previous evidence that mature neurons, including TH neurons, can indeed be viably isolated from ES cell cultures [11, 24, 25], we deemed Pitx3 a good midbrain candidate for the selection of dopamine neurons from ES cell cultures. Toward this goal, our aim was first to carefully characterize Pitx3-eGFP KI mES cells in vitro and subsequently to enrich for these cells by FACS for eGFP expression and perform transplantation to 6-hydroxydopamine-lesioned rats.
Analysis of intact cultures showed that Pitx3-eGFP expression colocalized almost completely with Pitx3, Nurr1, engrailed, and AADC, confirming that eGFP+ cells generated in the culture were of a midbrain dopamine neuron type. The majority of cells were also positive for Lmx1a and TH. The earlier developmental expression of En [44, 45] and AADC than of TH [22] might explain the better overlap in expression among En, AADC, and Pitx3 compared with TH. Furthermore, ventrolateral midbrain dopamine neurons express Pitx3 prior to TH [30], further explaining why TH and Pitx3 would show a lower level of coexpression compared with markers that are normally expressed several days prior to Pitx3. Lmx1a is an early marker of mesencephalic progenitors and is also expressed in postmitotic midbrain dopamine neurons [8]. Careful analysis of Lmx1a expression in midbrain dopamine neurons has been done through E13.5 in the mouse [8, 46]. The lower overlapping expression between Lmx1a and Pitx3-eGFP in our cultures could be due to a restricted temporal expression of Lmx1a in midbrain dopamine neurons, although this remains to be further investigated.
The progressive restriction of the ES cell differentiation protocol used, toward mesencephalic and metencephalic fates [2, 7, 47], appears to limit the expression of Pitx3 in the culture to dopamine neurons. Consequently, Pitx3-eGFP expression did not overlap with markers other than those of dopamine neurons, indicating that Pitx3-based sorting would be a good choice for removing all other cell types.
FACS for Pitx3-eGFP expression showed that eGFP+ neurons could be viably enriched to constitute >90% of the cells while maintaining a midbrain dopamine neuron phenotype. This is the first time that midbrain dopamine neurons have been isolated and cultured from differentiating ES cell cultures. Sorted Pitx3-eGFP cells showed a higher overlapping expression between eGFP and TH than cells in intact cultures. This increased overlap in expression after sorting could simply be due to the fact that the ventro-lateral mesencephalic cells, which are known to express Pitx3 prior to TH [30], have now had time to initiate the expression of TH, 1 day post-FACS. Another explanation could be that the increased overlap is due to the coculture of sorted Pitx3-eGFP+ cells with astrocytes, which are known to be important for maturation/differentiation, morphology, and survival of dopamine neurons [48–52]. Since it is currently not possible to culture purified midbrain dopamine neurons without astrocytes or astrocyte-conditioned medium [11, 32], it is difficult to distinguish between these two possibilities.
The cell population enriched for Pitx3-eGFP expression could efficiently reverse amphetamine-induced rotational behavior as well as significantly reducing apomorphine-induced rotational behavior. Previous analyses using primary sorted mesencephalic TH-eGFP cells showed partial recovery of clockwise amphetamine-induced rotations [32, 38] but never demonstrated a recovery large enough to induce counter-clockwise rotations such as those displayed in this study or a reduction in apomorphine-induced behavior. Improvement in the behavioral assays in our study was well correlated with the presence of grafted dopamine neurons that innervated the host striatum.
Previous ES cell transplantation studies have, through the engraftment of relatively immature unsorted cultures, intentionally or unintentionally, relied mainly on in vivo generation of midbrain dopamine neurons for behavioral recovery [4 –7, 47]. We and others have previously used TH promoter-driven eGFP expression in ES cells to enrich for cathecholaminergic neurons prior to transplantation [11, 53]. However, due to the expression of eGFP in cells of non-neuronal morphology [11, 53], the grafts were composed of a majority of nondopaminergic cells, and most of the dopamine neurons in the resulting grafts were generated in vivo after grafting rather than prior to the sorting procedure [11, 53]. In this current study, we demonstrate for the first time that mature ES cell-derived midbrain dopamine neurons can survive enrichment by FACS and that these cells can reverse drug-induced behavior. Furthermore, in a double eGFP sorting procedure, we were able to show that these mature dopamine neurons can in fact be sorted twice by FACS and remain viable, ensuring that an almost pure midbrain dopamine neuron population is recovered.
It has previously been shown that primary midbrain dopamine neurons isolated by FACS can survive transplantation [32, 38]. However, the survival postgrafting appears to be dependent on the purity of the cell population transplanted, with a 60% enrichment for TH-eGFP expression resulting in a survival rate similar to that of TH neurons in nonenriched transplants [38], whereas an almost homogeneous TH-eGFP preparation (99%) resulted in a much reduced survival [32]. In this study, we grafted our sorted cells in a cocktail of trophic factors and also used a pan-caspase inhibitor, which increased the survival of Pitx3-eGFP midbrain dopamine neurons after transplantation. The combination of inhibition of apoptosis and blocking of complement activation can additionally increase survival of grafted primary midbrain dopamine neurons [33] and could perhaps be a future approach to increasing the survival of ES cell-derived midbrain dopamine neurons after transplantation.
In addition, our experiments demonstrate that it might be necessary to purify ES cell-derived midbrain dopamine neurons beyond an enrichment of 80% to ensure the generation of homogeneous grafts. Our double-sorting procedure for eGFP expression showed that the dopamine neurons could be viably enriched to 95.4% ± 2.7%, and this population could perhaps be used to generate grafts consisting almost solely of midbrain dopamine neurons. Furthermore, if the percentage of Pitx3+ cells in the culture could be increased by additional (nongenetic) modifications to the in vitro differentiation protocol, it is possible that a single sorting step for eGFP could yield a high proportion of Pitx3+ cells similar to the current yield after a double FACS procedure. Another approach to ensuring the generation of more homogeneous grafts could be the use of a selective elimination of cells with proliferative capacity. Our current sorting strategy reduced the number of proliferating cells 10-fold compared with unsorted suspensions, as measured by the reduction in SSEA-1+ cells [54–57]. Complete removal of cell-to-cell contact between ES cells results in a default dopaminergic differentiation [4]. Therefore, a future transplantation strategy could involve combining a positive selection for Pitx3 with a negative selection step for the cell surface antigen SSEA-1 [11] or other neuronal surface markers [25].
Even if the resulting cell population transplanted was not entirely homogeneous for Pitx3 after the second sorting step, the removal of SSEA-1+ cells would reduce the risk of tumor formation while providing the cells needed to recover from loss of dopamine. Previous studies aimed at eliminating the risk of tumor formation used Sox1 to positively select for neuronal precursors [9, 10]. Although this selection strategy was successful in preventing tumor formation, Sox1 precursors do not efficiently generate dopamine neurons [10, 58], and therefore their use might not be an optimal strategy for ultimately selecting dopamine neurons.
CONCLUSION
Our study shows for the first time that midbrain dopamine neurons derived from ES cells can be viably isolated using FACS. These sorted midbrain dopamine neurons could survive both further in vitro culturing and transplantation into the adult rat striatum. Furthermore, this midbrain dopamine neuron-enriched population could also reverse drug-induced rotational behavior in an animal model of Parkinson’s disease.
Supplementary Material
ACKNOWLEDGMENTS
We thank Michaela Patterson and Casper Reske-Nielsen for excellent technical assistance. We are grateful to Dr M. Li for providing the Pitx3-eGFP mES KI cells. This work was supported by Udall Parkinson’s Disease Center of Excellence Grant P50 NS39793; National Institute of Mental Health Grant 48866; the Orchard, Anti-Aging, and Stern foundations; and the Harold and Ronna Cooper family. E.H. is currently affiliated with the Ludwig Institute for Cancer Research, Department of Cell and Molecular Biology, Karolinska Institute, Stockholm, Sweden.
Footnotes
DISCLOSURE OF POTENTIAL CONFLICTS OF INTEREST
The authors indicate no potential conflicts of interest.
REFERENCES
- 1.Deacon T, Dinsmore J, Costantini LC, et al. Blastula-stage stem cells can differentiate into dopaminergic and serotonergic neurons after transplantation. Exp Neurol. 1998;149:28–41. doi: 10.1006/exnr.1997.6674. [DOI] [PubMed] [Google Scholar]
- 2.Lee SH, Lumelsky N, Studer L, et al. Efficient generation of midbrain and hindbrain neurons from mouse embryonic stem cells. Nat Biotechnol. 2000;18:675–679. doi: 10.1038/76536. [DOI] [PubMed] [Google Scholar]
- 3.Kawasaki H, Mizuseki K, Nishikawa S, et al. Induction of midbrain dopaminergic neurons from ES cells by stromal cell-derived inducing activity. Neuron. 2000;28:31–40. doi: 10.1016/s0896-6273(00)00083-0. [DOI] [PubMed] [Google Scholar]
- 4.Bjorklund LM, Sanchez-Pernaute R, Chung S, et al. Embryonic stem cells develop into functional dopaminergic neurons after transplantation in a Parkinson rat model. Proc Natl Acad Sci U S A. 2002;99:2344–2349. doi: 10.1073/pnas.022438099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barberi T, Klivenyi P, Calingasan NY, et al. Neural subtype specification of fertilization and nuclear transfer embryonic stem cells and application in parkinsonian mice. Nat Biotechnol. 2003;21:1200–1207. doi: 10.1038/nbt870. [DOI] [PubMed] [Google Scholar]
- 6.Roy NS, Cleren C, Singh SK, et al. Functional engraftment of human ES cell-derived dopaminergic neurons enriched by coculture with telomerase-immortalized midbrain astrocytes. Nat Med. 2006;12:1259–1268. doi: 10.1038/nm1495. [DOI] [PubMed] [Google Scholar]
- 7.Kim JH, Auerbach JM, Rodriguez-Gomez JA, et al. Dopamine neurons derived from embryonic stem cells function in an animal model of Parkinson’s disease. Nature. 2002;418:50–56. doi: 10.1038/nature00900. [DOI] [PubMed] [Google Scholar]
- 8.Andersson E, Tryggvason U, Deng Q, et al. Identification of intrinsic determinants of midbrain dopamine neurons. Cell. 2006;124:393–405. doi: 10.1016/j.cell.2005.10.037. [DOI] [PubMed] [Google Scholar]
- 9.Fukuda H, Takahashi J, Watanabe K, et al. Fluorescence-activated cell sorting-based purification of embryonic stem cell-derived neural precursors averts tumor formation after transplantation. STEM CELLS. 2006;24:763–771. doi: 10.1634/stemcells.2005-0137. [DOI] [PubMed] [Google Scholar]
- 10.Chung S, Shin BS, Hedlund E, et al. Genetic selection of sox1GFP-expressing neural precursors removes residual tumorigenic pluripotent stem cells and attenuates tumor formation after transplantation. J Neurochem. 2006;97:1467–1480. doi: 10.1111/j.1471-4159.2006.03841.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hedlund E, Pruszak J, Ferree A, et al. Selection of embryonic stem cell-derived enhanced green fluorescent protein-positive dopamine neurons using the tyrosine hydroxylase promoter is confounded by reporter gene expression in immature cell populations. STEM CELLS. 2007;25:1126–1135. doi: 10.1634/stemcells.2006-0540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Björklund A, Stenevi U, Dunnett SB, et al. Cross-species neural grafting in a rat model of Parkinson’s disease. Nature. 1982;298:652–654. doi: 10.1038/298652a0. [DOI] [PubMed] [Google Scholar]
- 13.Schmidt RH, Ingvar M, Lindvall O, et al. Functional activity of substantia nigra grafts reinnervating the striatum: Neurotransmitter metabolism and [14C]2-deoxy-D-glucose autoradiography. J Neurochem. 1982;38:737–748. doi: 10.1111/j.1471-4159.1982.tb08693.x. [DOI] [PubMed] [Google Scholar]
- 14.Brundin P, Nilsson OG, Strecker RE, et al. Behavioural effects of human fetal dopamine neurons grafted in a rat model of Parkinson’s disease. Exp Brain Res. 1986;65:235–240. doi: 10.1007/BF00243848. [DOI] [PubMed] [Google Scholar]
- 15.Brundin P, Strecker RE, Widner H, et al. Human fetal dopamine neurons grafted in a rat model of Parkinson’s disease: Immunological aspects, spontaneous and drug-induced behaviour, and dopamine release. Exp Brain Res. 1988;70:192–208. doi: 10.1007/BF00271860. [DOI] [PubMed] [Google Scholar]
- 16.Clarke DJ, Brundin P, Strecker RE, et al. Human fetal dopamine neurons grafted in a rat model of Parkinson’s disease: Ultrastructural evidence for synapse formation using tyrosine hydroxylase immunocytochemistry. Exp Brain Res. 1988;73:115–126. doi: 10.1007/BF00279666. [DOI] [PubMed] [Google Scholar]
- 17.Mendez I, Sanchez-Pernaute R, Cooper O, et al. Cell type analysis of functional fetal dopamine cell suspension transplants in the striatum and substantia nigra of patients with Parkinson’s disease. Brain. 2005;128:1498–1510. doi: 10.1093/brain/awh510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smidt MP, van Schaick HS, Lanctot C, et al. A homeodomain gene Ptx3 has highly restricted brain expression in mesencephalic dopaminergic neurons. Proc Natl Acad Sci U S A. 1997;94:13305–13310. doi: 10.1073/pnas.94.24.13305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nunes I, Tovmasian LT, Silva RM, et al. Pitx3 is required for development of substantia nigra dopaminergic neurons. Proc Natl Acad Sci U S A. 2003;100:4245–4250. doi: 10.1073/pnas.0230529100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van den Munckhof P, Luk KC, Ste-Marie L, et al. Pitx3 is required for motor activity and for survival of a subset of midbrain dopaminergic neurons. Development. 2003;130:2535–2542. doi: 10.1242/dev.00464. [DOI] [PubMed] [Google Scholar]
- 22.Smidt MP, Smits SM, Bouwmeester H, et al. Early developmental failure of substantia nigra dopamine neurons in mice lacking the homeodomain gene Pitx3. Development. 2004;131:1145–1155. doi: 10.1242/dev.01022. [DOI] [PubMed] [Google Scholar]
- 23.Jacobs FM, Smits SM, Noorlander CW, et al. Retinoic acid counteracts developmental defects in the substantia nigra caused by Pitx3 deficiency. Development. 2007;134:2673–2684. doi: 10.1242/dev.02865. [DOI] [PubMed] [Google Scholar]
- 24.Schmandt T, Meents E, Gossrau G, et al. High-purity lineage selection of embryonic stem cell-derived neurons. Stem Cells Dev. 2005;14:55–64. doi: 10.1089/scd.2005.14.55. [DOI] [PubMed] [Google Scholar]
- 25.Pruszak J, Sonntag KC, Aung MH, et al. Markers and methods for cell sorting of human embryonic stem cell-derived neural cell populations. STEM CELLS. 2007;25:2257–2268. doi: 10.1634/stemcells.2006-0744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao S, Maxwell S, Jimenez-Beristain A, et al. Generation of embryonic stem cells and transgenic mice expressing green fluorescence protein in midbrain dopaminergic neurons. Eur J Neurosci. 2004;19:1133–1140. doi: 10.1111/j.1460-9568.2004.03206.x. [DOI] [PubMed] [Google Scholar]
- 27.Sakazume S, Sorokina E, Iwamoto Y, et al. Functional analysis of human mutations in homeodomain transcription factor PITX3. BMC Mol Biol. 2007;8:84. doi: 10.1186/1471-2199-8-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zetterstrom RH, Solomin L, Mitsiadis T, et al. Retinoid X receptor heterodimerization and developmental expression distinguish the orphan nuclear receptors NGFI-B, Nurr1, and Nor1. Mol Endocrinol. 1996;10:1656–1666. doi: 10.1210/mend.10.12.8961274. [DOI] [PubMed] [Google Scholar]
- 29.Zetterström RH, Williams R, Perlmann T, et al. Cellular expression of the immediate early transcription factors Nurr1 and NGFI-B suggests a gene regulatory role in several brain regions including the nigrostriatal dopamine system. Brain Res Mol Brain Res. 1996;41:111–120. doi: 10.1016/0169-328x(96)00074-5. [DOI] [PubMed] [Google Scholar]
- 30.Maxwell SL, Ho HY, Kuehner E, et al. Pitx3 regulates tyrosine hydroxylase expression in the substantia nigra and identifies a subgroup of mesencephalic dopaminergic progenitor neurons during mouse development. Dev Biol. 2005;282:467–479. doi: 10.1016/j.ydbio.2005.03.028. [DOI] [PubMed] [Google Scholar]
- 31.Teitelman G, Jaeger CB, Albert V, et al. Expression of amino acid decarboxylase in proliferating cells of the neural tube and notochord of developing rat embryo. J Neurosci. 1983;3:1379–1388. doi: 10.1523/JNEUROSCI.03-07-01379.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Donaldson AE, Marshall CE, Yang M, et al. Purified mouse dopamine neurons thrive and function after transplantation into brain but require novel glial factors for survival in culture. Mol Cell Neurosci. 2005;30:108–117. doi: 10.1016/j.mcn.2005.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cicchetti F, Costantini L, Belizaire R, et al. Combined inhibition of apoptosis and complement improves neural graft survival of embryonic rat and porcine mesencephalon in the rat brain. Exp Neurol. 2002;177:376–384. doi: 10.1006/exnr.2002.8007. [DOI] [PubMed] [Google Scholar]
- 34.Wallén A, Zetterstrom RH, Solomin L, et al. Fate of mesencephalic AHD2-expressing dopamine progenitor cells in NURR1 mutant mice. Exp Cell Res. 1999;253:737–746. doi: 10.1006/excr.1999.4691. [DOI] [PubMed] [Google Scholar]
- 35.Chung S, Hedlund E, Hwang M, et al. The homeodomain transcription factor Pitx3 facilitates differentiation of mouse embryonic stem cells into AHD2-expressing dopaminergic neurons. Mol Cell Neurosci. 2005;28:241–252. doi: 10.1016/j.mcn.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 36.di Porzio U, Rougon G, Novotny EA, et al. Dopaminergic neurons from embryonic mouse mesencephalon are enriched in culture through immunoreaction with monoclonal antibody to neural specific protein 4 and flow cytometry. Proc Natl Acad Sci U S A. 1987;84:7334–7338. doi: 10.1073/pnas.84.20.7334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kerr CW, Lee LJ, Romero AA, et al. Purification of dopamine neurons by flow cytometry. Brain Res. 1994;665:300–306. doi: 10.1016/0006-8993(94)91351-x. [DOI] [PubMed] [Google Scholar]
- 38.Sawamoto K, Nakao N, Kobayashi K, et al. Visualization, direct isolation, and transplantation of midbrain dopaminergic neurons. Proc Natl Acad Sci U S A. 2001;98:6423–6428. doi: 10.1073/pnas.111152398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Timmer M, Grosskreutz J, Schlesinger F, et al. Dopaminergic properties and function after grafting of attached neural precursor cultures. Neurobiol Dis. 2006;21:587–606. doi: 10.1016/j.nbd.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 40.Lindeberg J, Usoskin D, Bengtsson H, et al. Transgenic expression of Cre recombinase from the tyrosine hydroxylase locus. Genesis. 2004;40:67–73. doi: 10.1002/gene.20065. [DOI] [PubMed] [Google Scholar]
- 41.Rothman TP, Specht LA, Gershon MD, et al. Catecholamine biosynthetic enzymes are expressed in replicating cells of the peripheral but not the central nervous system. Proc Natl Acad Sci U S A. 1980;77:6221–6225. doi: 10.1073/pnas.77.10.6221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Semina EV, Reiter RS, Murray JC. Isolation of a new homeobox gene belonging to the Pitx/Rieg family: Expression during lens development and mapping to the aphakia region on mouse chromosome 19. Hum Mol Genet. 1997;6:2109–2116. doi: 10.1093/hmg/6.12.2109. [DOI] [PubMed] [Google Scholar]
- 43.Smidt MP, Smits SM, Burbach JP. Homeobox gene Pitx3 and its role in the development of dopamine neurons of the substantia nigra. Cell Tissue Res. 2004;318:35–43. doi: 10.1007/s00441-004-0943-1. [DOI] [PubMed] [Google Scholar]
- 44.Wurst W, Auerbach AB, Joyner AL. Multiple developmental defects in Engrailed-1 mutant mice: An early mid-hindbrain deletion and patterning defects in forelimbs and sternum. Development. 1994;120:2065–2075. doi: 10.1242/dev.120.7.2065. [DOI] [PubMed] [Google Scholar]
- 45.Danielian PS, McMahon AP. Engrailed-1 as a target of the Wnt-1 signalling pathway in vertebrate midbrain development. Nature. 1996;383:332–334. doi: 10.1038/383332a0. [DOI] [PubMed] [Google Scholar]
- 46.Ono Y, Nakatani T, Sakamoto Y, et al. Differences in neurogenic potential in floor plate cells along an anteroposterior location: Midbrain dopaminergic neurons originate from mesencephalic floor plate cells. Development. 2007;134:3213–3225. doi: 10.1242/dev.02879. [DOI] [PubMed] [Google Scholar]
- 47.Rodríguez-Gómez JA, Lu JQ, Velasco I, et al. Persistent dopamine functions of neurons derived from embryonic stem cells in a rodent model of Parkinson disease. STEM CELLS. 2007;25:918–928. doi: 10.1634/stemcells.2006-0386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Denis-Donini S, Glowinski J, Prochiantz A. Specific influence of striatal target neurons on the in vitro outgrowth of mesencephalic dopaminergic neurites: A morphological quantitative study. J Neurosci. 1983;3:2292–2299. doi: 10.1523/JNEUROSCI.03-11-02292.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Denis-Donini S, Glowinski J, Prochiantz A. Glial heterogeneity may define the three-dimensional shape of mouse mesencephalic dopaminergic neurones. Nature. 1984;307:641–643. doi: 10.1038/307641a0. [DOI] [PubMed] [Google Scholar]
- 50.O’Malley EK, Sieber BA, Black IB, et al. Mesencephalic type I astrocytes mediate the survival of substantia nigra dopaminergic neurons in culture. Brain Res. 1992;582:65–70. doi: 10.1016/0006-8993(92)90317-3. [DOI] [PubMed] [Google Scholar]
- 51.Wagner J, Akerud P, Castro DS, et al. Induction of a midbrain dopaminergic phenotype in Nurr1-overexpressing neural stem cells by type 1 astrocytes. Nat Biotechnol. 1999;17:653–659. doi: 10.1038/10862. [DOI] [PubMed] [Google Scholar]
- 52.Castelo-Branco G, Sousa KM, Bryja V, et al. Ventral midbrain glia express region-specific transcription factors and regulate dopaminergic neurogenesis through Wnt-5a secretion. Mol Cell Neurosci. 2006;31:251–262. doi: 10.1016/j.mcn.2005.09.014. [DOI] [PubMed] [Google Scholar]
- 53.Yoshizaki T, Inaji M, Kouike H, et al. Isolation and transplantation of dopaminergic neurons generated from mouse embryonic stem cells. Neurosci Lett. 2004;363:33–37. doi: 10.1016/j.neulet.2004.03.074. [DOI] [PubMed] [Google Scholar]
- 54.Fox N, Damjanov I, Martinez-Hernandez A, et al. Immunohistochemical localization of the early embryonic antigen (SSEA-1) in postimplantation mouse embryos and fetal and adult tissues. Dev Biol. 1981;83:391–398. doi: 10.1016/0012-1606(81)90487-5. [DOI] [PubMed] [Google Scholar]
- 55.Solter D, Knowles BB. Monoclonal antibody defining a stage-specific mouse embryonic antigen (SSEA-1) Proc Natl Acad Sci U S A. 1978;75:5565–5569. doi: 10.1073/pnas.75.11.5565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Eggens I, Fenderson B, Toyokuni T, et al. Specific interaction between Lex and Lex determinants. A possible basis for cell recognition in preimplantation embryos and in embryonal carcinoma cells. J Biol Chem. 1989;264:9476–9484. [PubMed] [Google Scholar]
- 57.Capela A, Temple S. LeX/ssea-1 is expressed by adult mouse CNS stem cells, identifying them as nonependymal. Neuron. 2002;35:865–875. doi: 10.1016/s0896-6273(02)00835-8. [DOI] [PubMed] [Google Scholar]
- 58.Barraud P, Thompson L, Kirik D, et al. Isolation and characterization of neural precursor cells from the Sox1-GFP reporter mouse. Eur J Neurosci. 2005;22:1555–1569. doi: 10.1111/j.1460-9568.2005.04352.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.